Abstract
Posttranslational activation of nitrate reductase (NR) in Arabidopsis (Arabidopsis thaliana) and other higher plants is mediated by dephosphorylation at a specific Ser residue in the hinge between the molybdenum cofactor and heme-binding domains. The activation of NR in green leaves takes place after dark/light shifts, and is dependent on photosynthesis. Previous studies using various inhibitors pointed to protein phosphatases sensitive to okadaic acid, including protein phosphatase 2A (PP2A), as candidates for activation of NR. PP2As are heterotrimeric enzymes consisting of a catalytic (C), structural (A), and regulatory (B) subunit. In Arabidopsis there are five, three, and 18 of these subunits, respectively. By using inducible artificial microRNA to simultaneously knock down the three structural subunits we show that PP2A is necessary for NR activation. The structural subunits revealed overlapping functions in the activation process of NR. Bimolecular fluorescence complementation was used to identify PP2A regulatory subunits interacting with NR, and the two B55 subunits were positive. Interactions of NR and B55 were further confirmed by the yeast two-hybrid assay. In Arabidopsis the B55 group consists of the close homologs B55α and B55β. Interestingly, the homozygous double mutant (b55α × b55β) appeared to be lethal, which shows that the B55 group has essential functions that cannot be replaced by other regulatory subunits. Mutants homozygous for mutation in Bβ and heterozygous for mutation in Bα revealed a slower activation rate for NR than wild-type plants, pointing to these subunits as part of a PP2A complex responsible for NR dephosphorylation.
Nitrate reductase (NR), a key enzyme in nitrogen assimilation, reduces nitrate to nitrite in the cytosol. The nitrite formed is further reduced to ammonium in the plastids, whereafter ammonium is incorporated into amino acids. Regulation of NR takes place at the transcriptional as well as posttranslational level. At both levels signals from the chloroplasts are involved in initiating an increase in NR activity levels, but these signals and their processing are still unknown (Jonassen et al., 2008; Lillo, 2008; Krouk et al., 2010; Nunes-Nesi et al., 2010). At the posttranslational level NR is rapidly activated by dephosphorylation, and active photosynthesis is necessary for this activation (Kaiser and Brendle-Behnisch, 1991; Huber et al., 1994; Douglas et al., 1995; Bachmann et al., 1996; Campbell, 1999; Lea et al., 2006). The phosphorylation status of NR, or any other protein, is the result of the action of both kinases and phosphatases. Recently, protein phosphatases have been brought into focus because it is recognized that protein phosphatases are themselves subjected to activation or inactivation by posttranslational mechanism, hence they are not passive partners of protein kinases, but have dynamic regulatory roles (DeLong, 2006; Janssens et al., 2008; Virshup and Shenolikar, 2009).
Microcystin-LR and okadaic acid, which are known inhibitors of the protein phosphatase families protein phosphatase 1 (PP1), PP2A, PP4, PP5, PP6, and PPP-Kelch, prevented light activation of NR in spinach (Spinacia oleracea) leaves and barley (Hordeum vulgare) protoplasts (MacKintosh, 1992; Lillo et al., 1996). Okadaic acid and microcystin also prevented activation of NR in extracts of leaves, whereas inhibitor 2 (known inhibitor of the PP1 family) had no effect. Mammalian PP2A, but not PP1, could activate NR in vitro, which further supported the importance of PP2A and related phosphatases, except PP1, in this process (MacKintosh, 1992). PP6 is inhibited by okadaic acid, but is predominantly expressed in flowers, and PP6 (from pea [Pisum sativum]) is known to require addition of Fe2+ or Zn2+ in vitro for performing dephosphorylation (Kim et al., 2002). The protein phosphatase(s) that activates NR is present in leaves and does not require addition of divalent cations to extracts for activity (MacKintosh, 1992). This makes PP6 an unlikely candidate for regulation of NR. Arabidopsis (Arabidopsis thaliana) has two, weakly expressed, genes encoding PP4 (Farkas et al., 2007). Furthermore, the two PP4 phosphatases (PPX1 and PPX2) are localized inside plastids (Pujol et al., 2000), therefore not likely to dephosphorylate NR, which is localized in the cytosol. Protein phosphatases with Kelch-repeat domains (BSU1, BSL1, 2, and 3) are also inhibited by okadaic acid, although higher concentrations are needed than for PP2A. BSU1 is localized in the nucleus and present in young tissue. BSL1, 2, and 3 are expressed in leaves and older tissue (Mora-García et al., 2004), hence can be considered candidates for dephosphorylating NR. PP5 is known to promote expression of the light-regulated genes, CHALCONE SYNTHASE, CAB2, and RBCS (Ryu et al., 2005), and is a putative candidate for regulating also NR.
