Abstract
A number of APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) genes have been shown to function in abiotic and biotic stress responses, and these genes are often induced by multiple stresses. We report here the characterization of an AP2/ERF gene in Arabidopsis (Arabidopsis thaliana) that is specifically induced during hypoxia. We show that under normoxic conditions, the expression of AtERF73/HRE1 can be induced by exogenous addition of 1-aminocyclopropane-1-carboxylic acid and that a combination of hypoxia and 1-aminocyclopropane-1-carboxylic acid results in hyperinduction of AtERF73/HRE1 expression. In addition, hypoxic induction of AtERF73/HRE1 is reduced but not completely abolished in ethylene-insensitive mutants and in the presence of inhibitors of ethylene biosynthesis and responses. These results suggest that, in addition to ethylene, an ethylene-independent signal is also required to mediate hypoxic induction of AtERF73/HRE1. To assess the role of AtERF73/HRE1, we generated three independent RNA interference (RNAi) knockdown lines of AtERF73/HRE1. Under normoxic conditions, the AtERF73/HRE1-RNAi seedlings displayed increased ethylene sensitivity and exaggerated triple responses, indicating that AtERF73/HRE1 might play a negative regulatory role in modulating ethylene responses. Gas chromatography analyses showed that the production of ethylene was similar between wild-type and RNAi lines under hypoxia. Quantitative reverse transcription-polymerase chain reaction analyses showed that hypoxia-inducible genes could be affected by AtERF73/HRE1-RNAi lines in two different ways: hypoxic induction of glycolytic and fermentative genes was reduced, whereas induction of a number of peroxidase and cytochrome P450 genes was increased. Taken together, our results show that AtERF73/HRE1 is involved in modulating ethylene responses under both normoxia and hypoxia.
Oxygen deficiency (hypoxia) is an abiotic stress encountered by plants during flooding in soil. The consequences of hypoxia, such as a decrease in cellular energy charge, drop in cytoplasmic pH, and accumulation of toxic end products from anaerobic respiration and of reactive oxygen species during recovery, are responsible for the slowed growth and reduced yield of many agriculturally important crops in the event of flooding (Subbaiah and Sachs, 2003). Plants have developed adaptive mechanisms to sense oxygen deficiency in their environments and make coordinated physiological and structural adjustments to enhance their hypoxic tolerance (Liu et al., 2005; Huang et al., 2008).
Several microarray studies showed that genes coding for enzymes of sugar metabolism, glycolysis, and fermentation are up-regulated in Arabidopsis (Arabidopsis thaliana) under low oxygen concentrations (Klok et al., 2002; Liu et al., 2005; Loreti et al., 2005; van Dongen et al., 2009). Hypoxia-induced genes can be controlled at transcriptional, posttranscriptional, and translational levels (Manjunath et al., 1998; Conley et al., 1999; Peng et al., 2005). In particular, the aerobic-anaerobic switch results in the induction of two genes: ALCOHOL DEHYDROGENASE (ADH) and PYRUVATE DECARBOXYLASE (PDC). Studies of ADH genes in Arabidopsis and maize (Zea mays) have identified a cis-acting element, designated anaerobic-responsive element, that is essential for hypoxia induction (Dolferus et al., 1994; Kyozuka et al., 1994; Hoeren et al., 1998). It was shown that ethylene is necessary but not sufficient for hypoxic induction of ADH in Arabidopsis (Peng et al., 2001, 2005). It was also reported that ethylene regulates aerenchyma formation in root tips of maize plants exposed to hypoxic conditions (He et al., 1996). These observations suggested that ethylene plays an essential role in hypoxia signaling pathways.
The APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) gene family encodes plant-specific transcription factors, which are often involved in regulating plant responses to biotic and abiotic stresses (Dubouzet et al., 2003; Gutterson and Reuber, 2004; Nakano et al., 2006). Expression of many AP2/ERF genes has been shown to be regulated by a variety of external stimuli, such as wounding, jasmonic acid (JA), salicylic acid (SA), ethylene, and infection by pathogens (McGrath et al., 2005; Pré et al., 2008). ERF proteins that bind to the GCC box, an ethylene-responsive element, have been identified from several plant species (Gu et al., 2000; Ohta et al., 2000; Zhang et al., 2004). Constitutive overexpression of Arabidopsis ERF1 (At3g23240) activates the expression of PLANT DEFENSIN1.2 (PDF1.2) and BASIC CHITINASE (ChiB) genes (Lorenzo et al., 2003). ORA59 (At1g06160), another member of the Arabidopsis AP2/ERF family, could also activate PDF1.2 gene expression and was shown to be involved in the cross talk between the JA and ethylene signal transduction pathways (Pré et al., 2008). In addition to positive regulatory roles, some AP2/ERF factors have negative regulatory functions. For example, ERF4 (At3g15210) down-regulates the expression of PDF1.2 (McGrath et al., 2005).
AP2/ERF genes have been reported to be involved in signaling pathways associated with abiotic stresses such as cold and drought; however, studies relating to their roles in hypoxia are very limited. In rice (Oryza sativa), the Sub1 locus contains two or three ERF-like genes whose transcripts are regulated by submergence and ethylene (Xu et al., 2006; Perata and Voesenek, 2007). The cultivars with Sub1A-1 are tolerant of submergence. In deepwater rice, a pair of ERF factors, SNORKEL1 (SK1) and SK2, also play a key role in allowing rice to adapt to flooding terrain (Hattori et al., 2009). In this study, we have investigated the functional roles of ethylene in hypoxia response in Arabidopsis. To mimic natural flooding conditions, we have adopted an “open system” treatment (Drew, 1997), in which only roots are subjected to hypoxia treatment. Using microarray analysis, we identified a number of AP2/ERF genes in Arabidopsis that are induced at different stages of hypoxia treatment. One of these genes, AtERF73/HRE1 (At1g72360), which is induced strongly by hypoxia but not by cold or dehydration, was chosen for further studies. Very recently, a study of the same gene was reported (Licausi et al., 2010). The authors showed that overexpression of this gene resulted in increases of PDC and ADH expression during hypoxia but not under normoxia, suggesting a positive regulatory role of AtERF73/HRE1 during hypoxia. In addition, it was shown that another member in the same subfamily, RAP2.2, might also be important for survival of Arabidopsis under hypoxia and that its expression is mainly affected by darkness and ethylene (Hinz et al., 2010). However, we show here that AtERF73/HRE1 was involved in modulating ethylene responses under both normoxia and hypoxia. In addition, our results also indicate that two pathways, one ethylene dependent and the other ethylene independent, are involved in hypoxia induction of AtERF73/HRE1.
