Abstract
Grass plants develop unique floral patterns that determine grain production. However, the molecular mechanism underlying the specification of floral organ identities and meristem determinacy, including the interaction among floral homeotic genes, remains largely unknown in grasses. Here, we report the interactions of rice (Oryza sativa) floral homeotic genes, OsMADS3 (a C-class gene), OsMADS13 (a D-class gene), and DROOPING LEAF (DL), in specifying floral organ identities and floral meristem determinacy. The interaction among these genes was revealed through the analysis of double mutants. osmads13-3 osmads3-4 displayed a loss of floral meristem determinacy and generated abundant carpelloid structures containing severe defective ovules in the flower center, which were not detectable in the single mutant. In addition, in situ hybridization and yeast two-hybrid analyses revealed that OsMADS13 and OsMADS3 did not regulate each other’s transcription or interact at the protein level. This indicates that OsMADS3 plays a synergistic role with OsMADS13 in both ovule development and floral meristem termination. Strikingly, osmads3-4 dl-sup6 displayed a severe loss of floral meristem determinacy and produced supernumerary whorls of lodicule-like organs at the forth whorl, suggesting that OsMADS3 and DL synergistically terminate the floral meristem. Furthermore, the defects of osmads13-3 dl-sup6 flowers appeared identical to those of dl-sup6, and the OsMADS13 expression was undetectable in dl-sup6 flowers. These observations suggest that DL and OsMADS13 may function in the same pathway specifying the identity of carpel/ovule and floral meristem. Collectively, we propose a model to illustrate the role of OsMADS3, DL, and OsMADS13 in the specification of flower organ identity and meristem determinacy in rice.
Studies in two model eudicot plants, Arabidopsis (Arabidopsis thaliana) and Antirrhinum majus, have suggested that MADS box genes play critical roles in regulating flower development. The proposed genetic ABC model explains how three classes of genes (A, B, and C) work together in specifying floral organ identities (Coen and Meyerowitz, 1991). In Arabidopsis, A (APETALA1 [AP1] and AP2) alone determines sepals, A and B (AP3 and PISTILLATA [PI]) together specify petals, B and C (AGAMOUS [AG]) define stamens, and C alone defines carpels (Coen and Meyerowitz, 1991). Subsequently, two additional classes of genes (D and E) have been included in the modified ABC model. The D-class genes specify ovules (Angenent et al., 1995), while E-class genes (SEPALLATA1/2/3/4 [SEP1/2/3/4]; formerly named AGL2/4/9/3) specify the identity of all four whorls of floral organs and floral meristem determinacy (Pelaz et al., 2000, 2001a, 2001b; Ditta et al., 2004; Liu et al., 2009).
As one of the largest families in flowering plants, the grass family (Poaceae) contains many economically important crops, such as rice (Oryza sativa), barley (Hordeum vulgare), and maize (Zea mays; Linder and Rudall, 2005). These crops have unique floral organization and morphology, which are distinct from those of eudicots and even other monocots (Grass Phylogeny Working Group, 2001; Rudall et al., 2005; Whipple et al., 2007). The spikelet is the structural unit of grass flowers, and each spikelet consists of a varied number of bract-like organs, glumes, and florets. A rice spikelet consists of two pairs of sterile glumes (i.e. rudimentary glumes and empty glumes) and one floret that contains one lemma, one palea in whorl 1, two lodicules in whorl 2 interior to the lemma, six stamens in whorl 3, and a carpel in whorl 4 (Yuan et al., 2009; Zhang and Wilson, 2009).
Although grass flowers are essential for producing grains, the underlying molecular basis that specifies grass floral organs still remains less understood (Clifford, 1987; Whipple et al., 2007). While the ABCDE model is thought to be partially applicable in explaining grass floral development, grasses have diversified genetic components in specifying the identity of floral organs and meristem (Thompson and Hake, 2009). For example, loss-of-function mutants of the orthologs of Arabidopsis AP3 in maize (Silky1) and in rice (SUPERWOMEN1 [SPW1] or OsMADS16) display homeotic transformations of stamens to carpels and of lodicules to lemma- or palea-like structures, suggesting the conservation of class B genes between grasses and Arabidopsis (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2007). Grasses have duplicated and subfunctionalized C-class genes (Kramer et al., 2004; Zahn et al., 2006). For example, rice has two AG homologs, OsMADS3 and OsMADS58 (Kramer et al., 2004). OsMADS3 plays key roles in both stamen identity specification and late anther development, while OsMADS58 is crucial for specifying floral meristem determinacy and carpel architecture (Yamaguchi et al., 2006; Hu et al., 2011). Similarly, there is a pair of AG homologs in maize: Zea agamous1 (zag1) and Zea mays mads2 (zmm2). The zag1 gene has been shown to determine floral meristem determinacy, while the biological function of zmm2 remains unclear (Mena et al., 1996).
