Abstract
DNA polymerases play a central role in the process of DNA replication. Yet, the proteins in charge of the replication of plant organelle DNA have not been unambiguously identified. There are however many indications that a family of proteins homologous to bacterial DNA polymerase I (PolI) is implicated in organelle DNA replication. Here, we have isolated mutant lines of the PolIA and PolIB genes of Arabidopsis (Arabidopsis thaliana) to test this hypothesis. We find that mutation of both genes is lethal, thus confirming an essential and redundant role for these two proteins. However, the mutation of a single gene is sufficient to cause a reduction in the levels of DNA in both mitochondria and plastids. We also demonstrate that polIb, but not polIa mutant lines, are hypersensitive to ciprofloxacin, a small molecule that specifically induces DNA double-strand breaks in plant organelles, suggesting a function for PolIB in DNA repair. In agreement with this result, a cross between polIb and a plastid Whirly mutant line yielded plants with high levels of DNA rearrangements and severe growth defects, indicating impairments in plastid DNA repair pathways. Taken together, this work provides further evidences for the involvement of the plant PolI-like genes in organelle DNA replication and suggests an additional role for PolIB in DNA repair.
Faithful replication of DNA is one of the most crucial processes in living organisms. Plant cells are particular in this regard because of their need to maintain genomes in three separate compartments: the nucleus, the plastid, and the mitochondria. While nuclear DNA replication and repair have been extensively studied, little is known about these processes in plant organelles. For example, the DNA polymerases responsible for the replication of the genome of mitochondria and plastids have not yet been clearly identified.
In higher plants, DNA polymerase activity has been purified from chloroplast extracts of spinach (Spinacia oleracea; Spencer and Whitfeld, 1969; Sala et al., 1980) and pea (Pisum sativum; McKown and Tewari, 1984), while a mitochondrial DNA polymerase activity has been purified from wheat (Triticum aestivum; Castroviejo et al., 1979) and the flowering plant Chenopodium album (Meissner et al., 1993). The DNA polymerase activities from these various sources have similar characteristics, including insensitivity to aphidicolin, a known inhibitor of nuclear eukaryotic alpha DNA polymerases, a broad alkaline pH optimal activity and the requirement for a divalent metal cation (Mg2+ or Mn2+). This suggests that plant organelles, like animal mitochondria, possess their own specialized enzymes, with distinct characteristics, for the replication of their genome (Sala et al., 1980).
Isolation of DNA polymerases from both the chloroplast and mitochondria of soybean (Glycine max) and tobacco (Nicotiana tabacum) cell cultures allowed further comparison of the two enzymes contained in these compartments (Heinhorst et al., 1990; Sakai et al., 1999). Interestingly, the biochemical properties and Mrs of these enzymes were shown to be very similar. They reach maximal activity under the same experimental conditions and are sensitive to the same chemical agents, suggesting that they are at least homologous proteins. These enzymes were also shown to share characteristics with the γ DNA polymerases (Polγ) found in the mitochondria of animals (Heinhorst et al., 1990; Sakai et al., 1999), such as insensitivity to aphidicolin, requirement for KCl, and template preference. Another important characteristic shared by these purified enzymes and Polγ is that they possess a 3′ to 5′ exonuclease activity, as demonstrated for the DNA polymerases from spinach chloroplasts (Keim and Mosbaugh, 1991) and C. album mitochondria (Meissner et al., 1993).
More recently, genes encoding proteins targeted to organelles and with similarity to the gene coding for Escherichia coli DNA polymerase I (PolI) were identified in rice (Oryza sativa; Kimura et al., 2002), Arabidopsis (Arabidopsis thaliana; Mori et al., 2005), and tobacco (Ono et al., 2007). Full-length cDNAs were cloned and regions corresponding to the DNA polymerase domains of these enzymes were produced in bacteria and tested for template preference, biochemical properties, and sensitivity to inhibitors. Like the enzymes purified from plants, the recombinant proteins are insensitive to aphidicolin and dideoxynucleotides (Kimura et al., 2002; Mori et al., 2005; Ono et al., 2007), suggesting that these DNA PolI-like enzymes are involved in DNA replication in plants’ organelles.
It therefore appears that a conserved family of eubacteria-like DNA polymerases is responsible for the replication of DNA inside plant organelles. Indeed, every plant genome that has been sequenced to date possesses two PolI-like genes which, in Arabidopsis and tobacco, encode proteins that localize to both organelles, thus suggesting an important conserved function (Elo et al., 2003; Christensen et al., 2005; Ono et al., 2007; Wamboldt et al., 2009). In addition, the recombinant proteins have a high processivity, which would allow them to replicate complete organelle genomes (Kimura et al., 2002; Mori et al., 2005; Ono et al., 2007). All these characteristics make the PolI proteins ideal candidates for a role in organelle DNA replication. However, this function has not yet been confirmed in vivo. Here we report the isolation of mutant lines for the two Arabidopsis PolI-like genes At1g50840 and At3g20540 (named PolIA and PolIB henceforth). While no visible phenotype was observed with two mutant alleles of PolIA, a strong mutant allele of PolIB yielded plants that grew more slowly than wild-type plants. More importantly, we were unable to recover plants bearing homozygous mutations in both genes, indicating that they play an essential, at least partially redundant role in Arabidopsis. All mutants tested showed reduced levels of organelle DNA relative to nuclear DNA, suggesting that they are involved in genome replication. Interestingly, only the mutants for PolIB showed increase sensitivity to the organelle-specific DNA double-strand break (DSB)-inducing agent ciprofloxacin, suggesting that one of the two PolI proteins has specialized in DNA repair.
