Abstract
The ontogeny of seed structure and the accumulation of seed storage substances is the result of a determinant genetic program. Using RNA interference, the synthesis of soybean (Glycine max) glycinin and conglycinin storage proteins has been suppressed. The storage protein knockdown (SP−) seeds are overtly identical to the wild type, maturing to similar size and weight, and in developmental ontogeny. The SP− seeds rebalance the proteome, maintaining wild-type levels of protein and storage triglycerides. The SP− soybeans were evaluated with systems biology techniques of proteomics, metabolomics, and transcriptomics using both microarray and next-generation sequencing transcript sequencing (RNA-Seq). Proteomic analysis shows that rebalancing of protein content largely results from the selective increase in the accumulation of only a few proteins. The rebalancing of protein composition occurs with small alterations to the seed’s transcriptome and metabolome. The selectivity of the rebalancing was further tested by introgressing into the SP− line a green fluorescent protein (GFP) glycinin allele mimic and quantifying the resulting accumulation of GFP. The GFP accumulation was similar to the parental GFP-expressing line, showing that the GFP glycinin gene mimic does not participate in proteome rebalancing. The results show that soybeans make large adjustments to the proteome during seed filling and compensate for the shortage of major proteins with the increased selective accumulation of other proteins that maintains a normal protein content.
Seed crops are propagated for the stored protein, oil, and carbohydrates that specifically accumulate in seeds. Seeds accumulate protein as a source of carbon, nitrogen, and sulfur as well as triglycerides and carbohydrate reserves, which are used as a source of carbon and ultimately energy (for review, see Herman and Larkins, 1999). Seeds can be characterized as storing primarily either protein and triglyceride or protein and carbohydrate. Soybean (Glycine max) seed storage proteins are encoded by a few conserved gene families (Schuler et al., 1982a, 1982b; Harada et al., 1989; Nielsen et al., 1989). Most seed proteins are members of the cupin superfamily (e.g. legumins and vicilins), but in some dicotyledonous seeds, lectins are abundant, whereas in cereal grains, the prolamins and to a lesser extent the legumins are abundant (Herman and Larkins, 1999).
During seed filling, the accumulation of seed storage proteins is regulated by an integrated genetic and physiological network (Golombek et al., 2001; Brocard-Gifford et al., 2003; Elke et al., 2005; Fait et al., 2006). However, the tight genetic control of seed development is modifiable within a certain range by maternal nutritional and environmental effects (Weber et al., 2005). The composition of seeds can be broadly defined as a developmental genetic program that is modified by nutrient source availability and the demands of the forming storage substance sink. Viewed from a systems biology perspective, the regulation and cross talk between the underlying developmental program modified by extrinsic conditions and nutrient source flux and nutrient sink formation determines the final content of the mature seed. Broadly, the mature seed can be considered the product of a combinatorial output of the interaction of program, source, and sink that yields a specific seed composition constrained within genetically defined limits.
The seed’s genetic program specifies a series of stage-specific gene expression patterns and regulons that provide a determinant framework for the formation and development of the seed (Hill and Breidenbach, 1974; Goldberg et al., 1981a, 1981b, 1983; Mienke et al., 1981; Walling et al., 1986; Naito et al., 1988; Harada et al., 1989; Perez-Grau and Goldberg, 1989; Nielsen and Nam, 1999; To et al., 2006). Seed-specific DNA-binding proteins have been discovered using sequence-specific probes and promoter-deletion analysis with reporter gene constructs (Chen et al., 1986; Jofuku et al., 1987; Allen et al., 1989; Lessard et al., 1991; Bäumlein et al., 1992; Kwong et al., 2003; Wang et al., 2007). A primary role of transcription factors in seed development is the primary control of protein and oil and possibly other classes of storage substance accumulation (for review, see Kroj et al., 2003; Gutierrez et al., 2007; Santos-Mendoza et al., 2008) and the developmental processes that support the accumulation of storage substances. Forward and reverse genetics experiments have defined regulators of seed development that consist of four master regulator transcription factors, LEC1, LEC2, ABI3, and FUS3, and at least four other proteins (for review, see Kroj et al., 2003; Gutierrez et al., 2007). The four master regulators have a hierarchal relationship and an interactive relationship that further regulate other transcription factors, forming a control network. For instance, LEC1 and LEC2 positively regulate themselves and the others (To et al., 2006). The master regulators form additional regulatory networks with other transcriptional regulators; for example, the AP2/wrinkled family is responsive to Suc source flux, and its expression alters seed size (Ohto et al., 2005) and triglyceride content (Cernac and Benning, 2004; Baud et al., 2007; Maeo et al., 2009; Liu et al., 2010).
The nutrient status of the maternal plant provides a further regulatory control that modulates the output of the seed genetic program in forming the seed sink (Gayler and Sykes, 1985). In this program, the plant couples the nutrient source (the maternal tissue) to the nutrient sink (the seed propaqule) on the assumption that there is a proportional linkage of the strength of the seed sink to the input of the nutrient source, perhaps regulated at the transport level. Viewed from this perspective, the maturing seed would maximize utilization of the source nutrients (Hernández-Sebastià et al., 2005; Gu et al., 2010). Sulfur availability is one example of a nutrient source regulating seed protein composition (Beach et al., 1985). Legumes such as soybean possess storage proteins with low sulfur amino acid content. The β-subunit of conglycinin exhibits plasticity in response to available sulfur (Holowach et al., 1986; Hirai et al., 1995; Tabe et al., 2002; Hagan et al., 2003). There is also feedback regulation of protein filling in the seed sink in response to nitrogen availability (Biermann et al., 1998; Ohtake et al., 2002). Another source of regulation is the overexpression of seed-specific amino acid permeases that increase the nutrient flux into the seed, resulting in an increase in seed sink protein content (Rolletschek et al., 2005). The plasticity induced in seed protein composition by altered nutrient source availability modulates the genetic developmental program, changing, for instance, the expression of transcripts as a result of sulfur availability (Rolletschek et al., 2005).
Because the stored seed sink will become a source in the following life cycle, plants maintain a proportional and species-specific defined inventory of protein, triglyceride, and carbohydrate for use by the germinating seedling. Breeders have long known that in soybean, the two major reserve substances, protein and triglyceride, are metabolically linked, and their level can be selected as a trait. A shortfall of accumulation of a major reserve substance limits the availability of critical nutrients for the postgermination seedling. Suppression of seed protein sink production results in compensating protein accumulation, shown by mutation-induced suppression or genetic modification of storage protein synthesis in maize (Zea mays), for example in opaque2 (Geetha et al., 1991; Hunter et al., 2002), and in soybean (Kinney et al., 2001; Takahashi et al., 2003), all resulting in rebalancing protein content by increased accumulation of other seed proteins. Moreover, seed protein and other constituents, most notably triglyceride, have an inverse relationship, where selection for increased protein or oil content results in a compensating decrease in the other reserve substance. The variability in protein and triglyceride content is maintained within relatively narrow limits of about 3% to 5% (w/w), with seeds that would function to maintain a relatively defined inventory of storage substances to provide nutritional reserves for the plant’s next generation.
One way to view the genetic, source, and sink regulation of seed protein fill is as a hierarchal series of controls, regulation, cross talk, and feedbacks from the genetic to the physiological level. In such a systems-oriented model, there is a determinant genetic framework that dictates the overall development of the seed, including its morphology, the developmental timing of gene expression, and reserve substance accumulation. But the genetic program through regulatory controls and feedback modulates the composition and balance of constituents, resulting in some plasticity that serves to maintain a relatively defined ratio of storage substances and composition in the mature seed.
In this paper, we demonstrate that the response to posttranscriptional silencing of soybean storage proteins results in a control that maintains the size of the seed protein sink by remodeling the proteome by greatly increased accumulation of a few proteins. Proteome rebalancing to maintain seed protein content occurs with minor collateral changes in the ontogeny of soybean seed development, including the transcriptome, metabolome, gross and ultrastructural morphology, and viability.
RESULTS
Creation of Storage Protein Knockdown Seeds
An RNA interference (RNAi) construct designed to suppress glycinin accumulation was transferred to soybean using biolistic transformation protocols (Parrott and Clemente, 2004). A FAD2 RNAi was also included in this construct to provide a marker for additional screening for a high-oleic acid phenotype and to maintain consistency with a prior conglycinin knockdown that also included the FAD2 knockdown (Kinney et al., 2001). Comparisons of the RNAi sequence show a component with homology to the glycinin family of storage proteins and little homology to the conglycinin family of storage proteins (Supplemental Fig. S1A). The FAD component of the RNAi has homology with the FAD family (Supplemental Fig. S1B). The regenerated somatic embryos and T0 seeds were screened for total protein distribution by one-dimensional SDS-PAGE and with immunoblot assays using anti-glycinin and anti-conglycinin antibodies. The recovered transgenic lines not only exhibited suppressed glycinin content but also an essentially complete knockdown of α/α′- and β-subunits of conglycinin. These lines that exhibited knockdown of both glycinin and conglycinin, termed SP− (for storage protein minus), were regenerated into plants. The growth morphology of the resulting plants, their seed set, and the mature seeds were overtly identical to wild-type controls (cv Jack). A total of five lines from two transformation experiments were created that all exhibited the same storage protein suppression.
