Abstract
Embryogenesis is central to the life cycle of most plant species. Despite its importance, because of the difficulty associated with embryo isolation, global gene expression programs involved in plant embryogenesis, especially the early events following fertilization, are largely unknown. To address this gap, we have developed methods to isolate whole live Arabidopsis (Arabidopsis thaliana) embryos as young as zygote and performed genome-wide profiling of gene expression. These studies revealed insights into patterns of gene expression relating to: maternal and paternal contributions to zygote development, chromosomal level clustering of temporal expression in embryogenesis, and embryo-specific functions. Functional analysis of some of the modulated transcription factor encoding genes from our data sets confirmed that they are critical for embryogenesis. Furthermore, we constructed stage-specific metabolic networks mapped with differentially regulated genes by combining the microarray data with the available Kyoto Encyclopedia of Genes and Genomes metabolic data sets. Comparative analysis of these networks revealed the network-associated structural and topological features, pathway interactions, and gene expression with reference to the metabolic activities during embryogenesis. Together, these studies have generated comprehensive gene expression data sets for embryo development in Arabidopsis and may serve as an important foundational resource for other seed plants.
Embryogenesis represents an important phase in the life cycle of flowering plants; it begins with the zygote, the product of fertilization of female gamete (egg cell) with male gamete (sperm cell). The developmental events that culminate in the production of a mature embryo from a single-cell zygote are precisely coordinated and relatively conserved in the majority of angiosperms (Goldberg et al., 1994). During this phase of development, the embryonic body plan is laid out, starting with apical-basal polarity, followed by partitioning of the apical domain into cotyledons and the shoot apical meristem, and differentiation of the basal domain to form the hypocotyl and root apical meristem. This overall theme is conserved among diverse plants although some species-specific differences also exist. This embryonic program, in coordination with the developing endosperm and seed coat, plays a central role in defining many of the key aspects of seed development and diversity, and this impacts many agronomic traits in crops (Huh et al., 2007; Braybrook and Harada, 2008).
Significant advances have been made in studying and understanding early embryogenesis in animal model systems because of the ease of isolating embryos (Carroll et al., 2005). In plants, fertilization and further embryo development occurs within the ovule and this makes it difficult to isolate embryos at very early stages (Wei and Sun, 2002). Thus the molecular events associated with early stages of plant embryogenesis are largely unknown. Without such information, it is not possible to advance the knowledge to a level comparable to that obtained by system-level integrative approaches in yeast (Saccharomyces cerevisiae) and animal models (Ihmels et al., 2004; Cui et al., 2007). Arabidopsis (Arabidopsis thaliana) is the best-studied model angiosperm, with several genetic resources and databases (Alonso and Ecker, 2006). Exploiting these resources requires overcoming the challenges in accessing the diminutive embryos. In this study, we report successful isolation and generation of global gene expression data sets of Arabidopsis embryos at key stages of development, determination of the metabolic networks and their dynamics during embryo development, and experimental validation of inferred key nodes. Furthermore, cases of higher-order regulation, chromosomal regiocentric coordination of gene expression, and maternal and paternal gamete contribution to early stages of embryo development are highlighted. Together, the results provide a comprehensive view of gene expression patterns, new regulatory insights, and metabolic network models for embryogenesis in Arabidopsis and these lay the foundation for further dissection of individual aspects of embryogenesis.
RESULTS AND DISCUSSION
Isolation of Developing Embryos from Arabidopsis Seeds
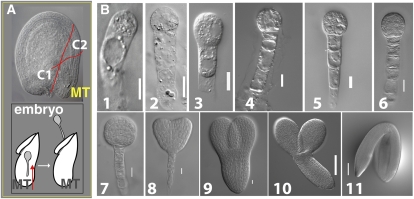
Isolation of zygote and early stage embryos has been a major challenge in plants. This is particularly the case in Arabidopsis because of the small size of the ovules at fertilization (160 × 140 μm; Fig. 1A) and the embryos within (i.e. elongated zygote size is 40 × 13 μm; Fig. 1B, 1). We developed a method for isolation of whole, live embryos from the fertilized ovules at various key stages including the zygote. Briefly, two incisions were made at the micropylar end of the ovule followed by careful microdissection of intervening cells exposing the central part of the micropylar tube where the transparent embryo is seated; the embryo is then released intact by gentle manipulation of the micropylar tube with fine forceps (Fig. 1, A and B). Following this procedure, embryos at the other early stages could also be obtained. Embryos at torpedo stage and thereafter were isolated by slicing the ovule and releasing the embryo with forceps. Structural integrity of the embryos was verified by Nomarski microscopy of randomly selected individual embryos following the retrieval of embryos from ovules (Fig. 1B). As expected, it was difficult to separate rapidly dividing early stage embryos into exclusively discrete stages; for example, the zygote (Z) stage would have some one-cell and two-cell embryos (Fig. 1B, 1–3).
Figure 1.
Embryo isolation from developing seeds of Arabidopsis. A, Nomarski image of ovule at fertilization showing two incisions (C1 and C2) made with forceps to detach the micropylar tube (MT; A, top). Isolated MT with the embryo inside is gently manipulated with forceps in the direction shown (arrow) to release the embryo (A, bottom). B, Nomarski images of dissected Arabidopsis embryos (B1–B11). Elongated zygote (B1); one-cell (B2); two-cell (B3); quadrant (B4); octant (B5); dermatogen (B6); globular (B7); heart (B8); torpedo (B9); bent (B10); and mature (B11) embryos. Bar = 0.01 mm (B1–B9); 0.1 mm (B10–B11).