RESULTS
PolIA and PolIB Share a Common Essential Function in Organelles
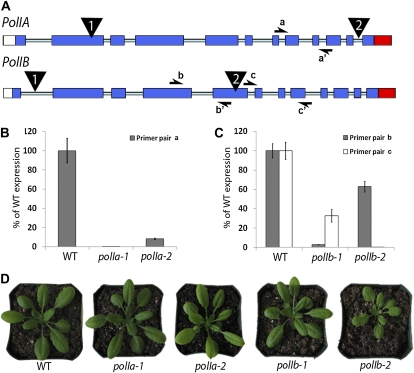
To provide genetic evidence for the implication of the Arabidopsis PolIA and PolIB genes in organelle DNA replication, we have obtained two T-DNA insertion mutant lines for each gene. Figure 1A shows the position of the insertion in each line. Figure 1B shows PolIA mRNA levels, measured by quantitative reverse transcription (qRT)-PCR, in the polIa lines. The two mutations in PolIA caused a marked reduction in the expression level of the gene. The polIa-1 line (SALK_022638) showed a 536-fold reduction and the polIa-2 line (SALK_065221) a 12-fold reduction in mRNA level as compared to the wild type. Mutations in the PolIB gene had a different effect depending on the region of the gene that was amplified. Figure 1C shows the results of two qRT-PCR reactions using primers located either in the middle of the gene or at the 3′ end (see Fig. 1A). The polIb-1 line (SALK_134274) showed a 33-fold reduction in expression in the midgene section but only a 3-fold reduction near the end of the gene. The polIb-2 line (WiscDsLoxHs021_09D) resulted in a 1.5-fold reduction of the midgene section and a 365-fold reduction at the 3′ end. This difference in signal level between two parts of the same messenger could be due to differential processing of the mRNA caused by the T-DNA insertion, or to leaky transcription from the 35S promoter inside the T-DNA insert. This could lead to different results when using probes placed on either side of the insertion site, as it is the case for polIb-2.
Figure 1.
Isolation of PolIA and PolIB mutant lines. A, Schematic representation of PolIA and PolIB genes. Exons are denotated by blue boxes, the 3′ untranslated region by a red box, and the 5′ untranslated region by a white box. The pale-blue lines represent introns. The position of the T-DNA insertions is indicated by black triangles. Half-arrows stand for the primers used for the qRT-PCR experiments. B, Histogram presenting the results of the qRT-PCR experiments measuring the level of expression of the PolIA gene inside the polIa mutant plants grown on Murashige and Skoog medium for 14 d. Expression levels are represented as percentages of wild-type (WT) level. The experiments were done in technical and biological triplicates. The error bars represent the sem on the biological triplicates. C, Histogram presenting the results of the qRT-PCR experiments measuring the level of expression of the PolIB gene inside the polIb mutant plants grown on Murashige and Skoog medium for 14 d. Expression levels are represented as percentages of wild-type level. The experiments were done in technical and biological triplicates. The error bars represent the sem on the biological triplicates. D, Representative photographs of 4-week-old Arabidopsis plants for wild-type and PolI mutant lines.
The phenotype of the two polIa lines did not differ from the wild type (Fig. 1D). Interestingly, while the polIb-1 mutation also yielded no differential phenotype, the polIb-2 line showed growth retardation (Fig. 1D). This difference in phenotype compared to polIb-1 might be attributed to the lower expression of PolIB (maximal reduction of 365-fold versus 33-fold for the polIb-1 line) or to the site of the insertion (fifth exon versus first intron). We then attempted to cross polIa-1 with polIb-1, and polIa-2 with polIb-2 to obtain mutant plants devoid of PolI proteins. We were unable to isolate double homozygous mutant plants in the progeny of double heterozygous plants. By testing the germination of seeds from the cross on Murashige and Skoog medium, we did not observe any aborted seed and we concluded that the double homozygous individuals do not produce mature seeds. We went a step further for the polIa-1/polIb-1 cross and isolated one plant homozygous for polIb-1, but heterozygous for polIa-1. We then tested 48 plants from the progeny of this plant and again could not recover any double homozygous plant. The significance of this result was confirmed by a χ2 test (P = 0). These results indicate that at least one intact PolI gene is essential for plant viability. Therefore, in the rest of our work we focused on the analysis of single-mutant plants.
