Abstract
Mutagenized populations have become indispensable resources for introducing variation and studying gene function in plant genomics research. In this study, fast neutron (FN) radiation was used to induce deletion mutations in the soybean (Glycine max) genome. Approximately 120,000 soybean seeds were exposed to FN radiation doses of up to 32 Gray units to develop over 23,000 independent M2 lines. Here, we demonstrate the utility of this population for phenotypic screening and associated genomic characterization of striking and agronomically important traits. Plant variation was cataloged for seed composition, maturity, morphology, pigmentation, and nodulation traits. Mutants that showed significant increases or decreases in seed protein and oil content across multiple generations and environments were identified. The application of comparative genomic hybridization (CGH) to lesion-induced mutants for deletion mapping was validated on a midoleate x-ray mutant, M23, with a known FAD2-1A (for fatty acid desaturase) gene deletion. Using CGH, a subset of mutants was characterized, revealing deletion regions and candidate genes associated with phenotypes of interest. Exome resequencing and sequencing of PCR products confirmed FN-induced deletions detected by CGH. Beyond characterization of soybean FN mutants, this study demonstrates the utility of CGH, exome sequence capture, and next-generation sequencing approaches for analyses of mutant plant genomes. We present this FN mutant soybean population as a valuable public resource for future genetic screens and functional genomics research.
The release of whole genome sequences in crops such as soybean (Glycine max; Schmutz et al., 2010) marks a new era for genomics research in crop species. Soybean is one of the most valued crops for its ability to fix nitrogen and provide seed protein and oil. Resources to study gene function in this important species are needed, and using mutagenesis to develop population resources has long proven to be a key step for identifying gene function in many organisms.
A number of mutagen sources exist for introducing genomic variation. These include chemical, radiation, and transformation-induced mutagenesis of plant genomes (Østergaard and Yanofsky, 2004; Waugh et al., 2006; Kuromori et al., 2009). Each of these methods results in a signature footprint of structural variation across the genome (Alonso and Ecker, 2006). Fast neutron (FN) radiation is a particularly promising source of mutagenesis due to the potential to create deletions in a wide range of sizes (Li and Zhang, 2002) for gene knockouts and disruptions.
FN radiation has been used to induce mutations for many decades and has been shown to be an effective mutagen in plants (Koornneef et al., 1982). The majority of mutations that result from FN bombardment are DNA deletions that range in size from a few base pairs to several megabases (Li et al., 2001; Men et al., 2002). Precedence exists in many species, including Arabidopsis (Arabidopsis thaliana; Alonso et al., 2003), Medicago truncatula (Oldroyd and Long, 2003), Glycine soja (Searle et al., 2003), barley (Hordeum vulgare; Zhang et al., 2006b), and Lotus japonicus (Hoffmann et al., 2007), for the use of FN mutagenesis in forward genetic screens. Many phenotype-associated genes have been successfully identified and cloned through such screens (Meinke et al., 2003).
Recent years have seen renewed interest in FN mutagenesis, and the advent of whole genome technologies has increased the capacity for mutant screening. Comparative genomic hybridization (CGH) microarrays and high-throughput next-generation sequencing (NGS) platforms are powerful tools for the analysis of copy number changes, polymorphisms, and structural variation in the genome. CGH compares DNA hybridization intensities from each genomic sample against a set of fixed probes and is capable of detecting regional copy number changes, such as deletions and duplications (Carter, 2007). NGS can be used to map deletions and insertions, polymorphisms, inversions, and translocations (Medvedev et al., 2009). Coupling whole exome sequence capture with high-throughput NGS can target genes for selective resequencing. Previous studies have shown the utility of each of these approaches to identify and locate genomic changes (Sebat et al., 2004; Hodges et al., 2007; Korbel et al., 2007; Choi et al., 2009). Application of these technologies to a FN mutant population brings detection of gene deletions and phenome-based genetic screens to the whole genome level.
In this study, we describe the development of a FN mutant population resource in soybean. This population was used to screen for seed composition, maturity, morphology, pigmentation, and nodulation mutants. Phenotypes observed in the FN mutant population are described, and deletions within mutated soybean genomes of interest are characterized. We show the promise of the FN mutant population as a community resource and demonstrate the utility of CGH and NGS methods for performing informative genome analyses on such a population.
RESULTS
Soybean FN Population Resource Development
Approximately 60,000 soybean seeds of cv M92-220 were irradiated with FNs in the first round of mutagenesis. One-quarter (15,000) of the seeds were irradiated at each of the following doses: 4, 8, 16, and 32 Gray units (Gy). In the first season, 20,000 M1 seeds, 5,000 from each dose, were planted in two locations and harvested by single-seed descent. An additional 60,000 seeds were irradiated in a second round of mutagenesis, half at 16 Gy and half at 32 Gy, for greater representation in the higher doses, and seeds were also harvested by single-seed descent. Independent M2 plants were propagated in three locations, and seed was harvested from individual plants. Statistics on dose comparisons for emergence, survival, and propagation percentages were compiled (Table I). As expected, seeds exposed to greater FN radiation doses resulted in fewer emerged M1 plants in the field and fewer individuals that produced viable seed at the end of the season. In the M2 generation, radiation dose also correlated inversely to the number of mutant lines that produced seed but did not affect plant emergence in the same manner. As the number of loci that are involved in overall plant development and reproduction are far greater than those limited to plant emergence, this result was not surprising. Since mutant phenotypes in the M2 generation are more likely due to heritable effects, as opposed to mutant phenotypes in the M1 generation that may be due to physical damage from FN bombardment, M2 plant leaf tissue and phenotypes were collected and cataloged. Corresponding M3 seed was collected from independent M2 individuals to form the basis of a FN mutant library resource.
Table I. Summary statistics for soybean mutant viability as a function of FN radiation dose.
The M2 generation was grown in Santiago, Chile, as single-seed descent from M1 plants grown in St. Paul, Minnesota.
| Radiation Dose | M1 Plants Emerged | M1 Plants Producing M2 Seed | M2 Plants Emerged | M2 Plants Producing M3 Seed |
| % | ||||
| 4 Gy | 76.24 | 48.98 | 88.00 | 80.00 |
| 8 Gy | 73.14 | 33.80 | 69.67 | 57.33 |
| 16 Gy | 69.22 | 23.36 | 66.58 | 56.83 |
| 32 Gy | 61.04 | 15.48 | 74.40 | 54.80 |
Forward Screen for Seed Composition Mutants
An immediate function for the FN soybean population was to serve as a forward genetics resource for seed composition mutants. To this end, 10,000 M3 lines were screened by near infrared (NIR) spectroscopy for seed protein or oil percentage of total seed composition. Additional preliminary data on carbohydrates and fatty acid and amino acid composition were also determined for a subset of samples. The mean value and detected range for seed composition components were assayed on seed harvested from three different locations (Supplemental Table S1) and showed significant increases and decreases in seed protein and oil within the FN mutant population.
A subset of mutant families consistently displayed increased or decreased seed protein or oil composition in subsequent generations. Eight seed protein and oil mutants, designated as “PO” mutants, were chosen primarily based on prior repeat performances in year-to-year rankings. PO1, PO3, and PO8 are mutants that displayed high seed protein phenotypes and ranked fourth, second, and first, respectively, among mutants in the M4 generation. PO3 also exhibited chimeric leaf pigmentation. PO2 possessed the second highest seed oil content detected in the M4 seed. PO4 and PO5 exhibited the lowest seed oil content, and PO4 also showed high seed raffinose and Suc (0.82% and 8.29% of total seed carbohydrate, respectively) content. PO6 exhibited the second lowest seed protein content in the M3 generation and the lowest seed protein content in the M4 generation. PO7 exhibited the highest seed oil content in the M3 and M4 generations. PO3 showed an increase in combined seed protein and oil content compared with M92-220, and PO7, PO2, and PO8 also showed minor increases in combined seed protein and oil content.