Majority of the Nuclear-Encoded Genes Are Expressed during Embryogenesis
After establishing a reproducible method for embryo isolation, we obtained embryos of the Z, quadrant (Q), globular (G), heart (H), torpedo (T), bent (B), and mature (M) stages (Fig. 1B). We then performed microarray-based global gene expression profiling using four biological replicates from each of the seven stages. Each microarray was hybridized with two embryo probe samples from different developmental stages that were labeled separately with Cy3 and Cy5. For each embryo stage, there were four biological replicates: two labeled with Cy3 and the other two labeled with Cy5 to control any dye bias. The experiments were performed in a manner that allowed direct comparison of the seven embryo developmental stages in 10 combinations: Z vs Q, Q vs G, G vs H, H vs T, T vs B, B vs M, Z vs H, Z vs M, Q vs T, and G vs B. Thus, all possible sequential stages were hybridized on individual slides, in addition to four combinations of stages that were not sequential. In addition, the data sets obtained from these microarray experiments were used to perform further comparative analysis of other 11 possible combinations (Z vs G, Z vs T, Z vs B, Q vs H, Q vs B, Q vs M, G vs T, G vs M, H vs B, H vs M, and T vs M) for the seven embryo developmental stages in Arabidopsis (Fig. 1; Supplemental Fig. S1a; Supplemental Table S22). These large data sets were used to analyze the global gene expression levels and the differentially expressed genes at all stages of embryo development (Fig. 2; Supplemental Fig. S4; Supplemental Table S1). A searchable digital gene expression database of our data sets for Arabidopsis embryo development is available at http://www2.bri.nrc.ca/plantembryo/ (Supplemental Fig. S1b). The analysis suggested that cumulatively, approximately 78% of the genes in the Arabidopsis genome were expressed during the course of embryogenesis. Prolific genome activity was evident even at the earliest zygote stage where 58% of genes were expressed (Supplemental Table S7a). Just prior to the developmentally important transition from globular to heart stage (G → H) where the primordial body plan is established, the largest fraction (62%) of the genes were expressed. Near maturity, fewer genes (55%) were expressed despite the embryo containing more cells (Supplemental Table S7a). Studies using laser-capture microdissection and microarray approaches have also estimated a comparable number of genes being expressed during embryogenesis in Arabidopsis and soybean (Glycine max; Casson et al., 2005; Le et al., 2007, 2010; Spencer et al., 2007). Representative genes selected in the high, medium, and low expression categories at different stages of embryo development in our data set shows good correlation with the published laser-capture microdissection data (Spencer et al., 2007; Supplemental Table S7b). Cluster analyses of differentially expressed genes between stages revealed unique patterns associated with biological activities operating during sequential developmental stages of Arabidopsis embryogenesis. For example, the Z and Q stages clustered closer than with other stages that are temporally distal (Fig. 2). This also provided an independent validation of the method used in isolating the embryos.
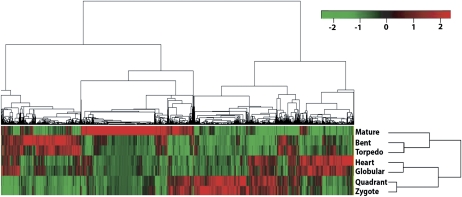
Figure 2.
. Hierarchical cluster analysis of gene expression patterns in Arabidopsis embryo development. Microarray analysis of seven embryo developmental stages identified 10,409 (shown in the top tree) differentially expressed genes at one or more stages referred to as modulated genes. We identified sets of modulated genes for each transition stage (namely, Z → Q, Q → G, G → H, H → T, T → B, B → M) using Limma software, then extracted their corresponding gene expression values for all seven stages (using single-channel normalization of Limma, see “Materials and Methods”). Z scores (statistical measure) were calculated for each of these genes and then used for hierarchical clustering. The analyses clustered Z and Q stages as phase I, G and H as phase II, T and B as phase III, and M stage as a distinct phase IV. Red indicates up-regulated genes whereas green indicates down-regulated genes. Scale bar represents fold change (log2 value).
Gene Expression Patterns Correlate Timing of Developmental Events and Transcription Factor Genes Are Robustly Modulated during Early Embryogenesis
On examining our data sets for genes that function during cell cycle, primary metabolism, storage reserve synthesis, auxin and abscisic acid biosynthesis, and signaling, we found discrete expression patterns. These are summarized in Supplemental Figure S2 and the highlights of these results are presented in the sections below. Within each stage, we found some expression patterns that were less shared with adjacent stages and therefore deemed unique to the given stage. Additionally, overlapping functional gene groups were also identified within Z and Q stages (phase I), G and H stages (phase II), T and B embryo stages (phase III), and M as distinct stage (phase IV; Fig. 2; Supplemental Fig. S4). These unique gene expression signatures reflect the biologically distinct programs associated with different phases. For example, in phase I where postfertilization sporophytic program is initiated, auxin stimulus and signaling events associated gene activities are more prevalent, and the Z and Q stages of this phase also cluster together (Supplemental Fig. S2b; Supplemental Table S5). Meristem and morphogenesis (Supplemental Fig. S4) genes are more active in phase II when the body plan is established and elaborated; carbohydrate, fatty acid, and storage protein synthesis activities are evident during phase III, reflecting deposition of storage reserves (Supplemental Figs. S2a and S4); finally, genes associated with abscisic acid response and dehydration are active in phase IV when the embryo undergoes desiccation to become a dormant and fully mature embryo (Supplemental Fig. S2c; Supplemental Table S5). Our study with isolated embryos revealed a clear separation between the early and late stages of embryogenesis (Fig. 2), and this is consistent with a previous study that clustered globular and heart stages together while later stages showed divergent clustering (Spencer et al., 2007).
Although ribosomal proteins are largely considered as constitutively expressed and housekeeping in function, our data show dynamic regulation (Supplemental Fig. S3A) with relatively high expression levels of ribosomal genes during Z, Q, and G stages that coordinated with the embryo growth rate (Supplemental Fig. S3, A, B, and D). Interestingly, the proteasomal genes are highly expressed at G stage, which coincides with key transition phase in embryogenesis (Supplemental Fig. S3, C–E).