Small chips were removed from the T0 seeds and used to assay for seed protein phenotype, and SP− seeds were regrown and reselected for two self-propagated generations to produce a population homozygous for the RNAi transgene. The SP− phenotype was stable through each of these generations, with α/α′- and β-conglycinin subunits being undetectable and glycinin levels being greatly reduced. The SP− phenotype has proven stable, with the plants and seeds appearing overtly normal in growth and development through subsequent greenhouse expansion generations and a field test. The oleic acid level in the SP− seeds was greater than 94% of the fatty acids, indicating that the FAD2 marker knockdown was also expressed. The size and dry weight of the greenhouse-grown SP− dormant seeds averaged 146 mg, which is similar to dormant seeds of the wild-type control, at 163 mg. The protein and oil contents of the SP− (40.2% and 19.1%) and the wild type (37.5% and 20.5%) are also similar. Together, these data show that the knockdown of the storage proteins (glycinin and conglycinin) that constitute the majority of the seed’s protein results in a soybean that rebalances its protein composition to a nearly identical protein and oil content and maintains its normal seed size. The SP− seeds germinate with about 100% frequency, and the initial stages of seedling growth appear overtly identical to the conventional Jack seeds. The SP− trait has been stable through more than eight generations under greenhouse growth conditions. Whether the SP− trait was maintained was further assessed in a field grow-out in the 2010 season in Wooster, Ohio. Both the proteome rebalancing of suppression of the glycinin and conglycinin storage proteins and their replacement by other intrinsic seed proteins were apparently identical in both greenhouse- and field-grown seeds (Supplemental Fig. S2). The FAD2 suppression cotrait was also maintained in the field-grown seeds. Although this grow-out was not a true field test, the overall productivity of the SP− plants was comparable to that of conventional soybean cultivars.
Proteomic Analysis of the SP− Seeds Shows That Other Seed Proteins Compensate for the Absence of Storage Protein Polypeptides
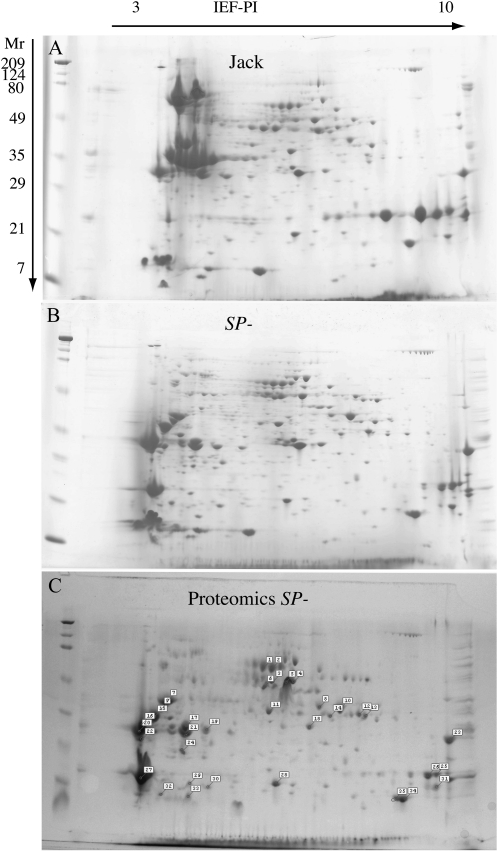
Two-dimensional (2D) isoelectric focusing/SDS-PAGE fractionation of protein extracts from SP− seeds in comparison with the Jack seed shows a large change in the spot distribution of the proteins that results from the knockdown of glycinin and conglycinin (Fig. 1, A and B; for soybean seed proteome maps, see Herman et al., 2003; Hajduch et al., 2005; for seed proteomics review, see Miernyk and Hajduch, 2011). The protein gel stained with Coomassie blue shows that there is a large-scale change in the protein distribution in SP− seeds, with the absent storage proteins being replaced by other abundant proteins. The knockdown of the glycinin and conglycinin storage proteins was confirmed by immunoblot probing a replicate gel with antibodies specific for these storage proteins. The identification of the induced proteins in the SP− seeds was achieved by mass spectrometric analysis of excised protein gel spots. Triplicate 2D gels were evaluated by visual examination with the assistance of gel spot size-scanning software. By comparing the protein spot sizes between SP− and wild-type seeds, significantly altered spots were excised, subjected to trypsin digestion, and analyzed by tandem mass spectroscopy. The map of the protein spots selected for mass spectroscopy is shown in Figure 1C.
Figure 1.
A and B, The two-dimensional gel fractionation of the total proteins of mature Jack (A) and SP− (B) seeds is shown. Note that abundant storage proteins in Jack seeds are absent in the SP− seeds, which accumulate a few alternative proteins that account for a large fraction of the total seed proteins. C, The spot selection for excision from the SP− seeds is shown to identify protein spots with changes in comparison with Jack. The spots were excised, digested with trypsin, and processed by tandem mass spectroscopy to identify the proteins. The spot numbers on the gel correspond to the identification and mass spectroscopy data outlined in Supplemental Table S1. IEF, Isoelectric focusing.
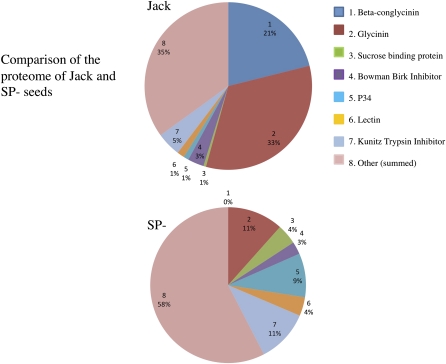
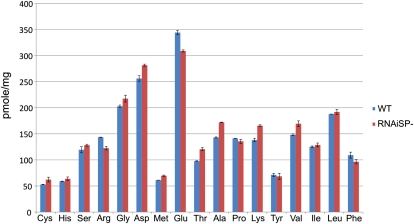
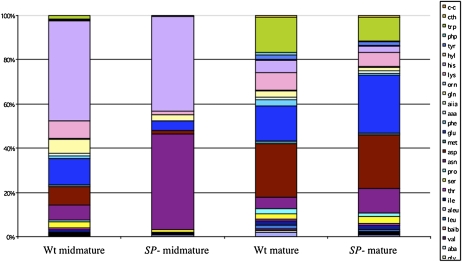
The compiled proteomic data (Supplemental Table S1) shows that most of the protein content rebalancing is due to greatly increased accumulation of only a few proteins and global increase of myriad other proteins that produce the seed proteome. The major proteins exhibiting an increase in abundance with the posttranscriptional silencing of SPs are Kunitz trypsin inhibitor (KTI; Fig. 1C, spots 2 and 7), soybean lectin (Fig. 1C, spots 17, 1, and 24), and the immunodominant soybean allergen P34 or Gly m Bd 30k (Fig. 1C, spot 16). Less prominent increases in abundance were also observed in Glc-binding protein (Fig. 1C, spots 1 and 2) and seed maturation-associated protein (Fig. 1C, spots 4 and 5). This is similar to the same set of proteins whose accumulation is greatly increased in a cross between two soybean lines that carry conglycinin and glycinin null alleles, respectively (Takahashi et al., 2003). The relative contribution of each major protein to the total proteome is shown in a pie chart (Fig. 2) comparing the conventional cv Jack with SP−. In Jack, the integrated spot volume of the 7S and 11S storage proteins from 2D broad-pH isoelectric focusing/SDS-PAGE is 54% of the total protein compared with 11% in SP−, which is primarily glycinin A4 with the SP− trait suppressing the more abundant group 1 glycinins. The silencing of storage proteins in SP− resulted in increased accumulation of Suc-binding protein from 1% in the conventional to 4% in total protein, of P34 from 1% to 9%, of lectin from 1% to 4%, and of KTI from 5% to 11%. These four proteins together constitute 28% of the total seed proteome, replacing over half of the silenced SP sink. In addition, there is a global increase in all other proteins, from 35% in the conventional to 58%, that together constitute 23% of the rebalanced SP− proteome. Not all proteins increase in abundance: the Bowman-Birk protease inhibitor (BBI) is the major sulfur amino acid sink in soybean, constituting 3% of the conventional seed proteome and remaining at 3% in the rebalanced proteome of SP−. Similarly, the major oil body protein, oleosin, constitutes about 2% of the total proteome, and it is also unchanged in SP−, consistent with the observation that total seed triglyceride content is unchanged. The impact of the change in protein composition on the total amino acid composition of the rebalanced soybean proteome was examined by digestion of SP− and Jack mature seed samples followed by fractionation and quantifying the resulting amino acid distribution. Figure 3 shows a bar graph comparing the amino acids and demonstrates that although the SP− phenotype and resulting protein composition rebalancing changes a majority of the protein content of the seed, there is little impact on total amino acid content, which is largely conserved in the rebalancing process.
Figure 2.
Shown are pie chart representations of the relative abundance of proteins in the proteomes of mature SP− and Jack determined by fractional spot volume. This illustrates the suppression of storage protein accumulation in SP− and its partial replacement by increased lectin, KTI, and P34 accumulation. In contrast, BBI, which is a major sulfur sink of soybean seeds, remains unchanged in SP− compared with Jack, indicating the selectivity of the proteins that rebalance the shortage of seed storage proteins.