The protein and oil composition of M6 seed from the 2010 season for mutants PO1 to PO8 are displayed (Table II). High seed protein and low seed protein as well as low seed oil phenotypes were confirmed in 2010. However, the selected high-seed oil phenotypes were not observed in the 2010 season, and we attribute this variation to growing season conditions. Several seed protein and oil content mutants displayed maturity differences; however, maturity date did not correlate with a fixed trend in high or low seed protein or oil.
Table II. Seed protein and oil composition data by NIR spectroscopy on M6 seed from eight soybean FN mutant lines (PO1–PO8) compared with the wild-type M92-220 in 2010.
Mean and sd values are shown. Maturity rankings are on a five-point scale from earliest (5) to latest (1) dates of plant maturity.
| Identifier | Percentage Protein | Percentage Oil | Percentage (Protein + Oil) | Maturity |
| M92-220 | 39.16 ± 0.52 | 21.20 ± 0.39 | 60.36 ± 0.65 | 4 |
| PO1 | 46.99 ± 1.18 | 12.92 ± 1.19 | 59.91 ± 1.68 | 2 |
| PO2 | 40.42 ± 0.36 | 20.59 ± 0.11 | 61.01 ± 0.38 | 3 |
| PO3 | 49.31 ± 0.62 | 16.54 ± 0.32 | 65.84 ± 0.70 | 3 |
| PO4 | 43.63 ± 0.35 | 10.93 ± 0.47 | 54.57 ± 0.59 | 3 |
| PO5 | 42.37 ± 1.07 | 11.45 ± 0.54 | 53.82 ± 1.20 | 4 |
| PO6 | 35.73 ± 0.29 | 21.68 ± 0.30 | 57.41 ± 0.42 | 1 |
| PO7 | 42.39 ± 0.91 | 21.24 ± 0.22 | 63.63 ± 0.93 | 5 |
| PO8 | 46.72 ± 1.42 | 14.95 ± 0.30 | 61.66 ± 1.45 | 2 |
FN Mutants with Visual Phenotypes
Visual phenotypes were recorded for the FN mutant population. Over 500 independent individuals were observed to display an abnormal visual phenotype. Altered phenotypes were observed for approximately 2% of the soybean FN mutant population. Visual phenotypes observed in the field that were not attributed to disease were categorized under the areas of morphology, pigmentation, or maturity. Morphological phenotypes accounted for over 70% of the observed abnormal phenotypes. Pigmentation abnormalities were noted for over 30% of observed phenotypes, and maturity differences were recorded for around 15% of recorded observations. Some of the mutant lines displayed more than one documented phenotype. Most visual phenotypes were observed for mutants derived from seed treated with 16-Gy (51.7%) or 32-Gy (38.8%) FN radiation doses.
A subset of mutants representing a few of the visual phenotypes observed in the FN population are shown in Figure 1. These include a short trichome mutant (Fig. 1A), chimeric and yellow pigmentation mutants (Fig. 1, B and C), and a short-petiole mutant with crinkled, curled leaves (Fig. 1D). A separate screen for root and nodulation mutants was also conducted within the growth chamber using duplicate M2 seed. Among observed phenotypes during the root and nodulation phenotype screen were a nonnodulating mutant (Fig. 1F), a robust mutant with early pod set (Fig. 1G), and a hypernodulating mutant (Fig. 1H).
Figure 1.
Selected soybean FN mutants with visual phenotypes. A, A short-trichome mutant, VP1 (top), displays short trichomes compared with wild-type soybean (bottom). B, A chimeric mutant, VP3, with altered leaf pigmentation patterns is visible among a field of other soybean mutants with normal pigmentation. C, A row of petite yellow-tinged mutants (VP4). D, The short-petiole and curled, crinkled leaf mutant (VP5). E, Wild-type root and nodules are shown at 3 weeks post germination. F, Mutant RN1 displays abnormal shoot-root connection and does not appear to nodulate. G, Robust mutant RN2 displays elongated internodes compared with the wild type and later begins pod set precociously. H, Hypernodulating mutant RN3 displays increased nodulation compared with the wild type in E.
CGH Validation and Marker Development for a Known FAD2 Gene Deletion in the M23 Mutant
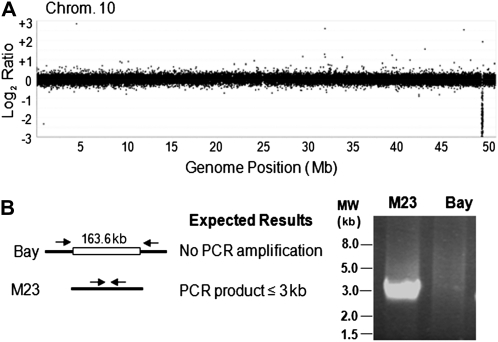
CGH possesses the potential to map deletions in mutant genomes. Accordingly, a custom NimbleGen soybean 700K-feature CGH microarray containing 696,139 unique soybean probes was designed. This design incorporated unique probes that were spaced approximately every 1,100 bp along the reference soybean genome sequence (www.phytozome.org). To validate the CGH method, genomic DNA from the M23 line was analyzed to verify a known ω-6 fatty acid desaturase (FAD2-1A) gene deletion. Increased oleate content in soybean oil improves its oxidative stability, thereby reducing the risk of producing trans-fatty acids from chemical hydrogenation of processed oil. Several soybean cultivars with increased oleate content are available, one of which is the midoleate mutant line M23 (Rahman et al., 1994) derived from x-ray mutagenized seed of the Bay soybean cultivar. Genetic studies indicated that the midoleate phenotype of M23 is associated with the deletion of FAD2-1A (Alt et al., 2005; Sandhu et al., 2007; Anai et al., 2008).
Given the agronomic importance of M23 in current breeding programs (Scherder et al., 2008) for increased seed oleate content, CGH could also be utilized to define the presence of other genetic lesions in the M23 genome, in addition to the previously reported FAD2-1A deletion. Based on the normalized log2 ratio of M23 to control (cv Bay) CGH data, a 163.6-kb copy number variation (CNV) event was detected on chromosome 10 where the M23 hybridization signal was approximately 4-fold less than in Bay, indicating a deletion (Fig. 2A). According to the reference soybean genome sequence, 20 annotated genes are predicted within the chromosome 10 deletion (Supplemental Table S2), including FAD2-1A (Glyma10g42470). No CNV was detected in the other characterized soybean ω-6 fatty acid desaturase genes FAD2-1B, FAD2-2A, FAD2-B, and FAD2-C (Schlueter et al., 2007; Pham et al., 2010; data not shown).
Figure 2.
Detection of a genomic deletion region encompassing a known deleted gene in M23. A, CGH analysis identifies a deletion at the end of chromosome 10. The y axis represents unaveraged log2 ratios of M23 to Bay hybridization signals. B, Confirmation of the predicted deletion by PCR. Arrows represent flanking amplification primers located 1.5 kb from the predicted 163.6-kb deletion.
To confirm the chromosome 10 deletion, the genomic region encompassing the deletion junction was amplified by PCR. Flanking primers used for amplification were approximately 1.5 kb from the predicted deletion. As expected, no amplification was detected when genomic DNA from wild-type Bay was used as a template, whereas a product of approximately 3.0 kb was obtained with M23 genomic DNA (Fig. 2B). Sequencing of the PCR product precisely mapped the deletion to base positions 49,369,546 to 49,533,559 of chromosome 10. Except for an extra nucleotide at the deletion junction, no sequence change was detected in the DNA regions immediately flanking the deletion. The PCR primers designed to amplify this region may be used as a molecular marker to select for the midoleate phenotype in segregating breeding populations.