Mutants for the PolI Genes Have Less Organelle DNA
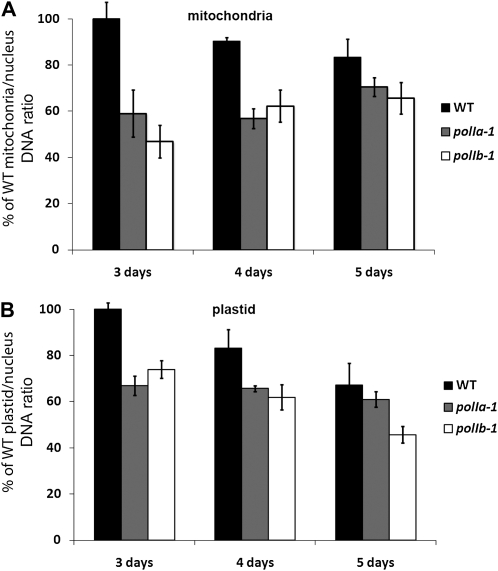
Mutants for DNA polymerase genes are expected to have a reduced capacity to synthesize and accumulate DNA. We therefore measured the relative level of organelle to nuclear DNA in the PolI mutants to evaluate organelle genome copy number in the PolI mutants (Zoschke et al., 2007; Rowan et al., 2009). Wild-type, polIa-1, and polIb-1 mutant lines were grown on Murashige and Skoog medium, whole seedlings (including the roots) were harvested at 3, 4, and 5 d after germination, and their total DNA isolated. The level of DNA was measured by qPCR using primers specific for a given DNA sequence inside the nucleus, the mitochondria, or the plastid. The levels for the mitochondria and plastid DNA (mtDNA and ptDNA) are reported as a ratio to the nuclear DNA level. Figure 2A shows that, in 3- and 4-d-old seedlings, the relative level of mtDNA is lower in both polIa-1 and polIb-1 as compared to the wild-type plants, but that this difference is less pronounced by day 5. A similar trend is observed when testing ptDNA, although in this case the difference between the wild type and polIb-1 is still pronounced at day 5 (Fig. 2B). Similar results were obtained with the polIa-2 line (Supplemental Fig. S1). However, it was not possible to test the polIb-2 line at these early time points due to its growth retardation defect. These results show that PolIA and PolIB are both required for maximal DNA accumulation in Arabidopsis plastids and mitochondria.
Figure 2.
Organelle DNA is less abundant in seedlings of polIa-1 and polIb-1 mutants. A, Histogram presenting the results of the qPCR analysis of mitochondrial genome copy number relative to nuclear genome copy number for 3-, 4-, and 5-d-old seedlings. The experiments were done in technical and biological triplicates. The error bars represent the sem on the biological triplicate. B, Histogram presenting the results of the qPCR analysis of plastid genome copy number relative to nuclear genome copy number for 3-, 4-, and 5-d-old seedlings. The experiments were done in technical and biological triplicates. The error bars represent the sem on the biological triplicate. WT, Wild type.
Mutants for the PolIB But Not PolIA Gene Are Impaired in Their Capacity to Repair DNA DSBs
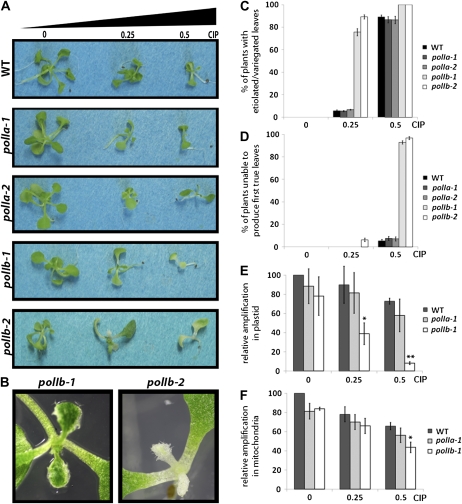
DNA-synthesis activity is required for the repair of DNA damages by mechanisms such as homologous recombination (HR). To test for a role in DNA repair, the PolI mutants were treated with ciprofloxacin, a molecule that induces DNA DSBs specifically in plant organelle DNA (Wall et al., 2004) and was recently shown to affect ptDNA conformation (Rowan et al., 2010). Interestingly, only the polIb mutants showed increased sensitivity to ciprofloxacin when compared to wild-type plants (Fig. 3, A and B). At 0.25 μm ciprofloxacin, polIb-2 plants were more sensitive to the drug than polIb-1 plants. In this condition, the polIb-1 plants were able to produce small first true leaves with white regions while polIb-2 plants produced abnormal first true leaves that were completely white (Fig. 3B). At 0.5 μm ciprofloxacin very few true leaves were observed for either polIb alleles. Figure 3, C and D, presents, for each line and condition tested, the percentage of plants showing first true leaves with white regions or no first true leaves, respectively.
Figure 3.