Zygote and quadrant stages represent the earliest events in embryogenesis. In these two stages, 3,162 genes are modulated, of which 1,630 were up-regulated and 1,532 were down-regulated, respectively, in the Z stage relative to the Q stage. Further analysis of the genes expressed in the Z stage revealed that transcription factors (TFs) represented 7.5% of the modulated genes (Supplemental Table S8a). Notable among these is the zinc-finger group. Only 4% to 5% of the modulated genes are TFs in the later stages of embryo development. Regulation of gene expression at the chromatin level, via DNA methylation/demethylation, DNA acetylation/deacetylation, and Polycomb/Trithorax complexes, is critical for epigenetic control of developmental programs in both animals and plants (Goldberg et al., 2007; Henderson and Jacobsen, 2007). Our data sets reveal several of the Arabidopsis genes that are involved in chromatin modification and regulation (Supplemental Table S8b). These genes include MOM1, CHR34, RPD3A, VRN2, and ATX1 that are modulated during the early stages of embryogenesis, suggesting their potential key roles in postfertilization.
Maternal and Paternal Gene Expression Programs in Zygote and Quadrant Stage Embryos
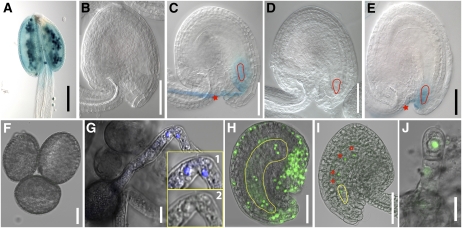
The gene expression programs of the parental gametes play important roles in fertilization and zygote development. The state of chromatin (Schaefer et al., 2007), activation of the zygote-specific gene expression (Stitzel and Seydoux, 2007), involvement of maternal transcripts in the initiation and maintenance of the zygote, and maternal to zygote transition (Baroux et al., 2008) are all important. Our knowledge of these processes in plants is still at infancy. We used published gametophyte-enriched microarray data sets (Becker et al., 2003; Honys and Twell, 2003; Yu et al., 2005) to discern any patterns of gene expression common to the zygote and the male or female gametophytes. Unless parents with expressed sequence polymorphisms are used and the hybridization platform is allele specific, parent of origin cannot be determined in such analyses. With this limitation in mind, our analysis showed that gamete and zygote expression shared a large number of genes at the two early stages (Z and Q) and such an overlap receded in subsequent embryo stages. We found that 56% (712) of the genes expressed in a female gametophyte-enriched manner are also expressed in the early stages (Z and Q). Notably, the corresponding number for male gametophyte expressed genes was 51% (239; Supplemental Table S2). If allele-specific expression was the underlying cause, the female genome would be contributing more than the male genome (Vielle-Calzada et al., 2000) but the contribution of the male genome would also be significant. Obviously, the data available above do not permit resolution of preexisting and de novo transcripts of parental genome origin. To address the question of parent-of-origin gene expression pattern in the zygote, we performed quantitative reverse transcription (qRT)-PCR analysis for 14 gene targets (nine pollen enriched, five embryo-sac enriched). The results confirmed their predicted specific expression in paternal or maternal parent as well as in the zygote (Supplemental Table S8e). Among the genes that show enriched expression in the ovule or pollen (Supplemental Table S8e), we selected two representative samples, one from the pollen (At3g28780) and the other from the embryo sac (At4g07410) for further expression analysis. The GUS reporter construct for the At3g28780 gene was made using its 1.98-kb putative promoter and introduced into Arabidopsis Columbia to generate transgenic lines. Analysis of these lines showed pollen-specific expression that continued in the pollen tube and after fertilization in the zygote (Fig. 3, A–C). Pollen enrichment of this gene is consistent with the findings of a previous pollen proteome study (Holmes-Davis et al., 2005). Reciprocal crosses of the GUS line with wild type confirmed that the GUS expression in the zygote is paternally derived (Fig. 3, B–E). The functional significance and implications of paternal contribution is evident from a recent finding of paternally controlled embryo patterning as determined by SHORT SUSPENSOR expression in Arabidopsis (Bayer et al., 2009). A GFP reporter assay with a maternally expressed (embryo-sac enriched) gene (At4g07410) selected from this study showed expression in the female gametophyte but not in the pollen (Fig. 3, F–H). The GFP reporter construct represents a translational fusion at the C terminus of At4g07410 open reading frame (1.95 kb putative promoter of this gene is used). When the GFP reporter was introduced paternally, expression was observed in the zygote but not in the pollen, suggesting de novo transcription after fertilization (Fig. 3, I and J). Together, these observations suggest that some of the predominantly or specifically expressed genetic programs in the contributing gametes are retained after fertilization in the zygote, indicating expression state of the corresponding genes after fertilization is likely influenced by contributing gametic programs. The gene expression patterns of zygote and quadrant stage embryos inferred from this study will likely include maternally and paternally inherited transcripts and/or de novo expression. These findings likely have implications in reprogramming of the parental genomes of egg and sperm cell to adopt the zygotic/embryonic program as suggested in animal studies (Tadros and Lipshitz, 2009). The observations from this study will likely provide important leads to investigate further biological implications of reprogramming of parental genomes after fertilization in plants.
Figure 3.