RESULTS
AtERF73/HRE1 mRNA Accumulation Is Controlled by Hypoxia and Ethylene Signal Transduction Pathways
By comparing our microarray data with published microarray data, we found that AtERF73/HRE1, corresponding to At1g72360, could only be induced during hypoxia but not by other abiotic stresses, including cold, drought, and dehydration. Recently, it was shown that among group VII ERF genes, AtERF73/HRE1 and AtERF71/HRE2 could be induced by hypoxia treatment, in which the entire seedlings were subjected to low-oxygen conditions (Licausi et al., 2010). Similarly, under our hypoxia treatment conditions, AtERF73/HRE1 and AtERF71, but not RAP2.2, RAP2.12, and RAP2.3, could be induced in roots (Supplemental Fig. S1). However, no significant change of AtERF73/HRE1 transcripts was observed in the shoots (Supplemental Fig. S2).
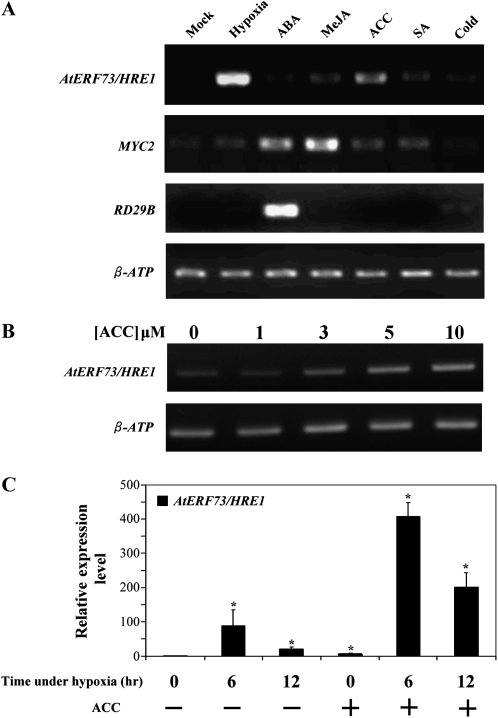
To investigate the effects of various signaling molecules, we used reverse transcription (RT)-PCR to compare the transcript levels of AtERF73/HRE1 from roots of Arabidopsis plants under hypoxia, abscisic acid (ABA), methyl jasmonate (MeJA), 1-aminocyclopropane-1-carboxylic acid (ACC; a precursor of ethylene), SA, or cold treatment. The data showed that AtERF73/HRE1 was highly induced during hypoxia, moderately induced by ACC treatment, and weakly induced upon MeJA or SA treatment (Fig. 1A, top panel). By contrast, RD29B was highly induced by ABA, while MYC2 could be induced by ABA and MeJA (Fig. 1A, middle panels). The expression patterns for RD29B and MYC2 are consistent with published results (Nakashima et al., 2006; Dombrecht et al., 2007). Next, we examined the dosage effect of ACC on AtERF73/HRE1 expression. The results showed that the mRNA accumulation of AtERF73/HRE1 reached a maximal level at ACC concentrations between 5 and 10 μm (Fig. 1B). To investigate whether there is a synergistic effect of hypoxia and ethylene, we determined by quantitative RT-PCR the level of AtERF73/HRE1 transcript after different combinations of hypoxia and ACC treatment. The AtERF73/HRE1 mRNA level was induced after 6 h of hypoxia treatment and decreased after treatment for 12 h. Plants subjected to both hypoxia and ACC treatments displayed a significant increase in the level of AtERF73/HRE1 transcripts as compared with those treated with hypoxia alone (Fig. 1C). These results demonstrate that hypoxia and ethylene have additive effects on AtERF73/HRE1 mRNA accumulation.
Figure 1.
Effects of abiotic stresses and hormones on AtERF73/HRE1 expression. A, RT-PCR analysis of AtERF73/HRE1 expression. The mRNA levels of AtERF73/HRE1, MYC2, and RD29B in roots of 14-d-old seedlings were determined after mock treatment (lane 1) and after treatment with hypoxia (lane 2), 5 μm ABA (lane 3), 50 μm MeJA (lane 4), 5 μm ACC (lane 5), 50 μm SA (lane 6), or cold (4°C; lane 7) for 6 h. β-ATP was used as a control. Experiments were carried out in triplicate with similar results. B, RT-PCR analysis of the effects of ACC. mRNA levels of AtERF73/HRE1 in 14-d-old seedlings were determined after treatment with 0, 1, 3, 5, or 10 μm ACC for 6 h. C, Quantitative RT-PCR analysis of the effects of combined ACC and hypoxic treatment. Levels of AtERF73/HRE1 mRNA in 14-d-old seedlings were determined after treatment with hypoxia for 6 or 12 h in the presence (+) or absence (−) of 5 μm ACC, respectively. The relative amounts of transcripts were calculated and normalized with β-ATP mRNA. Values represent means ± sd from three biologically independent experiments. * P < 0.05 versus the wild type (one-way ANOVA with Dunnett’s t test).
Hypoxic Induction of AtERF73/HRE1 mRNA Accumulation Is Affected in ein2-5 and etr1-1
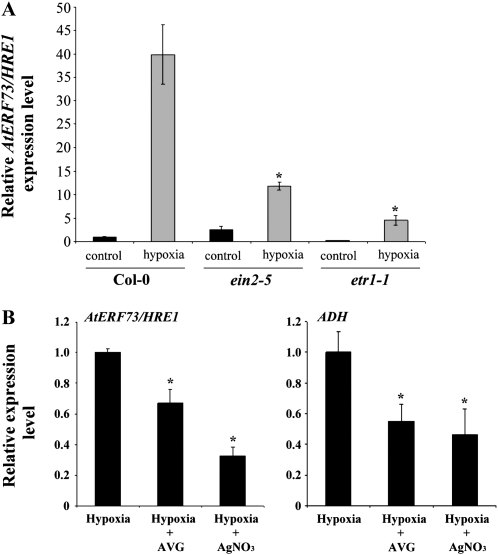
We have previously shown that ethylene is essential for hypoxic induction of ADH in Arabidopsis (Peng et al., 2001). To further investigate the role of ethylene in hypoxic induction of AtERF73/HRE1, we examined the expression pattern of AtERF73/HRE1 during hypoxia in two ethylene signaling mutants, ethylene insensitive2-5 (ein2-5) and ethylene resistant1-1 (etr1-1). EIN2 was shown to be a bifunctional transducer and may mediate the cross talk between ethylene and stress responses (Alonso et al., 1999). The ETR1 gene encodes an ethylene receptor protein with homology to two-component regulators (Chang et al., 1993). As shown in Figure 2A, the level of AtERF73/HRE1 transcript in the wild type increased 40-fold after 6 h of hypoxia treatment. By contrast, the levels of AtERF73/HRE1 in ein2-5 and etr1-1 increased only 3-fold and 8-fold, respectively, as compared with the levels under control conditions. We also examined the effects of aminoethoxyvinylglycine (AVG), an inhibitor of ethylene biosynthesis, and silver nitrate (AgNO3), an inhibitor of ethylene responses, on the expression of AtERF73/HRE1. The levels of AtERF73/HRE1 and ADH mRNA accumulation were reduced under hypoxia in the presence of either AVG or AgNO3 (Fig. 2B). These results indicate that ethylene has a positive role in regulating hypoxic induction of AtERF73/HRE1 mRNA accumulation. However, the fact that hypoxic induction of AtERF73/HRE1 in both ethylene-insensitive mutants and in the presence ethylene inhibitors is only reduced, but not completely abolished, suggests that an additional ethylene-independent signaling pathway is also involved in the induction of AtERF73/HRE1 during hypoxia.