Genomic Analyses of FN Mutants by CGH
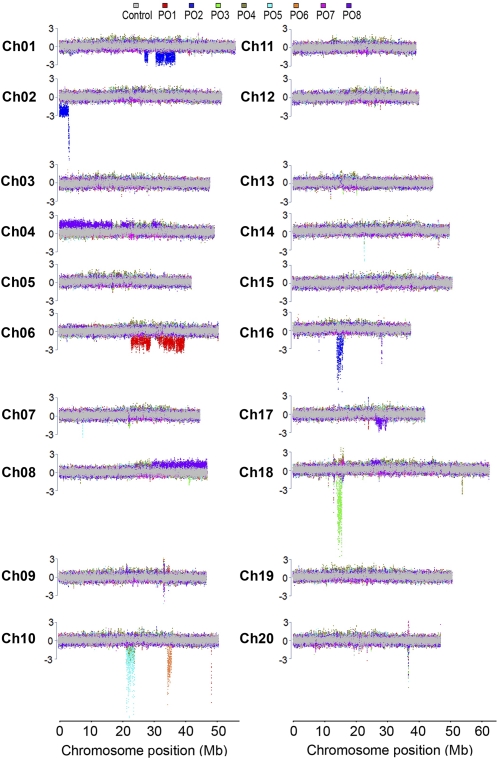
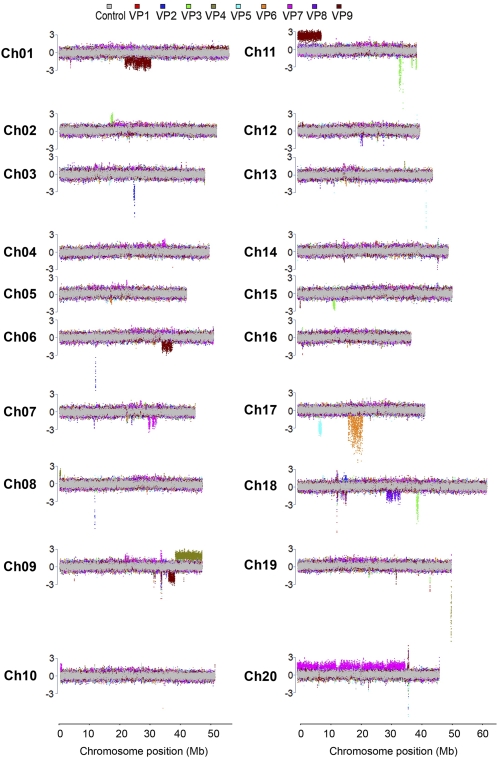
Many aberrant phenotypes were observed in the soybean FN mutant population and found to be heritable. These phenotypes were hypothesized to result from genomic changes within the mutant after exposure to FN radiation. To confirm and characterize mutations at the genomic level, we performed CGH using the custom NimbleGen soybean 700K-feature CGH microarray. Thirty microarray hybridizations were performed using genomic DNA from a subset of soybean FN mutants. These mutants were categorized under the following classes: eight seed protein and oil (PO), seven late maturity (LM), three early maturity (EM), three root and nodule (RN), and nine aboveground visual phenotype (VP) mutants. DNA copy number changes were readily detected in these lines. Full chromosome views of the normalized CGH log2 hybridization ratios for mutant versus control are shown (Figs. 3 and 4; Supplemental Figs. S1–S3).
Figure 3.
CNV events detected by CGH in soybean FN seed protein and oil mutants. Full chromosome views of CNV events are depicted for all 20 soybean chromosomes. The normalized log2 ratios of sample to control data are plotted as the median across 11 probe data points across chromosome positions. Results from each array are color coded for mutants PO1 through PO8. PO1 to PO3 and PO8 = high seed protein; PO4 = low seed oil and high seed raffinose and Suc; PO5 = low seed oil; PO6 = low seed protein; PO7 = high seed oil. The gray overlay represents CGH data from control versus control hybridization. Colored regions above and below the control regions potentially represent copy number change differences. The y axis scale is in terms of the number of sd from average, with the segment threshold for deletions or duplications at ±3.
Figure 4.
CNV events detected by CGH in soybean FN visual phenotype mutants. Full chromosome views of CNV events are depicted for all 20 soybean chromosomes. The normalized log2 ratios of sample to control data are plotted as the median across 11 probe data points across chromosome positions. Results from each array are color coded for mutants VP1 through VP9. VP1 and VP2, short trichomes; VP3, chimeric leaf pigmentation; VP4, petite and yellow leaf; VP5, short petiole and crinkled leaf; VP6, copper leaf; VP7, abnormal floral meristem development; VP8, fused trifoliates; VP9, thick, twisted petioles. The gray overlay represents CGH data from control versus control CGH. Colored regions above and below the control regions potentially represent copy number change differences. The y axis scale is in terms of the number of sd from average, with the segment threshold for deletions or duplications at ±3.
Eight seed protein and oil composition mutants were chosen for CGH analyses based on high or low seed protein or oil composition across multiple environments and generations (Table II). The M4 generation was used for CGH analyses, and high/low phenotypes were confirmed on the M5 seed harvested from an M4 plant. Genomic regions that exhibited changes in DNA copy number were detected in these soybean FN mutants and are collectively displayed in Figure 3.
Soybean FN mutants with aboveground visual phenotypes were also assayed by CGH to detect and map genomic changes. VP1 (Fig. 1A) and VP2 mutants were independently recovered, and both exhibited a short trichome phenotype. VP3 to VP5 pigmentation and short-petiole mutants (Fig. 1, B–D) were also assayed. In addition, a mutant with copper-colored leaves (VP6), a mutant with abnormal floral meristem development (VP7), a mutant showing fused trifoliates (VP8), and a mutant exhibiting thick, twisted petioles (VP9) were subjected to CGH for genomic analysis. DNA copy number changes detected by CGH for these visual phenotype mutant lines are displayed in Figure 4. Also shown are genomic regions with DNA copy number changes detected by CGH for late maturity, early maturity, and root and nodulation mutants (Supplemental Figs. S1–S3).
Microarray hybridizations were performed using genomic DNA from M2, M3, or M4 plant tissue. Of the 30 hybridization results, a subset of M4 seed composition and maturity mutant results were derived from a common M2 or M3 plant. Thus, several detected DNA copy number change regions coincided (Fig. 3; Supplemental Fig. S1) due to shared pedigree. These regions were on chromosome 10, at approximately 48.7 Mb (PO1, LM1, LM5) and approximately 23 Mb (PO4, PO5) . These results refer only to the relationship of specific mutant plants that were assayed by CGH and not to mutant lines in general that are unique.
Screening for False-Positive CNV Events in the Soybean FN Population
A number of genomic locations consistently exhibited CNV across many FN mutant genotypes. These can be observed in the multicolored groupings above or below the axis (Figs. 3 and 4; Supplemental Figs. S1–S3), for example, on chromosomes 9, 18, and 20. To further characterize and delineate these regions, we performed single-nucleotide polymorphism (SNP) genotyping across a subset of the soybean FN mutant population using the Illumina Goldengate SNP genotyping platform with 1,536 universal soybean SNP markers (Hyten et al., 2010). Twenty-four markers detected SNPs within the population on the following chromosomes: 3, 6, 9, 14, 16, 18, and 20 (Supplemental Table S3; Supplemental Fig. S4). These data indicate that there were several regions of genomic heterogeneity maintained among the M92-220 individuals within the population (bulked seed) that was exposed to FN mutagenesis. This type of intracultivar heterogeneity appears to be typical of many soybean accessions (Haun et al., 2011). Segregation of regions of genomic heterogeneity that include natural copy number polymorphisms may appear as false-positive FN-induced duplications or deletions in CGH CNV analyses.