In rice, OsMADS13 is a D-class gene that is orthologous to Arabidopsis SEEDSTICK (STK) and to FLORAL BINDING PROTEIN7 (FBP7) and FBP11 in petunia (Petunia hybrida). Cosuppression of FBP7 and FBP11 causes the conversion of ovules into carpelloid organs (Colombo et al., 1995). The osmads13 mutants are associated with homeotic transformation of ovules into carpelloid structures and indeterminate flowers (Dreni et al., 2007; Yamaki et al., 2011). This is in contrast to the mutation of the Arabidopsis STK gene, which does not display altered ovule identity (Pinyopich et al., 2003). In Arabidopsis, AG, STK, SHATTERPROOF1 (SHP1), and SHP2 are grouped in the monophyletic AG-like clade and have been shown to be involved in ovule identity specification. STK is the only D-lineage gene and is expressed in the ovule. stk single mutants develop a slightly abnormal ovule with a defect in funiculus development, while the stk shp1 shp2 triple mutant demonstrates the conversion of ovules into leaf-like or carpel-like organs (Favaro et al., 2003; Pinyopich et al., 2003). Furthermore, STK, SHP1, SHP2, and AG were shown to form multimeric complexes in yeast in the presence of SEP MADS box factors, and the defect of ovule development in sep1/SEP1 sep2 sep3 is similar to that in the shp1 shp2 stk triple mutant (Favaro et al., 2002), suggesting the role of Arabidopsis SEP genes participating in ovule identity specification. In addition, AG was shown to be involved in specifying ovule identity by affecting the expression of SHP1 and SHP2 (Brambilla et al., 2007).
The rice DROOPING LEAF (DL) gene, which is orthologous to Arabidopsis CRABS CLAW (CRC), encodes a YABBY domain protein and plays a crucial role in specifying the carpel identity and floral meristem determinacy (Yamaguchi et al., 2004). Severe dl mutants display complete homeotic transformation of carpels into stamens, while mutations of CRC cause abnormal carpel development (Alvarez and Smyth, 1999; Yamaguchi et al., 2004). Moreover, DL/CRC interacts antagonistically with class B genes (Alvarez and Smyth, 1999; Yamaguchi et al., 2004), suggesting that DL and CRC play a conserved and diversified role in controlling carpel identity in rice and Arabidopsis, respectively.
Grasses have diversified SEP-like genes, with at least five SEP-like members (OsMADS1, OsMADS5, OsMADS7, OsMADS8, and OsMADS34) in rice (Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007). OsMADS1 (also called LEAFY HULL STERILE1) has been characterized as a SEPALLATA (SEP)-like gene in rice, which is required for specifying the lemma/palea identity and the meristem of inner floral organs (Jeon et al., 2000; Prasad et al., 2001, 2005; Malcomber and Kellogg, 2004; Agrawal et al., 2005; Chen et al., 2006b). Knockdown of both OsMADS7 and OsMADS8 results in late flowering, homeotic transformations of lodicules, stamens, and carpels into palea/lemma-like organs, and a loss of floral determinacy. Simultaneous reduction of the expression of four rice SEP-like genes, OsMADS1, OsMADS5, OsMADS7, and OsMADS8, causes homeotic transformation of all floral organs except the lemma into leaf-like organs (Cui et al., 2010). OsMADS34 (also called PANICLE PHYTOMER2) plays a key role in controlling the development of inflorescences and spikelets in rice (Gao et al., 2010; Kobayashi et al., 2010). Moreover, investigation of the double mutant osmads34 osmads1 indicates that OsMADS34 and OsMADS1 redundantly specify the identities of floral organs, including the lemma/palea, lodicules, stamens, and carpel (Gao et al., 2010). All these data suggest the conserved and diversified functions of rice SEP-like genes in specifying flower organ identity. More recently AGAMOUS-LIKE6 (AGL6) genes in monocots and dicots have been also shown to play key roles in specifying floral organ and meristem identity (Hsu et al., 2003; Fan et al., 2007; Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Rijpkema et al., 2009; Thompson et al., 2009; Li et al., 2010; Viaene et al., 2010). AGL6-like genes are ancient and widely distributed in gymnosperms and angiosperms and form a sister clade to SEP-like genes (Purugganan et al., 1995; Theissen et al., 2000; Becker and Theissen, 2003; Zahn et al., 2005). Mutations in AGL6 homologous genes in grasses result in defective floral organ identity and meristem determinacy (Ohmori et al., 2009; Thompson et al., 2009; Li et al., 2010).
Although several genes are reported to play roles in specifying flower development in rice, their genetic interactions remain largely unknown. In this study, we characterized the genetic interaction of OsMADS3, DL, and OsMADS13 in specifying floral organs and floral meristem determinacy and provided new insights into the molecular mechanisms that regulate flower development in rice.
RESULTS
Identification of New Alleles of OsMADS13, OsMADS3, and DL
To identify rice mutants with floral defects, we screened a population of rice mutants for defective flowers in the japonica subspecies 9522 background treated by 60Co γ-ray (280 Gy; Chen et al., 2006a). One mutant line displaying complete female sterility was identified. Genetic analysis and map-based cloning indicated that this mutant has a one-base deletion in the fifth exon in OsMADS13 (Os012g10540; Supplemental Fig. S1A), causing a frameshift at amino acid 132 and the formation of a premature stop codon. OsMADS13 expression was specifically reduced in pistils of the mutant (Supplemental Fig. S1B). As the first two mutants of OsMADS13 (osmads13-1 and osmads13-2) have been reported (Dreni et al., 2007; Yamaki et al., 2011) and a genetic analysis indicated that our mutant is allelic to the reported osmads13-1, we named this mutant osmads13-3. This mutation is not associated with obvious alteration of the outer three whorl organs, although some osmads13-3 flowers (31%) displayed three or four stigmas (n = 121; Fig. 1, A and Q) instead of two stigmas in wild-type flowers. Like the osmads13-1 mutant, osmads13-3 showed complete female sterility with aborted ovule development (Fig. 1, B and Q) and carpelloid structures (Supplemental Fig. S1, F and G). In addition, the ectopic expression of DL was observed in the carpelloid structure of osmads13-3 (Fig. 2, A–F), suggesting that these ectopic structures have the carpel identity.