Figure 2.
Effects of ethylene-responsive mutations and inhibitors of ethylene biosynthesis or response on the hypoxic induction of AtERF73/HRE1. A, Quantitative RT-PCR analysis of AtERF73/HRE1expression in ein2-5 and etr1-1 mutants under hypoxic conditions. Total RNAs were isolated from roots of Col-0, ein2-5, and etr1-1 after hypoxia treatment for 6 h. Levels of AtERF73/HRE1 gene expression were determined. B, Expression levels of AtERF73/HRE1 and ADH in Arabidopsis seedlings after 6 h of hypoxia in the presence of 10 μm AVG and 10 μm AgNO3 were determined by quantitative RT-PCR, and the relative amounts of transcripts were calculated and normalized with β-ATP mRNA. Values represent means ± sd from three biologically independent experiments. * P < 0.05 versus the wild type (one-way ANOVA with Dunnett’s t test).
Generation of AtERF73/HRE1 RNA Interference Transgenic Plants
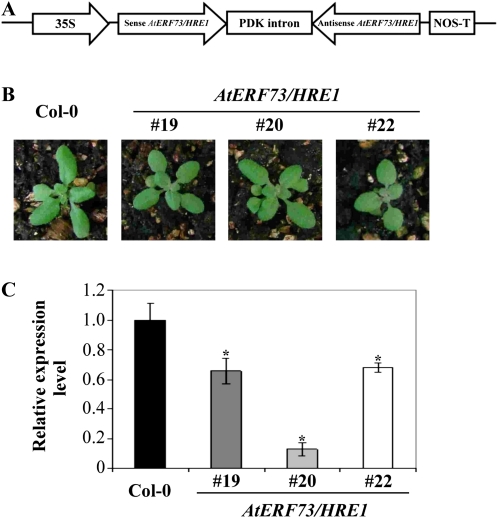
To assess the role of AtERF73/HRE1, we decided to use RNA interference (RNAi) to generate transgenic plants with reduced AtERF73/HRE1 mRNA accumulation. We generated an RNAi construct that contains the cauliflower mosaic virus 35S promoter fused to an inverted repeat of the AtERF73/HRE1 open reading frame (Fig. 3A). T1 and T2 kanamycin-resistant lines harboring this construct were recovered. We selected three T3 homozygous AtERF73/HRE1-RNAi lines, designated RNAi-19, RNAi-20, and RNAi-22, for further analysis. Under normal growth conditions, these AtERF73/HRE1-RNAi lines show slightly smaller rosette leaves compared with 14-d-old Arabidopsis wild-type (ecotype Columbia [Col-0]) plants (Fig. 3B). Quantitative RT-PCR showed that the AtERF73/HRE1 mRNA levels in roots of hypoxia-treated plants were reduced by approximately 2-fold in RNAi-19 and RNAi-22 lines and by approximately 10-fold in the RNAi-20 line in comparison with the wild type (Fig. 3C). AtERF73/HRE1 belongs to the group VII ERF subfamily, which has five members in the Arabidopsis genome (Licausi et al., 2010). We used quantitative RT-PCR to determine whether our RNAi construct also resulted in the reduction in expression of other members in the VII ERF subfamily in transgenic lines. The results show that the expression of four other genes in the VII subfamily is not affected in the three RNAi lines (Supplemental Fig. S3).
Figure 3.
AtERF73/HRE1 gene expression levels in transgenic AtERF73/HRE1-RNAi lines. A, Structure of the AtERF73/HRE1-RNAi construct. B, Phenotypes of 2-week-old seedlings of wild-type Col-0 and transgenic AtERF73/HRE1-RNAi lines. C, Quantitative RT-PCR analysis. The mRNA levels of AtERF73/HRE1 in roots of 2-week-old seedlings of Col-0 and AtERF73/HRE1 lines RNAi-19, RNAi-20, and RNAi-22 were determined after hypoxia treatment for 6 h by quantitative RT-PCR, and the relative amounts of transcripts were calculated and normalized with β-ATP mRNA. Values represent means ± sd from three biologically independent experiments. * P < 0.05 versus the wild type (one-way ANOVA with Dunnett’s t test).
AtERF73/HRE1-RNAi Transgenic Lines Display Increased Ethylene Sensitivity
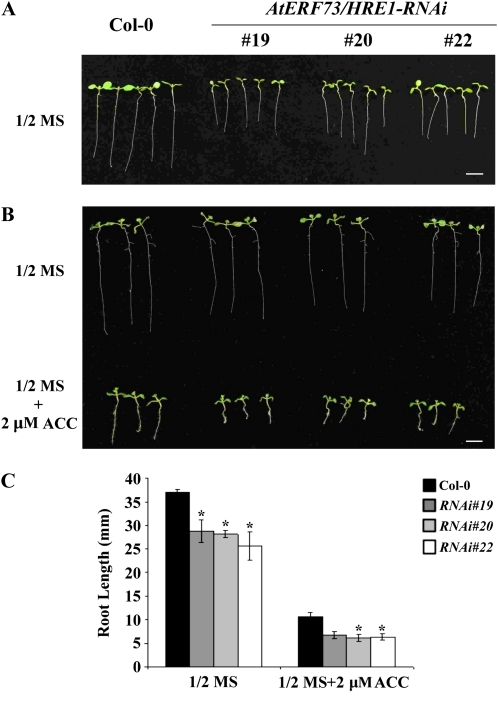
To determine whether AtERF73/HRE1 functions as a regulator of ethylene signaling pathways, seeds from the wild type and the three AtERF73/HRE1-RNAi lines were germinated in the presence or absence of ACC. We found that the root lengths in 7-d-old seedlings, grown on one-half-strength Murashige and Skoog (MS) medium in the absence of ACC, were significantly shorter in the AtERF73/HRE1-RNAi lines in comparison with the wild type (Fig. 4A). When seedlings were grown for 10 d, transgenic plant lines developed roots with average lengths of 28.7, 28.2, and 26.0 mm, respectively, while the average root length of wild-type plants was 37.0 mm (Fig. 4C). Addition of ACC resulted in inhibition of root elongation in both wild-type and transgenic lines; however, the root lengths in AtERF73/HRE1-RNAi lines were shorter than those in the wild type (Fig. 4, B and C).