Examples of paternal- and maternal-enriched gene expression in early Arabidopsis embryogenesis. A to E, Expression of a paternally enriched gene (At3g28780) using GUS reporter after fertilization in the zygote. The At3g28780 promoter:GUS line showed pollen-specific expression (A), no detectable expression in the ovule before fertilization (B), and expression in the zygote after fertilization (C). No GUS expression was detected in the zygote (red outline) when this GUS reporter line was used as female parent and crossed with pollen from wild-type male parent (D). In the reciprocal cross (pollen from the GUS reporter line as male parent and wild-type female), the GUS expression was observed after fertilization in the zygote (E). The red star and red outline in C and E indicate GUS expression in the pollen tube and zygote, respectively. F to J, De novo expression of a maternally enriched gene (At4g07410) tagged with GFP in the zygote. The GFP reporter line showed no detectable expression in the pollen (F) or in the pollen tube (G) but showed expression in the female gametophyte (H). The inserts in G (1 and 2) showed 4′,6-diamino-phenylindole staining of sperm nuclei (1) but no GFP signal (2). The GFP signal was also detected in the unfertilized ovule and specifically in the embryo sac nuclei, namely, egg cell, two synergids, two polar nuclei, and three antipodals (embryo sac, yellow outline; H). When the pollen from the GFP reporter line (no detectable expression of GFP) was crossed with wild type as female, the GFP signal was observed in the zygote (yellow outline) and the endosperm nuclei (red stars; I). In the reciprocal cross where the pollen from wild type was crossed into transgenic reporter line as female, GFP expression was observed in the zygote (J). Bar = 0.05 mm (A–E, H, and I) and 0.01 mm (F, G, and J).
Higher Order Gene Expression Patterns
To explore the dynamic gene expression patterns across embryo stages, we compared the modulated genes between adjacent embryonic stages. We found that most of the modulated genes (44%) are embryo stage transition-specific genes, implying that those differentially expressed genes are significant to that stage and then remain at similar expression levels for the rest of embryogenesis (Supplemental Table S9). To further analyze gene expression patterns across embryogenesis, as described previously (Yu et al., 2007), we classified the expressed genes into seven groups according to the number of embryo stage samples analyzed. We found that most of the genes were expressed across multiple embryonic stages, 73% of the genes were expressed in more than five stages (Supplemental Tables S3 and S4). These results suggest that many genes and their associated biological processes are shared by different embryo stages. Similar observations were made in a recent independent study using seeds at different developmental stages (Le et al., 2010). To test if the observed gene expression patterns were associated with specific chromosomes, we further examined the distribution of these seven groups on five Arabidopsis chromosomes. This analysis suggests that broadly expressed genes during embryogenesis were enriched on chromosomes 1 and 2, whereas, stage-specific genes were predominantly distributed on chromosomes 3, 4, and 5 (Supplemental Fig. S5a). Consistently, more modulated genes can be found on chromosomes 3, 4, and 5 compared to chromosomes 1 and 2 (Supplemental Fig. S5b). To test if these expression patterns observed in Arabidopsis embryos is a general phenomenon across organisms, similar analysis was performed using microarray data sets available for Caenorabditis elegans embryo development (Hill et al., 2000). Interestingly, a similar chromosome-specific distribution of higher-order gene expression was also observed in C. elegans (Supplemental Fig. S5c). We next investigated the highly expressed gene clusters and their neighbors on chromosomes in Arabidopsis. Statistical analysis of the highly expressed gene clusters (see “Materials and Methods”) showed that chromosome 5 is enriched for such clusters (Supplemental Tables S10 and S11). Furthermore, comparing these clusters across stages showed that zygote and quadrant stage embryos shared most of these clusters compared to the later embryo stages, suggesting that a significant number of these clusters (6/15) are stage specific (Supplemental Table S11). Together, these analyses suggest coregulation at higher-order level, namely, the chromosomal or chromosomal segment level. The significance of chromosomal context for coregulated gene clusters observed in this study may have similar implications to gene expression as highlighted for animal systems in a recent review (Cremer and Cremer, 2010).
Validation and Functional Analysis
We validated the microarray data by qRT-PCR for 20 selected genes and the results showed good correlation with the microarray studies (Supplemental Table S8, f and g). We conducted similar correlation analysis using virtual data sets of Genevestigator (Zimmermann et al., 2004) and the eFP browser (Winter et al., 2007). Unlike our data set, these data sets were generated from developing whole seeds (Hennig et al., 2003; Schmid et al., 2005; Winter et al., 2007). For the majority of the genes analyzed, especially those active in late embryogenesis, the expression patterns were comparable (Supplemental Fig. S2a). In addition to the above analyses, we isolated putative promoters of 12 differentially expressed genes and tested their expression patterns with a GUS reporter. Ten of the 12 selected genes showed GUS expression in the embryos consistent with the corresponding microarray data sets (Fig. 4; Supplemental Table S8c). Together, these validation studies provided experimental evidence for the microarray data sets generated in this study.
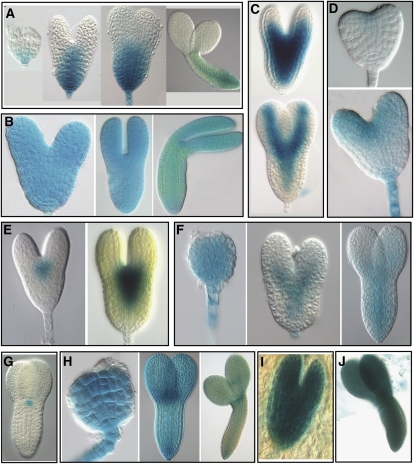
Figure 4.
Expression patterns of GUS reporter constructs during Arabidopsis embryo development. Promoter:GUS transcriptional fusion constructs were generated with 10 selected embryo-expressed genes: At4g17800 (A); At1g67320 (B); At5g20885 (C); At5g43250 (D); At5g66070 (E); At1g27470 (F); At2g27250 (G); At5g41880 (H); At5g37478 (I); and At5g63780 (J). The expression values of these genes are shown in Supplemental Table S8c. These 10 reporter constructs were introduced into Arabidopsis and the corresponding transgenic lines were analyzed for GUS expression during embryo development. A range of expression patterns were observed including broad expression in the embryo (B, H–J), expression in the basal region (A), predominant expression in the apical region of the embryo and suspensor (D), expression in the axis and provascular domain (C and F), and expression in the shoot apical meristem (E and G) that includes CLAVATA3 (At2g27250)-based GUS reporter with a consistent expression pattern as previously reported (G). Overall, these GUS reporter expression patterns are consistent with the corresponding microarray results.