Figure 1.
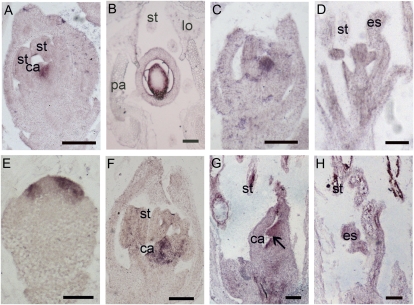
Flower phenotypes of osmads13-3 osmads3-4. A, One osmads13-3 flower with the removal of the lemma and the palea showing normal lodicules and stamens but its pistil with three stigmas. B, Longitudinal section of one osmads13-3 flower showing abnormal development of the ovule. C, One osmads3-4 flower with the removal of the lemma and the palea. D, Longitudinal section of one osmads3-4 flower showing normal development of the carpel and the ovule. E, One osmads13-3 osmads3-4 flower with the removal of the lemma and the palea showing inner organs. F, Longitudinal section of one osmads13-3 osmads3-4 flower showing the formation of higher order carpels. G, SEM observation of one osmads13-3 osmads3-4 flower primordium with the removal of the lemma and the palea. H, SEM observation of one osmads13-3 osmads3-4 flower primordium displaying the loss of determinacy in the center. I, SEM observation of one osmads13-3 flower primordium at stage Sp6. J, SEM observation of one osmads3-4 flower primordium at stage Sp6. K, SEM observation of one osmads13-3 osmads3-4 flower primordium at stage Sp6. As in J, the primordium of one ectopic organ is emerged. L, SEM observation of a wild-type flower at early stage Sp8 showing the termination of the floral meristem activity. M, SEM observation of one osmads13-3 osmads3-4 flower at the early stage Sp8 showing the remaining activity of the floral meristem. N, Closeup of M. The primordium of the primary carpel, the secondary carpel, and the floral meristem are indicated. O, Expression pattern of OSH1 in a wild-type flower at stage Sp8. P, Expression pattern of OSH1 in one osmads13-3 osmads3-4 flower at stage Sp8. Q to S, Floral diagrams of osmads13-3 (Q), osmads3-4 (R), and osmads13-3 osmads3-4 (S). ca, Capel; ii, inner integument; l-a, lodicule-anther mosaic organs; ll, lodicule-like structure; lo, lodicules; mtp, marginal tissue of the palea; oi, outer integument; ov, ovule; pc, primary carpel; sc, secondary carpel; st, stamen; tc, tertiary carpel. Bars = 1 mm in A, C, E, and G; 100 μm in B, D, F, and I to L; and 500 μm in H.
Figure 2.
Expression pattern of DL in flowers. A to C, In situ hybridization of DL mRNA in wild-type flowers. A, At stage Sp6, when the stamen primordia are forming, DL transcripts were detected in the midrib of the lemma. B, At stage Sp7, when the carpel primordia are forming, DL mRNA was observed in the wild-type carpel primordium. C, At stage Sp8, when the ovule is forming, the expression of DL was still observed in the wild-type carpel. D to F, In situ hybridization of DL mRNA in osmads13-3 flowers. D, At stage Sp7, DL transcripts were detected in the osmads13-3 carpel primordium. E, At stage Sp8, DL expression signal was observed in the osmads13-3 carpel (indicated by the arrow). F, At late stage Sp8, DL expression was strongly detectable in the indeterminate organ within the carpel in one osmads13-3 flower. The signal is indicated by the arrow. G, Normal expression of DL in osmads3-4 at stage Sp7. H, Ectopic expression of DL in osmads3-4 osmads13-3 at stage Sp8. ca, Carpel; fm, floral meristem; le, lemma; lo, lodicule; pa, palea; st, stamen. Bars = 50 μm in A and 100 μm in B to H.
Subsequently, we identified a new null mutant of DL, called dl-sup6, which was allelic with the reported d1-2 mutant (Nagasawa et al., 2003; Yamaguchi et al., 2004). Sequence analysis showed the insertion of one DNA fragment at the second intron of the DL gene (Supplemental Fig. S2A), which abolished the expression of DL in the mutant (Supplemental Fig. S2B). Because of five previously identified strong dl alleles (dl-sup1 to dl-sup5; Nagasawa et al., 2003; Yamaguchi et al., 2004), we named this mutant dl-sup6. Like the severe dl mutants, dl-sup6 displayed a phenotype of drooping leaves (Supplemental Fig. S2C) with ectopic stamens at the position of the carpel (Supplemental Figs. S2, E–H, and 4Q). Some flowers displayed a loss of floral meristem determinacy (Supplemental Fig. S2, F and J). In some cases, ectopic lodicule-like structures or fused anthers were observed in dl-sup6 (Supplemental Fig. S2, E–G). Scanning electronic microscopy (SEM) observation revealed that dl-sup6 flowers developed normally at stage Sp6 (Supplemental Fig. S2I). The staging of flowers refers to a previous report (Ikeda et al., 2004). At stage Sp7 or Sp8, dl-sup6 flowers generated ectopic stamen primordia (Supplemental Fig. S2, G and K). In addition, dl-sup6 lemmas displayed alternating numbers of vascular tissues, with three, four, or five vascular bundles (Supplemental Fig. S2L), while the wild-type lemma had the characteristic five vascular bundles (Yuan et al., 2009), suggesting an important role of DL in specifying lemma identify.