We used a combination of CGH and SNP genotyping data to mask regions of genomic heterogeneity from FN CNV analysis. Microarray probes located within confirmed polymorphic SNP regions were removed from consideration as FN-induced polymorphisms. The degree of variation at each probe position across 30 CGH microarrays was visualized by calculating and plotting values that crossed the 95th percentile log2 CGH ratio threshold (Supplemental Fig. S4). Major peaks of variation and SNP locations coincided to confirm regions of genomic heterogeneity within the population and to differentiate these regions from DNA CNV arising from other sources (i.e. FN bombardment).
Genomic CNV Events Detected in FN Mutants
Analysis of CGH data revealed a total of 61 genomic DNA regions with CNV among the 30 lines tested. This number was calculated using a stringent threshold of segments ±3 sd from the mean normalized log2 ratio for the sample versus control. Of the 61 CNV events that passed the threshold, 52 were putative deletion regions (85.25%) and nine were putative duplication regions (14.75%). The average number of regions with CNV per mutant was 2.03, and the average number of deletion regions detected per mutant was 1.73.
Our stringent analyses criteria were designed to identify homozygous deletions. Several heterozygous or hemizygous events were potentially observed. For example, CGH variation detected in RN1 and VP7 to -9 did not pass the threshold, possibly due to allelic differences in genomic DNA from M2 generation individuals. In VP9, deviations from background and control hybridizations appear (Fig. 4, Ch01, -06, -09, and -11) near the threshold. Other putative heterozygous or hemizygous events were observed, for example, in Figure 4: VP4 on Ch09, VP5 on Ch17, and VP7 on Ch20. In comparison, candidate homozygous genomic deletions reach a greater log2 ratio amplitude (Fig. 3, Ch10 and Ch18). A mixture of these two deletion types is observed in PO2 (Fig. 3, Ch02).
Among the 61 genomic regions exhibiting significant CNV, putative deletions and duplications ranged in size from 986 bp to almost 3 Mb, and the mean size of a DNA copy number change region was around 367 kb. The mean and median sizes of the total detected DNA copy number change regions per mutant were 777 and 319 kb, respectively. A summary of the detected CNVs from 30 CGH microarrays for seed composition, maturity, root and nodule, and other visual phenotypes is shown (Supplemental Table S4). Detected CNV regions were found on every chromosome and distributed as follows: Ch01, 3; Ch02, 5; Ch03, 2; Ch04, 1; Ch05, 2; Ch06, 5; Ch07, 2; Ch08, 1; Ch09, 4; Ch10, 8; Ch11, 2; Ch12, 1; Ch13, 1; Ch14, 2; Ch15, 2; Ch16, 4; Ch17, 7; Ch18, 2; Ch19, 6; Ch20, 1.
The number of genes located within each of the 61 significant CNV events ranged from zero to 145. On average, 17 genes were found within each CNV event. A total of 1,048 genes, including 634 high-confidence genes, were found within all CNV events defined across the 30 CGH microarrays (Supplemental Table S5). For 130 of these genes, putative paralogous genes were found elsewhere in the genome. Over half of the detected CNV events occurred in pericentromeric regions, with 29 of 52 deletions and five of nine duplications. This finding may be expected, as approximately half of the genome space consists of pericentromeric regions. Importantly, deletion regions that contained single genes and mutants with as few as four predicted genes within all detected deletions were recovered.
Genomic Analysis of Soybean FN Mutants by Exome Resequencing
To confirm CGH-detected deletions in FN mutants, exome capture and resequencing was performed on four of the 30 soybean FN mutants assayed by CGH. Mutant DNA libraries were constructed and hybridized to a NimbleGen capture array designed to cover the soybean exome. The design comprises 69% of the Glyma 4.0 annotated coding sequence features, with total capture space that represents 52.3 Mb and targets 226,207 coding sequence features (Haun et al., 2011). DNA captured by the array was amplified and sequenced by high-throughput Illumina NGS technologies to carry out exome resequencing of the mutant DNA.
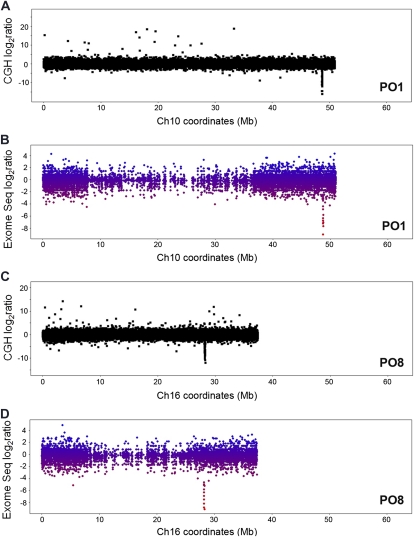
The exomes of four mutants, PO1, PO8, VP1, and VP5, were resequenced. Exon counts were normalized to the total counts in each sample and visualized to display deleted exons (Fig. 5). Single chromosome views for the main deletions detected by CGH are shown for PO1 and PO8 (Fig. 5, A and C). CGH analyses detected deletions on chromosome 10 at approximately 48.7 Mb in PO1 and on chromosome 16 at approximately 28.1 Mb in PO8. Exome resequencing confirmed these deletions, as there was no evidence for the existence of exon sequence in these respective regions in the mutant sample DNA, while the control sample did provide sequence reads that mapped to these regions (Fig. 5, B and D). Similarly, exome resequencing and whole genome paired-end mapping with NGS confirmed a CGH-detected deletion on chromosome 13 at approximately 42.3 Mb in VP5 (data not shown). Exome resequencing was performed on genomic DNA from a sibling of the short-trichome VP1 mutant assayed by CGH; this sibling also displayed the short-trichome phenotype. In addition, whole genome paired-end mapping using NGS was performed on VP1 (data not shown). The same deletion was detected on chromosome 5 at approximately 36.4 Mb by three different approaches, CGH, exome resequencing, and whole genome paired-end mapping through NGS, in two individuals with the VP1 short-trichome mutation.
Figure 5.
Exome resequencing confirms gene deletions detected by CGH. A, The corrected log2 ratios of sample PO1 to control intensities are shown for chromosome 10, where a deletion is detected at approximately 48.7 Mb. B, The normalized exome resequencing log2 ratios of sample PO1 to control exon counts are displayed for chromosome 10. Each colored dot represents an exon in a high-confidence gene call. The color gradient indicates the lowest (red) to highest (blue) amount of read count evidence for an exon in sample PO1 compared with the control. The absence of sequence evidence for exons at approximately 48.7 Mb is shown and parallels the deletion found by CGH in A. C, The corrected log2 ratios of sample PO8 to control intensities are shown for chromosome 16, where a deletion is detected at approximately 28.1 Mb. D, The normalized exome resequencing log2 ratios of sample PO8 to control exon counts are displayed for chromosome 10. Each colored dot represents an exon in a high-confidence gene call. The color gradient indicates the lowest (red) to highest (blue) amount of read count evidence for an exon in sample PO8 compared with the control. The absence of sequence evidence for exons at approximately 28.1 Mb is shown and parallels the deletion found by CGH in C.