PolIB mutants exhibit an increased sensitivity to ciprofloxacin (CIP) and are impaired in their ability to process DNA DSBs. A, Representative photographs of the different polI mutants grown for 21 d on Murashige and Skoog medium containing 0, 0.25, or 0.5 μm ciprofloxacin. B, Representative photographs of the first true leaves of polIb-1 and polIb-2 plants grown for 21 d on Murashige and Skoog medium containing 0.25 μm ciprofloxacin. C, Histogram representing the percentage of plants with white/variegated first true leaves after 21 d of growth on medium with different concentrations of ciprofloxacin. Error bars represent the sd of the counts made on different plates. D, Histogram representing the percentage of plants without any first true leaves after 21 d of growth on medium with different concentrations of ciprofloxacin. Error bars represent the sd of the counts made on different plates. E, Histogram presenting the results of the PCR experiments to evaluate the abundance of DNA lesions upon ciprofloxacin treatment in ptDNA. DNA lesions are significantly more abundant for the polIb-1 mutant when treated with both 0.25 and 0.5 μm ciprofloxacin. Error bars represent the sem of three separate experiments where wild-type signal at 0 μm ciprofloxacin is expressed as 100 for each repetition. One asterisk indicates a significant difference with a P < 0.05 in the Student’s t test and two asterisks, a difference with a P < 0.01. F, Histogram presenting the results of the PCR experiments to evaluate the abundance of DNA lesions in mitochondrial DNA upon treatment with ciprofloxacin. DNA lesions are significantly more abundant for the polIb-1 mutant when treated with 0.5 μm ciprofloxacin. Error bars represent the sem of three separate experiments where wild-type signal at 0 μm ciprofloxacin is expressed as 100 for each repetition. Asterisk indicates a significant difference with a P < 0.05 in the Student’s t test. WT, Wild type.
These results suggest that in addition to a role in DNA replication, PolIB is also required for the repair of DNA. In that case one can predict that the DSBs induced by ciprofloxacin will accumulate to higher levels in polIb-1 plants than in wild-type or polIa-1 plants. This was tested by measuring the level of DNA lesions in plastid and mitochondrial DNA using a PCR approach (Yakes and Van Houten, 1997). Figure 3, E and F, shows a clear, dose-dependent decrease in the relative amplification of a long DNA fragment when using polIb-1 DNA isolated from plants treated with ciprofloxacin as template. This indicates the presence of a higher level of DNA lesions in the organelle genomes of these plants as compared to wild-type and polIa-1 plants. Although a similar trend was observed when comparing the results for ptDNA (Fig. 3E) and mtDNA (Fig. 3F), the difference with wild-type DNA was less marked when testing mtDNA. This difference was nevertheless significant (P < 0.05 in Student’s t test) for the highest concentration of the antibiotic. To confirm that the DSBs induced by ciprofloxacin are specific to organellar DNA, we designed a similar experiment with nuclear probes and found, as expected, no increase in DNA lesions in wild-type and polIb-1 plants treated with 0.5 μm ciprofloxacin (Supplemental Fig. S2). Taken together, these results indicate a role for PolIB in DNA repair in both the mitochondria and the plastid.
Genetic Interaction between PolIB and Whirly Genes
The above results suggest that PolIB is specifically recruited at DSB sites and contributes to their repair. In prokaryotes, the recruitment of repair proteins to the DNA is orchestrated by the single-stranded DNA-binding proteins (for review, see Shereda et al., 2008). We have recently reported that the single-stranded DNA-binding proteins Whirly are required for accurate repair of DSBs in plant organelles and proposed that their function in this process is comparable to that of single-stranded DNA-binding proteins in prokaryotes (Maréchal et al., 2009; Cappadocia et al., 2010). To test whether a genetic interaction exists between the Whirly and the PolI genes, we crossed the polIa-1 and the polIb-1 mutants with the mitochondria (why2-1) or the plastid (why1why3) Whirly mutants reported previously (Maréchal et al., 2009; Cappadocia et al., 2010). All but one of the crosses yielded plants with no difference in phenotype when compared to wild-type plants, except for the occasional appearance of variegation in lines bearing the why1why3 mutations (Maréchal et al., 2009; Fig. 4A; Supplemental Fig. S3). Interestingly, the polIb-1/why1why3 homozygous triple mutant exhibited a severe growth defect and pale leaves, indicating that a synergistic interaction exists between plastid Whirlies and PolIB.
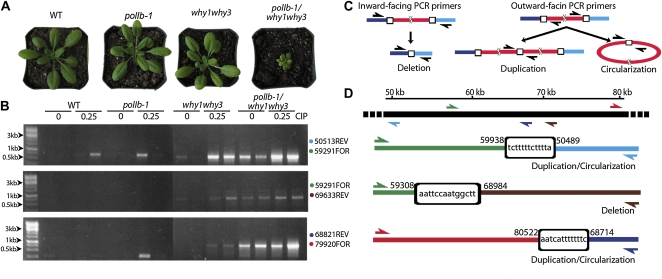
Figure 4.
A polIb-1/why1why3 triple mutant yields a pale-green dwarf phenotype and gives rise to abundant DNA rearrangements in plastids. A, Representative photographs of 4-week-old plants for the genotypes used in the cross and the triple-mutant progeny compared to the wild type (WT). B, Representative PCR reactions to evaluate the abundance of DNA rearrangements dependant on microhomology in plastids of the plants presented in A. The plants were grown for 21 d on Murashige and Skoog medium containing 0 or 0.25 μm ciprofloxacin. Two DNA samples were tested for each condition. PCR reactions were done using the primers indicated on the right of the gels. C, Schematic representation of DNA rearrangements evaluated by the PCR approach. Inward-facing primers placed far apart will yield a PCR product only if a deletion has occurred in the region that separates the primers. Outward-facing primers will yield a PCR product only if the template DNA is duplicated or circularized. Primers are represented by black half-arrows. D, Schematic representation of the DNA rearrangements amplified by PCR. The top section shows the position of the primers used to test the plastid genome of Arabidopsis. The bottom section gives a graphical representation of the rearranged DNA molecules. The sequences in the white boxes are the microhomologous sequences that were used to prime the MMBIR process. The original position of the microhomologous sequences in wild-type ptDNA is indicated.