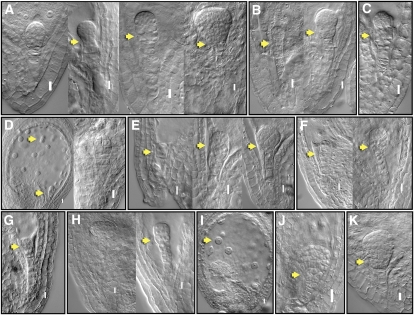
We selected 16 genes encoding TFs that were modulated at different stages of embryo development for functional characterization (Supplemental Table S8, d and h). Loss-of-function screens for T-DNA insertion lines for these genes did not produce detectable embryo phenotypes, presumably due to functional redundancy. In another approach, we used the Drosophila Engrailed (En) repressor domain that was shown to produce dominant-negative phenotypes in Arabidopsis (Markel et al., 2002): 11 of the 16 TF constructs showed a range of strong embryo phenotypes (Fig. 5; Supplemental Table S8, d and h), and the rest showed weaker embryo, endosperm, and in some cases postembryonic phenotypes (Supplemental Table S8, d and h). Among these, five putative zinc-finger-encoding genes that are modulated in Z and Q stages caused developmental arrest at early stages of embryogenesis. These include, developmental arrest at two-, four-, and eight-cell stages and additionally also displayed abnormal cell divisions further affecting basal lineage, suspensor, and endosperm development (Fig. 5). These phenotypes suggest that the putative zinc-finger TFs are likely redundantly involved in conferring important functions during early phases of embryogenesis in Arabidopsis. Interestingly, recent studies have also implicated zinc-finger TFs in the early embryo development in Drosophila (Liang et al., 2008). We have also observed mutant embryo phenotypes with BELL1 LIKE, WOX6, TCP16, and SCL7 TFs (Fig. 5; Supplemental Table S8, d and h). These dominant phenotypes observed for some of the TFs suggest that their expression and functions may be critical for embryo development.
Figure 5.
Functional analysis of selected TFs during Arabidopsis embryo development. Nomarski images of embryo-defective phenotypes observed with translational fusions of TFs with En repressor domain under the control of 35S promoter (“Materials and Methods”). Putative zinc-finger TFs: At3G24050 (A); At3G51080 (B); At5G66320 (C); At3G54810 (D); At4G32890 (E); HD-ZIP At4G37790 (F); BELL1 LIKE homeodomain, A4G34610 (G); WOX6, At2G28610 (H); TCP16, At3G45150 (I); GRAS family SCL7, At3G50650 (J); and MYB, At3G55730 (K). Embryo phenotypes (arrows) observed include arrest at two-cell stage (E), quadrant (G), and octant (I); defective hypophyseal cell division (J); abnormal divisions in the basal region of the embryo at later stages of development (A, B, E, F, H, and K); abnormal divisions in the upper suspensor cells (B and C); and abnormal endosperm cell division (D and I). Details of observed phenotypes summarized in Supplemental Table S8d. Bar = 0.01 mm.
Dynamic Regulation of Metabolic Networks
A hallmark of plant embryos is their accumulation of storage reserves and secondary metabolites (Vicente-Carbajosa and Carbonero, 2005). However, there is very little information on the dynamic aspects of metabolism vis-à-vis embryo development. Using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa and Goto, 2000) and our embryo-specific global gene expression data, we constructed stage-transition metabolic networks that contains 723 nodes and 1,568 links (Supplemental Fig. S6; Supplemental Table S6; “Materials and Methods”). The torpedo to bent stage (T → B) transition metabolic network that displayed most dynamic changes is shown in Figure 6. In this study, we focused on the genes associated with biochemical pathways in fatty acid, carbohydrate, amino acid, nucleotide, vitamin metabolism, as well as tricarboxylic acid cycle. The six-stage transition networks were constructed by mapping the modulated genes of adjacent stages onto the network. As shown in Supplemental Figure S6, the resulting networks suggest that gene regulation between adjacent stages is highly dynamic during embryogenesis (Supplemental Table S13). We studied the gene regulatory patterns in the networks by examining the network structure characteristics, i.e. upstream nodes, downstream nodes, hubs, and cutpoints (see “Materials and Methods”; Supplemental Table S12b, legend). We identified 71 network hubs that represent intersections of many pathways that are preferentially regulated in most stage transitions (Supplemental Tables S12b and S14) whereas many cutpoints and their downstream nodes are least preferentially regulated (Supplemental Table S15). The gene coexpression network modules, clusters of nodes that are either up- or down-regulated in the networks, were examined using modulated genes in stage-transition networks. The relatively larger up-regulated coexpression network modules were found in the Q → G, G → H, H → T, and T → B stage transition networks whereas the largest down-regulated network module was found in the B → M transition network (Supplemental Fig. S7a). These network modules reflect the turning on or off of different metabolic activities such as fatty acid, carbohydrate, and protein synthesis during stage transitions.
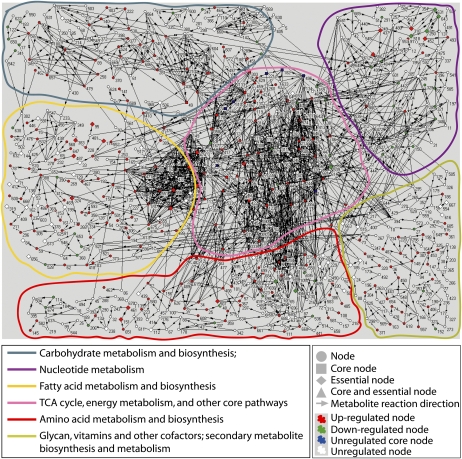
Figure 6.