Demarcation of FN Deletion Regions and Cosegregation of a Deletion with a Dominant Mutant Phenotype
A small deletion detected by both CGH and exome resequencing in VP1 contained a single gene (Glyma05g31280) encoding a tetratricopeptide repeat-containing chaperone-binding protein. This region was chosen for PCR confirmation. Primers were designed within the CGH probe sequences that flanked the deletion region, and PCR was performed under short extension conditions on VP1 versus wild-type M92-220 genomic DNA templates. A single 579-bp PCR product was obtained from VP1 and not seen in the wild-type control (Fig. 6A). Upon sequencing of the 579-bp product and alignment to the reference genome sequence, the exact break point sites for the deletion were determined to span chromosome 5 base positions 36,426,532 to 36,430,207 (Fig. 6B). A larger deletion of nearly 40 kb on chromosome 10 that was detected by CGH (Fig. 5A) and exome resequencing (Fig. 5B) was also confirmed by PCR in PO1 (Fig. 6, C and D).
Figure 6.
Demarcation and confirmation of deletion regions and cosegregating phenotypes by PCR. A, Agarose gel electrophoresis of the PCR product across a deletion region in short-trichome mutant VP1 next to 100-bp marker (M) and wild-type (WT) M92-220 template PCR control lanes. B, A diagram shows the reference sequence region length (4,254 bp) versus the VP1 mutant region length (579 bp) characterized by PCR amplification and sequencing of the region. C, Agarose gel electrophoresis of the PCR product across a deletion region in high-seed protein mutant PO1 next to 1-kb marker and wild-type M92-220 template PCR control lanes. D, A diagram depicts the reference sequence region length (39,806 bp) versus the PO1 mutant region length (approximately 1 kb) characterized by PCR amplification and sequencing of the region. E, A deletion region on chromosome 17 was confirmed by PCR and mapped in VP5. This genetic marker locus in VP5 cosegregates with progeny displaying the short-petiole phenotype (+) and is not found in the wild type or in progeny without the short-petiole phenotype (−) shown in F.
A putative heterozygous deletion on chromosome 17 detected by CGH and by whole genome paired-end mapping through NGS (data not shown) in VP5 was also confirmed by PCR. Sequencing of the PCR product (Fig. 6E) revealed an 837,919-bp deletion on chromosome 17. This deletion spans chromosome 17 base positions 7,770,585 to 6,932,666, encompasses 87 high-confidence genes, and interrupts a ubiquitin-specific proteinase gene at one end. M3 progeny of VP5 segregated approximately 3:1 for the short-petiole phenotype, indicative of a dominant mutant phenotype. The chromosome 17 deletion was found to cosegregate with the mutant phenotype (Fig. 6, E and F) in all 14 M3 progeny. We used CGH to genotype six of the M3 progeny (data not shown) and found three mutant individuals heterozygous for the chromosome 17 deletion. A total of 41 M4 individuals derived from the heterozygous M3 parents were scored for plant architecture and the chromosome 17 deletion. A perfect correlation was found between the presence of at least one copy of the chromosome 17 deletion and the short-petiole phenotype among the 30 mutant and 11 wild-type segregating individuals. These data suggest that the chromosome 17 hemizygous deletion may be sufficient to confer the mutant phenotype in a dominant fashion or is tightly linked to the dominant causative locus. Furthermore, these data provide evidence for the potential to detect associated loci and develop specific markers for mutant phenotypes within this soybean FN mutant population.
Soybean FN Mutant Database
The complete catalog of soybean FN M2 mutants from this study has been launched at http://www.soybase.org/mutants. FN mutant trait data, observed phenotype descriptors, and photographs are presented on this site along with parallel data from the unmutagenized wild-type M92-220. Currently, the soybean FN mutant database lists over 23,000 independent FN mutant lines. The original seed composition data on M3 seed from over 10,000 independent mutants are also displayed on the site.
Information compiled on the soybean FN mutant population is available for users to browse and search for recorded phenotypes of interest. A menu with key descriptors is provided to facilitate searches, and recorded trait ranges across the mutant population are displayed upon trait selection to assist in data filtering. Photographs of mutant plants and plant parts are presented in a user-friendly browser. Images of special-interest mutants chosen for further analysis may be accumulated during browsing to view multiple images on a single page, facilitating comparisons between mutants. Standardized soybean trait ontology tags are associated with each descriptor or measured feature. Seed availability is noted for each mutant line. Over 240 mutants with observed visual phenotypes have available seed. Bulk M3 and M4 seed from the soybean FN mutant population are also available.
FN mutant genomic analyses results, observations connected to the original M2 identifier, and seed status are continually updated in this dynamic database. Multichromosome diagrams and normalized log2 hybridization ratios for each CGH microarray experiment are available for viewing and download at the soybean FN population database. Sequence homology search capabilities through BLAST (Altschul et al., 1990) allow the user to find mutants with CNV events that cover a gene with nucleotide or protein sequence similarity to a sequence of interest. An added “mutant” track for FN mutants on the genome browser displays the locations of all CNV events defined in this study and leads to information on the genes within identified deletion and duplication regions.
DISCUSSION
In this study, we established a soybean FN mutant population and performed phenotypic and genomic analyses on mutants of phenotypic interest. We identified soybean FN mutants with seed composition phenotypes and mapped gene deletions within these mutants. Additional phenome analyses were performed by selecting mutants with observed visual phenotypes and identifying genomic CNV events. Notably, this study combines complementary genome-wide microarray-based and NGS technologies to map FN-induced deletions. This population is a resource for studying gene function, and the soybean FN mutant database provides open access to this resource.
Existing Mutant Resources
A number of resources currently exist for phenome analysis in plant species. Open databases are available for Arabidopsis (Sundaresan et al., 1995; Martienssen, 1998; Tzafrir et al., 2003, 2004; Kuromori et al., 2006), rice (Oryza sativa; Zhang et al., 2006a; Miyao et al., 2007; Larmande et al., 2008), maize (Zea mays; Fernandes et al., 2004; Lawrence et al., 2007), sorghum (Sorghum bicolor; Xin et al., 2008), barley (Caldwell et al., 2004), and tomato (Solanum lycopersicum; Menda et al., 2004), among others. A number of mutant population resource databases exist for legume species, including Tnt-1 (Tadege et al., 2008) and FN bombardment (Wang et al., 2006; Rogers et al., 2009) populations in M. truncatula, ethyl methanesulfonate TILLING populations in soybean (Cooper et al., 2008) and L. japonicus (Kuromori et al., 2009), and now a soybean FN mutant database composed of the mutants developed in this study. In terms of recovering viable M2 plants with observable mutant phenotypes, the most successful FN dose rates were 16 to 32 Gy for our soybean population. This was comparable to the optimized FN doses of 18 to 20 Gy reported for rice (Li et al., 2001) and 32.5 Gy reported for M. truncatula (Rogers et al., 2009). This study describes, to our knowledge, the assembly and characterization of the largest collection of soybean FN mutants to date accompanied by the only FN mutant database to display genome-wide coverage of deletion events in addition to recorded phenotypic traits.
Forward Genetic Screening and Prospects for Reverse Genetic Screening
Mutant populations are sources of increased genetic and phenotypic diversity. Three phenotypic categories, physical, chemical, and biological, have been proposed for the application of phenome analysis to mutant resources (Kuromori et al., 2009). In this study, we describe visual or physical phenotypes observed in the soybean FN mutant population as well as chemical phenotypes profiled by NIR spectroscopy of mutant seed. Biological conditional phenotypes, involving screens in the presence of different stresses or growth conditions, are also possible. Forward screens for drought resistance, soybean cyst nematode resistance, and yield have been initiated in the soybean FN mutant population. Additional seed composition and root and nodulation screens are also in progress.