Figure 4.
Comparison of root growth in Col-0 and AtERF73/HRE1-RNAi lines with or without ACC treatment. A, The phenotypes of seedlings grown on one-half-strength MS medium for 7 d. Bar = 5 mm. B, The phenotypes of seedlings grown for 10 d on one-half-strength MS medium in the absence or presence of ACC (2 μm). Bar = 5 mm. C, Average root length for seedlings grown on one-half-strength MS or medium containing ACC (2 μm). The data represent average values ± sd from 50 seedlings of each genotype obtained from three biologically independent experiments. * P < 0.05 (Student’s t test).
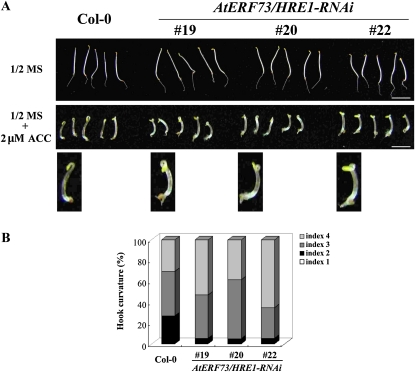
To further assess the ethylene sensitivity of AtERF73/HRE1-RNAi lines, we measured the degrees of triple response to ethylene in etiolated seedlings. No difference in the phenotype between 4-d-old etiolated seedlings of the wild type and three AtERF73/HRE1-RNAi lines was observed in the absence of ACC (Fig. 5A, top panel). However, in the presence of 2 μm ACC, etiolated seedlings of AtERF73/HRE1-RNAi lines showed exaggerated curvature of the apical hook compared with wild-type seedlings (Fig. 5A, middle and bottom panels). The level of apical curvature was analyzed for more than 100 seedlings each and classified according to an index scale ranging from 1 to 4, where index 1 and 2 represent curvatures of 90° and 180°, respectively, and index 3 and 4 represent the beginning of hook formation and the presence of a full hook, respectively (Leclercq et al., 2002). As shown in Figure 5B, about 95% of the seedlings of AtERF73/HRE1-RNAi lines displayed a phenotype with hook formation and full hook (index 3 and 4), whereas a significant fraction of wild-type seedlings (27%) lacked hook formation but displayed 180° apical curvature (index 2). The root and hypocotyl elongation in the wild type and AtERF73/HRE1-RNAi lines did not show any apparent difference (Supplemental Fig. S4). Taken together, these results suggest that AtERF73/HRE1 negatively regulates a subset of ethylene responses in Arabidopsis seedlings under normoxic conditions.
Figure 5.
Effects of ethylene on the apical hook curvature in AtERF73/HRE1-RNAi lines. A, The phenotypes of seedlings grown on one-half-strength MS medium with or without ACC (2 μm) for 4 d in the dark. Bars = 5 mm. B, The level of apical curvature was estimated visually for at least 100 seedlings using a scale ranging from 1 to 4 (1, 90° curvature; 2, 180° curvature; 3, beginning of hook formation; 4, full hook).
Effects of AtERF73/HRE1 Knockdown Lines on Hypoxia-Inducible Genes
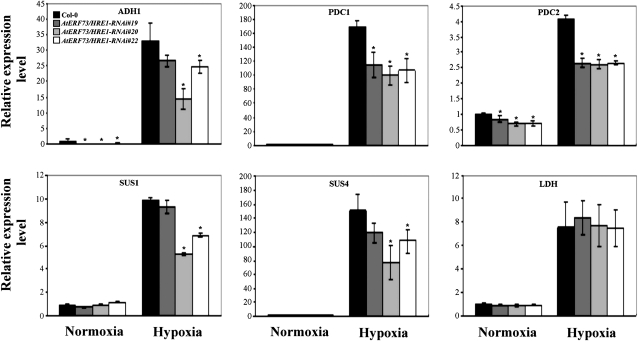
To determine how a decrease in AtERF73/HRE1 expression affects hypoxia responses, we examined the expression of glycolytic and fermentative genes, including ADH, PDC1, PDC2, SUS1, SUS4, and LDH, in AtERF73/HRE1-RNAi lines. Quantitative RT-PCR analyses showed that, under hypoxic conditions, the mRNA levels for all of these genes, with the exception of LDH, were reduced significantly in AtERF73/HRE1-RNAi lines (Fig. 6). The levels of reduction in three AtERF73/HRE1-RNAi lines roughly correlated with the degrees of AtERF73/HRE1 knockdown, with RNAi-20 having the most pronounced effect on hypoxic induction of these genes. The data suggest that AtERF73/HRE1 positively regulates a subset of glycolytic and fermentative genes during hypoxia.
Figure 6.
Effects of AtERF73/HRE1 lines on fermentative and related carbohydrate metabolic genes under normoxic or hypoxic conditions. Total RNAs were extracted from roots of 14-d-old seedlings exposed to normoxic or hypoxic conditions for 6 h. The mRNA levels of ADH, PDC1, PDC2, SUS1, SUS2, and LDH genes were determined by quantitative RT-PCR, and the relative amounts of transcripts were calculated and normalized with β-ATP mRNA. Data represent means ± sd from three biologically independent experiments. * P < 0.05 versus the wild type (one-way ANOVA with Dunnett’s t test).