Illustration of the dynamic metabolic network for torpedo-bent stages of Arabidopsis embryo development. The network model was generated in Pajek using the microarray data sets and KEGG database (Supplemental Table S6; “Materials and Methods”). Numbers represent the metabolites and the lines (with arrows) that connect the metabolites represent the genes encoding enzymes that catalyze their reactions (Supplemental Table S6). The pathways that represent six key biochemical reactions for carbohydrates, nucleotides, fatty acids, tricarboxylic acid cycle, amino acids, and vitamins are outlined in color (details in the bottom left box; see Supplemental Fig. S6 for metabolic networks of other embryo stages).
The top 10% of the highly expressed genes across all embryo stages involved in metabolism was mapped onto the network to identify the network core (“Materials and Methods”). The network core contains 28 connected nodes (reactions), most of which are involved in glycolysis, pentose phosphate pathway, pyruvate metabolism, and carbon fixation (Supplemental Table S16), indicating the essential role of these nodes in core metabolic activities that operate during embryogenesis (Supplemental Table S17). A significantly larger number of links was found between nodes in the network core (Supplemental Table S18) that is consistent with the overrepresentation of hubs in the core (i.e. 46% of the core nodes are hubs; hubs are highly connected nodes representing reactions that share metabolites, see Supplemental Table S12b). These characteristics of the network core indicate that metabolites can be easily converted in both directions in the core. Interestingly, the stage transitions between Q and T stages showed higher number of up-regulated nodes than the others (Supplemental Table S19), highlighting the coordinated elaboration of metabolic pathways during embryo development.
Gene-Regulation Relationships between the Network Core and Its Periphery
Examination of the fractions of differentially regulated (modulated) nodes in nth-order neighbors from the network core in the transition networks (“Materials and Methods”) revealed a positive correlation between the increase of modulated nodes in the core and their neighbors (Supplemental Table S20), indicating that they are coordinately regulating metabolic activities. Closer look at the up- and down-regulated pathways in these transition networks revealed that fatty acid biosynthesis and other metabolism-related genes were significantly up-regulated in the transition networks, namely, H → T and T → B (Supplemental Table S21, a and b). Furthermore, we observed that carbohydrate metabolism and biosynthesis genes were up-regulated relatively earlier than fatty acid biosynthesis, whereas the protein synthesis genes were up-regulated relatively later than fatty acid biosynthesis (Supplemental Table S21, a and b). However, genes involved in the metabolism of fatty acids and carbohydrates were all significantly up-regulated from the G and H to B embryo stages consistent with previous reports (Girke et al., 2000; Schmid et al., 2005).
Embryo-Defective Mutations Maps to End Nodes
To gain biological insights into the metabolic network models, we selected 52 of the known Arabidopsis embryo-defective mutants (EMBs; 358 EMBs listed at www.seedgenes.org; Tzafrir et al., 2003) associated with metabolic pathways that cause embryo lethality (Tzafrir et al., 2003, 2004) and mapped them onto the metabolic networks (Supplemental Table S12a). By calculating the enrichment of different node types (Supplemental Table S12b), we found that the end nodes (i.e. the last reaction in a pathway) are enriched for EMBs. These EMBs are associated with biosynthetic and metabolic pathways of fatty acids, carbohydrates, nucleotides, and amino acids. Therefore, lesions in the genes associated with the end nodes are not compensated by alternative feedback from neighboring pathways and can result in embryo lethality and perturb embryo development (Supplemental Fig. S8; www.seedgenes.org; Kajiwara et al., 2004). However, interestingly EMBs associated with hub nodes display patterning defects (e.g. acc1), suggesting that these metabolic lesions also likely impact on embryo developmental programs.
CONCLUSION
In this study, we have successfully isolated live zygote to late-stage embryos of Arabidopsis and studied global gene expression patterns that regulate embryogenesis. To our knowledge, this represents the first genome-wide gene expression profiling of embryogenesis in Arabidopsis and in plants, and the data presented here will serve as foundational resource for future studies addressing fundamental molecular and developmental mechanisms that govern plant embryogenesis. The digital expression of individual genes and entire data sets for Arabidopsis embryo development can be viewed and downloaded (Supplemental Table S1; Supplemental Fig. S1b) at http://www2.bri.nrc.ca/plantembryo/. The highlighted in-depth analyses clearly show the dynamics of embryo-specific gene expression and metabolic pathways as the embryo progresses from a zygote to a physiologically mature embryo. The metabolic networks developed in this study provide an integrated view of modulated and progressively elaborated biochemical pathways. Further, mapping of critical genes in embryogenesis on the networks illustrates the validity and utility. Implications of the findings from this study of Arabidopsis embryogenesis will contribute to future plant embryo research.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. Plants were grown under 16-h light/8-h dark photoperiod with constant temperature of 22°C at 120 μE m−2 s−1 light intensity.
Embryo Dissection, Total RNA Isolation, and Microarray Experiments
Embryo isolations were performed as described in Figure 1 using the dissecting microscope and fine forceps (Dumont 55 forceps, catalog no. 11295-55, Fine Science Tools) in a 5% Suc solution that contained 0.1% RNALater (Ambion, catalog no. AM7021) solution. Total RNA was extracted from each stage embryo sample following the protocol of RNAqueous-micro kit (Ambion, catalog no. 1927). These RNAs were used to make probes for the microarray experiments as well as for qRT-PCR analysis (Supplemental Table S22).