Figure 3.
The distribution and abundance of total amino acids in Jack and SP− seeds is shown. Note the relative conservation of amino acid content in the SP− phenotype compared with the Jack control (WT, wild type). Of particular note is the lack of substantial change in the sulfur amino acids Cys and Met, which corresponds to the lack of change in the abundance of the main sulfur amino acid-containing protein, BBI (Fig. 2).
SP− Seeds Form Protein Storage Vacuoles in a Developmentally Correct Morphology and Pattern
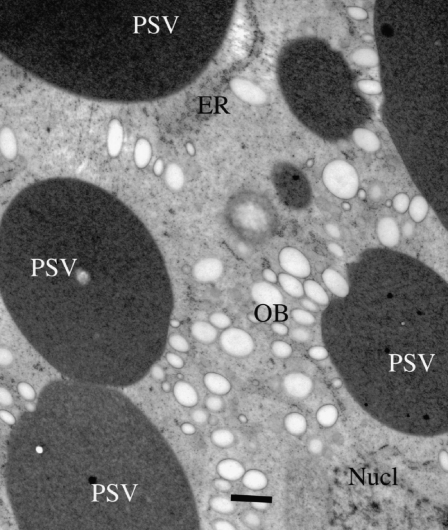
Protein storage vacuoles (PSVs) of dicotyledonous seeds, such as soybean, are formed by the subdivision of the central vacuole, which occurs coordinately with the synthesis and deposition of the storage proteins (Herman and Larkins, 1999). This results in protein-filled PSVs that fill much of the cytoplasm of seeds accumulating seed proteins. In order to examine the cellular structure of the SP− seeds, maturing cotyledons prepared by high-pressure cryofixation and the resulting samples were freeze substituted with acetone/OsO44 and then embedded in Epson plastic. In comparison with the wild-type cotyledons, the PSVs of the SP− lines are overtly similar in size and appearance (Fig. 4), and they possess a protein-filled amorphous matrix typical of soybean. (Note that soybean seed PSVs do not form protein-specific domains, or crystalloids, in the matrix.) In some SP− seed PSVs, there appears to be an excess of autophagic inclusions (for examples in soybean, see Melroy and Herman, 1991) in some of the PSVs of the maturing seeds as compared with the wild type, although this was not quantified or examined further.
Figure 4.
Transmission electron microscopy of SP− seed cotyledon storage parenchyma cells shows that the alternative set of proteins induced by proteome rebalancing in SP− are sequestered in filled PSVs. OB, Oil body. Bar = 1 μm.
The knockdown of conglycinin, whether by directed genetic engineering (Kinney et al., 2001) or as a consequence of a spontaneously arising null allele (Mori et al., 2004), results in the retention of a large fraction of normally soluble transport-competent glycinin in the precursor proglycinin form, which is accreted in endoplasmic reticulum (ER)-derived protein bodies. In contrast, in response to the knockdown of both glycinin and conglycinin, the storage parenchyma cells contain only PSVs, indicating that the compensating PSV-associated proteins remain vacuolar, without redirection of the proteins into ER-derived protein bodies (for discussion, see Herman, 2008). The structure and distribution of all other subcellular organelles and structures appear to be identical in SP− and Jack.
Maturing SP− Soybean Seeds Exhibit Limited Changes in the Global Transcript Profile
The global transcript profile of mid- to late-maturation SP− seeds were compared with the wild type using the Affymetrix DNA GeneChip platform, incorporating both biological and technical replicates in the design of the experiment. The resulting transcriptome data indicate that few transcripts show any significant change in accumulation, using a relatively stringent 3-fold up/down cutoff with a t test value of P < 0.05. Only those transcripts that had a positive correlation in both technical and biological replicates were scored as valid. The data are retrievable as accession number GSE12314. The DNA array results were annotated using Brandon et al. (2007). The transcriptome data showed RNAi suppression of all of the glycinin- and conglycinin-related transcripts. Seventy sequences exhibit decreased abundances of more than 3.0-fold, and 45 sequences are increased in abundance, as summarized in the scatterplot in Figure 5, with the expression data shown in Supplemental Table S2. Some of the transcripts represent the suppressed cupin superfamily storage proteins. The other transcript changes are a diverse set. Among the down-regulated transcripts is the AP2 gene domain-related transcription factor, which has the largest fold decrease of the transcripts assayed by DNA arrays and has been associated with the deposition of storage products, particularly seed oil (Kwong et al., 2003; Cernac and Benning, 2004). Other transcripts of note include decreased abundance of pre-mRNA processing-related proteins, proteins related to a variety of oxygen or oxygenase responses, and ferritin.
Figure 5.
Scatterplot of 3-fold up- and down-regulated transcript abundance of midmaturation SP− and Jack cotyledons assayed with the Affymetrix soybean DNA array using technical and biological replicates. The data displayed represent transcript hits filtered to be present in all replicates.
To further analyze the possible changes in transcriptome resulting from the SP− phenotype, replicate RNA samples converted to cDNA were subjected to Illumina sequencing. One half-plate of cDNA sequence was obtained on the Illumina Genome Analyzer for both SP− and Jack, which resulted in more than 12 million and more than 10 million 36mer sequences, respectively. Each sequence set was aligned to the soybean gene index (Quackenbush et al., 2001) Glycine max Gene Index version 14 (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=soybean), counts were normalized to RPKM values (reads per kilobase of target, per million mappable reads; Mortazavi et al., 2008), and only alignments where both SP− and Jack were represented with a minimum RPKM value of 10.0 were examined. This yielded 12,568 distinct sequence elements, of which 997 were down-regulated and 151 were up-regulated more than 3-fold in SP− compared with Jack. Overall, the results of the Illumina sequencing confirmed those observed with the Affymetrix array but provided greater breadth and depth of analysis as well as actual representation of individual transcripts in the transcriptome. Since the soybean gene index that the Illumina reads were aligned against a more comprehensive collection of soybean transcript sequences than represented on the Affymetrix array provides additional depth of information that extends as well as confirms the more preliminary array data set. Table I displays a summary of selected transcript abundance data comparing the Affymetrix and Illumina data for the transcripts of the silenced proteins and the proteins that compensate as well as a few other seed proteins such as the oil body-localized oleosin. The Illumina data enabled some transcript signals identified in the Affymetrix analysis to be resolved to specific members of gene families. The Illumina data set is available as Supplemental Table S3, with the data available for download in Excel format and the raw data set retrievable as accession number GSE21116 at the National Center for Biotechnology Information. The transcripts down-regulated include those encoding the proteins with suppressed accumulation in the SP− phenotype shown in the proteomic assay (Figs. 1 and 2; Table I) and in the Affymetrix transcript assay (Supplemental Table S2). The Illumina data set, as a quantitative assessment of transcript abundance, shows a much greater breadth of down-regulation of 40- to 175-fold compared with the Affymetrix results, confirming and extending the array results (Supplemental Table S2). KTI, lectin, and P34 transcripts that are the proteins that rebalance the protein content in SP− do not exhibit changes in transcript abundance of more than 3-fold. This confirms the Affymetrix array results, which also showed that the increased abundance of KTI, lectin, and P34 in SP− occurs without a large increase in transcription (Table I). The Illumina results also showed that other major seed proteins, for instance oil body oleosin, PSV aquaporin α-TIP, and BBI, do not show significant changes in transcript abundance (Table I). Among the metabolic enzymes with increased steady-state expression in SP− is asparaginase (transcripts 12528 and 12529, 6.3-fold up-regulation, plus two other transcripts, 7131 and 11879, with less than 2-fold down- and up-regulation), an enzyme that has been associated with postgermination storage protein mobilization when the amino acid flux generated by proteolysis becomes the nutrient source for the growing embryo. The free amino acid and metabolic profile of SP− shows a 5.98-fold excess of free Asn compared with the Jack control (Figs. 6 and 7; Supplemental Table S4), suggesting that the resulting increase in asparaginase expression is a feedback regulation.
Table I. Illumina transcriptome results of selected transcripts encoding proteins altered by storage protein suppression in SP− along with selected additional seed proteins shown as quantitative data.