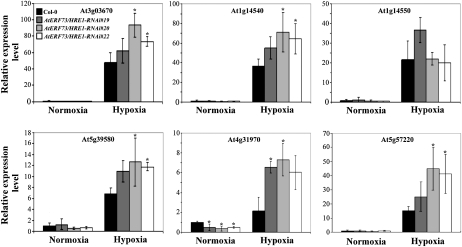
In order to identify additional genes whose expression is regulated by AtERF73/HRE1, we performed microarray analysis to compare gene expression profiles of the wild type and AtERF73/HRE1-RNAi20, in which AtERF73/HRE1 expression was severely knocked down. Arabidopsis Affymetrix GeneChip arrays were probed with RNAs isolated from roots of untreated plants (controls) and plants treated with hypoxia for 6 h. Up-regulated genes in wild-type plants were identified from four independent experiments at a 5-fold change cutoff and a P value cutoff of P ≤ 0.05. The results showed that there were 285 genes up-regulated in the wild type upon hypoxic treatment (Supplemental Table S1). Among these hypoxia-inducible genes, 13.3% had higher induction levels in the wild type, 7.4% had higher induction levels in the AtERF73/HRE1-RNAi20 line, and 79.3% had about the same induction level (Supplemental Table S1). A list of 24 genes, which exhibited a change in raw intensity above 1.5-fold and displayed different levels of hypoxic induction between the wild type and the AtERF73/HRE1-RNAi20 line, was generated (Table I). Among these 24 genes, most genes had higher levels of hypoxic induction in the AtERF73/HRE1-RNAi20 line than in the wild type. We noticed that, among the genes that had higher induction levels in the AtERF73/HRE1-RNAi20 line, peroxidase and cytochrome P450 genes were overrepresented. These genes, including At3g03670 (putative peroxidase), At1g14540 (putative anionic peroxidase), At1g14550 (putative anionic peroxidase), At5g39580 (putative peroxidase), At4g31970 (CYP82C2), At5g57220 (CYP81F2), and At2g30750 (CYP71A12), were selected to confirm their expression patterns by quantitative RT-PCR analysis (Fig. 7). Consistent with the microarray data, five of the genes were induced at significantly higher levels in the AtERF73/HRE1-RNAi20 line, whereas the induction level of At1g14550 did not differ significantly between the wild type and AtERF73/HRE1-RNAi20. These results show that AtERF73/HRE1 could negatively modulate hypoxic induction of these peroxidase and cytochrome P450 genes in Arabidopsis.
Table I. Microarray-derived expression of selected genes with differential expression between the wild type and AtERF73/HRE1-RNAi upon hypoxia.
| Accession No. | Affymetrix No. | Gene Description | Wild-Type Fold Changea | Pb | AtERF73/HRE1-RNAi Fold Changec | Pb |
| AT4G30380 | 253616_at | Encodes a plant natriuretic peptide (PNP) | 180.8 | 2.48E | 28.4 | 1.43E |
| AT4G37710 | 253060_at | VQ motif-containing protein | 101.8 | 2.59E | 68.3 | 1.65E |
| AT3G03670 | 259197_at | Peroxidase | 51.7 | 1.19E | 334.6 | 4.49E |
| AT1G14540 | 261474_at | Peroxidase | 48.5 | 1.03E | 239.6 | 2.82E |
| AT4G31970 | 253505_at | Cytochrome P450 (CYP82C) | 36.5 | 2.03E | 414.4 | 3.24E |
| AT5G57220 | 247949_at | Cytochrome P450 (CYP81F2) | 30.5 | 2.71E | 201.0 | 3.50E |
| AT1G26410 | 261006_at | FAD-binding domain-containing protein | 26.3 | 1.21E | 66.5 | 1.93E |
| AT5G13320 | 250286_at | Encodes PBS3 (avrPphB susceptible) | 21.1 | 3.81E | 65.2 | 8.77E |
| AT2G23270 | 245082_at | Unknown protein | 19.6 | 2.23E | 74.5 | 2.16E |
| AT1G57630 | 246405_at | Disease resistance protein (TIR class), putative | 19.2 | 2.51E | 23.6 | 1.16E |
| AT4G19520 | 254587_at | Disease resistance protein (TIR-NBS-LRR class) | 16.4 | 5.86E | 18.1 | 2.36E |
| AT5G26920 | 246821_at | Calmodulin-binding protein CBP60g | 15.9 | 2.16E | 28.1 | 1.31E |
| AT2G26560 | 245038_at | Lipid acyl hydrolase | 15.8 | 2.78E | 42.1 | 4.11E |
| AT5G25250 | 246927_s_at | Membrane-associated protein | 15.4 | 4.36E | 66.4 | 2.18E |
| AT5G57510 | 247913_at | Unknown protein | 15.2 | 1.25E | 9.4 | 4.81E |
| AT2G26150 | 266841_at | Heat shock transcription factor A2 (ATHSFA2) | 14.3 | 2.13E | 3.14 | 5.74E |
| AT1G14550 | 261475_at | Peroxidase | 14.1 | 4.38E | 38.3 | 9.65E |
| AT1G19250 | 256012_at | Flavin-dependent monooxygenase 1 (FMO1) | 13.8 | 9.24E | 7.9 | 1.23E |
| AT2G44800 | 266875_at | Oxidoreductase, 2OG-Fe(II) oxygenase family | 13.2 | 2.24E | 1.62 | 8.11E |
| AT1G26380 | 261021_at | FAD-binding domain-containing protein | 12.8 | 8.30E | 130.0 | 7.63E |
| AT3G55790 | 251755_at | Unknown protein | 12.5 | 5.31E | 64.4 | 2.34E |
| AT1G05880 | 261249_at | ARIADNE12 (ARI12) | 12.1 | 5.82E | 34.8 | 1.48E |
| AT5G64905 | 247215_at | Elicitor peptide 3 precursor (PROPEP3) | 11.4 | 1.39E | 28.2 | 3.85E |
| AT1G53620 | 260971_at | Unknown protein | 11.1 | 2.15E | 21.7 | 6.64E |
Relative expression of wild-type fold change upon normoxia conditions compared with hypoxia conditions.
The P value was determined by t test with fold change of the wild type and AtERF73/HRE1-RNAi upon hypoxia conditions.
Relative expression of AtERF73/HRE1-RNAi line fold change upon normoxia conditions compared with hypoxia conditions.
Figure 7.
Effects of AtERF73/HRE1 lines on hypoxia-inducible peroxidase and cytochrome P450 genes under normoxic or hypoxic conditions. Total RNAs were extracted from roots of 14-d-old seedlings exposed to normoxic or hypoxic conditions for 6 h. The mRNA levels of peroxidase (At3g03670, At1g14540, At1g14550, and At5g39580) and cytochrome P450 (At4g31970 and At5g57220) genes were determined by quantitative RT-PCR, and the relative amounts of transcripts were calculated and normalized with β-ATP mRNA. Data are means ± sd from three biologically independent experiments. * P < 0.05 versus the wild type (one-way ANOVA with Dunnett’s t test).