RNA Amplification and Labeling
The quantity of RNA isolated from the embryos was insufficient for preparation of probes for the microarray experiments. Therefore the mRNA was amplified prior to labeling. The first round of mRNA amplification was conducted according to the protocol provided in the MessageAmp aRNA kit (Ambion, catalog no. 1750). During the second round of amplification, aminoallyl-UTP was incorporated into the newly synthesized aRNA; 3 μL of aminoallyl-UTP (50 mm) plus 2 μL of UTP (75 mm) instead of 4 μL of UTP were added. The purpose of incorporating aminoallyl-UTP is to provide a reactive chemical group to which the fluorescent dyes can be attached. After purification of the aRNA, the NHS-ester dyes were coupled to the modified bases of aRNA in a chemical reaction.
Microarray Experimental Design and Hybridization
The Arabidopsis 70-mer oligo array slides prepared by University of Arizona were used in all the microarray experiments (http://ag.arizona.edu/microarray/). Antisense RNA labeling was performed following the protocol of Wellmer et al. (2004). The aRNA samples representing four biological replicates from each of the seven embryo stages were labeled (two cy3 and two cy5) and hybridized to these slides following the protocol described in http://ag.arizona.edu/microarray and the experimental design shown in Supplemental Figure S1a. Hybridized slides were scanned sequentially for Cy3- and Cy5-labeled mRNA targets with a ScanArray 4000 laser scanner at a resolution of 10 μm. The image analysis and signal quantification were performed using the QuantArray program (GSI Lumonics).
qRT-PCR
Embryo isolation was performed as described in Figure 1 and total RNA samples were isolated from different embryo stages as described in RNAqueous-micro kit (Ambion, catalog no. 1927) and the respective double-strand cDNAs were produced using amplified aRNA following the protocol of MessageAmpTM II aRNA kit (Ambion, catalog no. 1751). Gene-specific primers were designed using Primer 3 software. qRT-PCR reactions were performed using the protocol and equipment of Applied Biosystem Step One.
Construction of En-TF Plasmids
PCR fragments were amplified with Phusion Taq polymerase (Biolabs) and subcloned into pSTBlue-1 (Clontech) or pENTR/D-TOPO (Invitrogen) before cloning in the binary vectors. A new multiple cloning site (MCS) including a C-terminal E-tag (5′-GTTTAAACCAACTAGTAAAGATCTACAAGTTTGTACAAAGTGGTTCCGGGTGCGCCGGTGCCGTATCCGGATCCGCTGGAACCGCGTGCTCGAGCATCGCGAGCTCTAGA-3′) was generated by overlapping primers and cloned in the PmeI and XbaI sites of the binary Gateway destination vector pK7WG2 (VIB-Ghent University). The t35S terminator was amplified with primers 5′-CACCTCGCGATGACGGCCATGCTAGAGTCCGCA-3′ and 5′-TCTAGAGTCACTGGATTTTGGTTTTAGG-3′, and cloned as an NruI/XbaI fragment in the respective sites of the new MCS. The BsrGI Gateway cassette (GW) fragment and the PmeI/SpeI p35S promoter fragment were reintroduced from pK7WG2 by cloning in the respective sites of the new MCS resulting in pER310. The En repressor domain was PCR amplified from pLD16125 (Drosophila Genomics Resource Centre) with primers 5′-CACCACTAGTATGGCCCTGGAGGATCGCTG-3′ and 5′-AGATCTGGATCCCAGAGCAGATTTCTC-3′ and inserted at the N-terminal side of the GW site in pER310 resulting in pER311. To accommodate different reading frames of a collection of TFs in relation to the upstream Gateway attL1 site, the intermittent BglII site was filled in with Klenow enzyme and self ligated (pER311A). The entry clones containing the TF genes were recombined with the pER311A using LR clonase (Invitrogen) and the resulting expression T-DNA vectors were sequenced and after confirmation shuttled into Agrobacterium strain MP90 by three-parental mating.
Microarray Analysis
Limma Software (Smyth, 2004) was used to normalize and to determine the modulated genes from microarray data. Signal background correction was carried out using normexp method and offset = 50 as suggested by Limma. To determine which genes are expressed in each embryo stage, we used the single-channel normalization. Within-array normalization was carried out using loess method while between-array normalization was performed using a quantile method (Smyth, 2004). For individual genes from each embryo stage, we performed t tests using the expression values of the gene and the black spots (background spots) on the chips. To estimate the expressed genes in a conserved manner, we multiplied the values of black spots by 1.05. False-positive discovery rate was applied to correct raw P values. If a corrected P value is less than 0.05, the gene is counted as expressed. To determine the modulated genes between embryo stages, we applied the two-channel normalization. The methods applied for within and between array normalizations, were the same as described above. Empirical Bayes statistics was applied to determine the differential expression of the genes (Smyth, 2004).
Microscopy
Isolated embryos representing different key stages of Arabidopsis embryogenesis were cleared in chloral hydrate solution (8:1:2, chloral hydrate:glycerol:water; w/v/v) and viewed under Leica DMR compound microscope with Nomarski optics. Images were captured using the Magnafire camera (Optronics) and were edited in Abobe Photoshop CS.
Network and Statistical Analysis
Microarray data were processed using Limma package (Smyth, 2004). Metabolic network construction, analysis, and randomization tests were followed as described previously (Wang and Purisima, 2005; Tibiche and Wang, 2008). Details of these analyses are provided in “Materials and Methods.”
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Microarray experimental design and snapshot of the embryo-specific gene expression Web site.
Supplemental Figure S2. Expression patterns of biosynthesis- and development-related genes, auxin-related genes, and abscisic acid-related genes.
Supplemental Figure S3. Expression patterns of ribosomal and proteasomal genes.
Supplemental Figure S4. Pathway-associated genes are coordinately expressed.
Supplemental Figure S5. Gene distribution on Arabidopsis and Caenorabditis elegans chromosomes.
Supplemental Figure S6. Dynamic regulation of metabolic networks during embryo development.
Supplemental Figure S7. Number of genes expressed at each of the seven embryo stages and comparison of gene expression levels in our data versus the laser capture microdissection study.
Supplemental Figure S8. Metabolic lesions affect embryo development.