In Arabidopsis, as well as in other eukaryotes, PP2As are oligomeric proteins composed of a 36- to 38-kD catalytic subunit (C) bound to a scaffolding subunit (A) of 65 kD. A number of variable regulatory subunits of molecular mass 48 to 74 kD associate with the core dimer to form a trimeric PP2A. Presently three B families are identified in plants (B/B55, B’, B’’; Farkas et al., 2007; Wang et al., 2007). The A subunit is required as a scaffold for the formation of the heterotrimeric PP2A complex. In mammals, two different A subunits are present, whereas Arabidopsis has three such scaffolding subunits, and yeast (Saccharomyces cerevisiae) has one. The A subunits are composed of 15 imperfect HEAT repeats, each of 39 amino acids, that form a hook-shaped molecule. The repeats consist of two α-helices connected by an intrarepeat loop. B subunits bind to repeats 1 to 10, whereas C subunits bind to repeats 11 to 15 (Li and Virshup, 2002). The Arabidopsis A subunits have been studied by different research groups, and are known to be important for root morphology (Zhou et al., 2004) and auxin transport (Michniewicz et al., 2007). They show overlapping functions, and the triple knockout is most likely lethal (Zhou et al., 2004; Michniewicz et al., 2007). One of the A subunits, ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1 (RCN1), was suggested to have a cardinal role in regulation of phosphatase activity, whereas functions of the other two, PP2AA2 and PP2AA3, were only unmasked when RCN1 was absent (Zhou et al., 2004). By using these different loss-of-function mutants and a transgenic line with tamoxifen-inducible expression of artificial microRNAs (amiRNAs) targeting all three A subunits of PP2A (Michniewicz et al., 2007), we here investigated the importance of A subunits for activation of NR. A clear effect on NR by inducible knockdown of all three A subunits established the involvement of PP2A for activation of NR in vivo.
The PP2A regulatory B subunits play an important role in conferring substrate specificity and determine subcellular localization of PP2As in mammals (Sontag, 2001; Eichhorn et al., 2009). It is assumed that also in plants the different B subunits have such functions (Matre et al., 2009). In Arabidopsis, the TON2/FASS is the only B (a B’’) regulatory subunit that has been firmly characterized so far, and was found to be involved in cortical microtubule organization and important for cell shape (Camilleri et al., 2002; Wang et al., 2007). Very recently it was shown that silencing of TAP46 in Arabidopsis, the ortholog of the yeast TAP42 protein, a PP2A catalytic subunit interacting protein, affects NR expression (Ahn et al., 2011). When we tested different B subunits for interaction with NR, positive results were found for the B55 group. In other eukaryotes B55 has a regulatory role in mitosis (Mochida et al., 2009; Schmitz et al., 2010). Little is known about the functions of the two 55-kD B isoforms in plants. This type of subunit is almost entirely composed of β-sheets and turns, whereas B’ and B’’ are mostly α-helical. B55 subunits contain four to seven degenerate WD40 repeats. In mammals the B55 subunit family consists of four members, α, β, δ, and γ (Eichhorn et al., 2009). The mammalian isoforms are localized to nucleus, cytosol, or associated with the cytoskeleton, and are involved in reactions with different substrates (Eichhorn et al., 2009). The mammalian Bγ is the form most similar to the plant B55. Evolutionarily, plants and mammals divided before the various isoforms were developed, and their isoforms are named independently (Terol et al., 2002). A necessary step for dissecting the signal transduction chain connecting processes in the chloroplasts and activation of NR in the cytosol is to identify phosphatase(s) of physiological importance for NR activation, and this is the subject of the present report.
RESULTS
PP2A-Like Protein Phosphatases
PP5 is a protein phosphatase susceptible to okadaic acid inhibition, and a possible candidate for activation of NR. NR activity state following dark-to-light transitions was tested for a PP5 loss-of-function mutant (pp5ko) and also a PP5 overexpressor (pp5ox; papp5-1 and PAPP5-OX1 in Ryu et al., 2005). No effects on NR activation during dark-to-light transitions were revealed for these mutants (see Supplemental Fig. S2). Mutants homozygous for T-DNA insert in PPP-KELCH motif protein phosphatase genes BSL1, BSL2, and BSL3 were tested, however no indications of effects on NR activation were seen. From these experiments it can be concluded that none of these phosphatases had a major effect on NR activation (Supplemental Fig. S2), however they cannot be excluded from having some effect on NR, that would be overtaken by another phosphatase in the mutants.
