Abstract
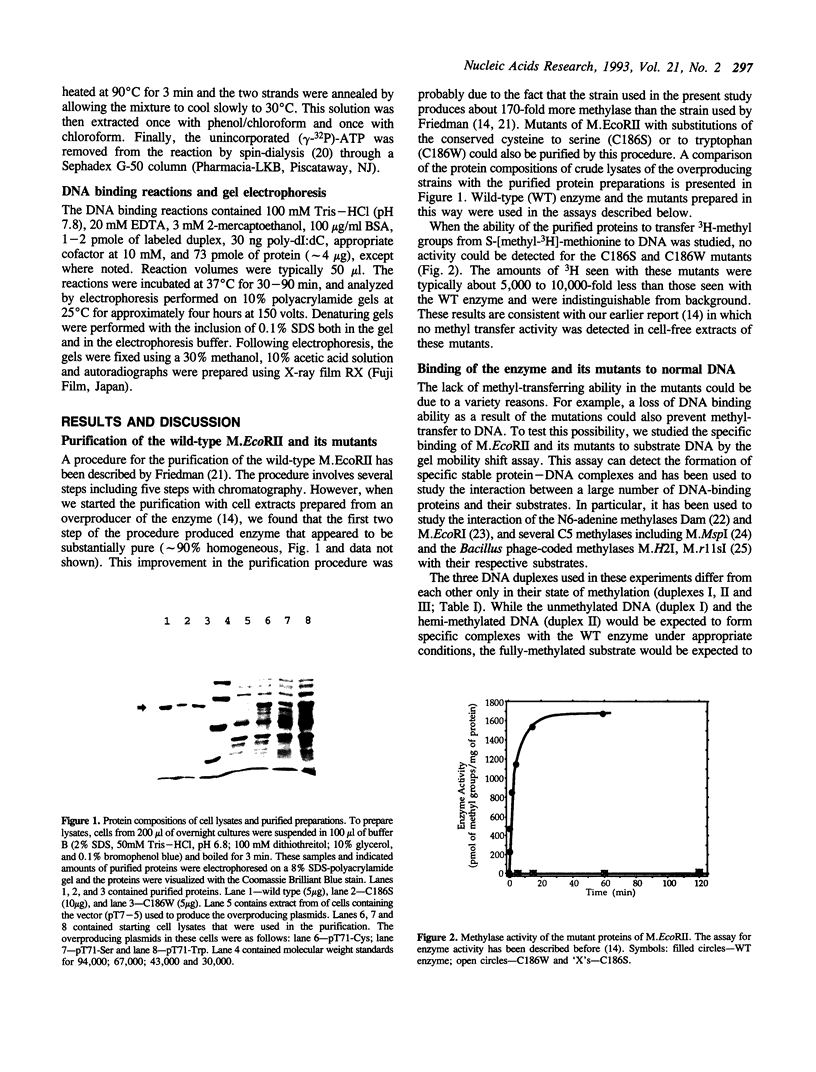
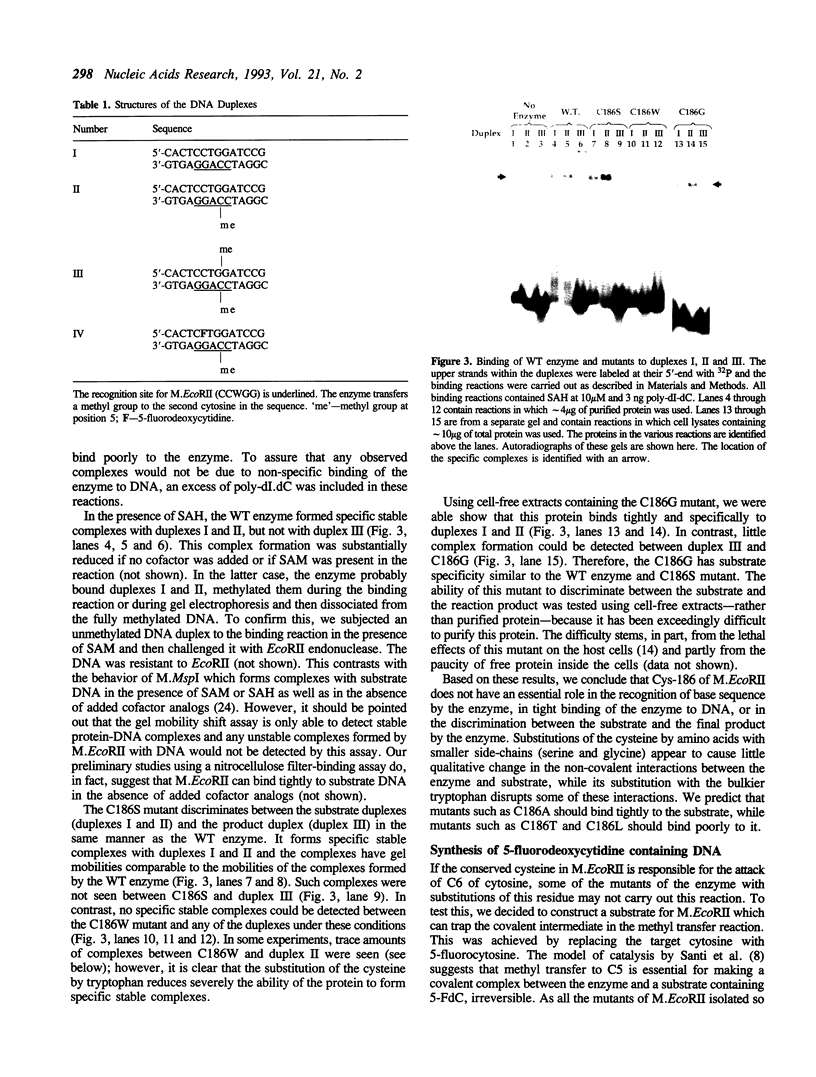
All DNA (cytosine-5)-methyltransferases contain a single conserved cysteine. It has been proposed that this cysteine initiates catalysis by attacking the C6 of cytosine and thereby activating the normally inert C5 position. We show here that substitutions of this cysteine in the E. coli methylase M. EcoRII with either serine or tryptophan results in a complete loss of ability to transfer methyl groups to DNA. Interestingly, mutants with either serine or glycine substitution bind tightly to substrate DNA. These mutants resemble the wild-type enzyme in that their binding to substrate is not eliminated by the presence of non-specific DNA in the reaction, it is sensitive to methylation status of the substrate and is stimulated by an analog of the methyl donor. Hence the conserved cysteine is not essential for the specific stable binding of the enzyme to its substrate. However, substitution of the cysteine with the bulkier tryptophan does reduce DNA binding. We also report here a novel procedure for the synthesis of DNA containing 5-fluorocytosine. Further, we show that a DNA substrate for M. EcoRII in which the target cytosine is replaced by 5-fluorocytosine is a mechanism-based inhibitor of the enzyme and that it forms an irreversible complex with the enzyme. As expected, this modified substrate does not form irreversible complexes with the mutants.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Patel L., Rauscher F. J., 3rd, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990 Sep 7;249(4973):1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- Barone A. D., Tang J. Y., Caruthers M. H. In situ activation of bis-dialkylaminophosphines--a new method for synthesizing deoxyoligonucleotides on polymer supports. Nucleic Acids Res. 1984 May 25;12(10):4051–4061. doi: 10.1093/nar/12.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellisario R. L., Maley G. F., Galivan J. H., Maley F. Amino acid sequence at the FdUMP binding site of thymidylate synthetase. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1848–1852. doi: 10.1073/pnas.73.6.