Supplemental Table S1. Gene expression values of different embryo developmental stages.
Supplemental Table S2. Shared expressed genes between the pollen or female with the zygote.
Supplemental Table S3. Overlap of expressed genes during seven embryo stages.
Supplemental Table S4. List of embryo stage-specific genes.
Supplemental Table S5. Expression levels of genes related to plant hormone pathways.
Supplemental Table S6. Genes and their respective KEGG/Pajek identifications used for networks.
Supplemental Table S7. Number of genes expressed at each of the seven embryo stages and comparison of gene expression levels in our data versus the LCM study.
Supplemental Table S8. Percentage of modulated TFs, expression levels of chromatin modification genes and data related to promoter-GUS analysis, qRT-PCR analysis and dominant-negative studies.
Supplemental Table S9. Modulated genes overlap between transition stages.
Supplemental Table S10. Number of observed gene domains in Arabidopsis chromosomes.
Supplemental Table S11. Highly expressed gene domains in Arabidopsis embryogenesis.
Supplemental Table S12. Arabidopsis EMBs mapped on the metabolic network and their distribution on the network.
Supplemental Table S13. Modulated genes and nodes represented on metabolic networks.
Supplemental Table S14. Enrichment of modulated network hubs in embryo stage transitions.
Supplemental Table S15. Network cut points and input nodes are nonpreferentially modulated.
Supplemental Table S16. Core nodes on the metabolic networks.
Supplemental Table S17. Pathway enrichment in the network core.
Supplemental Table S18. Modulated nodes represented as links in the network core.
Supplemental Table S19. Up- and down-regulated nodes in the network core.
Supplemental Table S20. Fraction of modulated nodes in core and its nth order neighbors.
Supplemental Table S21. Modulated pathways in transition networks.
Supplemental Table S22. Arabidopsis embryos dissected for microarray analysis.
Acknowledgments
This is National Research Council of Canada publication number 50156. We thank Sandra Stone and Don Palmer for critical comments on the manuscript.
D.X., P.V., E.W., and R.D. designed and performed the experiments. C.T., E.W., and M.C. analyzed the microarray data. H.Y. and V.B. contributed to microarray experiments and GUS reporter assays in transgenics. E.R. contributed to engrailed gene constructs and their analysis in transgenic plants. Y.C. performed qRT-PCR experiments. W.K. and G.S. contributed valuable advice and reagents. D.X., P.V., E.W., and R.D. wrote the manuscript with contributions from G.S.
References
- Alonso JM, Ecker JR. (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7: 524–536 [DOI] [PubMed] [Google Scholar]
- Baroux C, Autran D, Gillmor CS, Grimanelli D, Grossniklaus U. (2008) The maternal to zygotic transition in animals and plants. Cold Spring Harb Symp Quant Biol 73: 89–100 [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ. (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Carroll S, Grenier J, Weatherbee S. (2005) From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, Ed 2. Blackwell Science, Malden, MA [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K. (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42: 111–123 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. (2010) Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Ma Y, Jaramillo M, Bari H, Awan A, Yang S, Zhang S, Liu L, Lu M, O’Connor-McCourt M, et al. (2007) A map of human cancer signaling. Mol Syst Biol 3: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128: 635–638 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. (2007) Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Hennig L, Menges M, Murray JA, Gruissem W. (2003) Arabidopsis transcript profiling on Affymetrix GeneChip arrays. Plant Mol Biol 53: 457–465 [DOI] [PubMed] [Google Scholar]
- Hill AA, Hunter CP, Tsung BT, Tucker-Kellogg G, Brown EL. (2000) Genomic analysis of gene expression in C. elegans. Science 290: 809–812 [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S. (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5: 4864–4884 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh T-F, Fischer R. (2007) Endosperm gene imprinting and seed development. Curr Opin Genet Dev 17: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Levy R, Barkai N. (2004) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22: 86–92 [DOI] [PubMed] [Google Scholar]
- Kajiwara T, Furutani M, Hibara K, Tasaka M. (2004) The GURKE gene encoding an acetyl-CoA carboxylase is required for partitioning the embryo apex into three subregions in Arabidopsis. Plant Cell Physiol 45: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Wagmaister JA, Kawashima T, Bui AQ, Harada JJ, Goldberg RB. (2007) Using genomics to study legume seed development. Plant Physiol 144: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. (2008) The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel H, Chandler J, Werr W. (2002) Translational fusions with the engrailed repressor domain efficiently convert plant transcription factors into dominant-negative functions. Nucleic Acids Res 30: 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CB, Ooi SKT, Bestor TH, Bourc’his D. (2007) Epigenetic decisions in mammalian germ cells. Science 316: 398–399 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Spencer MWB, Casson SA, Lindsey K. (2007) Transcriptional profiling of the Arabidopsis embryo. Plant Physiol 143: 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. (2007) Regulation of the oocyte-to-zygote transition. Science 316: 407–408 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. (2009) The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Tibiche C, Wang E. (2008) MicroRNA regulatory patterns on the human metabolic network. Open Systems Biology Journal 1: 1–8 [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. (2003) The Arabidopsis seedgenes project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J-P, Baskar R, Grossniklaus U. (2000) Delayed activation of the paternal genome during seed development. Nature 404: 91–94 [DOI] [PubMed] [Google Scholar]
- Wang E, Purisima E. (2005) Network motifs are enriched with transcription factors whose transcripts have short half-lives. Trends Genet 21: 492–495 [DOI] [PubMed] [Google Scholar]
- Wei J, Sun M. (2002) Embryo sac isolation in Arabidopsis thaliana: a simple and efficient technique for structure analysis and mutant selection. Plant Mol Biol Rep 20: 141–148 [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-J, Hogan P, Sundaresan V. (2005) Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol 139: 1853–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Jian Z, Shen S-H, Purisima E, Wang E. (2007) Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res 35: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]