PP2A A1, A2, and RCN1 Subunits and NR Activity
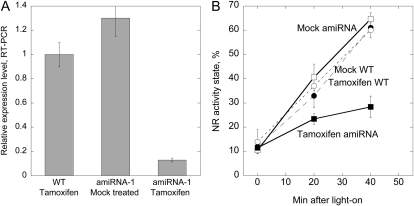
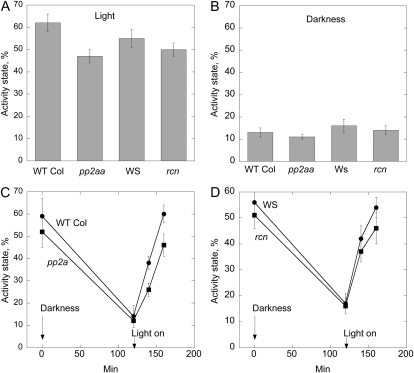
A triple knockout for all three A subunits is to our knowledge not available, and this is most likely due to lethality of such a mutant (Michniewicz et al., 2007). Knockdown of all three A subunits of PP2A was achieved by using tamoxifen-inducible microRNA overexpression with amiRNA-1 and amiRNA-2 lines (Michniewicz et al., 2007). Three-week-old plants were treated with tamoxifen every day for 1 week, which led to a strong decrease (90%) in expression of the A subunit examined (Fig. 1A). A marked decrease in NR activation was observed in these amiRNA plants (Fig. 1B). The NR activity state gives the amount of nonphosphorylated (active) NR as a percentage of the total amount of NR (the nonphosphorylated plus phosphorylated forms; MacKintosh et al., 1995). Only the phosphorylated form of NR is inhibited by Mg++. When Mg++ is present in the assay only nonphosphorylated NR is active, but when EDTA is added to the assay also the phosphorylated form becomes active because of sequestering of cations and release of the 14-3-3 inhibitor (Huber et al., 2002). The differential Mg++/EDTA assays, therefore, enable an efficient and precise way of calculation of activity state and also adjust for possible variations in total NR in each extract (Supplemental Fig. S3). Within 40 min of illumination, NR activity state increased from 10% to 66% in mock-treated amiRNA-1 plants, and from 11% to 28% in tamoxifen-treated amiRNA-1 plants (Fig. 1). Wild-type (Columbia [Col]) plants treated with tamoxifen confirmed that NR was still activated as in nontreated wild-type plants (Fig. 1B). The low PP2AA expression seemed to have no effect on NR inactivation during the dark period since the NR activation state was the same in mock- and tamoxifen-treated darkened plants (time zero in Fig. 1B). NR activity was also measured in A-subunit loss-of-function mutants (Zhou et al., 2004), i.e. rcn1 (Wassiljevskaja [WS] background) and double mutant pp2aa2 pp2aa3 (Col background; Fig. 2). Activity state for samples harvested during the photoperiod was 47% ± 3% for the pp2aa2 pp2aa3 double mutant and significantly higher, 62% ± 4%, for the wild-type Col control (Fig. 2A). NR activity state in the rcn1 mutant was not significantly different from the control (WS; Fig. 2A). Samples harvested after 2 h of darkness showed activity state between 11% and 16%, but no significant differences between mutants and controls (Fig. 2B). Change in activity state in response to light on/off was also tested, and showed that for the pp2aa2 pp2aa3 double-mutant activation of NR was lower than in wild type. After 20 min of light, wild type showed activity state of 38% ± 7% and the double mutant showed activity state of 26% ± 5% (Fig. 2C). For the rcn1 mutant the difference from the WS control was not significant (Fig. 2D). In summary these experiments showed that PP2A is required for rapid NR activation in vivo, and that the structural A subunits have overlapping functions in this process.
Figure 1.
The scaffolding A subunits of PP2A are required for light activation of NR. A, Relative transcript levels for the PP2AA subunit in tamoxifen-treated wild-type and amiRNA-1 plants, and mock-treated plants. Expression levels were tested with RT-PCR using SYBR green with plant samples also used for testing NR activation. se for the RT-PCR assay is given, n = 3. B, Activity state of NR following a dark-to-light transition of wild-type (dots) and amiRNA-1 (squares) plants treated with tamoxifen (black symbols), and mock-treated plants (white symbols). Plants were grown in 12-h light/12-h darkness for 3 weeks. Two hours into the photoperiod, when NR activity was at a high level, plants were transferred to darkness and samples harvested after 1 h of darkness (time zero), then after 20 and 40 min of white light (150 μmol m−2 s−1). Data are means of three samples, se is given.
Figure 2.
NR activity state in light or darkness in different A subunit mutant plants. Wild-type Col, the double-mutant pp2aa2 pp2aa3, WS, and rcn1 mutant were tested. Leaves of rosette stage plants were harvested daily during 6 d, 1.5 to 3 h into the light period, and after approximately 12 h of darkness. A, Light samples. B, Dark samples. n = 13, se is given. To test changes in activity stage during light/dark transitions samples were harvested from rosette stage plants 3 h into the daily light period (time 0), light was turned off, and samples were harvested after 2 h of darkness. Light was then switched on, and samples taken after 20 and 40 min of light. C, Double-mutant pp2aa2 pp2aa3 and control wild-type Col. D, rcn1 mutant and control WS. The data are means of four independent experiments, se is given.