The transcript number corresponds to its number in the Excel data set (Supplemental Table S3). The RPKM is an adjusted transcript count that was adjusted for transcript length, and fold values are for Jack RPKM/SP− RPKM, reflecting differential transcript abundance. The glycinin (G1–G3 isoforms) and α-, α′-, and β-subunits of conglycinin exhibit nearly complete silencing. The transcripts of the major proteins that rebalance the storage protein shortfall (lectin, P34, and KTI) exhibit 2-fold or less increase in transcript abundance.
| Transcript No. | Name | Jack RPKM | SP− RPKM | Fold Jack/SP− |
| 1527 | Glycinin G1 | 23,402 | 133 | −175.0 |
| 1526 | Glycinin G2 | 20,923 | 134 | −155.0 |
| 0379 | Glycinin G3 | 7,547 | 47 | −75.0 |
| 1525 | Glycinin G4 | 12,601 | 14,459 | 1.14 |
| 644 | Glycinin A3B4 | 6,257 | 15,885 | 2.55 |
| 5230 | Conglycinin α/α′ | 8,642 | 57 | −150 |
| 1365 | Conglycinin α | 3,277 | 81 | −40 |
| 4119 | Conglycinin β | 1,173 | 2,971 | 2.53 |
| 5734 | Conglycinin β | 1,244 | 6,095 | 4.90 |
| 4080 | Lectin | 3,639 | 8,435 | 2.32 |
| 7124 | KTI | 6,399 | 6,478 | 1.01 |
| 382 | KTI | 2,935 | 2,031 | −1.44 |
| 3256 | Suc-binding protein | 2,970 | 3,851 | 1.29 |
| 10331 | P34 allergen | 12,047 | 16,089 | 1.33 |
| 6408 | BBI | 13,024 | 8,228 | −1.58 |
| 8253 | 7S-28k allergen | 764 | 1,418 | 1.85 |
| 9370 | 2S albumin | 4,033 | 4,579 | 1.13 |
| 9393 | 2S albumin | 3,996 | 6,465 | 1.61 |
| 1002 | α-TIP | 508 | 946 | 1.86 |
| 2622 | P24 oleosin | 6,060 | 8,251 | 1.36 |
| 2933 | Low-Mr Cys-rich inhibitor | 2,364 | 3,792 | 1.66 |
| 1041 | FAD2 | 569 | 19 | −28 |
Figure 6.
A graphic distribution of the relative free amino content of midmaturation and mature Jack and SP− is shown. Note that the divergence in free amino acid content of the midmaturation seeds narrows in upon seed maturation, resulting in little difference in Jack and SP− mature seeds. Wt, Wild type.
Figure 7.
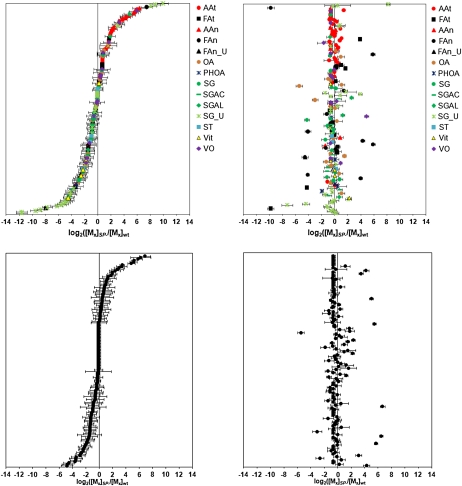
. Log2 ratios of soybean metabolites of midmaturation seeds and mature seeds. A, Known metabolites in midmaturation seeds. B, Known metabolites in mature seeds. C, Unknown metabolites in midmaturation seeds. D, Unknown metabolites in mature seeds. Data are sorted by log2 ratios in midmaturation seeds on the values from the smallest to the largest. Red circles, free amino acids extracted by the EZ:fast kit; black squares, fatty acids extracted by barium hydroxide; red triangles, amino acids from total metabolites; black circles, fatty acids; black triangles, unknown fatty acids; brown circles, organic acids; blue stars, phenolic acids; green circles, sugars; green dashes, sugar acids; green diamonds, sugar alcohols; light green stars, unknown sugars; sky blue squares, sterols; yellow triangles, vitamins; purple diamonds, volatiles. se bars were calculated on three replicates.
The posttranscriptional knockdown of storage proteins resulted in up and down changes in the abundance of selected transcriptional factors. The Affymetrix array showed that the transcription factor AP2 was one of the transcripts exhibiting the largest change in abundance (Supplemental Table S2). The Illumina results confirmed and extended this observation, showing that several different transcripts encoding AP2/wrinkled/ethylene-binding factor (EBF) are greatly decreased in abundance, as shown in Supplemental Table S3 (transcript 19, 11.8-fold down-regulated; transcripts 343 and 344, 3.94-fold down-regulated; transcripts 600–602, 3.43-fold down-regulated; and transcript 937, 3-fold down-regulated). Each of these individual genes presumably contributed to the overall observation of the AP2 domain transcription down-regulation results from the DNA array. Transcript 33 (8.25-fold down-regulated) is a member of the bzip family of transcription factors. Members of this family have been shown to be involved in a regulon-related Suc allocation to storage compounds. Breeders have long asserted that there is a negative correlation between oil and protein content in soybean, where either one decreases in abundance in response to increases, likely reflecting carbon flux allocation. In the SP− phenotype, were if not for the observed protein rebalancing based on breeding experience, oil accumulation would be expected to increase, but it does not. What, if any, role the decrease in the bzip transcription factor has in maintaining the balance of protein and oil accumulation will require further investigation. Among the other 1,000-plus transcripts up- and down-regulated comparing SP− and Jack are a wide variety of proteins representing many presumably unrelated pathways representing primary, secondary, and tertiary effects, showing that suppressing SP causes broad adjustments to the expression of low-abundance message in the seed transcriptome.
Metabolite Profiling of SP− Seeds Indicates That the Metabolic Differences from the Wild-Type Seeds at Midmaturation Stage Are Narrowed by the Onset of Seed Maturation
The alteration of protein accumulation in SP− seeds would be expected to impose an altered dynamic on the metabolism of the influx carbon and nitrogen nutrients, as they are converted to the protein and oil seed reserves. Total free amino acid analysis showed that the SP− trait resulted in the accumulation of excess free Asn largely at the expense of other amino acids (Fig. 6). Asn is one of the major nitrogen transport vehicles of plants, including soybean. Additional insights into these metabolic changes were obtained by conducting a combination of nontargeted metabolomics analyses and metabolite profiling that was targeted for amine-containing metabolites and fatty acids. In combination, these analyses measured the relative abundance of about 320 metabolite analytes (Fig. 7; Supplemental Table S4). These analyses were conducted on extracts made from midmaturation seeds and mature seeds (approximately 200 mg of tissue per extract). Seed samples were quickly frozen in liquid N2 to rapidly quench metabolism, and three parallel analytical procedures were used to assess the metabolite pool sizes in the samples. All three procedures used gas chromatography-mass spectrometry (GC-MS) as the analytical platform for gathering this data set, adapting procedures that have been developed for assessing metabolite pools in Arabidopsis (Arabidopsis thaliana; Bais et al., 2010; www.plantmetabolics.org). Two of these analytical procedures targeted the analysis of amine-containing metabolites (enriched in amino acids) and fatty acids, respectively. The rationale for targeting the analysis of these metabolites is that they are intermediates and products of the major seed reserves (i.e. proteins and oils, respectively). The third analytical procedure assessed the abundance of metabolites irrespective of their chemical class (i.e. nontargeted metabolite profiling).
The identity of the metabolites was determined by a combination of chromatographic behavior (i.e. retention indices relative to a series of hydrocarbon standards) and matches with mass spectra using either authentic commercially available standards or chemical databases (i.e. National Center for Biotechnology Information, www.plantmetabolomics.org, and PubChem). In combination, the three analytical protocols identified about 320 metabolite entities, and of these, 180 are metabolites whose chemical identities were determinable via the above two criteria; the remaining 143 metabolite entities were not chemically defined but were labeled by a convention that has been defined by the plant metabolomics research community (Bino et al., 2004). These latter analytes were mainly detected by the nontargeted metabolite profiling platform.
Because the response of the MS detector used to assess the abundance of the metabolites is dependent on the chemical nature of each metabolite, it is not possible to generate absolute abundance data for those metabolite entities whose chemical nature was not determined. Moreover, even for those metabolites whose chemical identity was determinable, the response of the MS detector is variable for each metabolite. Therefore, in order to integrate all the metabolite data irrespective of whether the chemical identity of the metabolite is known, we assessed the effect of the SP− transgene on the relative abundances of all metabolite entities.
Figure 7 shows the relative abundances of the 320 metabolite entities in the two genotypes assessed at two different stages of seed development. Figure 7, A and B, plot the log ratio of the 180 metabolites whose chemical identity is known, and the order of these metabolites on the y axis is the same in both plots. Similarly, Figure 7, C and D, plot the log ratio of the 140 analytes whose chemical identity is unknown, and the order of these analytes on the y axis is the same in both of these plots. In these log-ratio plots, those metabolites that hyperaccumulate or hypoaccumulate as a consequence of the genetic manipulation plot to the extreme left or right of the zero ordinate of the x axis, respectively, whereas those metabolites whose abundance was minimally affected by the genetic manipulation plot near the zero ordinate. Cursory examination of these plots indicates that at the midmaturation stage of seed development, the relative abundance of more metabolites is affected by the SP− transgene than in seeds at near maturity. Specifically, of the 320 metabolites that were assessed, 95 either hyperaccumulate or hypoaccumulate more than 4-fold in the SP− lines relative to the wild type, whereas the equivalent number is 38 metabolites at seed maturation. Therefore, these data indicate that although the metabolic state of the seeds is altered by the transgenic event at the midmaturation stage, as the seeds develop to maturity compensatory mechanisms appear to be expressed that bring the metabolic status of the seeds to near the wild-type state.