Effects of AtERF73/HRE1 Knockdown Lines on Ethylene Production
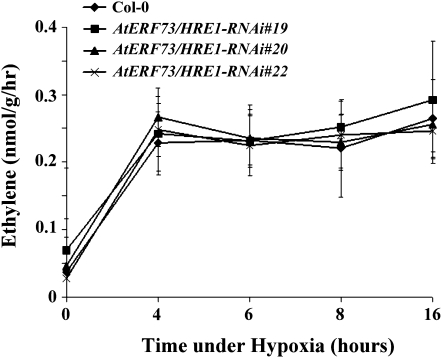
Since we have shown previously that ethylene production is increased during hypoxia in Arabidopsis (Peng et al., 2001), the negative effect of AtERF73/HRE1 on ethylene responses could be due to its inhibition of ethylene synthesis or its effects on ethylene signaling pathways. To distinguish between these two possibilities, we measured the level of ethylene production during hypoxia by gas chromatography. The results show that the wild type and AtERF73/HRE1-RNAi lines exhibited similar levels of ethylene production under normoxic and hypoxic conditions (Fig. 8).
Figure 8.
Production of ethylene in Col-0 and AtERF73/HRE1-RNAi lines during hypoxia. The Col-0 (diamonds) and AtERF73/HRE1-RNAi lines 19 (squares), 20 (triangles), and 22 (crosses) subjected to hypoxic treatment for different time periods were harvested and assayed for ethylene production as described in “Materials and Methods.” The means of six independent hypoxic treatments are plotted. Bars represent sd.
DISCUSSION
Effects of Hypoxia and Ethylene on AtERF73/HRE1 mRNA Accumulation
Microarray studies have shown that a number of Arabidopsis AP2/ERF genes are induced during hypoxia (Branco-Price et al., 2005; Gonzali et al., 2005; Liu et al., 2005). However, the regulatory functions of these transcription factors remained largely unknown. AtERF73/HRE1 transcription factor belongs to the AP2/ERF subfamily group VII of plant transcription factors. Members within this subfamily displayed differential expression patterns, with AtERF73/HRE1 and HRE2 (AtERF71) being induced at different stages of hypoxia (Licausi et al., 2010; Supplemental Fig. S1). It was reported that overexpression of HRE1 improved the tolerance of Arabidopsis plants to anoxia (Licausi et al., 2010). However, whether ethylene is involved in regulating AtERF73/HRE1 during hypoxia was not analyzed.
This report focused on the interplay between AtERF73/HRE1 and ethylene during hypoxia. When we tested the effects of other abiotic stresses and hormones, such as ABA, MeJA, ACC, SA, or cold treatment, our data showed that AtERF73/HRE1 expression could be induced not only by hypoxia but also by ACC treatment (Fig. 1, A and B). We further demonstrated that hypoxia and ACC treatments had an additive effect on the mRNA accumulation of AtERF73/HRE1 (Fig. 1C). However, we have shown previously that production of ethylene is increased in hypoxia-treated Arabidopsis seedlings (Peng et al., 2001). This raises the question of why exogenous addition of ethylene could further enhance the induction of AtERF73/HRE1 during hypoxia. Two possible explanations can be offered: the level of ethylene production has not reached maximum under our hypoxia treatments, or there is a feedback control to prevent the effects of ethylene reaching a maximal level. Results from our studies are consistent with the latter explanation, since AtERF73/HRE1-RNAi lines resulted in exaggerated ethylene responses under both normoxic and hypoxic conditions. Additionally, the wild type and AtERF73/HRE1-RNAi lines had the same ethylene production under normoxic and hypoxic conditions. This result established the AtERF73/HRE1 gene was involved in ethylene signaling but not in the biosynthesis pathway.
Interestingly, another member of the ERF subfamily VII, RAP2.2, is constitutively expressed at a high level in roots and at a lower level in shoots, but it is induced by dark and ethylene treatments (Hinz et al., 2010). Although RAP2.2 was not induced during hypoxia, its overexpression resulted in the improvement of plant survival under hypoxic stress (Hinz et al., 2010). In addition, transgenic lines containing T-DNA knockout mutations of RAP2.2 also have lower survival rates than the wild type. These results suggest that although RAP2.2 might not play a direct role in regulating hypoxic signaling pathways, it controls the expression of a set of genes whose products are involved in the survival of Arabidopsis under hypoxic stress.
Hypoxia-Responsive AtERF73/HRE1 Expression Is Mediated by Ethylene-Dependent and Ethylene-Independent Pathways
We have previously reported that the production of ethylene is increased during hypoxia and that ethylene is necessary but not sufficient in the hypoxic induction of ADH in Arabidopsis (Peng et al., 2001, 2005). We showed here that hypoxic treatment for 6 h could induce 12- and 5-fold increases in the expression of AtERF73/HRE1 in ein2-5 and etr1-1 mutants. However, the levels of induction in these mutants were much lower than that determined in the wild type (Fig. 4). These results indicate that ethylene is required for the mRNA accumulation of AtERF73/HRE1 during hypoxia. On the other hand, our results also show that hypoxic induction of AtERF73/HRE1 is not completely abolished in both ein2-5 and etr1-1. Indeed, the obtained patterns are similar to those observed for the hypoxic induction of ADH in ein mutants (Peng et al., 2005). Similarly, addition of inhibitors of ethylene (AVG and AgNO3) resulted in only partial reduction in hypoxic induction of AtERF73/HRE1, suggesting that ethylene is necessary but not sufficient for the hypoxic induction of AtERF73/HRE1.
AtERF73/HRE1 Functions as a Positive and Negative Regulator
Ethylene has been shown to regulate a number of cellular responses, including inhibition of root elongation and elicitation of triple responses in dark-grown seedlings (Huang et al., 2003; Swarup et al., 2007). The triple-response phenotype, which consists of shortening of the hypocotyl and root, radial swelling of the hypocotyl, and exaggerated curvature of the apical hook, is a well-known effect of ethylene on etiolated dicotyledonous seedlings (Guo and Ecker, 2004). In our studies, seedlings of AtERF73/HRE1-RNAi lines displayed an ethylene-oversensitive phenotype under normoxic conditions (Figs. 4 and 5), suggesting that AtERF73/HRE1 might negatively regulate ethylene responses.