Subcellular Localization of B55 Subunits and Interactions with NR
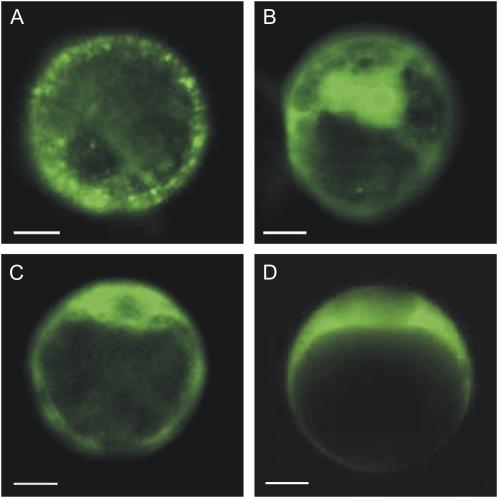
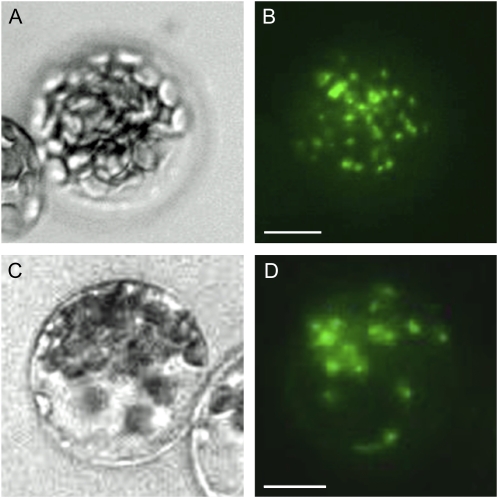
NR1 and NR2 linked by their C terminus to EYFP were transiently expressed in protoplasts of Arabidopsis suspension culture and mesophyll cells. The fusion proteins localized to the cytosol, with a tendency to form speckles (representative pictures are shown in Fig. 3, A and B). The B55α and B55β subunits both localized to the cytosol regardless of N- (Fig. 5, C and D) or C-terminal (not shown) fusion to the EYFP. Interaction of NR1 and B55α or B55β was tested in planta by bimolecular fluorescence complementation (BiFC). This assay is based on the reconstitution of EYFP when nonfluorescent N-terminal (NFP) and C-terminal (CFP) fragments of the EYFP are brought together by two interacting proteins fused to NFP and CFP (Hu et al., 2002; Maple et al., 2005). The NFP was fused to the C-terminal end of full-length NR1, and CFP was fused to the C-terminal end of full-length B55α and B55β. The results firmly established that NR1 and Bα interact in planta (Fig. 4, A and B). In experiments performed with Bβ, some interactions were detected, however, with lower occurrence. Several other B’ and B’’ subunits (At5g03470/B’α, At3g09880/B’β, At3g26030/B’δ, At1g13460/B’θ, At4g15410/B’γ, At3g26020/B’η, At3g54930/B’ε, At3g21650/B’ζ, At5g18580/TON2, and At5g53000/TAP46) were also tested for interaction with NR1 without positive results. Combined these results showed that specifically B55, but not other B subunits tested, interacted with NR1. To confirm NR interactions with B55α and to determine the interacting domains, we furthered the BiFC experiments by testing interaction between NR protein fragments and B55-type subunits. The experiments showed that B55α interacted with the domain containing the hinge1 flanked by dimerization domain and heme-binding domains (amino acid 340–621; Fig. 4, C and D). The N-terminal 100 amino acids containing an acidic domain, and the flavin-binding domain linked with hinge2 (amino acid 621–917), gave the same type of positive images (not shown). The molybdenum cofactor factor binding domain (amino acid 100–340) did not interact with Bα. For scheme of interacting domains see Supplemental Figure S4 (Buchanan et al., 2000).
Figure 3.
Fusion proteins of NR and B55 subunits localize to the cytoplasm in Arabidopsis protoplasts. A, NR1 (At1g77760) and B, NR2 (At1g37130) were fused at their C terminus to EYFP (NR-EYFP) and transiently expressed in protoplasts from Arabidopsis cell suspension culture. C, B55α (At1g51690) protein fused at its N terminus to EYFP and transiently expressed in protoplasts localized to cytosol. D, B55β (At1g17720) protein fused at its N terminus to EYFP and transiently expressed in protoplasts localized to cytosol.
Figure 5.
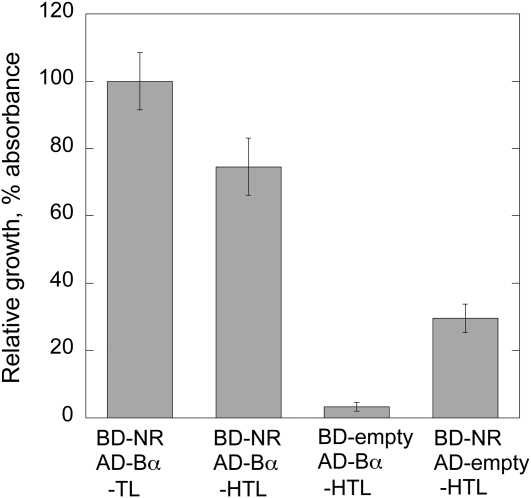
NR1 and B55α interact in yeast. HF7c cells were cotransformed with the indicated combination of bait (BD) and prey (AD) vectors and grown on selection plates at 30°C for 48 h. The growth on −HTL medium and −TL medium was assayed and the ratio calculated as an indicator of the strength of protein interaction. n = 3.
Figure 4.
Full-length and truncated forms of NR interact with Bα as tested by BiFC. A and B, Interaction of full-length NR1 and Bα in cytosol of Arabidopsis mesophyll protoplasts. C and D, Interaction of the NR1 fragment comprising the dimerization domain, hinge1, and heme domain with Bα in Arabidopsis mesophyll protoplasts. Scale bar = 10 μm.
To verify the BiFC interactions of NR and B55 the yeast two-hybrid assay was employed. To this end we decided to assay the specific PP2A regulatory B55 subunits by coexpression with NR1 in yeast. On selection medium lacking His, the yeast cells are able to grow only when BD-NR1 and an AD-B subunit interact and reconstitute the transcription factor needed for growth in the absence of His. His auxotrophy was restored in cells cotransformed with BD-NR1 and AD-B55α. Growth experiments were repeated in triplicate and showed reconstituted growth when the BD-NR1 and AD-B55α were coexpressed, whereas controls with empty AD vector showed nearly no growth, and controls with empty BD vector showed some retarded growth only. Relative growth was measured by incubating 5 μL of yeast cultures on the agar medium for 48 h, the colonies were resuspended in synthetic dropout medium lacking His, Trp, Leu (SD/−HTL), and the absorbance was read at 600 nm (Fig. 5). Similar results were obtained for BD-NR1 and AD-B55β, however, due to high autoactivating for the BD-empty control plus AD-B55β coexpression, the assay was nonconclusive for B55β.