Supplemental Table S4 lists the metabolites that show altered accumulation at midmaturation stage and at seed maturity, and these are listed in the order of magnitude difference in abundance, from those that hyperaccumulate (most red-shaded cell) to those that hypoaccumulate (most-blue shaded cell) in response to the SP− transgenic event, and the chemical nature of these metabolites is indicated in Figure 7, A and B. These data indicate that in response to the SP− transgenic event, at midmaturation stage of development, amino acid metabolites hyperaccumulate (as indicated by the enrichment of the red symbols at the top of Fig. 7A) and carbohydrate metabolites hypoaccumulate (as indicated by the enrichment of the green symbols at the bottom part of Fig. 7B). However, although source-free amino acids accumulate, the process of proteome rebalancing maintains a fixed protein and total amino acid content (Fig. 3), albeit with a very different proteome composition. The accumulation of carbohydrate likely results from the interplay between carbon allocation to amino acids and other polymer precursors, with the rebalancing process tipping the ratio of allocation to favor carbohydrates.
A Foreign Protein Introgressed into the SP− Background Does Not Contribute to Seed Protein Rebalancing
The impact of expressing a foreign protein gene was tested in the context of the protein rebalancing process occurring in the SP− lines. The expression of the extrinsic gene product consisted of the G2 glycinin promoter sequence, a glycinin terminator, and a modified GFP that included a chitinase signal sequence and a C-terminal ER retention sequence. This mimics a G2 glycinin allele with an exchanged open reading frame (GFP). This transgene is designed to accrete in the ER, forming ER-derived protein bodies, which are inert, de novo-created organelles (Schmidt and Herman, 2008a) that are not normally found in soybean. Because the GFP-tagged protein bodies stably accumulate during seed maturation (Schmidt and Herman, 2008a), the fluorescence associated with this protein/organelle can be quantified to measure its accumulation throughout seed development.
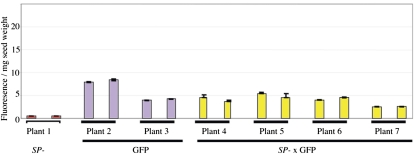
The GFP construct was introduced into soybean by biolistic transformation followed by selection and regeneration of the plants. GFP-positive seeds were regrown for T1 and T2 generations, producing a homozygous line of seed-specific GFP-HDEL-expressing seeds. Accumulation of the GFP-HDEL protein was assayed using a fluorometer assay with a standard curve control using commercially obtained GFP (Schmidt and Herman, 2008a). These assays showed that the GFP-HDEL protein in the parental homozygous seeds accumulates to 1.6% to 2% GFP of seed protein (Fig. 8). GFP-HDEL homozygous plants were then used as pollen donors in a cross to homozygous SP− seeds. Successful crosses were obtained and subjected to recurrent selection, producing homozygous GFP-HDEL/SP− seeds that were assayed for GFP fluorescence. GFP fluorescence in the SP− background was found to be approximately equal to that of the GFP-HDEL parental line, indicating that the GFP-HDEL glycinin allele mimic does not appear to participate in proteome rebalancing in the SP−.
Figure 8.
The fluorometric analysis of GFP concentration resulting from the introgression of the GFP-HDEL glycinin gene mimic (Schmidt and Herman, 2008a) into the SP− background. The measurement of the GFP accumulation shows that the GFP-HDEL abundance is slightly reduced when expressed in the SP− background, indicating that it is not recruited to participate in the proteome rebalancing of the SP− phenotype.
DISCUSSION
Protein Rebalancing Remodels the Seed into an Alternative Variant of Its Conventional Developmental Ontogeny
These results show that there is a remarkable adaptive response to the shortage of SP accumulation in soybean. A fascinating question concerns the selection process that led to the development of a physiological process to precisely compensate for the lesion of a shortage of most of the conserved SPs maintaining protein content, the ratio of protein to oil content, and the seed’s amino acid content. That seeds possess compensatory mechanism(s) by which an alternative maturation program results in an overtly normal conventional soybean, albeit with a greatly altered proteome, may require the utilization of unrecognized feedback and control mechanisms. That the soybean SP− and Jack soybean content diverges in the metabolome during maturation but all of these differences narrow by seed maturation shows that even with the large perturbation in composition, the seed strives to rebalance to a variation of normal composition. The overt development of the SP− soybean seeds is indistinguishable from that of Jack. Our results suggest that soybean seeds and likely other seeds possess far more plastic developmental programs than is inferred by plant breeding experience. The capacity to absorb large perturbations and to rebalance composition and the developmental program to reach a specific composition end point indicates that the interplay of all the various metabolic and synthetic pathways is capable of adjusting to a new program leading to an overtly normal seed. The 11S and 7S SP families are conserved in seeds from their first evolution. That the soybean seed proteome rebalancing process functions equally well in field-grown as well as greenhouse-grown seeds exchanging SPs for non-SPs raises many interesting questions about the selective factors that have maintained SPs throughout plant evolution. The results shown here show that soybean seeds have the capacity to possess an extensively remodeled protein composition without overt change in the seed’s developmental pattern of maturation and content in plants that appear to be equally productive to conventional soybean.
Suppression of Seed Storage Protein Accumulation Induces Proteome Rebalancing to Maintain Protein Content and Amino Acid Balance within a Fixed Box
The results presented here provide a new perspective on the developmental process of protein filling of soybean seeds and, perhaps by implication, seeds in general. The results of this study indicate that there is an intrinsic process(es) that evaluates the progress of protein filling during seed development and can alter the mix of proteins synthesized to rebalance the system to produce a mature seed with the correct protein and oil content with a greatly altered protein composition. The results reported here extend a prior investigation in which α/α′-conglycinin is suppressed (Kinney et al., 2001), resulting in transgenic soybeans that rebalance protein content by increasing the level of glycinin to compensate. This results in soybean seeds that possess an identical total protein content but a different proteome dominated by glycinin. A similar rebalancing of glycinin for conglycinin shortage occurs in a conglycinin null obtained from screening a germplasm collection (Mori et al., 2004). A cross of the conglycinin mutant with a glycinin mutant also results in rebalancing, with several PSV proteins along with an increase in free amino acids replacing the SP shortage (Takahashi et al., 2003). The posttranscriptional RNAi knockdown of all three SP proteins, α/α′-conglycinin, β-subunit of conglycinin, and glycinin results in a rebalancing of protein distribution to maintain total protein content with a complex alteration of the proteome, resulting in maintaining the seed’s protein content. Taken together, the results presented here and in prior publications (Kinney et al., 2001; Takahashi et al., 2003; Mori et al., 2004) show that soybean seeds have the capacity to compensate for a protein accumulation shortage and to rebalance that shortage by accumulating additional quantities of other intrinsic seed proteins to attain a predetermined protein content of approximately 40%. The rebalancing process appears to function whether the SP shortage is generated by mutation (Mori et al., 2004) or by directed knockdown in transgenic soybeans (Kinney et al., 2001). This indicates that seed size and composition are regulated so that the seed rebalances the proteome to maintain overall protein content.
The introduced SP− lesion is posttranscriptional RNAi-mediated suppression, implying that the mechanism(s) that mediates proteome rebalancing has components that must recognize the protein shortage and induce sufficient alternative protein synthesis to compensate for the protein shortfall and maintain the amino acid balance. Such mechanisms could involve primarily translational control; alternatively, the shortage of storage protein may be recognized by a signal transduction system that induces changes in gene expression. The RNAi used in this study was designed to target glycinin; that the accumulation of both glycinin and conglycinin was suppressed is surprising. This may support a role for a signaling mechanism that functions to repress transcription of the mRNA of other related proteins when the RNAi targets one of the gene families, perhaps by an epigenetic process. Proteomic analysis shows that in SP−, no other cupin superfamily SP protein compensates in the rebalancing of seed protein composition. Instead, other PSV proteins normally produced are accumulated at higher proportional levels, resulting in seeds with a normal protein content but a different protein composition. The use of PSV proteins KTI, SBA, and P34 as major contributors to the rebalancing process is similar to the rebalancing previously shown to occur in the cross of glycinin and conglycinin null mutants (Takahashi et al., 2003).
Protein Content Rebalancing Occurs without Significant Collateral Changes in the Transcriptome
The Affymetrix arrays show comparatively few changes in transcript abundance in SP− compared with conventional Jack seeds. In contrast, the Illumina cDNA sequencing of the same samples showed 997 transcripts down-regulated and 151 transcripts up-regulated comparing SP− and Jack. There are several reasons for the apparent increase in sensitivity, including the only partial representation of the soybean transcriptome on the Affymetrix array, that DNA array hybridization is at best a compromise average of binding conditions for the sequences, and that the DNA array poorly assesses the abundance of low-abundance transcripts. The Illumina transcriptome results point to more complex and subtle changes in transcription resulting from silencing SPs. The Illumina results confirmed and extended the Affymetrix array results by quantifying the transcript populations, and the SP silencing and resulting proteome rebalancing does not result from a large increase in transcription of the genes encoding the compensating proteins. But the apparent changes in many unrelated pathways indicate that there is the potential for complex feedback control, regulation, and cross talk, indicating that overt proteome rebalancing has components that are primary, secondary, and tertiary events resulting from the SP suppression. How these unrelated pathways intersect and have regulatory connections remains to be determined, given the complications in both data gathering and interpretation necessary. The complexities of regulatory cross talk in seed development are beginning to be established. Gu et al. (2010) recently showed that changes in the amino acid catabolism of seeds result in cross talk in what they described as a complex network of regulation of accumulation of amino acids generated by unrelated pathways.