FN mutant populations for reverse genetic screening have been created for Arabidopsis (Li et al., 2001), M. truncatula (Rogers et al., 2009), and rice (Wu et al., 2005). The resources established in this study provide the basis for future reverse genetic screening of genes of interest. A library of DNA isolated from unique M2 individuals is under development and will allow for the screening and recovery of mutants with DNA copy number changes at specific locations. Previous estimates for the number of mutants required for saturation mutagenesis of a plant genome were based on estimates calculated through the success rate of screening for single gene mutations (Li et al., 2001; Rogers et al., 2009) and prior estimated correlations with observed albino frequencies (Koornneef et al., 1982; Rogers et al., 2009). Through the use of a genome-wide CGH platform, we obtained a more comprehensive view of the number and size of deletions that are present within our soybean FN mutant lines. In a simplified calculation, if approximately 1,000 genes are deleted per 30 mutants, as seen in this study, our collection of over 20,000 mutants may provide 10× coverage of the soybean genome.
Large-scale reverse genetic screening is now feasible with the availability of the soybean genome sequence. PCR-based strategies, such as Deleteagene (Li et al., 2002) and De-TILLING (Rogers et al., 2009), designed to screen FN mutant populations for mutations at individual genes of interest may be facilitated in the future by screening pools of mutant DNA through paired-end NGS to first identify the location and size of potential deletions within the population. Through paired-end NGS of multiplexed mutant genomic DNA samples, we were able to detect deletions confirmed by CGH (data not shown). Once identified by NGS and deposited as putative deletions in a database, deletions of interest may then be chosen for confirmation by PCR. Such a strategy, combined with the pooling methods described previously (Rogers et al., 2009), may facilitate high-throughput reverse genetic screening. With a population size of over 23,000 mutants and the tools launched in this study for mutant genome characterization, the development of a tiled set of identified deletions and duplications within the mutants may be possible. The establishment of such a resource would be of great value for future research in soybean functional genomics.
Genomic Mutation Detection Methods
The NimbleGen 700K soybean CGH microarray was designed with unique probes spaced approximately every 1,100 bp along the soybean genome sequence, for a total of 696,139 probe sequences. This coverage allowed for preliminary whole genome information to be gained regarding the effect of FN radiation exposure on soybean genomic DNA. The resolution of the microarray platform and analysis method was limited in many regions to approximately 2 kb, due to the requirement for a CNV event to cover at least two adjoining probes. In reality, we were able to detect a 986-bp deletion because the spacing between probes in this case was closer than average at approximately 450 bp. It is also possible that a single deviant data point on the CGH array could represent a true deletion; however, such a region would not be identified using our stringent filtering criteria, and the exact break points for fragmented deletion borders may not be resolved using CGH alone. The size of FN-induced deletions defined in this study range from less than 1 kb to almost 3 Mb and expands the range of FN-induced deletions reported in plants (Li et al., 2001; Men et al., 2002). However, an underestimation of structural variation events, particularly of smaller deletions and duplications, is likely to occur.
To validate the use of CGH for detecting gene deletions, we performed CGH on the midoleate x-ray mutant M23 with a known FAD2-1 gene deletion. Through CGH, we delineated an x-ray-induced genomic deletion to an estimated size of 163.6 kb on chromosome 10, encompassing the FAD2-1A gene, and subsequent sequencing of the region showed that the deletion covered 164.01 kb. This control provided a proof of concept that CGH analysis can be reliably used to identify and develop markers for important deletion break points in the soybean genome.
The use of paired-end mapping NGS technologies to detect structural variations in the genome has been reported previously (Korbel et al., 2007). In this study, we combined paired-end mapping NGS with array-capture technologies to detect the presence and absence of gene exons in mutant versus control genomic DNA. Our use of NGS combined with exome capture and whole genome paired-end mapping confirmed CGH-detected deletions. Hybridization-based platforms are subject to many sources of error that may reduce the power of detecting polymorphisms, particularly for small features. The addition of NGS approaches allowed us to impose stringent parameters that, in conjunction with CGH, resulted in high-confidence calls, particularly for polymorphic genomic regions. The use of NGS paired-end technologies also adds the utility of mapping the insertion location of genomic duplications and translocations. As NGS costs decrease in the future, the cost per genotype will likely become comparable or preferable to CGH and enable the high-throughput genotyping of lesion-induced mutant populations.
Connecting Gene to Function
Genes detected in copy number change regions are candidate genes for the associated traits of interest. Assessment of existing annotations reveals potential genes that may play a role in the observed phenotype. For example, genes involved in regulation of protein metabolism (Glyma10g18310, encoding ubiquitin-conjugating enzyme) and proteolysis (Glyma10g19260, encoding Ser carboxypeptidase) are deleted in low-seed-oil mutants (PO4 and PO5; Supplemental Table S5). Transcripts for a gene encoding a ubiquitin-conjugating enzyme were previously observed to accumulate during seed development of a high-seed oil soybean cultivar (Wei et al., 2008). In addition, a gene involved in protein biosynthesis (Glyma02g03750, encoding threonyl-tRNA synthetase) is absent in a high-seed oil mutant (PO2). Examination of differentially accumulated transcripts in soybean near-isogenic lines also showed higher accumulation of a gene encoding a tRNA synthetase in the higher seed oil line LoPro (Bolon et al., 2010). Furthermore, a gene involved in lipid metabolism (Glyma10g26510, encoding lipase [class 3]) is deleted in a low-seed protein mutant (PO6). These observations may reflect changes in flux and relate to the inverse correlation between protein and oil levels observed in the soybean seed (Bolon et al., 2010).
Likewise, a detected deletion region in late maturity mutant LM6 contains genes (Glyma04g36620 and Glyma04g36630) encoding jumonji domain transcription factors with homology to Arabidopsis Relative of Early Flowering6 (REF6), a suppressor for the expression of a gene that delays flowering called Flowering Locus C (Noh et al., 2004). Deletion of the REF6 gene may explain or contribute to the late maturity phenotype observed in LM6. At least 15 genes involved in the regulation of transcription were found in deletion regions identified in this study. These include AP2, Dof, WRKY, helix-loop-helix, NAM, bZIP, and B3 domain transcription factors (Supplemental Table S5). Previously identified quantitative trait locus regions for seed protein, seed oil, and pod maturity exist on chromosomes with detected deletions. Additional mapping and follow-up studies will be required to determine whether these regions coincide.
Greater evidence for the existence of functional genes within FN mutant deletion regions may be found through the use of complementary genome resources. For example, examination of genes within deletion regions that possess minimum transcript accumulation evidence of at least 10 read counts in at least one tissue of the recently reported RNA-Seq soybean gene expression atlas (Libault et al., 2010; Severin et al., 2010) reduces the gene list from 1,048 to just over 200 genes. Of these genes, only approximately 150 do not possess paralogs elsewhere in the genome. Using such deduction methods, the potential pool of candidate genes may be condensed. The genome browser function at SoyBase (www.soybase.org/gbrowse; Grant et al., 2009) allows for direct comparison of the locations of soybean FN deletions and duplications detected in this study with soybean gene models, gene expression evidence, and duplicated gene segments.
The assembly of a FN mutant population in soybean facilitates the study of genes with loss-of-function phenotypes. Our findings support the utility of FN radiation as a mutagen to delete gene regions with tandem duplications. For example, 18 genes with F-box and WD domain annotations (Glyma15g19120–Glyma15g19290) were found within a deletion region detected in EM3 (Supplemental Table S5). Multiple Leu-rich repeat genes (Glyma16g27520–Glyma16g27560), aminotransferase-related genes (Glyma09g07320–Glyma09g07340), and pentatricopeptide repeat genes (Glyma16g27780–Glyma16g27800) are tandemly arrayed within deletion regions detected in EM1. The use of FN bombardment to delete genes facilitates the identification of genes that are not functionally redundant. Additional genomic and cytogenetic studies involving this population may also provide insight into the minimal necessary soybean genome.