Mutations in B55 Subunits Affect NR Activity State
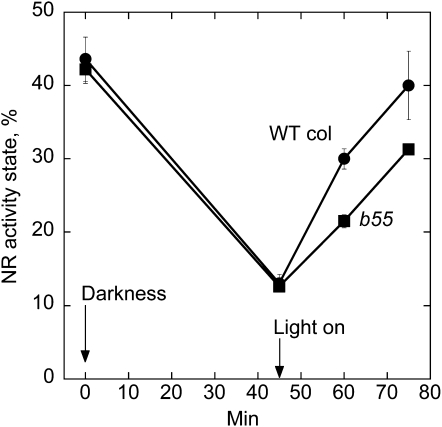
The T-DNA single-mutant lines of B55α or B55β were not different from wild type regarding NR activation (Supplemental Fig. S5). We did not succeed in obtaining a homozygous double mutant, most likely due to lethality. An additional complicating factor was that the Bα and Bβ genes are situated on the same chromosome. The homozygous single-mutant lines were crossed, and the F1 progeny was selfed. When two genes are linked it is easier to find double mutants in the F3 generation after suitable selection of crossover between the two mutated alleles (Weigel and Glazebrook, 2002). First two plants homozygous for the Bβ mutation and heterozygous for the Bα mutation were selected in the F2 progeny. In these two plants a crossing over had taken place, leading to mutated Bβ and Bα on one chromosome, whereas the sister chromosome was mutated in Bβ only. Selfing of such plant should give 25% homozygous double-mutant plants in the F3 generation (if viable), but none appeared among 30 plants first tested. Therefore, plants heterozygous for T-DNA insert in B55α and homozygous for T-DNA insert in B55β were propagated, and identified in each generation by PCR. In total approximately 100 plants were tested, but a homozygous double mutant never appeared, which confirmed the assumption that such homozygous mutations would be lethal. See Supplemental Figure S1 for details. All seeds from the selfed plants germinated and seedlings were heterozygous, showing that lethality would have occurred prior to seed formation, i.e. a homozygous double mutant would likely be embryo lethal. To test for a possible dose effect of B55 on NR activation, plants heterozygous for B55α insert and homozygous for Bβ insert were identified and used to test for NR activation (Fig. 6; Supplemental Fig. S6). In these plants a reproducible and significant decreased NR activation rate was found. NR activity state increased from 13% to 30% in wild type (four intact B55 alleles), and from 13% to 21% in the mutant (one intact B55 allele).
Figure 6.
NR activation rate is reduced in low B55 dosage plants. Plants were grown in 16-h light/8-h darkness for 3 weeks. Three hours into the photoperiod when NR activity was at a high level samples were harvested before subjected to darkness. Samples were then harvested after 45 min of darkness. Plants were transferred back to light for activation of NR, and sampled after 15 and 30 min of light (150 μmol m−2 s−1). Data are means of five samples at 0, 45, and 60 min, and three samples at 75 min. se is given. The plants used were first determined by PCR to be heterozygous for the mutation T-DNA insert in Bα and homozygous for insert in Bβ (homozygosis for insert in both genes appeared lethal, see text).
DISCUSSION
Based on previous studies with inhibitors, it was anticipated that PP2A would be important for dephosphorylation and hence activation of NR in response to dark/light transitions of plants (MacKintosh, 1992; Huber et al., 2002). In agreement with this hypothesis knockdown of all three A (scaffolding) subunits by amiRNA strongly impaired NR activation hence the involvement of PP2A in NR activation in situ was firmly established (Fig. 1). Complete abolishment of expression is, however, difficult to achieve by inducible amiRNA, and a low background activity of subunit A expression could therefore possibly be responsible for creating the slow NR activation observed (Fig. 1). Some residual activity due to a complex of the catalytic PP2A subunits with B55, but devoid of A subunits, as has been found for yeast PP2A (Koren et al., 2004) cannot be excluded. Alternatively, a different protein phosphatase, not yet identified, may additionally be involved. It was previously found that PP2A activity, as measured by phosphohistone dephosphorylation, in extracts from the rcn1 mutant was reduced by about 50% (Zhou et al., 2004). However, NR activation was not impaired in the rcn1 mutant. NR activation was lowered in the pp2aa2 pp2aa3 double mutant, showing that the PP2AA2 and PP2AA3 subunits were necessary for full NR activation, but since activation of NR was still taking place to a high degree, RCN1 could apparently substitute for PP2AA2 and PP2AA3. In conclusion the results are in agreement with NR being activated by PP2A complexes containing different structural A subunits.
Clearly, the B55α subunit interacted with NR as shown by BiFC and the yeast two-hybrid assay, and is thereby a candidate for targeting a PP2A complex to NR. B55α interacted with different domains of NR. Two of the domains tested, i.e. the N-terminal 100 amino acids, and the FAD-binding domain, do not contain the known regulatory, phosphorylation site. This implies that interaction between B55 and NR does not require the conserved motif in hinge1. This is in agreement with the knowledge that short motifs are recognized by protein kinases, whereas phosphatases do not appear to recognize consensus motifs within their substrates (Virshup and Shenolikar, 2009). Interestingly, the N-terminal end, which interacted with B55α in the BiFC test, was previously pointed out as important for regulation of NR from Nicotiana plumbaginifolia (Nussaume et al., 1995; Provan et al., 2000).