Our analyses showed that AtERF73/HRE1-RNAi lines could affect hypoxia-regulated gene expression in two ways. For one group of genes, including ADH and a subset of glycolytic and fermentative genes, levels of hypoxic induction were significantly reduced in three AtERF73/HRE1-RNAi lines (Fig. 7), indicating that AtERF73/HRE1 positively regulates the induction of these genes during hypoxia. However, it has to be pointed out that in the report by Licausi et al. (2010), hypoxic induction of ADH was affected by the double mutation of AtERF73/HRE1 and AtERF71/HRE2 but not by the single mutation of AtERF73/HRE1 in the single knockout line. In contrast, the expression of ADH was increased in the AtERF73/HRE1 overexpression line. The difference in the effects of AtERF73/HRE1-RNAi lines might reflect differences in hypoxia treatment conditions, in which an open system was used in our experiments. Our analyses also showed that hypoxic induction of several peroxidase and cytochrome P450 genes was enhanced in the AtERF73/HRE1-RNAi lines, suggesting that AtERF73/HRE1 could inhibit the hypoxic induction of these genes. The Arabidopsis genome contains as many as 286 different cytochrome P450 genes, some of which play crucial roles in the biosynthesis of a variety of endogenous lipophilic compounds and cross talk in the responses to abiotic and biotic stresses (Paquette et al., 2000; Noordermeer et al., 2001; Narusaka et al., 2004). Peroxidases are known to be involved in oxidative cross-linking of cell wall (e.g. lignification) and in hydrogen peroxide (H2O2) detoxification. Numerous studies have shown that hypoxia stress triggers the formation of reactive oxygen species and induces oxidative stress in plants (Geigenberger, 2003; Garnczarska and Bednarski, 2004). It was reported that the level of H2O2 increased in response to oxygen deprivation (Baxter-Burrell et al., 2002) and that there is a mechanism to maintain the homeostasis of H2O2. Therefore, it might be possible that AtERF73/HRE1 plays a role in maintaining the homeostasis of H2O2 by regulating the expression of genes involved in antioxidation. Consistent with this hypothesis, we found that expression of AtERF73/HRE1 could also be induced by H2O2 (data not shown).
Potential Roles of AtERF73/HRE1 and Ethylene in Hypoxia Signal Pathways
The results we have reported here indicate that AtERF73/HRE1 plays an essential role in hypoxia signaling pathways. We showed that hypoxia triggers ethylene-dependent and ethylene-independent pathways, both of which are required for the accumulation of AtERF73/HRE1 transcripts. The induced AtERF73/HRE1 in turn participates in the activation of ADH and other genes related to fermentation and carbohydrate metabolism. In addition, we showed that AtERF73/HRE1 could modulate ethylene responses under both normoxic and hypoxic conditions. We also showed that AtERF73/HRE1 negatively regulated a group of peroxidase and cytochrome P450 genes through its inhibitory effects on ethylene signaling responses. The functions of AtERF73/HRE1 are very similar to those of the rice Sub1A and SK genes, which play a central role in submergence tolerance of lowland and deepwater rice, respectively (Xu et al., 2006; Hinz et al., 2010). In rice, the Sub1 locus encodes three submergence-inducible ERF genes, Sub1A, Sub1B, and Sub1C (Fukao et al., 2006; Xu et al., 2006; Voesenek and Bailey-Serres, 2009). Sub1B and Sub1C exist in all rice cultivars that have been examined. The Sub1A has two alleles among indica cultivars. The submergence-tolerant cultivars, such as FR13A, carry the Sub1A-1 allele, whereas the submergence-sensitive cultivars carry the Sub1A-2 allele. In FR13A, induction of Sub1A-1 resulted in the inhibition of Sub1C expression as well as the retardation of root and shoot elongation during the submergence of rice plants (Fukao et al., 2006; Xu et al., 2006). It has been demonstrated that ethylene is involved in the induction of Sub1A-1 during submergence and that Sub1A in turn suppresses ethylene production and GA responses, which allow rice plants maintain a low metabolic state to survive long periods of submergence (Fukao et al., 2006; Perata and Voesenek, 2007). In deepwater rice, which does not carry Sub1A, a pair of ERF factors, SK1 and SK2, are induced under submergence conditions through the action of ethylene signaling pathways (Hattori et al., 2009). In contrast to the lowland rice, the activation of SK1 and SK2 resulted in enhanced GA responses to stimulate shoot elongation, which allows the deepwater rice to adapt to flooding conditions (Hattori et al., 2009). Despite the similarity of the proposed functions of AtERF73/HRE1, Sub1A, and SK, induction of Sub1A and SK genes results in submergence tolerance in rice, whereas induction of AtERF73/HRE1 probably results in only temporary survival of Arabidopsis. We are currently determining the downstream targets of both AtERF73/HRE1 and Sub1A to identify transcriptional regulatory pathways mediated by these two factors to determine the molecular basis of the difference in hypoxia sensitivity between Arabidopsis and rice.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatment
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used in this study. Two ethylene-insensitive mutants, ein2-5 and etr1-1, were obtained from Dr. L.-C. Wang, and their phenotypes were confirmed before use. All seeds were sterilized and kept for 3 d at 4°C in the dark to break dormancy. Seeds were germinated on 0.8% (w/v) agar plates containing one-half-strength MS medium supplemented with 0.5% (w/v) Suc at pH 5.7. The plates were incubated at 22°C under a 16-h-light (81 μmol s−1 m−2)/8-h-dark cycle in a growth chamber for 7 to 10 d. For morphological observations, seeds were sown on a 2:1:6 mixture of vermiculite, perlite, and peat moss and irrigated with water for 2 weeks.
For stress treatments, 14-d-old seedlings were treated with 5 μm ABA, 50 μm MeJA, 5 μm ACC, or 50 μm SA at 4°C for 6 h in the dark. For hypoxia treatment, 14-d-old Arabidopsis seedlings were placed on filter paper presoaked with one-half-strength MS solution supplemented with 0.5% (w/v) Suc at pH 5.7 for 1 h in the dark and then transferred onto a floating platform with roots dipped into one-half-strength MS solution containing 0.5% (w/v) Suc at pH 5.7, which was constantly bubbled with 3% (v/v) oxygen and 97% (v/v) nitrogen, in the dark for the times indicated. For hypoxia treatment with the presence of AVG and AgNO3, Arabidopsis seedlings were treated in the same way except that 10 μm AVG or 10 μm AgNO3 was added to the MS solution. To measure root lengths, Arabidopsis seeds were germinated on 0.8% (w/v) agar plates containing one-half-strength MS and 0.5% (w/v) Suc with or without 2 μm ACC at pH 5.7. The plates were kept at 4°C in the dark for 3 d and then transferred to 22°C under a 16-h-light (81 μmol s−1 m−2)/8-h-dark cycle in a growth chamber for 7 to 10 d. The root lengths were measured for 50 seedlings of each genotype. The measurements were performed three times with independently germinated seedlings. Photographs were taken at 7 or 10 d of treatment. For apical hook observations, seeds were grown on 0.8% (w/v) agar plates containing one-half-strength MS and 0.5% (w/v) Suc supplemented with or without 2 μm ACC at pH 5.7. The plates were kept at 4°C for 3 d and then transferred to 22°C for 4 d in the dark. The level of apical curvature was estimated visually for at least 100 seedlings. Photographs were taken at the end of the treatment.