In addition, we recently characterized a new weak allele of OsMADS3 called osmads3-4, which is allelic to osmads3-1 (Hu et al., 2011). In osmads3-4, a two-base deletion was observed in the fifth exon of OsMADS3, leading to premature translational termination at amino acid 137 within the K domain. osmads3-4 flowers developed ectopic lodicule-like structures in whorl 2 and lodicule-like or lodicule-anther mosaic organs in whorl 3 (Fig. 1, C, J, and R). Unlike severe allele osmads3-3 (Yamaguchi et al., 2006), most osmads3-4 flowers displayed normal pistil development in the forth whorl (Fig. 1D).
OsMADS3 and OsMADS13 Synergistically Specify Ovule Identity and Floral Meristem Determinacy
To investigate the genetic interaction between OsMADS13 and OsMADS3 in determining rice flower development, we constructed the double mutant osmads13-3 osmads3-4. osmads13-3 osmads3-4 flowers displayed similar developmental defects in the second and third whorls to osmads3-4 (Fig. 1, E and S). Surprisingly, osmads13-3 osmads3-4 flowers displayed indeterminate floral development with supernumerary whorls of carpelloid structures without detectable ovule morphology in the flower center (Fig. 1, E–H), which was not observed with the corresponding single mutant. SEM observation showed that osmads13-3 osmads3-4 floral meristem was similar to that of osmads3-4 at stage Sp6 during the formation of stamen primordia (Fig. 1, J and K). At early stage Sp8, when the wild-type flower displays one carpel primordium in the fourth whorl and the floral meristem terminates (Fig. 1L), osmads13-3 osmads3-4 generated both primary and secondary carpel primordia and the floral meristem still persisted (Fig. 1, M and N), suggesting that floral stem cells are not terminated in a timely manner in the double mutant (Fig. 1S). In support of this, the expression of OSH1, a maker gene of rice floral meristem (Yamaki et al., 2005), was detectable in the indeterminate floral meristem of osmads13-3 osmads3-4 at stage Sp8, while the floral meristem in the wild-type flowers had been consumed at the same stage (Fig. 1, O and P). These observations suggest that OsMADS13 and OsMADS3 play synergistic roles in ovule development and determinacy of the floral meristem.
To further elucidate the mechanism of OsMADS13 and OsMADS3 in floral development, a yeast two-hybrid experiment was performed, and we observed no interaction of these two proteins, as judged by the growth condition in selective culture medium (Supplemental Fig. S3). RNA in situ hybridization analysis indicated that the OsMADS13 expression pattern was not obviously reduced in osmads3-4 at stage Sp8, when the ovule forms (Fig. 3, A–C), and the OsMADS3 mRNA signal was not obviously changed in osmads13-3 (Fig. 3G). Thus, OsMADS13 and OsMADS3 do not seem to influence each other at the transcriptional level.
Figure 3.
In situ hybridization of OsMADS13 and OsMADS3. A, Longitudinal section of a wild-type flower at early stage Sp8 showing the specific expression of OsMADS13 in ovule primordium. B, Transverse section of one wild-type flower at late stage Sp8 showing the expression of OsMADS13 in the ovule. C, The expression of OsMADS13 in osmads3-4 at stage Sp8. D, No detectable expression of OsMADS13 in dl-sup6. E, The expression of OsMADS3 in the wild-type stamen primordia at stage Sp6. F, At stage Sp8, there is detectable expression of OsMADS3 in the wild-type ovule. G, OsMADS3 transcripts were observed in the abnormal ovule in osmads13-3 at stage Sp8. H, OsMADS3 transcripts in ectopic stamens in dl-sup6 at stage Sp8. ca, Capel; es, ectopic stamen; fm, floral meristem; lo, lodicules; st, stamen. Bars = 100 μm except for 50 μm in E.
OsMADS3 and DL Synergistically Terminate the Floral Meristem
To further characterize the potential interaction between OsMADS3 and DL in controlling rice flower development, the double mutant osmads3-4 dl-sup6 was constructed. Morphological observations indicated that osmads3-4 dl-sup6 flowers had an altered vascular pattern in the lemma, resembling that of dl-sup6 (data not shown), suggesting that DL controls lemma identity independent of OsMADS3. This is consistent with the lack of expression of OsMADS3 in whorl 1 (Yamaguchi et al., 2006). The floral organs in the second and third whorls of osmads3-4 dl-sup6 appeared similar to those of osmads3-4 (Fig. 4, A, B, Q, and R, compared with Fig. 1). Furthermore, osmads3-4 dl-sup6 developed ectopic floral organ primordia that were similar to those of osmads3-4 at stage Sp6 (Fig. 4F, compared with Fig. 1G), suggesting that OsMADS3 functions in lodicule and stamen development independent of DL. This is in agreement with the fact that DL is not expressed in lodicules and stamens (Fig. 2; Yamaguchi et al., 2004). Strikingly, osmads3-4 dl-sup6 flowers generated supernumerary whorls of undifferentiated lodicule-like organs in the position of the pistil, which seemed to be arranged in bilateral symmetry along the elongated axis (Fig. 4, C–F). In addition, the floral meristem was observed on the top of the axis (Fig. 4E). This phenotype implies a severe loss of floral meristem determinacy, which was further confirmed by the in situ hybridization of OSH1 mRNA (Fig. 4H). SEM observation showed that at the early stage Sp8, the osmads3-4 dl-sup6 flower violated the normal development process and formed an indeterminate floral meristem in the flower center (Fig. 4G). Transverse section analysis indicated that these underdeveloped tissues were morphologically close to those of lodicules, with the characteristic pattern of vascular bundles (Fig. 4, I–K). Also, this indication was confirmed by the SEM observation that the morphology of epidermal cells of these underdeveloped tissues appeared similar to that of lodicules (Fig. 4, L and M). Meanwhile, the mRNA of the rice B-class gene SPW1 (OsMADS16), which accumulates in wild-type lodicules and stamens (Fig. 4N; Nagasawa et al., 2003), was detectable in these undifferentiated organs (Fig. 4O). This was combined with the presence of transcripts of the putative class A gene OsMADS15 (also called Degenerative Palea [DEP]; Wang et al., 2010a) in the undifferentiated tissues within the flower center of osmads3-4 dl-sup6 (Fig. 4P). In addition, the normal expression pattern of DL was detectable in osmads3-4 (Fig. 2G) and OsMADS3 expression was detected in ectopic stamens of dl-sup6 (Fig. 3H), suggesting that OsMADS3 and DL do not affect the expression of each other at the transcriptional level. These results suggest that OsMADS3 and DL may define the floral meristem in parallel during rice flower development.