We did not find any effect on NR activity when comparing B55α or B55β single mutant with wild-type plants. Since B55α and B55β are close homologs, i.e. 87% similar, they may have overlapping functions. The single mutants were crossed, but interestingly a mutant homozygous for T-DNA inserts in both genes could not be obtained and is most likely lethal. This showed that the B55 subunits are essential and not replaceable by the other B (B’ and B’’) subunits. By propagating the double mutant through plants homozygous for insertion in Bβ and heterozygous for insertion in Bα, and identifying these plants in every generation a dosage effect of the B55 genes on NR activation was shown, confirming that B55 is indeed involved in activation of NR. Single mutants of several B’ and B’’ had been tested without finding effects on NR activity, thereby excluding each for being solely responsible for NR activation. However, a contribution from some of the B’ and B’’ subunits in targeting PP2A to NR cannot be excluded. By testing several PP2A-like phosphatases it was also recognized that none of these were exclusively responsible for PP2A interaction with NR, but again some contribution to NR activation is possible.
In conclusion, the results showed that PP2A structural genes are essential for activation of NR in situ after a dark-to-light transition. Furthermore, the regulatory B55 subunit genes are required for full activation rate of NR. Further work will include analysis of the posttranslational modifications of the subunits in response to light and photosynthesis.
MATERIALS AND METHODS
Plant Growth
Seeds for inducible knockdown of all three A subunits of PP2A were provided by Professor Jiri Friml (Ghent University, Belgium). Two different amiRNAs had been used to construct lines with inducible overexpression of amiRNA (amiRNA-1, amiRNA-2) that simultaneously targeted all three A subunit genes of PP2A (Schwab et al., 2006; Michniewicz et al., 2007). Seeds with loss-of-function mutations in the RCN1, PP2AA2, and PP2AA3 genes were provided by Professor Alison DeLong (Brown University, RI). The rcn1 and pp2aa2-1 pp2aa3-1 double mutants were used (Zhou et al., 2004). Arabidopsis (Arabidopsis thaliana) mutant lines SALK_062514 (B55β), SALK_095004 (B55α), SALK_055335 (BSL2), SALK_088206C (BSL1), and SALK_071689C (BSL3) were from the Ecker collection (Alonso et al., 2003) and obtained through the European Arabidopsis Stock Centre (Nottingham). Homozygous mutant selection was done by PCR using primers for T-DNA insertion lines recommended by the SALK institute Web site SIGnAL (http://signal.salk.edu/tdnaprimers.2.html). For the B55α × B55β double mutant, plants heterozygous for the B55α mutation and homozygous for the B55β mutation were identified (Supplemental Fig. S1). To confirm that B55 mutant lines are true knockouts they were verified with reverse transcription (RT)-PCR using gene-specific primers (Supplemental Fig. S1).
Seeds from PP5 overexpressor plants (PAPP5-OX1) and loss-of-function mutant (papp5-1) were provided by Professor Hong Gil Nam (Pohang University, South Korea).
Small (2 weeks after sowing) amiRNA plants were induced by tamoxifen/estradiol treatment. Plants were sprayed every day for 1 week with 10 μm tamoxifen and 50 μm estradiol (Sigma-Aldrich).
The seeds were stratified at 4°C for 1 to 3 d and then transferred to the green house. During germination and growth plants were placed at 20°C under artificial light in short days (8-h light/16-h dark) or long days (16-h light/8-h dark) or 12-h light/12-h dark regimens.
Cell Culture
Arabidopsis (Col) cell suspension was cultured in JPL medium as described by Droillard et al. (2000) at 22°C in constant light. Cells were subcultured every week and harvested 3 to 4 d after subculture. Protoplasts were prepared using 1% w/v cellulase RS (Yakult) and 0.2% Macerozyme R-10 (Yakult; Boudsocq et al., 2007).
Assay of NR Activity
Plants were grown in soil. Rosette leaves (0.2–0.3 g) were homogenized in a mortar with cold, 800 to 1,000 μL extraction buffer 100 mm HEPES-KOH, 1 mm EDTA, 7 mm cystein. The extract was centrifuged for 2 min, and the supernatant, 50 μL, was assayed in 700 μL assay volume with 200 μm NADH and generally 2 mm EDTA to assay total NR activity (nonphosphorylated plus phosphorylated NR). To measure actual activity (nonphosphorylated NR) NR was tested in the presence of 5 mm MgCl2. The assay was run at 25°C for 10 min. Nitrite formed was determined by addition of 700 μL 1% sulphanilamide and 0.02% N-(naphtyl)-ethylene-diamine dihydrochloride in 1.5 n HCl, and read spectrophotometrically at 540 nm. Absorbance of 1 corresponds with 27 μmol NO2− formed per g fresh weight per h. NR activity state was defined by the ratio of the actual activity (Mg assay) to the total NR activity (EDTA assay) multiplied by 100 (MacKintosh et al., 1995).