The large-scale changes in that SP− proteome that occur with minimal changes in transcription shown with the Affymetrix array were confirmed by Illumina cDNA sequencing. KTI, lectin, and P34 transcript abundances of SP− were within 2-fold of what was observed in conventional cv Jack and represent examples of the primary proteins that compensate for the SP shortfall. BBI abundance provides another contrasting observation, where BBI protein accumulation is not altered in SP− seeds and its transcript level is essentially the same as in the control. That BBI does not exhibit altered transcript levels and/or protein accumulation likely indicates another separate proteome regulation within the conserved protein content box. BBI contains a large fraction of the total sulfur inventory of the seed, and its regulation is tied to sulfur availability; therefore, its regulation might be expected to be separated from other seed proteins that do contribute to the protein rebalancing in SP−. This indicates that there is a source availability component to proteome rebalancing where the amino acid balance of the seed is conserved along with its protein content.
For the SP silencing, the transcript changes represent the repercussion of silencing a large fraction of the total seed protein and therefore a change in the majority of the seed protein sink. Some of the transcript changes are likely significant to the physiological changes imposed by the SP− lesion, for instance, the change in transcript level of the wrinkled/AP2/ethylene response protein transcription factors that have been shown to regulate processes resulting from changes in sugar source and the formation of the triacylglycerol (TAG) sink (Cernac and Benning, 2004; Baud et al., 2007; Maeo et al., 2009; Liu et al., 2010) and seed size (for review, see Ohto et al., 2005; Okamuro et al., 2007). Similarly, the bzip transcription factor (Wiese et al., 2004) that is highly down-regulated in SP− is related to Suc regulation of transcription. Although the SP− maintains the same protein content as the wild-type cv Jack, the regulatory processes and cross talk between pathways that occur presumably impact the process of control of the other storage substance accumulation even if, in the final seed, the proportions of the storage substances in the mature seed remain fixed. One speculative role for the decrease of AP2/wrinkled/EBF-related transcripts in SP− seeds is if the SP− trait would induce a reduction of seed size that is then mitigated by a decrease in AP2, this through interaction and cross talk could maintain normal seed protein content and seed size in SP− seeds. This should be testable by combining SP− and an overexpression of AP2/wrinkled/EBF transcripts, experiments currently in progress. Similarly, the excess of free Asn in SP− may be an example of feedback gene regulation in SP−, where the excess of Asn is sensed and induces the increase of asparaginase expression.
Protein Rebalancing Does Not Significantly Impact the Metabolome
The metabolomic analysis does show some minor changes occurring as a consequence of the SP− trait and proteome rebalancing. However, the remodeling of protein composition within a fixed protein content box would not be expected to require major metabolic changes exchanging like sink. The free amino acids do show differences when comparing immature green seeds during the process of protein fill. Some of these may be due to differential composition of the now dominant proteins that present a different amino acid sink for their synthesis. It is likely that the seed is capable of making minor adjustments in amino acid availability to compensate for the altered amino acid sink requirements. The amino acid differences in maturing seeds between SP− and Jack narrow but are not entirely eliminated in the mature seed. Similarly, many of the other metabolites, including sugars and oligosaccharides, exhibit some variance between the Jack and SP− in immature seeds during protein filling, but the differences narrow and often are eliminated in the mature seed. This again shows that rebalancing occurs, with the mature SP− seed possessing a composition that is similar to that of Jack. This indicates that although there may be some perturbations of the metabolome by altering the SP protein sink, the rebalancing process adjusts the metabolome by the end point of seed maturation to produce seeds close to the wild-type metabolome configuration. The most significant difference between SP− and Jack in the metabolomics assays is the greater abundance of Man and various oligosaccharides, which may reflect a minor redistribution of reserve carbon in favor of carbohydrate. This again indicates that protein rebalancing is largely a physiological process contained within the context of protein and in particular vacuolar protein accumulation that has little collateral impact on the remaining cellular processes. Although the metabolomics analysis did not analyze TAG precursor synthesis, the SP− seeds possess an essentially wild-type content of TAG, so the inference, even if not demonstrated, is that rebalancing protein has little impact on the carbon flow to TAG. However, although there is no apparent change in TAG accumulation, the changes in transcription factors and metabolic alterations may be adjustments to maintain the differential flux of source carbon into the various reserve substances. Reversing the changes in expression in transcription factors in the SP− is one approach under way to examine how the rebalancing process leads to a fixed content of seed protein with a very different composition.
The Introgression of Foreign Protein into the Protein Content Rebalancing Background Indicates That Rebalancing Is Specific
In a prior study, it was shown that suppression of conglycinin results in the increased accumulation of proglycinin/glycinin to compensate (Kinney et al., 2001). Foreign proteins introduced into a conglycinin-suppressed background as a mimic of the glycinin gene will participate in the proteome rebalancing of glycinin exchanging for a conglycinin shortage (Schmidt and Herman, 2008a). The introgression of a reporter foreign protein, GFP-HDEL, into the SP background does not result in a large change, either up or down, of the GFP-HDEL accumulation (Fig. 8). The proteomics assays of the rebalancing show that the selection of intrinsic proteins is selective. The lack of an increase in a foreign protein’s expression introgressed into the SP− background substantiates the selective choices used in protein rebalancing. Although our experiment to test the potential of increased abundance of a foreign protein by the rebalancing was unsuccessful, if the underlying process can be exploited, this would open the potential to obtain high yields of foreign proteins in soybean.
MATERIALS AND METHODS
Construction of a Storage Protein Suppression RNAi Cassette
An RNAi cassette specific for the simultaneous suppression of both endogenous soybean (Glycine max) storage proteins, β-conglycinin and glycinin, and FAD-2 was produced as described by Schmidt and Herman (2008b), with the inverted arms of the construct produced by gene-specific amplifications as follows. cDNA from 150 mg of soybean cotyledon was amplified using SuperScript II (Invitrogen) and primers specific to glycinin subunit A1bB2 (GenBank accession no. AB030495) and FAD-2 (GenBank accession no. AB188250) open reading frames in two separate PCRs. Primer pair 5′Gly (5′-TTCTAGACTCGAGTATATTGACGAGACCATTTGCACA-3′) and 3′Gly (5′-CAGTGGCGGATATCGAGCTCCAGCCAACCGCAAAGTTTTGT-3′), including restriction sites XbaI and XhoI (underlined and boldface, respectively) on the 5′ primer located at 939 bp and homologous FAD-2 (underlined), EcoRV (italics), and SacI (boldface) on the 3′ primer located at 1,270 bp, were used to amplify a 331-bp region of the glycinin A1bB2 gene. Similarly, primer pair 5′FAD2 (5′-GGAGCTCGATATCCGCCACTGCTGTTTCTCTTCTCGTCACA-3′) and 3′FAD2 (5′-TAAGCTTACTAGTTCACGGTTAGAATATATGGG-3′), including restriction sites SacI and EcoRV (boldface and underlined, respectively) on the 5′ primer located at 618 bp and restriction sites HindIII and SpeI (underlined and boldface, respectively) on the 3′ primer located at 746 bp, were used to amply a 128-bp region of the FAD-2 gene. The two resultant amplification products, 331-bp glycinin and 128-bp FAD2, were gel purified (Qiagen gel extraction kit) and used as a template in a single PCR using the 5′Gly and 3′FAD2 primers as above. The single 459-bp amplicon, consisting of a 5′ glycinin region with XbaI and XhoI restriction sites and a 3′ FAD2 region flanked by HindIII and SpeI restriction sites, was cloned into TOPOvector (Invitrogen) and subjected to two separate double digestions, XbaI/HindIII and XhoI/SpeI, which would in turn be placed on either side of an intron to make up the inverted repeats in the hairpin RNAi cassette, as described previously (Schmidt and Herman, 2008b). The cassette with the hairpin for both storage proteins and FAD-2 in tandem was then placed into a vector described previously (Moravec et al., 2007; Schmidt and Herman, 2008a) under the regulatory control of the glycinin promoter and terminator and also containing the hygromycin resistance gene; this vector is hereby referred to as pRNAiSP−.
Transgenic Seeds and Proteomic Analysis
Transgenic soybean plants expressing the pRNAiSP− cassette were produced as described previously (Schmidt and Herman, 2008a, 2008b), and the resultant seeds were analyzed by proteomic analysis. Initial screening to identify phenotype-positive seeds was preformed by both one-dimensional SDS-PAGE stained for total soluble protein by 0.1% Coomassie Brilliant Blue stain and a replicate immunoblot probed using a mixture of polyclonal antibodies, one specific to glycinin and another to β-conglycinin, produced previously in this laboratory. Nontransformed soybean seed was used as a positive control. Seeds whose corresponding chips were shown to have the desired phenotype were grown into the next generation. Two generations were grown and screened in this manner until homozygosity was obtained.