Figure 4.
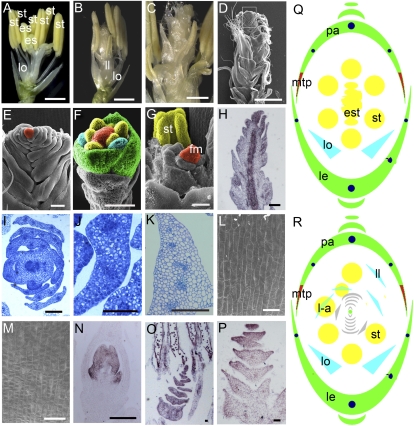
Flower phenotype of dl-sup6 osmads3-4. A, One dl-sup6 flower showing ectopic stamens in the center. B, One dl-sup6 osmads3-4 flower showing the phenotypes in the second and third whorls similar to osmads3-4. C, Closeup of one dl-sup6 osmads3-4 flower showing mosaic organs and indeterminate organs in the center. D, SEM observation of one dl-sup6 osmads3-4 flower showing the supernumerary whorls of indeterminate undifferentiated organs in the floral center. E, Closeup of D. F, SEM observation of one dl-sup6 osmads3-4 flower at stage Sp6. G, SEM observation of one dl-sup6 osmads3-4 flower after stage Sp7 showing ectopic organs and indeterminate meristem. H, Expression pattern of OSH1 in the dl-sup6 osmads3-4 flower at stage Sp8. I and J, Transverse sections of one dl-sup6 osmads3-4 flower showing the ectopic organs in the flower center. K, Transverse section of one wild-type lodicule. L and M, SEM analysis of epidermal cells of osmads3-4 du-sup6 ectopic organs and wild-type lodicules, respectively. N, Expression of SPW1 in wild-type lodicules and stamens. O, Transcripts of SPW1 detectable in lodicule-like organs of one dl-sup6 osmads3-4 flower center. P, OsMADS15 is expressed in lodicule-like organs in one dl-sup6 osmads3-4 flower center. Q and R, Floral diagrams of dl-sup6 (Q) and osmads3-4 dl-sup6 (R). est, Ectopic stamen; fm, floral meristem; l-a, lodicule-anther mosaic organs; ll, lodicule-like structure; lo, lodicules; mtp, marginal tissue of the palea; st, stamen. Bars = 1 mm in A, B, and D; 500 μm in C; 100 μm in E, H to K, and N to P; 50 μm in F; and 20 μm in L and M.
Analysis of the Interaction between OsMADS13 and DL
To determine the relationship between OsMADS13 and DL, we constructed the osmads13-3 dl-sup6 double mutant, and osmads13-3 dl-sup6 displayed flower defects similar to those of dl-sup6 (Fig. 5; Supplemental Fig. S2). Moreover, in situ analysis showed that OsMADS13 transcripts were not obviously detected in dl-sup6 flowers (Fig. 3D). In contrast, DL expression was ectopically observed in the indeterminate organ within the carpel in osmads13-3 flowers (Fig. 2, D–F). Therefore, we proposed that OsMADS13 and DL may function in the same pathway in specifying carpel/ovule identity and floral determinacy, and DL may act upstream of OsMADS13, positively regulating OsMADS13 expression, while OsMADS13 may repress the ectopic expression of DL in the ovule.
Figure 5.
Flower phenotypes of osmads13-3 dl-sup6. A, One osmads13-3 dl-sup6 flower with a weak phenotype, in which several ectopic stamens formed. B, One osmads13-3 dl-sup6 flower with a severe phenotype.
DISCUSSION
Rice Has a Conserved and Diversified Mechanism Controlling Ovule Identity
Ovule development is of importance in the plant life cycle. The ovule is the source of the megagametophyte and the precursors of seeds, consisting of the nucleus, integument(s), and funiculus (Reiser and Fischer, 1993; Colombo et al., 2008). Previous studies in petunia, Arabidopsis, and rice revealed that the MADS box genes belonging to the AG clade are necessary for specifying ovule identity.