Gene Cloning for in Planta Expression
All cDNAs were cloned into pWEN18, pWEN 25, pWEN-NY, and pWEN-CY in the same way: Full-length coding sequences of NR1, NR2, Bα, Bβ, and other B’ and B’’ subunits were PCR amplified from the cDNAs in pCRScript using the primer pairs listed in Supplemental Table S1. The PCR fragments were then cloned into pCRScript (Stratagene) before digestion with the restriction enzymes (Supplemental Table S1) and ligation into the corresponding restriction sites of pWEN18, pWEN-CY, or pWEN-NY as N-terminal fusions or pWEN25 as a C-terminal fusion. The vectors pWEN18 and pWEN25 have previously been described by Kost et al. (1998), and these vectors were modified by Fujiwara et al. (2004). pWEN18 provided in planta expression of B-EYFP (free N terminus of B), and pWEN25 provided expression of EYFP-B (free C terminus of B). The vectors pWEN-CY or pWEN-NY used for the BiFC have previously been described by Maple et al. (2005).
Transformation and Fluorescence Microscopy
Arabidopsis (Col) protoplasts, prepared from suspension culture, were transiently transformed using the polyethylene glycol transformation protocol as described by Droillard et al. (2000). For the BiFC in Arabidopsis mesophyll protoplasts plasmids for interacting proteins as well as negative control plasmids were cotransformed into protoplasts using polyethylene glycol transformation protocols described by Sheen (http://genetics.mgh.harvard.edu/sheenweb/). Microscopy was carried out on a Nikon TE-2000U fluorescence microscope equipped with an Exfo X-cite 120 fluorescence illumination system and filters and a special red chlorophyll autofluorescence filter set (exciter HQ630/39, emitter HQ680/40; Chroma Technologies). Images were captured using a Hamamatsu Orca ER 1394 cooled CCD camera and using Openlab and Velocity II (Improvision). Images were subsequently processed for optimal presentation with Adobe Photoshop version 7.0 (Adobe Systems).
Yeast Two-Hybrid Assay
The assay was performed according to Maple et al. (2005). Yeast (Saccharomyces cerevesiae) strain AH109 (MATa, trp-901, leu2-112, ura3-52, i, gal4D, gal80D), and plasmids pGADT7 and pGBKT7 encoding the Gal4 activation domain and the Gal4 DNA-binding domain, respectively, were derived from MATCHMAKER two-hybrid system version 3 (Clontech Laboratories).
The plasmids pGADT7 and pGBKT7 encoding Gal4 AD (activating domain) and Gal4 DNA-BD (binding domain), were used for protein-protein interaction studies in yeast (MATCHMAKER two-hybrid system version 3, Clontech Laboratories). The NR1 construct was cloned into pGBKT7 by the primers: forward ATCATATGGCGACCTCCGTCGATAACCG, reverse ATCCCGGGTTACTAGAAGATTAAGAGATCCTCC; and Bα was cloned into pGADT7 using primers, forward TACATATGAACGGTGGTGAT GAGGTCGTC, and reverse ATATCCCGGGTTAAGCATAGTACATGTACAAGC. Yeast HF7c cells were cotransformed with the resulting plasmids according to the manufacturer’s instructions (Clontech). After 5 d of incubation at 30°C single colonies were inoculated in SD/−TL and grown at 30°C in a shaking incubator for one night. Cultures were grown to OD600 of 1.0 and 5 μL spotted onto SD/−TL plates, or SD/−HTL to monitor HIS3 reporter expression by cell growth. Plates were incubated at 30°C for 48 h, each spot was resuspended in SD/−HTL medium, and the OD600 measured. This OD was taken as a measure of relative cell growth.
RT-PCR
Total RNA was isolated using RNeasy plant mini kit (Qiagen), and cDNA synthesized using the high-capacity cDNA archive kit (Applied Biosystems). Real-time PCR reactions were assayed using an ABI 7300 fast real-time PCR system. SYBR green real-time RT-PCR was carried out in triplicate with SYBR green PCR master mix (Applied Biosystems Inc.) using RCN1 primers, At1g25490LP1 and RP1, and βTUBULIN4 primers described in Supplemental Table S1. The relative expression (arbitrary units) of the RCN1 gene was obtained by dividing by the reference βTUBULIN4 gene expression. No-reverse-transcriptase and no-template controls were included as negative controls for each set of reactions. Two independent biological repeats gave similar results.
RT-PCR on amiRNA mutants was performed on tamoxifen-treated mutant and wild-type plants using primers for all three A subunits using primers At1g13320LP and RP, At3g25800LP and RP, At1G25490LP and RP (see Supplemental Table S1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Selecting double mutant At1g51690 × At1g17720.
Supplemental Figure S2. NR activity state after a darkness to light transition of plants with mutations in various PP2A-like protein phosphatases.
Supplemental Figure S3. Assays of NR in the presence of EDTA or Mg2+ used to calculate NR activity state.
Supplemental Figure S4. NR domains and B55 tested for interaction by bimolecular fluorescence complementation.
Supplemental Figure S5. NR activation rate in the single mutants b55α and b55β.
Supplemental Figure S6. Assays of NR in the presence of EDTA or Mg2+ used to calculate NR activity state for wild type and b55.
Supplemental Table S1. Primers used for cloning and RT-PCR.