Constructs and Plant Transformation
For the AtERF73/HRE1-RNAi lines, the AtERF73/HRE1 (At1g72360) open reading frame was amplified by PCR from the Col-0 ecotype using the following primer pair: 5′-GTAGAGGAGAATCATGAGGCTGATC-3′ (forward) and 5′-CTCGAGGGTACCCAAAGAGACTACTCAGAGACGTTGA-3′ (reverse). The reverse primer contains the recognition sites for KpnI and XhoI. The amplified product was cloned as an inverted repeat with a pyruvate-orthophosphate dikinase artificial intron into the binary vector pBI121. The recombinant plasmid was then transformed into Agrobacterium tumefaciens strain GV3101. The resulting Agrobacterium was used to generate transgenic Arabidopsis by vacuum infiltration. The seeds were planted on one-half-strength MS agar plates containing kanamycin (50 μg mL−1) to identify T1 transgenic plants. T1 seeds were generated on the kanamycin plates to select homozygous T2 lines.
RT-PCR and Quantitative RT-PCR Analyses
RNA samples were isolated from frozen tissues of Arabidopsis using the TRIzol reagent (Invitrogen). For RT-PCR analysis of Arabidopsis RNAs, total RNA samples from roots (2 μg per reaction) were first treated with DNase I and then reverse transcribed into cDNAs by Moloney murine leukemia virus reverse transcriptase. Primers used for RT-PCR are listed in Supplemental Table S2. The RT-PCR products were separated on a 1.0% agarose gel by electrophoresis.
Quantitative RT-PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems) with the Power SYBR Green PCR Master Mix according to the manufacturer’s recommendations. Amplification of β-ATP cDNA under identical conditions was used as an internal control to normalize the level of cDNA. The data obtained were analyzed with the 7500 System SDS software. Amplification conditions were as follow: 95°C for 10 min, and then 40 cycles at 95°C for 15 s and 60°C for 1 min. The specificity of primer pairs (Supplemental Table S2) was based on an analysis of dissociation curves (75°C–99°C). Quantitative RT-PCR experiments were repeated three times independently, and the data were averaged.
Ethylene Measurements
The 14-d-old seedlings of the wild type and AtERF73/HRE1-RNAi were treated with hypoxia as described above. Ten plants were transferred to a 12- × 75-mm test tube and capped for 1 h at room temperature in the dark. Ethylene was measured using a gas chromatograph (6890N Agilent GC system). The experiments were repeated six times independently, each time in triplicate.
Microarray Analysis
All microarray experiments were carried out using the ATH1 Arabidopsis GeneChip microarray (Affymetrix) containing a set of 22,810 probes. Four independent biological replicates were performed using cDNA obtained from control and hypoxia-treated (6 h) samples from 14-d-old seedlings. Fluorescence labeling of cDNA probes starting from mRNA was performed by in vitro transcription followed by fragmentation following the manufacturer's suggestion (GeneChip Expression Analysis Technical Manual rev5; Affymetrix). Microarray experiments were performed at the DNA microarray core facility of the Institute of Plant and Microbial Biology at Academia Sinica in Taiwan. The raw microarray data from all hybridization assays were analyzed and normalized by the GeneSpring GX 7.3 software. Normalization was performed, as values below 0.01 were set to 0.01, per chip was divided by the 50th percentile, and per gene was normalized by the median of corresponding gene measurements in normoxic samples. GeneSpring was used with a 5-fold change and P ≤ 0.05 cutoffs to identify up- and down-regulated genes for further analysis.
The microarray Gene Expression Omnibus accession number is GSE27475.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The expression levels of group VII ERF genes after hypoxic treatment.
Supplemental Figure S2. The shoot mRNA levels of AtERF73/HRE1 after hypoxic treatment.
Supplemental Figure S3. The expression levels of group VII ERF genes in AtERF73/HRE1-RNAi lines after hypoxia treatment.
Supplemental Figure S4. The hypocotyl and root lengths on one-half-strength MS medium with ACC for 4 d in the dark.
Supplemental Table S1. List of up-regulated genes in the wild type upon hypoxic treatment.
Supplemental Table S2. Primers for PCR and quantitative RT-PCR.
Acknowledgments
We thank Dr. L.-C. Wang (Institute of Plant and Microbial Biology, Academia Sinica) for providing seeds of ein2-5 and etr1-1 mutants. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/), supported by Academia Sinica.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot 96: 647–660; erratum [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Ann Bot (Lond) 96: 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Conley TR, Peng HP, Shih MC. (1999) Mutations affecting induction of glycolytic and fermentative genes during germination and environmental stresses in Arabidopsis. Plant Physiol 119: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES. (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48: 223–250 [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnczarska M, Bednarski W. (2004) Effect of a short-term hypoxic treatment followed by re-aeration on free radicals level and antioxidative enzymes in lupine roots. Plant Physiol Biochem 42: 233–240 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Novi G, Poggi A, Alpi A, Perata P. (2005) The use of microarrays to study the anaerobic response in Arabidopsis. Ann Bot 96: 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. (1996) Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiol 112: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Olive M, Peacock WJ, Dennis ES, Shimamoto K. (1994) Promoter elements required for developmental expression of the maize Adh1 gene in transgenic rice. Plant Cell 6: 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq J, Adams-Phillips LC, Zegzouti H, Jones B, Latché A, Giovannoni JJ, Pech JC, Bouzayen M. (2002) LeCTR1, a tomato CTR1-like gene, demonstrates ethylene signaling ability in Arabidopsis and novel expression patterns in tomato. Plant Physiol 130: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath S, Lee CH, VanWinkle P, Bailey-Serres J. (1998) Molecular and biochemical characterization of cytosolic phosphoglucomutase in maize: expression during development and in response to oxygen deprivation. Plant Physiol 117: 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K. (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55: 327–342 [DOI] [PubMed] [Google Scholar]
- Noordermeer MA, Veldink GA, Vliegenthart JF. (2001) Fatty acid hydroperoxide lyase: a plant cytochrome p450 enzyme involved in wound healing and pest resistance. ChemBioChem 2: 494–504 [DOI] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R. (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307–317 [DOI] [PubMed] [Google Scholar]
- Peng HP, Chan CS, Shih MC, Yang SF. (2001) Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 126: 742–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HP, Lin TY, Wang NN, Shih MC. (2005) Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol Biol 58: 15–25 [DOI] [PubMed] [Google Scholar]
- Perata P, Voesenek LA. (2007) Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci 12: 43–46 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM. (2003) Molecular and cellular adaptations of maize to flooding stress. Ann Bot (Lond) 91: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J. (2009) Plant biology: genetics of high-rise rice. Nature 460: 959–960 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Zhang H, Huang Z, Xie B, Chen Q, Tian X, Zhang X, Zhang H, Lu X, Huang D, Huang R. (2004) The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220: 262–270 [DOI] [PubMed] [Google Scholar]