In rice, the AG clade contains four MADS box members: two C-lineage genes, OsMADS3 and OsMADS58, and two D-lineage genes, OsMADS13 and OsMADS21 (Kramer et al., 2004; Zahn et al., 2006). The expression of OsMADS13 is restricted in the ovule, which is very similar to that of STK, FBP7, and FBP11. In contrast, OsMADS21 is mainly expressed in developing seeds (Lee et al., 2003; Dreni et al., 2007) and was thought to play a minor role in controlling ovule development (Dreni et al., 2007). Grass species including maize, wheat (Triticum aestivum), barley, and rice have duplicated C-class genes (Mena et al., 1996; Kramer et al., 2004; Yamaguchi et al., 2006; Zahn et al., 2006). To date, there is no evidence indicating that class C genes are required for carpel identity in grasses (Thompson and Hake, 2009). In rice, analyses of mutations of OsMADS3 and knockdown of OsMADS58 suggested that the two C-class genes have subfunctionalized and redundant functions in rice flower development (Yamaguchi et al., 2006; Hu et al., 2011; M.M. Kater, personal communication; Fig. 6). osmads3-3 is a strong allele of OsMADS3, displaying homeotic transformation of nearly all stamens in whorl 3 into lodicule-like organs, suggesting a major role of OsMADS3 in stamen specification (Yamaguchi et al., 2006). The intermediate mutant osmads3-4 displays defective postmeiotic anther development with an abnormal accumulation of reactive oxygen species. OsMADS3 was also shown to directly regulate the expression of MT-1-4b, which encodes a type 1 small Cys-rich and metal-binding protein with superoxide anion- and hydroxyl radical-scavenging activity, suggesting that OsMADS3 is a key transcriptional regulator in rice male reproductive development, at least in part by regulating reactive oxygen species homeostasis through MT-1-4b (Hu et al., 2011). Previously, OsMADS58 was shown to play a key role in regulating floral meristem determinacy and normal carpel morphogenesis by the analysis of OsMADS58 RNA-silenced lines (Yamaguchi et al., 2006). However, a T-DNA insertion knockout mutant of OsMADS58 was recently identified and showed no obvious floral defects (M.M. Kater, personal communication). The osmads3-4 osmads58 double mutant displayed more severe defects of inner floral organs and meristem determinacy, suggesting that OsMADS58 and OsAMDS3 redundantly regulate inner floral organ identity and flower determinacy (M.M. Kater, personal communication). Therefore, it will be interesting to investigate the genetic interaction of OsMADS58 with OsMADS13 and DL in the future.
Figure 6.
Proposed model to illustrate the genetic interaction between OsMADS3, OsMADS13, and DL in rice flower development. A, Interactions between rice floral organ homeotic genes of A-function genes (such as OsMADS15), SPW1, OsMADS3, OsMADS13, and DL. Different colors represent the expression patterns of genes in lodicules, stamens, the carpel, and the ovule. OsMADS3 possibly represses the expression of A-function genes such as OsMADS15 in the inner floral organs; DL may antagonize the expression of SPW1 and OsMADS15. While OsMADS13 may indirectly limit the expression of DL in the ovule, DL may directly or indirectly positively regulate OsMADS13 expression. The broken arrow indicates the possibly indirect or direct regulation of the OsMADS13 expression by DL. B, Functions of OsMADS3, DL, and OsMADS13 in specifying floral organ identities and floral meristem termination. Green lines and red arrows indicate the functions of repression and promotion, respectively. OsMADS3 regulates the number of lodicules in whorl 2 by suppressing lodicule development, particularly near the palea (Yamaguchi et al., 2006), represses the formation of lodicules and determines the stamen identity in whorl 3, and specifies ovule identity in the floral center. DL represses the formation of stamens and specifies the carpel identity in the flower center, while OsMADS13 represses carpel formation and determines ovule identity. OsMADS13 may terminate floral meristem termination in parallel with OsMADS3, and DL may regulate the floral meristem determinacy in the same pathway of OsMADS13. OsMADS3 and DL can redundantly terminate the floral meristem.
Similarly, two duplicated AG homologs (zag1 and zmm2) are present in the maize genome, and mutations in zag1 cause loss of floral meristem determinacy in the ear, without obvious alteration of floral organ identity (Mena et al., 1996). Currently, no mutants of zmm2 have been identified, but the expression pattern of zmm2 is in agreement with that of class C function (Mena et al., 1996). Here, our genetic analysis of the double mutant osmads13-3 osmads3-4 indicated that OsMADS3 plays a critical role in ovule formation and floral meristem determinacy redundantly with OsMADS13 (Fig. 6). These data also support that the C-class and D-class genes probably retain their functions even though they underwent multiple subfunctionlization events and several neofunctionalizations after duplication within the AG clade (Rijpkema et al., 2010).
In rice, the YABBY domain gene DL was shown to be crucial for carpel specification (Nagasawa et al., 2003; Yamaguchi et al., 2004), which is different from the well-known ABC genes. In addition, the role of DL is distinct from the closely related YABBY gene CRC of Arabidopsis, which plays a mild role in carpel development (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Yamaguchi et al., 2004). Analysis of osmads3-4 dl-sup6 flowers indicated that DL and OsMADS3 play a redundant role in terminating floral meristem, but they may function in a distinct pathway (Fig. 6). The ectopic expression of SPW1 in the supernumerary whorls of lodicule-like organs of the double mutant flower may be explained by the antagonistic role of DL in reacting to class B genes in the flower center (Yamaguchi et al., 2004; this study). The ectopic expression of the putative class A gene OsMADS15 in the floral center may be caused by the mutation of OsMADS3. In Arabidopsis and Antirrhinum, A- and C-class genes were shown to be antagonistic to each other (Coen and Meyerowitz, 1991). Given the conserved role of the C gene in plant flower development, in combination with the ectopic formation of lodicule-like organs in some dl-sup6 flowers, we hypothesize that OsMADS3 and DL likely inhibit the expression of putative class A genes such as OsMADS15 in inner flower organs (Fig. 6).