References
- Ahn CS, Han JA, Lee H-S, Lee S, Pai H-S. (2011) The PP2A regulatory subunit Tap46, a component of the TOR signalling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo B-C, Harmon AC, Huber SC. (1996) Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell 8: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. (2000) Biochemistry & Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD [Google Scholar]
- Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D. (2002) The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell WH. (1999) Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol 50: 277–303 [DOI] [PubMed] [Google Scholar]
- DeLong A. (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol 9: 470–477 [DOI] [PubMed] [Google Scholar]
- Douglas P, Morrice N, MacKintosh C. (1995) Identification of a regulatory phosphorylation site in the hinge 1 region of nitrate reductase from spinach (Spinacea oleracea) leaves. FEBS Lett 377: 113–117 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Thibivilliers S, Cazalé AC, Barbier-Brygoo H, Laurière C. (2000) Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: two crossroad MAP kinases and one osmoregulation-specific protein kinase. FEBS Lett 474: 217–222 [DOI] [PubMed] [Google Scholar]
- Eichhorn PJA, Creyghton MP, Bernards R. (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795: 1–15 [DOI] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Fujiwara MT, Nakamura A, Itoh R, Shimada Y, Yoshida S, Møller SG. (2004) Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J Cell Sci 117: 2399–2410 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Huber JL, Redinbaugh MG, Huber SC, Campbell WH. (1994) Regulation of maize leaf nitrate reductase activity involves both gene expression and protein phosphorylation. Plant Physiol 106: 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, MacKintosh C, Kaiser WM. (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol Biol 50: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Janssens V, Longin S, Goris J. (2008) PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem Sci 33: 113–121 [DOI] [PubMed] [Google Scholar]
- Jonassen EM, Lea US, Lillo C. (2008) HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 227: 559–564 [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Brendle-Behnisch E. (1991) Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis. Plant Physiol 96: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Kang J-G, Yang S-S, Chung K-S, Song P-S, Park C-M. (2002) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren R, Rainis L, Kleinberger T. (2004) The scaffolding A/Tpd3 subunit and high phosphatase activity are dispensable for Cdc55 function in the Saccharomyces cerevisiae spindle checkpoint and in cytokinesis. J Biol Chem 279: 48598–48606 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay Y-F. (2010) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13: 1–8 [DOI] [PubMed] [Google Scholar]
- Lea US, Leydecker M-T, Quilleré I, Meyer C, Lillo C. (2006) Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol 140: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Virshup DM. (2002) Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur J Biochem 269: 546–552 [DOI] [PubMed] [Google Scholar]
- Lillo C. (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Lillo C, Smith LH, Nimmo HG, Wilkins MB. (1996) Regulation of nitrate reductase and phosphoenol carboxylase activities in barley leaf protoplasts. Planta 200: 181–185 [Google Scholar]
- MacKintosh C. (1992) Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochim Biophys Acta 1137: 121–126 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Douglas P, Lillo C. (1995) Identification of a protein that inhibits the phosphorylated form of nitrate reductase from spinach leaves. Plant Physiol 107: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Møller SG. (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Matre P, Meyer C, Lillo C. (2009) Diversity in subcellular targeting of the PP2A B’η subfamily members. Planta 230: 935–945 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. (2009) Regulated activity of PP2A-B55 δ is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J 28: 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Vincentz M, Meyer C, Boutin J-P, Caboche M. (1995) Post-transcriptional regulation of nitrate reductase by light is abolished by an N-terminal deletion. Plant Cell 7: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan F, Aksland L-M, Meyer C, Lillo C. (2000) Deletion of the nitrate reductase N-terminal domain still allows binding of 14-3-3 proteins but affects their inhibitory properties. Plant Physiol 123: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol G, Baskin TI, Casamayor A, Cortadellas N, Ferrer A, Ariño J. (2000) The Arabidopsis thaliana PPX/PP4 phosphatases: molecular cloning and structural organization of the genes and immunolocalization of the proteins to plastids. Plant Mol Biol 44: 499–511 [DOI] [PubMed] [Google Scholar]
- Ryu JS, Kim J-I, Kunkel T, Kim BC, Cho DS, Hong SH, Kim S-H, Fernández AP, Kim Y, Alonso JM, et al. (2005) Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120: 395–406 [DOI] [PubMed] [Google Scholar]
- Schmitz MHA, Held M, Janssens V, Hutchins JRA, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, et al. (2010) Live-cell imaging RNAi screen identifies PP2A-B55α and importin-β1 as key mitotic exit regulators in human cells. Nat Cell Biol 12: 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E. (2001) Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal 13: 7–16 [DOI] [PubMed] [Google Scholar]
- Terol J, Bargues M, Carrasco P, Pérez-Alonso M, Paricio N. (2002) Molecular characterization and evolution of the protein phosphatase 2A B’ regulatory subunit family in plants. Plant Physiol 129: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol Cell 33: 537–545 [DOI] [PubMed] [Google Scholar]
- Wang H, Chevalier D, Larue C, Cho SK, Walker JC. (2007) The protein phosphatases and protein kinases of Arabidopsis thaliana. The Arabidopsis Book 5: e0106, doi/10.1199/tab.0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Genetic analysis of mutants. Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 41–55 [Google Scholar]
- Zhou H-W, Nussbaumer C, Chao Y, DeLong A. (2004) Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]