Two-Dimensional Protein Analysis and Mass Spectroscopy Analysis
Total soluble protein was isolated from mature seeds as described previously (Schmidt and Herman, 2008a, 2008b). The soluble protein extract (150 μg) from both a nontransformed soybean seed and a homozygote pRNAiSP− seed was separated on first dimension 11-cm immobilized pH gradient gel strips (pH 3–10 nonlinear; Bio-Rad) and then on second dimension SDS-PAGE gels (8%–16% linear gradient), subsequently stained in 0.1% (w/v) Coomassie Brilliant Blue R250 in 40% (v/v) methanol, 10% (v/v) acetic acid overnight, and then destained for approximately 3 h in 40% methanol, 10% acetic acid. Individual spots of interest were excised and digested with trypsin, and the fragments were analyzed and identified by tandem mass spectroscopy as described previously (Schmidt and Herman 2008b).
DNA Array Analysis
Total RNA was extracted from 150 mg of cotyledons from both wild-type and pRNAiSP− plants, and microarray analysis was performed using Soybean Arrays (Affymetrix) at the Iowa State University Service Center. RNA used in the array experiment was from three biological samples, a nontransformed control and two individual pRNAiSP− cotyledons, with each sample performed in duplicate. The array data were analyzed by ArrayAssist (Stratagene), and only data with a stringent positive correlation coefficient and significant data in both technical and biological replicates were scored. The Affymetrix annotation of the GeneChip was supplemented with a search of additional annotations by Brandon et al. (2007).
Illumina Transcriptome Sequencing
A portion of the RNA isolated above (see DNA Array Analysis) was converted to cDNA; hence, the Illumina sequencing can be considered a technical replicate of the Affymetrix assay. Following second strand synthesis, end repair, and A-tailing, adapters complementary to sequencing primers were ligated to cDNA fragments (mRNA-Seq Sample Prep Kit; Illumina). Resultant cDNA libraries were size fractionated on agarose gels, and 250-bp fragments were excised and amplified by 15 cycles of PCR. Resultant libraries were quality assessed using a Bioanalyzer 2100 and sequenced for 36 cycles on an Illumina GA II DNA sequencing instrument using standard procedures.
All Illumina reads from the wild type and pRNAiSP− were aligned to the soybean gene index (Quackenbush et al., 2001) Glycine max Gene Index version 14 (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=soybean) with the BOWTIE aligner version 0.9.9.3 (Langmead et al., 2009) with default parameters, which results in only one alignment reported for each Illumina sequence. Counts on each gene index sequence were converted to RPKM values, which normalizes for transcript length and for the total read number in each experiment (Mortazavi et al., 2008). Target length was simply determined by the length of the gene index target sequence that was aligned. Since many of the gene index sequences are likely not to be full length, many target lengths should be considered as estimates of actual lengths.
Quantitation of Hydrolyzed Amino Acids and Free Amino Acids
Samples of midmaturation and mature Jack and SP− cotyledons were submitted for hydrolyzed and free amino acid analysis. For hydrolyzed amino acids, samples were hydrolyzed for 24 h at 116°C in 6 n HCl containing 0.5% phenol. Samples are dried down and resuspended in 20 mm HCl, derivatized with the AccQ-tag reagent (Waters), separated by a Waters Acquity ultraperformance liquid chromatography system, and quantified according to the manufacturer’s method. Samples for free amino acid analysis were extracted according to Hacham et al. (2002) and subjected to analogous derivitization, detection, and quantifying methods. Triplicates were assayed for each biological sample.
Metabolomics
Chemicals
Standards such as ribitol, nonadecanoic acid, n-hydrocarbon mix (C8–C40), and N,O-bis(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane (BSTFA/TCMS) were purchased form Sigma-Aldrich. The EZ:fast kit for free amino acid analysis was purchased from Phenomenex. All other chemicals were purchased from Fisher Scientific.
Soybean Sample Preparation
For nontargeted metabolite analysis, soybean seeds were pulverized in liquid nitrogen with a cryogenic grinder (SPEX CertiPrep). The samples (20 mg) were homogenized with 0.35 mL of hot methanol (60°C) and spiked with 25 μg of ribitol and 25 μg of nonadecanoic acid as internal standards. The mixture was immediately incubated at 60°C for 10 min and sonificated for 10 min. The mixture was homogenized with 0.35 mL of chloroform and 0.3 mL of water and centrifuged. Two hundred liters of the upper polar fraction and lower nonpolar fraction was transferred into 2-mL glass vials and dried with a SpeedVac concentrator. The extracts were derivatized using methoxyamine hydrochloride at 30°C for 90 min and BSTFA/TCMS at 60°C for 30 min. One sample was spiked with n-hydrocarbon standards mix for retention index.
For amino acid analysis, soybean samples (25 mg) were homogenized with 0.5 mL of 10% trichloroacetic acid spiked with 5 nmol of nor-Val as an internal standard.
Extracts were centrifuged, and the supernatants were transferred to a 2-mL glass vial. Purification of extracts and derivatization of amino acids were performed using the EZ:fast kit from Phenomenex according to the manual provided by the manufacturer. For fatty acid analysis, soybean samples (50 mg) were homogenized with 0.5 mL of 10% barium hydroxide and 0.55 mL of 1,4-dioxane containing 20 μg mL−1 nonadecanoic acid as an internal standard. The mixture was incubated at 110°C for 24 h. After cooling to room temperature, the mixture was acidified using 6 n HCl, and fatty acid analytes were recovered by extracting the aqueous phase with hexane. The recovered fatty acids were derivatized by methylation with 1 n HCl in methanol at 80°C for 60 min and by silylation with BSTFA/TCMS at 60°C for 30 min. The mixture was transferred to a 2-mL glass vial for GC-MS analysis.
GC-MS
GC-MS analyses were performed with an Agilent 6890N gas chromatograph interfaced to a 5973 MSD detector (Agilent Technologies). A HP5ms column (30 m × 0.25 mm, 0.25-m film thickness; Agilent Technologies) was used. For nontargeted metabolite analysis and fatty acid analysis, the temperature gradient was programmed from 80°C to 320°C at a rate of 5°C min−1 with helium flow rate at 2.2 mL min−1. Operating parameters were set to 70 eV for ionization voltage and 280°C for interface temperature. Collected GC-MS data were deconvoluted and analyzed using the Automated Mass Spectral Deconvolution and Identification System program (National Institute of Standards and Technology) with retention index information. Metabolites were identified based on their mass spectra by comparison with those of authentic standards in our laboratory’s standard compounds library and the National Institute of Standards and Technology 05 mass spectra library. Amino acid analysis was performed on an Agilent 6890 gas chromatograph equipped with a flame ionization detector. A Phenomenex ZB-AAA gas chromatograph column was used. The temperature gradient was programmed from 110°C to 290°C at a rate of 30°C min−1 with helium flow rate at 1.2 mL min−1. Analytes were identified based on comparison of their retention times with standard mixtures.
SP− × GFP Seeds
Homozygous seeds expressing an ER-targeted and retained GFP in a seed-specific manner were produced previously, analyzed, and described by Schmidt and Herman (2008a). Homozygous pRNAiSP− plants were cross-pollinated using homozygous GFP-HDEL plants as a pollen source. Putative seeds from the resultant cross were analyzed under blue light fluorescence. Crossed seeds were heterozygote for both traits of interest, SP− and GFP fluorescence, and were grown to homozygosity for both traits by simultaneously analyzing resultant seeds for fluorescence under blue light and SDS-PAGE analysis for storage protein suppression phenotype. Seeds homozygous for both traits, GFP and SP−, were obtained and analyzed for the amount of GFP by fluorescence on a spectrophotometer in triplicate, as described previously (Schmidt and Herman, 2008a), and calculated GFP (units mg−1 protein ± se). The quantity of GFP produced in seeds from the two parental lines (GFP-HDEL and SP−) and four homozygote cross lines was compared.
Electron Microscopy
Mid-maturation and late-maturation soybean cotyledons were excised and cryofixed in a Balzers high-pressure device. The cryofixed tissue was freeze substituted in the presence of osmium tetroxide. The fixed dehydrated tissue was embedded in epoxy resin, thin sectioned, counterstained with 5% (w/v) aqueous uranyl acetate, and visualized with a LEO electron microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GSE21115.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the RNAis with the targeted storage protein and FAD2 gene family sequences.
Supplemental Figure S2. Comparative polypeptide distribution of Jack with greenhouse- and field-grown SP−.
Supplemental Table S1. Proteomics data set of protein spots analyzed from Figure 1C.
Supplemental Table S2. Compiled Affymetrix data set comparing maturing Jack and SP− seeds.
Supplemental Table S3. Compiled Illumina RNA-Seq results comparing Jack and SP− seeds as a replicate assay of the Affymetrix results.
Supplemental Table S4. Compiled data sets for metabolomics assays of mature and midmature Jack and SP− seeds.
Acknowledgments
We thank Dr. Prachuab Kwanyuen (U.S. Department of Agriculture/Agricultural Research Service, North Carolina State University) for measuring the protein and oil composition of the SP− seeds. We also acknowledge the technical assistance of Dr. Howard Berg and Dr. Sophie Alverez of the Donald Danforth Plant Science Center’s Integrative Microscopy Facility and Mass Spectroscopy Facility. We greatly appreciate Dr. Rouf Mien (U.S. Department of Agriculture/Agricultural Research Service) for grow-out and harvest of the SP− seeds.