The rice genome contains four putative A-class genes encoding AP1/FRUITFULL-like proteins: OsMADS14, OsMADS15, OsMADS18, and OsMADS20 (Fornara et al., 2004; Kater et al., 2006; Preston and Kellogg, 2006). However, few class A mutants have been identified in addition to those in Arabidopsis, and the roles of class A genes in floral organ identity are not as clear as was hypothesized by the ABCDE model (Preston and Kellogg, 2006). Unfortunately, besides the dep mutant, no other single or double knockout mutant lines for these rice genes have been reported. The dep mutant containing a single nucleotide G-to-C substitution at position 94 of the first exon of OsMADS15 displayed shrunken paleas and slightly elongated lemmas and glumes (Wang et al., 2010a), which are different from the mutant phenotype of class A genes AP1 and AP2 in Arabidopsis, with the conversion of sepals into leaf- or bract-like structures and petals into stamen-like organs or loss of sepals (Mandel et al., 1992; Jofuku et al., 1994). Therefore, whether DEP functions as an Arabidopsis A-class gene in rice flower development remains to be investigated. AP2 transcription factors in maize and rice have been shown to regulate shoot apical meistem determinacy. In maize, indeterminate spikelet1 (ids1) and the paralog of ids1, sid1, are required for floral meristem determinacy, and ids1 sid1 double mutants have no floral meristem, which was replaced by the formation of many bract-like organs, terminating in an ovule-like structure (Chuck et al., 2008). Similarly, mutations in the ids1-like gene SUPERNUMERARY BRACT in rice result in a delayed transition of spikelet meristem to floral meristem, with additional bract-like organs (Lee et al., 2007).
Furthermore, in this study, our finding suggests that OsMADS13 and DL specify carpel/ovule and floral meristem identity in the same pathway. Besides the observation that osmads13-3 dl-sup6 displayed flower defects similar to that of dl-sup6, no obvious OsMADS13 expression was detectable in dl-sup6 flowers, and DL transcripts were ectopically detected in osmads13-3 flowers, suggesting that DL may directly or indirectly regulate OsMADS13 expression. In other words, loss of OsMADS13 expression in dl-sup6 may result from the altered carpel/ovule identity in dl-sup6, or DL regulates carpel/ovule and meristem identity by controlling OsMADS13 expression. Furthermore, the ectopic expression of DL in osmads13-3 is likely caused by the altered identities of ovule and meristem, and OsMADS13 may indirectly restrict the expression of DL in the ovule (Fig. 6).
Regulation of Rice Floral Meristem Termination
Floral organs are formed by a floral meristem, a pool of pluripotent and dividing cells (Prunet et al., 2009). The regulation of the floral meristem seems to be widely conserved among angiosperms (Ferrario et al., 2004; Prunet et al., 2009). In Arabidopsis, AG is a master regulator terminating the floral meristem by turning WUSCHEL (WUS) off (Sieburth et al., 1998; Sun et al., 2009). In addition to homeotic transformations of stamens into petals, strong ag alleles (ag-1–ag-3) showed a complete loss of floral meristem determinacy, and the carpel was replaced by a new flower (Bowman et al., 1989, 1991; Yanofsky et al., 1990). The genomes of both eudicot and monocot species, including Antirrhinum, rice, maize, and barley, contain duplicated and subfunctionalized AG homologs (Zahn et al., 2006). Recent analysis of the osmads3-4 osmads58 double mutant suggests that two rice C-class genes, OsMADS3 and OsMADS58, redundantly regulate floral meristem determinacy (M.M. Kater, personal communication). In Antirrhinum, the class C MADS box gene PLENA (PLE) specifies reproductive organ identity and floral meristem termination, and the phenotype of ple mutants is similar to ag mutants, with homeotic conversion of reproductive organs to perianth organs (with the exception of nested flowers appearing inside whorl 4 instead of whorl 3 in strong ag mutants) and a loss of floral determinacy. In contrast, the mutation of FARINELLI (FAR), the close paralog of PLE, displayed normal flower development only with partial male sterility (Bradley et al., 1993; Davies et al., 1999). Moreover, the B-class MADS box genes DEF and GLO, which are not normally expressed in the fourth whorl, appeared to be ectopically expressed in ple far double mutants, suggesting a distinct role of the C class in Antirrhinum genes from that in Arabidopsis in redundantly and negatively regulating the B-function MADS box genes.
It is known that AG regulates the floral meristem by indirectly repressing the expression of WUS (Lenhard et al., 2001). Recently, KNUCKLES (KNU) encoding a C2H2 zinc-finger protein was shown to serve as the mediator in this feedback loop (Sun et al., 2009). AG directly regulates the expression of KNU, which can negatively regulate WUS expression (Sun et al., 2009). It remains unclear whether there is a similar mechanism in grasses. In this work, our genetic analyses elucidate the role of OsMADS3, OsMADS13, and DL in floral meristem determinacy (Fig. 6). There are 13 WOX (for WUSCHEL-related homeobox gene family) members in the rice genome, and OsWUS was found to be closely related to the Arabidopsis WUS gene (Nardmann and Werr, 2006; Dai et al., 2007; Nardmann et al., 2007; Zhang et al., 2010). But the biological function of OsWUS remains unclear. Nardmann and Werr (2006) isolated two WUS homologs (ZmWUS1 and ZmWUS2) in maize and rice OsWUS and found that they were not expressed in the organizing center of the vegetative shoot apical meristem, as was the WUS gene in Arabidopsis.