Further analyses are required to fully characterize the phenotypes identified in this study and to confirm gene candidates. Map-based cloning and confirmation of candidate gene function by complementation have successfully been performed using FN legume mutants to characterize nodulation genes NSP2 (Oldroyd and Long, 2003; Searle et al., 2003; Kaló et al., 2005) and DMI1 (Ané et al., 2004) in M. truncatula and NTS-1 (Men et al., 2002) in G. soja. Microarray-based strategies together with bulked segregant analysis (Gong et al., 2004) or transcript profiling (Mitra et al., 2004) have also resulted in gene cloning from deletion mutants. Together with NGS technological advances, the utility of a FN mutant population may now be explored more rapidly and comprehensively than ever.
MATERIALS AND METHODS
Resource Development
Soybean (Glycine max) seeds of midmaturity group I cv M92-220, derived from the 2006 Crop Improvement Association seed stock of var MN1302 (Orf and Denny, 2004), were exposed to FN radiation doses of 4, 8, 16, and 32 Gy at the McClellan Nuclear Radiation Center at the University of California-Davis. M1 seed was planted and propagated by single-seed descent. M2 seed was planted in a grid format, and M2 plants were individually tagged with an assigned barcode identifier. Young leaf tissue was collected for each M2 plant, and M3 seed was single-plant harvested from more than 20,000 M2 plants. Seeds were analyzed for seed composition by NIR spectroscopy using a Perten DA7200 diode array instrument equipped with calibration equations developed by Perten in cooperation with the University of Minnesota.
SNP Genotyping
Genomic DNA samples from soybean leaf tissue were isolated using the Qiagen DNeasy kit protocol. DNA samples were then assayed on the Illumina Goldengate platform for genotyping using 1,536 soybean universal SNP BARC markers (Hyten et al., 2010). Polymorphic SNP markers across the soybean FN population were detected, and SNP markers were subject to BLAST (Altschul et al., 1990) analysis to recover the physical SNP position along the reference genome sequence.
CGH
CGH was performed as described previously (Haun et al., 2011) on the NimbleGen soybean CGH microarray, which consists of 696,139 unique oligonucleotide probes (50- to 70mers) designed from the reference Williams 82 sequence (Schmutz et al., 2010) and spaced at approximately 1.1-kb intervals (platform details can be found in Gene Expression Omnibus accession no. GPL11198 at http://www.ncbi.nlm.nih.gov/geo/). Mutant (Cy3 dye) and reference (Cy5 dye) labeling reactions were performed with 1 μg each of genomic DNA from mutant and pooled reference leaf tissue samples. Pooled reference samples consisted of DNA from 35 independent M92-220 mutant families. The same reference pool was used as the Cy5 dye in all hybridizations. Wild-type M92-220 and wild-type control pool samples were also labeled for comparison purposes against the pooled mutant reference. For the M23 control, M23 x-ray mutant DNA was labeled with Cy3 dye and cv Bay wild-type DNA was labeled with Cy5 dye. Labeled DNA was quantified and then hybridized for 72 h at 42°C on the CGH microarrays. PO1, LM1, and LM5 are related. PO6 and LM7 were M4 samples derived from the same M3 plant. PO4 and PO5 were M4 samples also derived from a common M3 individual. Supplemental Table S6 provides the database and pedigree key for the 30 FN mutant lines that were analyzed by CGH.
CGH Analysis of CNV Events
CGH data were analyzed using the Roche NimbleGen NimbleScan version 2.5 segMNT algorithm. Corrected log2 ratios were obtained for each probe data point. Probe segmentation and corrected log2 ratios were obtained for each probe data point essentially as described (Haun et al., 2011). Parameters were set for minimum segment lengths of two probes and segment log2 ratio differences of 0.1 between segments at the 0.999 acceptance percentile. Spatial correction and qspline normalization were applied. Significant copy number changes were determined by retrieving segments with an average corrected log2 ratio greater than the average plus 3 sd (increase) or less than the average minus 3 sd (decrease). If a gap between potential segments was less than half the size of the total distance covered by neighboring segments, then the entire region was considered a single CNV event. Final mutant deletion and addition regions were determined after filtering each CNV event against regions of natural heterogeneity determined through SNP genotyping and CGH analysis of mutants within the population. Genes (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v4.1/Gmax/annotation/Glyma1.gff2) that overlapped CNV events were determined using a custom Perl script. Paralogous genes in soybean were identified using BLAST (Altschul et al., 1990), DAGChainer (Haas et al., 2004), and selection of gene pairs from synteny blocks with average synonymous change rate values between 0.03 and 0.60.
CGH Analysis of Natural Genetic Heterogeneity within the Mutant Population
The sd from the average corrected log2 ratio of Cy3 (sample) to Cy5 (control) intensities was calculated at each probe position for each CGH array. After the average absolute value of the above was calculated for each probe position across 30 CGH arrays, the 95th percentile border (1.103589) was calculated across 696,139 unique probe positions, and the median value across each 11-probe sliding window was determined. Regions with median values that peaked above the 95th percentile border were candidate regions of genetic heterogeneity highlighted for further examination.
Exome Resequencing and Data Analysis
Genomic DNA preparations from four mutants, PO1, PO8, VP1, and VP5, were extracted using the Qiagen Plant DNeasy system. Library preparation, exome capture, amplification, and high-throughput sequencing of exome-captured libraries were performed as described previously (Haun et al., 2011). The Illumina Solexa 76-base paired-end short-read sequences were aligned to the soybean genome sequence version 4.1 (Gmax.main_genome.scaffolds assembly; ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v4.1/Gmax/assembly/sequences/) using software SOAP2. The unique alignment allowed for a maximum of two mismatches. The Glyma version 5.0 annotation file Glyma1_highConfidence.gff3 (February 8, 2010) was used for exon annotations. There are 55,787 mRNAs and a total of 345,213 exons in this high-confidence annotation. After unique alignment of paired read sequences to the reference soybean genome, the number of reads in any direction at each exon in each gene was counted using a custom Perl script. For read counts, a minimum of 70 out of the 76 bases of read sequence were required to overlap the reference exon sequence. The count number was globally normalized by dividing each number by the total counts in the sample. For visualization of exome differences, one count was added to each value, and the log2 ratio of the normalized sample over the control count number was calculated and plotted.
PCR Analysis
Select regions with detected deletions were chosen for confirmation by PCR. Primers were designed within CGH probes that flanked the detected deletion region for a PCR product that spanned the deletion site. The PCR product was gel extracted and sequenced. Alignment of the PCR product sequence to the reference soybean genome sequence assembly (www.phytozome.net) was performed to determine the exact break point borders. The following primers were used for PCR confirmation: VP1, 5′-GTAAGTAGCCTACGCATGACC-3′ (forward) and 5′-CAATGTGACCAAGCACTGACAC-3′ (reverse); PO1, 5′-CACTTTCCGGTAAGATTAAGGG-3′ (forward) and 5′-CAGTTTGCTTACACTCTGACT C-3′ (reverse); VP5, 5′-TATAAAGAGGGAAGGTTTGTGC-3′ (forward) and 5′-CATGGGCAAACTATTATGCTTG-3′ (reverse); and M23, 5′-CCACATCCTGAATATTCGGAATCTGTGAA-3′ (forward) and 5′-GTGAAGCAACATACCTTGATGGCTTCGAT-3′ (reverse).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CNV events detected by CGH in soybean FN late maturity mutants.
Supplemental Figure S2. CNV events detected by CGH in soybean FN early maturity mutants.
Supplemental Figure S3. CNV events detected by CGH in soybean FN root and nodule mutants.
Supplemental Figure S4. Genomic regions of high variability across the soybean FN population detected by CGH and SNP genotyping.
Supplemental Table S1. Mean, sd, and range of FN mutant M3 seed composition traits derived from three locations analyzed by NIR on a dry matter basis.