The why1why3 mutant line of Arabidopsis has been shown to promote the repair of DSBs through an error-prone microhomology-mediated break-induced replication (MMBIR) pathway (Cappadocia et al., 2010) that leads to major genome rearrangements (for review, see Hastings et al., 2009; Maréchal and Brisson, 2010). About 5% of the why1why3 plants also exhibit a variegated leaf phenotype that correlates with selectively amplified genomic regions (Maréchal et al., 2009). As the results in Figure 3E show that the amount of DSBs is increased in ptDNA from ciprofloxacin-treated polIb-1 plants, it suggests that the severe phenotype of the polIb-1/why1why3 plants is also linked to an accumulation of rearranged DNA products in the plastid genome. This was tested by monitoring the extent of genome rearrangements in the mutant lines using eight primer pairs employed in our previous study to survey ptDNA rearrangements in Whirly mutant plants (Cappadocia et al., 2010). These primers are either in an opposite orientation or placed far apart (more than 5 kb) from each other (Fig. 4C) and amplify, respectively, junctions of duplicated/circular or deleted DNA regions produced by the MMBIR pathway in plastids. Figure 4B shows the results of representative PCR reactions with three primer pairs. A graphic representation of the main rearranged molecule is illustrated for each reaction in Figure 4D. Untreated wild-type and polIb-1 plants show no detectable rearrangements, while the same genotypes treated with 0.25 μm ciprofloxacin show only few rearrangements (Fig. 4B). The why1why3 plants contain a constitutive level of rearranged DNA molecules and, as reported before (Cappadocia et al., 2010), this level increases noticeably after treatment with 0.25 μm ciprofloxacin. The level of DNA rearrangements is however markedly higher in the untreated triple mutant as compared to untreated why1why3 plants. This level is actually similar to that of the treated why1why3 plants and appears to further increase upon treatment with the drug. Therefore, the severe phenotype of the polbI-1/why1why3 plants correlates with increased rearrangements of the plastid genome.
DISCUSSION
An Essential Role for the PolI Genes in Higher Plants
Our understanding of plant organelle DNA replication is still very incomplete (Bendich, 2007; Nielsen et al., 2010). This is largely due to our ignorance concerning some of the most basic factors, such as DNA polymerases, that contribute to this fundamental process in plants. Many studies have suggested that the PolI-like DNA polymerases found in higher plants are responsible for most if not all the DNA-synthesis activity found in mitochondria and plastids (Kimura et al., 2002; Mori et al., 2005; Ono et al., 2007). This is based on the biochemical properties of the recombinant proteins, which indicate that these enzymes have sufficient processivity to be able to replicate both genomes. In addition, the measured in vitro characteristics of these enzymes correspond almost perfectly to those measured ex vivo from the purified DNA-synthesis activity of mitochondria and plastids (Spencer and Whitfeld, 1969; Sala et al., 1980; McKown and Tewari, 1984; Heinhorst et al., 1990; Meissner et al., 1993). All these elements, plus the fact that they are present in both organelles (Elo et al., 2003; Christensen et al., 2005; Mori et al., 2005; Ono et al., 2007; Wamboldt et al., 2009), suggest that the PolI DNA polymerases are responsible for DNA replication in plant organelles.
In this study, we bring further arguments to support a role for PolI proteins in the replication of ptDNA and mtDNA. Using Arabidopsis mutant plants for the PolIA and PolIB genes, we show that concurrent mutation of the two genes is lethal, indicating that they share an essential function in plants. Furthermore, each gene is able to partially compensate for the function of the other gene as all single PolI mutants tested are viable. However, the fact that the polIb-2 line displays a growth retardation phenotype suggests that PolIA cannot entirely compensate for the function of PolIB, or that PolIB carries another function not shared by PolIA.
A role for the PolI proteins in DNA replication is also supported by our results showing that, as compared to wild-type plants, the relative level of organelle DNA is reduced in plastids and mitochondria of both polIa-1 and polIb-1 plants. The reduction is most pronounced at 3 d postgermination, which is consistent with expression data showing that the PolI genes are most highly expressed in young, meristematic tissues (Kimura et al., 2002; Mori et al., 2005; Ono et al., 2007). These results are also in line with previous studies showing that DNA replication in organelles is highly active in 3-d-old Arabidopsis seedlings (Fujie et al., 1993, 1994). Our results therefore provide a functional link between the PolI genes and DNA accumulation inside plant mitochondria and plastids.
Interestingly, the ratios of organelle to nuclear DNA appear to decrease in wild-type plants between day 3 and 5 (Fig. 2; Supplemental Fig. S1). This could either mean that nuclear DNA ploidy is increasing or that organelle DNA is less abundant in cells of older seedlings. However, since wild-type and mutant plants of the same age are being compared, any variations in nuclear or organellar DNA levels should not affect our results. It is interesting that previous studies reported a similar trend for ptDNA (Zoschke et al., 2007) and mtDNA (Preuten et al., 2010) ratios in aging cotyledons of Arabidopsis, although different samples were used in these studies (cotyledon alone versus whole seedling with roots).