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W. The double role of methyl donor and allosteric effector of S-adenosyl-methionine for Dam methylase of E. coli. Nucleic Acids Res. 1990 Aug 11;18(15):4369–4375. doi: 10.1093/nar/18.15.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Dubey A. K., Roberts R. J. Sequence-specific DNA binding by the MspI DNA methyltransferase. Nucleic Acids Res. 1992 Jun 25;20(12):3167–3173. doi: 10.1093/nar/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Friedman S. The irreversible binding of azacytosine-containing DNA fragments to bacterial DNA(cytosine-5)methyltransferases. J Biol Chem. 1985 May 10;260(9):5698–5705. [PubMed] [Google Scholar]
- Lange C., Jugel A., Walter J., Noyer-Weidner M., Trautner T. A. 'Pseudo' domains in phage-encoded DNA methyltransferases. Nature. 1991 Aug 15;352(6336):645–648. doi: 10.1038/352645a0. [DOI] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Appelt K., Oatley S. J., Xuong N. H. Crystal structure of Escherichia coli thymidylate synthase containing bound 5-fluoro-2'-deoxyuridylate and 10-propargyl-5,8-dideazafolate. J Mol Biol. 1990 Aug 20;214(4):923–936. doi: 10.1016/0022-2836(90)90346-N. [DOI] [PubMed] [Google Scholar]
- McBride A. A., Klausner R. D., Howley P. M. Conserved cysteine residue in the DNA-binding domain of the bovine papillomavirus type 1 E2 protein confers redox regulation of the DNA-binding activity in vitro. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7531–7535. doi: 10.1073/pnas.89.16.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. O., Mashhoon N. Inhibition of EcoRI DNA methylase with cofactor analogs. J Biol Chem. 1990 May 25;265(15):8966–8970. [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Pein C. D., Fritz H. J., Cech D. Chemical synthesis of 2'-deoxyoligonucleotides containing 5-fluoro-2'-deoxycytidine. Nucleic Acids Res. 1992 May 25;20(10):2421–2426. doi: 10.1093/nar/20.10.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Bhagwat A. S. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992 Jan 25;20(2):319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen R. W., Vertino P. M., Nelkin B. D., Yu J. J., el-Deiry W., Cumaraswamy A., Lennon G. G., Trask B. J., Celano P., Baylin S. B. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992 May 11;20(9):2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]