Supplemental Table S2. List of deleted genes on chromosome 10 of M23.
Supplemental Table S3. Universal SNPs on the Illumina Goldengate platform with evidence for segregation within the soybean FN population.
Supplemental Table S4. DNA CNV segment borders detected in soybean FN mutants by CGH.
Supplemental Table S5. List of genes within detected CNV events in soybean FN mutants.
Supplemental Table S6. Identifiers and pedigree for soybean FN mutant lines assayed by CGH.
Acknowledgments
We thank Jeffrey Roessler, Dimitri Von Ruckert, Renee Schirmer, and Gabriel Bascur Bascur for technical and field support, Dr. Zheng Jin Tu for supercomputing support, and Mauricio Assuncao, Phil Schaus, Arthur Killam, Dhananjay Mani, Tracy O’Neil, Dr. Jill Miller-Garvin, and numerous students for contributions to the project. In addition, we thank Dr. Bruna Bucciarelli for photography and plant ontology support, Dr. David Hyten for Goldengate SNP genotyping, Dr. Steven Cannon for soybean paralog data, and Dr. Kristin Bilyeu for the M23 line. Special thanks to Kevin Feeley and Nathan Weeks for database and Web site support. We also acknowledge the use of resources at the Minnesota Supercomputing Institute at the University of Minnesota.
References
- Alonso JM, Ecker JR. (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7: 524–536 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR. (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JL, Fehr WR, Welke GA, Sandhu D. (2005) Phenotypic and molecular analysis of oleate content in the mutant soybean line M23. Crop Sci 45: 1997–2000 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Anai T, Yamada T, Hideshima R, Kinoshita T, Rahman SM, Takagi Y. (2008) Two high-oleic-acid soybean mutants, M23 and KK21, have disrupted microsomal omega-6 fatty acid desaturase, encoded by GmFAD2-1a. Breed Sci 58: 447–452 [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, et al. (2010) Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. (2004) A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). Plant J 40: 143–150 [DOI] [PubMed] [Google Scholar]
- Carter NP. (2007) Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet (Suppl) 39:S16–S21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, et al. (2009) Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 106: 19096–19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V. (2004) Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JM, Waner DA, Horie T, Li SL, Horie R, Abid KB, Schroeder JI. (2004) Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Nelson RT, Cannon SB, Shoemaker RC. (2009) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38: D843–D846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646 [DOI] [PubMed] [Google Scholar]
- Haun WJ, Hyten DL, Xu WW, Gerhardt DJ, Albert TJ, Richmond T, Jeddeloh JA, Jia G, Springer NM, Vance CP, et al. (2011) The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol 155: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ.et al (2007) Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527 [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Jiang Q, Men A, Kinkema M, Gresshoff PM. (2007) Nodulation deficiency caused by fast neutron mutagenesis of the model legume Lotus japonicus. J Plant Physiol 164: 460–469 [DOI] [PubMed] [Google Scholar]
- Hyten DL, Cannon SB, Song Q, Weeks N, Fickus EW, Shoemaker RC, Specht JE, Farmer AD, May GD, Cregan PB. (2010) High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LW, van der Veen JH. (1982) EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. (2007) Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. (2009) Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol 50: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al. (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47: 640–651 [DOI] [PubMed] [Google Scholar]
- Larmande P, Gay C, Lorieux M, Périn C, Bouniol M, Droc G, Sallaud C, Perez P, Barnola I, Biderre-Petit C, et al. (2008) Oryza Tag Line, a phenotypic mutant database for the Genoplante rice insertion line library. Nucleic Acids Res 36: D1022–D1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC. (2007) MaizeGDB’s new data types, resources and activities. Nucleic Acids Res 35: D895–D900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lassner M, Zhang Y. (2002) Deleteagene: a fast neutron deletion mutagenesis-based gene knockout system for plants. Comp Funct Genomics 3: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Song Y, Century K, Straight S, Ronald P, Dong X, Lassner M, Zhang Y. (2001) A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J 27: 235–242 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y. (2002) Reverse genetics by fast neutron mutagenesis in higher plants. Funct Integr Genomics 2: 254–258 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Martienssen RA. (1998) Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA 95: 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P, Stanciu M, Brudno M. (2009) Computational methods for discovering structural variation with next-generation sequencing. Nat Methods (Suppl) 6: S13–S20 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Meinke LK, Showalter TC, Schissel AM, Mueller LA, Tzafrir I. (2003) A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol 131: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men AE, Laniya ST, Searle IR, Iturbe-Ormaetxe I, Gresshoff I, Jiang Q, Carroll BJ, Gresshoff PM. (2002) Fast neutron mutagenesis of soybean (Glycine soja L.) produces a supernodulating mutant containing a large deletion in linkage group H. Genome Lett 3: 147–155 [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H. (2007) A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol 63: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR. (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orf JH, Denny RL. (2004) Registration of ‘MN1302’ soybean. Crop Sci 44: 693 [Google Scholar]
- Østergaard L, Yanofsky MF. (2004) Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J 39: 682–696 [DOI] [PubMed] [Google Scholar]
- Pham AT, Lee JD, Shannon JG, Bilyeu KD. (2010) Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol 10: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SM, Takagi Y, Kubota K, Miyamoto K, Kawakita T. (1994) High oleic acid mutant in soybean induced by x-ray irradiation. Biosci Biotechnol Biochem 58: 1070–1072 [Google Scholar]
- Rogers C, Wen J, Chen R, Oldroyd G. (2009) Deletion-based reverse genetics in Medicago truncatula. Plant Physiol 151: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D, Alt JL, Scherder CW, Fehr WR, Bhattacharyya MK. (2007) Enhanced oleic acid content in the soybean mutant M23 is associated with the deletion in the Fad2-1a gene encoding a fatty acid desaturase. J Am Oil Chem Soc 84: 229–235 [Google Scholar]
- Scherder CW, Fehr WR, Shannon JG. (2008) Stability of oleate content in soybean lines derived from M23. Crop Sci 48: 1749–1754 [Google Scholar]
- Schlueter JA, Vasylenko-Sanders IF, Deshpande S, Yi J, Siegfried M, Roe BA, Schlueter SD, Scheffler BE, Shoemaker RC. (2007) The FAD2 gene family of soybean. Crop Sci 47: S-14–S-26 [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. (2004) Large-scale copy number polymorphism in the human genome. Science 305: 525–528 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li G, Chen R. (2006) Fast neutron bombardment (FNB) mutagenesis for forward and reverse genetic studies in plants. Teixeira da Silva JA, , Floriculture, Ornamental and Plant Biotechnology: Advances and Special Issues, Vol 1. Global Science Books, pp 629–639 [Google Scholar]
- Waugh R, Leader DJ, McCallum N, Caldwell D. (2006) Harvesting the potential of induced biological diversity. Trends Plant Sci 11: 71–79 [DOI] [PubMed] [Google Scholar]
- Wei W-H, Chen B, Yan X-H, Wang L-J, Zhang H-F, Cheng J-P, Zhou X-A, Sha A-H, Shen H. (2008) Identification of differentially expressed genes in soybean seeds differing in oil content. Plant Sci 175: 663–673 [Google Scholar]
- Wu JL, Wu C, Lei C, Baraoidan M, Bordeos A, Madamba MR, Ramos-Pamplona M, Mauleon R, Portugal A, Ulat VJ, et al. (2005) Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 59: 85–97 [DOI] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J. (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. (2006a) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fetch T, Nirmala J, Schmierer D, Brueggeman R, Steffenson B, Kleinhofs A. (2006b) Rpr1, a gene required for Rpg1-dependent resistance to stem rust in barley. Theor Appl Genet 113: 847–855 [DOI] [PubMed] [Google Scholar]