A Specific Role for PolIB in DNA DSB Repair
DNA repair activity is essential for correcting DNA damage caused by exogenous stress or that occur naturally during replication. Many mechanisms used to repair DNA, such as HR and MMBIR, involve a DNA-synthesis step. Since, as mentioned above, PolIA does not fully compensate for the loss of PolIB in the polIb-2 line, there is a possibility that PolIB could be involved in an additional process, such as synthesis of DNA during repair. This was tested by inducing DSBs in the organelle genome of all the PolI mutants and by measuring the effect of this stress on plant phenotype and on the level of DSBs. The fact that the growth of the polIb lines, but not of the polIa lines, was more affected than wild-type plants by the genotoxic stress provides evidence for a role of PolIB in DNA repair. Similar increased sensitivity to ciprofloxacin was also reported for Whirly (Cappadocia et al., 2010) and cpRecA (Rowan et al., 2010) mutants, which are both implicated in the maintenance of plastid genome stability. A role in DNA repair for PolIB is further supported by the observation that the polIb-1 line accumulates higher levels of DNA lesions than the polIa-1 line or wild-type plants when treated with ciprofloxacin.
The higher levels of DNA lesions in plastids as compared to mitochondria suggest a specialized role for PolIB in this organelle. However, it could also simply reflect the presence of alternate repair mechanisms more active in the mitochondria. For example HR between short repeats (>100 bp and <1,000 bp; for review, see Maréchal and Brisson, 2010), a process very active in plant mitochondria, would provide an efficient mean for the repair of DSBs. Also, we cannot exclude the possibility that ciprofloxacin could be less active in mitochondria. The different response of the polIa and polIb mutant lines to the genotoxic stress is surprising, considering that the two PolI proteins have over 72% identity at the amino acid sequence level (Mori et al., 2005) and that they are nearly identical regarding their 3′ to 5′ exonuclease and DNA polymerase domains. Indeed, there does not seem to be any distinctive motif between PolIA and PolIB when comparing amino acid sequences of available plant proteins. Therefore our results indicate that Arabidopsis PolIB has either evolved a specific repair activity or that it is specifically recruited to damage sites. This additional function in DNA repair might also explain why one of the PolIB mutants shows a growth retardation phenotype while both PolIA mutants are undistinguishable from wild-type plants. However, we cannot exclude that PolIA is involved in some other DNA repair mechanisms that do not involve DSBs.
A Genetic Interaction between PolIB and Plastid Whirly Genes
Previous studies have proposed that Whirly proteins play a role in organelle DNA replication (Maréchal et al., 2008; Prikryl et al., 2008). More recently, these proteins were shown to be involved in the maintenance of plastid genome stability (Maréchal et al., 2009) and the repair of DSBs in plastids and mitochondria (Cappadocia et al., 2010). It is therefore interesting that a strong synergistic interaction exists between PolIB and the plastid Whirly genes. This suggests that PolIB and the plastid Whirlies function in separate, but complementary pathways. The large increase of MMBIR rearrangements in ptDNA observed in the polIb-1/why1why3 mutant in the absence of genotoxic stress supports this conclusion and indicates that PolIB is not required or essential for DNA repair by MMBIR. This is reminiscent of the situation in yeast (Saccharomyces cerevisiae) where break-induced replication as well as MMBIR are dependent on a nonessential subunit of Polδ called POL32 (Lydeard et al., 2007; Payen et al., 2008) and in Escherichia coli where it is dependent on DinB (Galhardo et al., 2009). Our results suggest that, in the absence of PolIB, conservative repair of DSBs is reduced, leading to the accumulation of substrates (DSBs) that are repaired by the error-prone MMBIR in absence of Whirlies. It also indicates that another DNA polymerase is responsible for the synthesis of rearranged DNA molecules, at least regarding the MMBIR process.
The fact that no phenotype is observed when polIb-1 is crossed with the mitochondrial Whirly mutant allele why2-1 supports once again the presence of an alternative DNA repair mechanism in this organelle. This is in agreement with our previous results where no visible or molecular phenotype was observed in the why2-1 mutant unless an exogenous stress was applied (Cappadocia et al., 2010). This contrasts with the situation with the why1why3 plastid Whirly mutant where all the plants showed DNA rearrangements and 4.6% were variegated (Maréchal et al., 2009).