References
- Allen RD, Bernier F, Lessard PA, Beachy RN. (1989) Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant Cell 1: 623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais P, Moon SM, He K, Leitao R, Dreher K, Walk T, Sucaet Y, Barkan L, Wohlgemuth G, Roth MR, et al. (2010) PlantMetabolomics.org: a Web portal for plant metabolomics experiments. Plant Physiol 152: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Bäumlein H, Nagy I, Villarroel R, Inzé D, Wobus U. (1992) Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J 2: 233–239 [PubMed] [Google Scholar]
- Beach LR, Spencer D, Randall PJ, Higgins TJV. (1985) Transcriptional and post-transcriptional regulation of storage protein gene expression in sulfur-deficient pea seeds. Nucleic Acids Res 13: 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann BJ, Debanzie JS, Handelsman J, Thompson JF, Madison JT. (1998) Methionine and sulfate increase a Bowman-Birk-type protease inhibitor and its messenger RNA in soybeans. J Agric Food Chem 46: 2858–2862 [Google Scholar]
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al. (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9: 418–425 [DOI] [PubMed] [Google Scholar]
- Brandon L, Wagmaister J, Bui A. (2007) Affymetrix soybean genome array design, annotation and analysis. http://estdb.biology.ucla.edu/seed/media/Soybean_Array_Annotation_Su.pdf (March 17, 2011)
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR. (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chen ZL, Schuler MA, Beachy RN. (1986) Functional analysis of regulatory elements in a plant embryo-specific gene. Proc Natl Acad Sci USA 83: 8560–8564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elke GR, Blackmore D, Offler CE, Patrick JW. (2005) Increased capacity for sucrose uptake leads to earlier onset of protein accumulation in developing pea seeds. Funct Plant Biol 32: 997–1007 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayler KR, Sykes GE. (1985) Effects of nutritional stress on the storage proteins of soybeans. Plant Physiol 78: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha KB, Lending CR, Lopes MA, Wallace JC, Larkins BA. (1991) opaque-2 modifiers increase gamma-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell 3: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Ditta GS, Breidenbach RW. (1981a) Developmental regulation of cloned superabundant embryo mRNAs in soybean. Dev Biol 83: 218–231 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Tam SH, Ditta GS, Breidenbach RW. (1981b) Abundance, diversity, and regulation of mRNA sequence sets in soybean embryogenesis. Dev Biol 83: 201–217 [DOI] [PubMed] [Google Scholar]
- Golombek S, Rolletschek H, Wobus U, Weber H. (2001) Control of storage protein accumulation during legume seed development. J Plant Physiol 158: 457–464 [Google Scholar]
- Gu L, Jones AD, Last RL. (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Avraham T, Amir R. (2002) The N-terminal region of Arabidopsis cystathionine gamma-synthase plays an important regulatory role in methionine metabolism. Plant Physiol 128: 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan ND, Upadhyaya N, Tabe LM, Higgins TJ. (2003) The redistribution of protein sulfur in transgenic rice expressing a gene for a foreign, sulfur-rich protein. Plant J 34: 1–11 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ. (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JJ, Barker SJ, Goldberg RB. (1989) Soybean β-conglycinin genes are clustered in several DNA regions and are regulated by transcriptional and posttranscriptional processes. Plant Cell 1: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman E. (2008) Endoplasmic reticulum bodies: solving the insoluble. Curr Opin Plant Biol 11: 672–679 [DOI] [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ. (2003) Genetic modification removes an immunodominant allergen from soybean. Plant Physiol 132: 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM, Larkins BA. (1999) Protein storage bodies and vacuoles. Plant Cell 11: 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sebastià C, Marsolais F, Saravitz C, Israel D, Dewey RE, Huber SC. (2005) Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low- and high-protein soybean lines. J Exp Bot 56: 1951–1963 [DOI] [PubMed] [Google Scholar]
- Hill JE, Breidenbach RW. (1974) Proteins of soybean seeds. II. Accumulation of the major protein components during seed development and maturation. Plant Physiol 53: 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Chino M, Naito S. (1995) Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol 36: 1331–1339 [PubMed] [Google Scholar]
- Holowach LP, Madison JT, Thompson JF. (1986) Studies on the mechanism of regulation of the mRNA level for a soybean storage protein subunit by exogenous l-methionine. Plant Physiol 80: 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter BG, Beatty MK, Singletary GW, Hamaker BR, Dilkes BP, Larkins BA, Jung R. (2002) Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14: 2591–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Okamuro JK, Goldberg RB. (1987) Interaction of an embryo DNA binding protein with a soybean lectin gene upstream region. Nature 328: 734–737 [DOI] [PubMed] [Google Scholar]
- Kinney AJ, Jung R, Herman EM. (2001) Cosuppression of the α subunits of β-conglycinin in transgenic soybean seeds induces the formation of endoplasmic reticulum-derived protein bodies. Plant Cell 13: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F. (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard PA, Allen RD, Bernier F, Crispino JD, Fujiwara T, Beachy RN. (1991) Multiple nuclear factors interact with upstream sequences of differentially regulated beta-conglycinin genes. Plant Mol Biol 16: 397–413 [DOI] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. (2010) Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem 48: 9–15 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K. (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Melroy DL, Herman EM. (1991) TIP, an integral membrane protein of the soybean seed protein storage vacuole, undergoes developmentally regulated membrane insertion and removal. Planta 184: 113–122 [DOI] [PubMed] [Google Scholar]
- Mienke DW, Chen J, Beachy RN. (1981) Expression of storage-protein genes during soybean seed development. Planta 153: 130–139 [DOI] [PubMed] [Google Scholar]
- Miernyk JA, Hajduch M. (2011) Seed proteomics. J Proteomics 74: 389–400 [DOI] [PubMed] [Google Scholar]
- Moravec T, Schmidt MA, Herman EM, Woodford-Thomas T. (2007) Production of Escherichia coli heat labile toxin (LT) B subunit in soybean seed and analysis of its immunogenicity as an oral vaccine. Vaccine 25: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Mori T, Maruyama N, Nishizawa K, Higasa T, Yagasaki K, Ishimoto M, Utsumi S. (2004) The composition of newly synthesized proteins in the endoplasmic reticulum determines the transport pathways of soybean seed storage proteins. Plant J 40: 238–249 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Naito S, Dube PH, Beachy RN. (1988) Differential expression of conglycinin alpha-subunit and beta-subunit genes in transgenic plants. Plant Mol Biol 11: 109–123 [DOI] [PubMed] [Google Scholar]
- Nielsen NC, Dickinson CD, Cho TJ, Thanh VH, Scallon BJ, Fischer RL, Sims TL, Drews GN, Goldberg RB. (1989) Characterization of the glycinin gene family in soybean. Plant Cell 1: 313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen NC, Nam YW. (1999) Soybean globulins. Shewry PR, Casey R, , Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 285–313 [Google Scholar]
- Ohtake N, Kawachi T, Okuyama I, Fujikake H, Sueyoshi K, Ohyama T. (2002) Effect of short-term application of nitrogen on the accumulation of beta-subunit of beta-conglycinin in nitrogen-starved soybean (Glycine max L.) developing seeds. Soil Sci Plant Nutr 48: 31–41 [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada J. (2005) Control of seed mass by APETLA2. Proc Natl Acad Sci USA 22: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. (2007) The AP2 domain of APETLA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott WA, Clemente TE. (2004) Transgenic soybean. Soybeans: Improvement, Production and Uses. American Society of Agronomy/Crop Science Society of America/Soil Science Society of America, Madison, WI, pp 265–302 [Google Scholar]
- Perez-Grau L, Goldberg RB. (1989) Soybean seed protein genes are regulated spatially during embryogenesis. Plant Cell 1: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Hosein F, Miranda M, Heim U, Götz KP, Schlereth A, Borisjuk L, Saalbach I, Wobus U, Weber H. (2005) Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol 137: 1236–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM. (2008a) Proteome rebalancing in soybean seeds can be exploited to enhance foreign protein accumulation. Plant Biotechnol J 6: 832–842 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM. (2008b) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Schuler MA, Ladin BF, Fryer G, Pollaco JG, Beachy RN. (1982a) Structural sequences are conserved in the genes coding for the {alpha}, {alpha}′ and {beta} subunits of the soybean seed storage protein and other seed proteins. Nucleic Acids Res 10: 8245–8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Schmitt ES, Beachy RN. (1982b) Closely related families of genes code for the alpha and alpha′ subunits of the soybean 7S storage protein complex. Nucleic Acids Res 10: 8225–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L, Hagan N, Higgins TJ. (2002) Plasticity of seed protein composition in response to nitrogen and sulfur availability. Curr Opin Plant Biol 5: 212–217 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Uematsu Y, Kashiwaba K, Yagasaki K, Hajika M, Matsunaga R, Komatsu K, Ishimoto M. (2003) Accumulation of high levels of free amino acids in soybean seeds through integration of mutations conferring seed protein deficiency. Planta 217: 577–586 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling L, Drews GN, Goldberg RB. (1986) Transcriptional and post-transcriptional regulation of soybean seed protein mRNA levels. Proc Natl Acad Sci USA 83: 2123–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52: 716–729 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]