Taken together, our results indicate that the action of PolIB in DNA repair most likely involves a classical repair mechanism, such as HR. It is thus expected that reduced expression or inactivation of PolIB in Arabidopsis would lead to an increase in unattended DSBs and/or to slower repairs, even in the absence of exogenous stress. These breaks or arrested replication forks would be more likely to be corrected by other mechanisms, such as break-induced replication. In the absence of Whirly proteins, however, the breaks would also be repaired through error-prone mechanisms such as MMBIR, leading to the accumulation of rearranged DNA molecules such as those observed in polIb-1/why1why3 and that could account for the severe phenotype of this line. It will be interesting to confirm this model in the future by further genetic crosses between the polIb alleles and other mutant alleles for genes involved in organelle DNA repair.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0) mutant lines polIa-1 (SALK_022638), polIa-2 (SALK_065221), polIb-1 (SALK_134274), and polIb-2 (WiscDsLoxHs021_09D) were obtained from the Arabidopsis Biological Resource Center. Seeds were either sown on soil or sterilized and placed on petri dishes containing Murashige and Skoog basal media (Sigma-Aldrich, www.sigmaaldrich.com) supplemented with 1% Suc and 0.8% agar with or without ciprofloxacin (Sigma-Aldrich). After 3 d of vernalization at 4°C, the seeds were placed under light (100 μmol m−2 s−1, 16 h day/8 h dark) at an average temperature of 22°C. Whole plants grown on Murashige and Skoog medium were harvested at 3, 4, 5 (for seedlings), 14 (five to six true leaves), or 21 (mature plants) d after germination by pulling the plants (including the roots) carefully from the agar and instantly frozen in liquid nitrogen. Plants were pooled to make 75-mg samples.
Nucleic Acids Isolation
DNA was isolated using a cetyl trimethylammonium bromide protocol (Weigel and Glazebrook, 2002). RNA was isolated using TRIzol reagent (Invitrogen, www.invitrogen.com) according to the manufacturer’s instructions. RNA pellets were resuspended in 30 μL of nuclease-free water and kept at −80°C. cDNA was synthesized using the first-strand cDNA synthesis kit from Fermentas (www.fermentas.com) according to the manufacturer’s instructions. cDNA reactions were diluted 10-fold and kept at −20°C.
qPCR
Real-time qPCR reactions were done using the SYBR green master mix (SABiosciences, www.sabiosciences.com) according to the manufacturer’s instructions. Primers used for qRT-PCR experiments are as follows: 5′Tubu-qRT, CCT TCT CGG TGT TCC CTT CAC C; 3′Tubu-qRT, GGG AAC CTC AGA CAG CAA GTC AC; 5′PolIA-qRTa, ACC GCG GTC GGG CTT TCT AGA; 3′PolIA-qRTa, GCA CGT GAC TTT GAC GCC GGA; 5′PolIB-qRTb, GCG GAG ATC GCA AAG GAC ATA GTT GT; 3′PolIB-qRTb, TCC TTC AAC TAC GGC CGG TCT CT; 5′PolIB-qRTc, GAC GCT TAT CGG CTA GAA GGC CA; and 3′PolIB-qRTc, CTT GAG TGG AAG TCT CCA CCA GCT. Primers used for qPCR experiments are as follows: 5′ncDNA-qPCR, GTT GAA GCC TCC GTT CCC TGC TA; 3′ncDNA-qPCR, CTC TTC CAC CGT GCA TGG CTT GT; 5′mtDNA-qPCR, CCT GAT TCT GCG CGT AGA AGA CCT; 3′mtDNA-qPCR, AGG CGT AAG CGC AGC AGT TAG A; 5′ptDNA-qPCR, CCC TCT CTC TCG TAG TGT GGG GAA; and 3′ptDNA-qPCR, TCG AAA GGG GTT ACC CCA TGA ATG G. The amplification efficiency of each reaction was determined using dilution of template cDNA or DNA. Calibration runs determined the use of 1/10 dilution for cDNA and 1/100 for DNA. Melting curves confirmed the presence of a unique PCR product in each case. Amplification was carried out by using the LightCycler480 (Roche, www.roche-applied-science.com) and all the data were analyzed using the LightCycler480 software version 1.5.
Long-Amplification PCR
DNA lesions were quantified using the technique described in Yakes and Van Houten (1997). Briefly, DNA was amplified using GeneAmp XL PCR kit (Applied Biosystems, www.appliedbiosystems.com) following the manufacturer’s recommendations. For mitochondria, plastids, and the nucleus we amplified a long (about 20 kb) and a short (about 200 bp) PCR fragment. It is expected that the long amplification will be more susceptible to breaks than the short amplification. The primers used for ptDNA were as follows: 5′ptLongAmp, CCG GGA CTC GAA CCC GGA ACT A; 3′ptLongAmp, CTT CCT CCC GAG TTG AGA CCC A; 5′ptShortAmp, CTC CAG TGC ATT TCG CCC TCT GA; and 3′ptShortAmp, ACG TAC ACG TCA CGG GCA TCC T. The primers used for mtDNA were as follows: 5′mtLongAmp, CCT ATC AGT AGC TAG GCC CTG GTC C; 3′mtLongAmp, CCT AAC TAC ACC GAC CTC CGA GTC C; 5′mtShortAmp, CCT GAT TCT GCG CGT AGA AGA CCT; and 3′mtShortAmp, AGG CGT AAG CGC AGC AGT TAG A. The primers used for ncDNA were as follows: 5′ncLongAmp, TCG CGG AGA TCT TTC TCT CGC AC; 3′ncLongAmp, CTG CCT CCA CCA ACT CGT TCA C; 5′ncShortAmp, GTT GAA GCC TCC GTT CCC TGC TA; and 3′ncShortAmp, CTC TTC CAC CGT GCA TGG CTT GT. DNA quantification of the reactions was done using PicoGreen reagent (Invitrogen) following the manufacturer’s instructions. Fluorescence was measured using a Perkin Elmer/Packard fusion α-FP microplate analyzer (PerkinElmer, www.perkinelmer.com). The level of PCR products for the long amplification was reported on the level of the short product for each sample and normalized to 100 with wild-type untreated signal.
Detection of DNA Rearrangements
Total DNA from Murashige and Skoog medium-grown 21-d-old plants was used to survey ptDNA rearrangements. DNA was equilibrated with a low-cycle PCR amplification of a Ycf2 DNA fragment. The PCR strategy and primers used were described in Cappadocia et al. (2010). PCR reactions products were migrated on ethidium bromide-stained agarose gels.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Organelle DNA is less abundant in seedlings of polIa-2.
Supplemental Figure S2. Ciprofloxacin does not generate DSB in the nuclear genome.
Supplemental Figure S3. Crosses between PolI and Whirly mutants.
Acknowledgments
The assistance of Alexandre Maréchal in isolating and crossing mutant lines is gratefully acknowledged. We thank B. Franz Lang and all the members of the Brisson laboratory for valuable discussions.
References
- Bendich AJ. (2007) The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. Bioessays 29: 474–483 [DOI] [PubMed] [Google Scholar]
- Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N. (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castroviejo M, Tharaud D, Tarrago-Litvak L, Litvak S. (1979) Multiple deoxyribonucleic acid polymerases from quiescent wheat embryos: purification and characterization of three enzymes from the soluble cytoplasm and one from purified mitochondria. Biochem J 181: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. (2005) Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo A, Lyznik A, Gonzalez DO, Kachman SD, Mackenzie SA. (2003) Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15: 1619–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Mutoh S, Kuroiwa T. (1994) Behavior of organelles and their nucleoids in the soot apical meristem during leaf development in Arabidopsis thaliana L. Planta 194: 395–405 [DOI] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Suzuki T, Kawano S, Kuroiwa T. (1993) Organelle DNA synthesis in the quiescent center of Arabidopsis thaliana (col.). J Exp Bot 44: 689–693 [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM. (2009) DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC, Weissbach A. (1990) Chloroplast and mitochondrial DNA polymerases from cultured soybean cells. Plant Physiol 92: 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim CA, Mosbaugh DW. (1991) Identification and characterization of a 3′ to 5′ exonuclease associated with spinach chloroplast DNA polymerase. Biochemistry 30: 11109–11118 [DOI] [PubMed] [Google Scholar]
- Kimura S, Uchiyama Y, Kasai N, Namekawa S, Saotome A, Ueda T, Ando T, Ishibashi T, Oshige M, Furukawa T, et al. (2002) A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res 30: 1585–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Sabar M, Véronneau-Lafortune F, Abou-Rached C, Brisson N. (2008) Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown RL, Tewari KK. (1984) Purification and properties of a pea chloroplast DNA polymerase. Proc Natl Acad Sci USA 81: 2354–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner K, Heinhorst S, Cannon GC, Börner T. (1993) Purification and characterization of a gamma-like DNA polymerase from Chenopodium album L. Nucleic Acids Res 21: 4893–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Kimura S, Saotome A, Kasai N, Sakaguchi N, Uchiyama Y, Ishibashi T, Yamamoto T, Chiku H, Sakaguchi K. (2005) Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem Biophys Res Commun 334: 43–50 [DOI] [PubMed] [Google Scholar]
- Nielsen BL, Cupp JD, Brammer J. (2010) Mechanisms for maintenance, replication, and repair of the chloroplast genome in plants. J Exp Bot 61: 2535–2537 [DOI] [PubMed] [Google Scholar]
- Ono Y, Sakai A, Takechi K, Takio S, Takusagawa M, Takano H. (2007) NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol 48: 1679–1692 [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. (2008) Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Cincu E, Fuchs J, Zoschke R, Liere K, Börner T. (2010) Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J 64: 948–959 [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. (2009) A multiple-method approach reveals a declining amount of chloroplast DNA during development in Arabidopsis. BMC Plant Biol 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. (2010) RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J Exp Bot 61: 2575–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Suzuki T, Nagata N, Sasaki N, Miyazawa Y, Saito C, Inada N, Nishimura Y, Kuroiwa T. (1999) Comparative analysis of DNA synthesis activity in plastid-nuclei and mitochondrial-nuclei simultaneously isolated from cultured tobacco cells. Plant Sci 140: 9–19 [Google Scholar]
- Sala F, Amileni AR, Parisi B, Spadari S. (1980) A gamma-like DNA polymerase in spinach chloroplasts. Eur J Biochem 112: 211–217 [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43: 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D, Whitfeld PR. (1969) The characteristics of spinach chloroplast DNA polymerase. Arch Biochem Biophys 132: 477–488 [DOI] [PubMed] [Google Scholar]
- Wall MK, Mitchenall LA, Maxwell A. (2004) Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci USA 101: 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WB, Mackenzie SA. (2009) Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Yakes FM, Van Houten B. (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA 94: 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]