Similar to the role of eudicot SEP-like genes in floral meristem determinacy, grass SEP- and AGL6-like genes are capable of regulating carpel/ovule development and floral meristem determinacy (Jeon et al., 2000; Prasad et al., 2001, 2005; Agrawal et al., 2005; Chen et al., 2006b; Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Thompson et al., 2009; Cui et al., 2010; Gao et al., 2010; Kobayashi et al., 2010; Li et al., 2010). However, how these genes regulate floral organ identity and meristem determinacy in grasses remains less understood. It is likely that SEP-like and/or AGL6-like proteins act as mediators that constitute multimeric complexes with MADS domain proteins from different clades to regulate flower development in grasses (Immink et al., 2009; Seok et al., 2010; Wang et al., 2010b). In maize, double mutants of the AGL6-like gene bearded-ear (bde) and the class C gene zag1 display a severe ear phenotype with the conversion of floral meristems to branch-like meristems, which is not detectable in either single mutant, suggesting that bde and zag1 redundantly specify floral meristem identity (Thompson et al., 2009). Moreover, BDE and ZAG1 can physically interact, suggesting that these two proteins act in complexes to control floral development in the maize ear (Thompson et al., 2009). OsMADS7 (also called OsMADS45) and OsMADS8 (also called OsMADS24) were shown to have a similar interaction profile to those of Arabidopsis SEP proteins (Kater et al., 2006; Cui et al., 2010). They can interact with the AG-like protein OsMADS13, which is similar to STK. OsMADS7 and OsMADS8 also interact with Arabidopsis STK and petunia FBP7 (Favaro et al., 2002, 2003).
In summary, this study reveals the genetic interaction of the floral homeotic genes OsMADS3, OsMADS13, and DL and describes an unknown model to illustrate the role of OsMADS3, DL, and OsMADS13 in the specification of flower organ identity and meristem determinacy in rice.
MATERIALS AND METHODS
Plant Materials
The mutants osmads13-3 and dl-sup6 were identified from an M2 population of rice (Oryza sativa subspecies japonica ’9522’) mutagenized with radiation of 60Co γ-ray (Chen et al., 2006a). The strong allele of OsMADS13 (osmads13-1) and the weak allele (dl-2) were kindly provided by Prof. Martin M. Kater (Universita degli Studi di Milano) and Prof. Hiro-Yuki Hirano (University of Tokyo), respectively. Prior to the analysis, osmads13-3, osmads3-4, and dl-sup6 were all crossed with wild-type 9522 three times. Double mutant plants were isolated by phenotype observation and verified by genotyping with primers 3TPF/3TPR and 13TPF/13TPR for osmads3-4 and osmads13-3, respectively (Supplemental Table S1). Mutant and wild-type rice plants were planted in paddy fields under normal conditions in Shanghai or in a greenhouse at Shanghai Jiao Tong University.
Histological Analysis and Microscopy Observation
Materials were fixed and dehydrated as described by Li et al. (2006). For histological analysis, tissues were substituted by xylene and embedded in Paraplast plus. Then, materials were sectioned to 8 μm thick, stained with toluidine blue, and photographed using a Nikon E600 microscope and a Nikon DXM1200 digital camera. SEM observation was performed with JSM-6360LV (JEOL) as described previously (Li et al., 2006). The dividing of the ovule stages refers to a previous report (Lopez-Dee et al., 1999).
In Situ Hybridization
Treatment of samples was as described previously (Li. et al., 2006). For the construction of specific probes for OsMADS13, SPW1/OsMADS16, and DL, gene-specific fragments of OsMADS13 cDNA (367–958 bp), OsMADS16 cDNA (211–686 bp), and DL cDNA (121–639 bp) were amplified by reverse transcription (RT)-PCR using primers 13PPF/13PPR, 16PPF/16PPR, and DLPPF/DLPPR, respectively (Supplemental Table S1) and cloned into pBluescript II KS+ phagemid vector (Stratagene). The probe construct of OSH1 was generated as described previously (Agrawal et al., 2005; Yamaki et al., 2005; Li et al., 2010). Construction of the OsMADS3 and OsMADS15 probes referred to previous reports (Kyozuka et al., 2000; Yamaguchi et al., 2006). Digoxigenin-labeled antisense and sense probes were transcribed in vitro as described previously (Chu et al., 2006). Images were obtained using the Olympus Nikon E600 microscope.
Yeast Two-Hybrid Analysis
The MATCHMAKER GAL4 Two-Hybrid System (Clontech) was used to detect the interaction between OsMADS3 and OsMADS13. cDNA fragments encoding the IKC domain of OsMADS3 and OsMADS13 were amplified by RT-PCR with primers 3YF/3YR and 13YF/13YR, respectively (Supplemental Table S1), and the cDNA fragment encoding the IKC14 domain of OsMADS6 was amplified by RT-PCR with primers 6YF/6YR (Supplemental Table S1). Then, these cDNA fragments were cloned into pGBKT7 and pGADT7 to fuse with the BD (bait domain) and AD (activation domain) of GAL4, respectively. Recombinant vectors were named AD-13, BD-13, AD-3, BD-3, AD-6, and BD-6 respectively. Self-activation was assayed on selective synthetic dropout medium plates (−Leu/−His/+3-amino-1,2,4-triazole [3-AT] or −Trp/−His/+3-AT). Then, combinations of AD-3/BD-13 and BD-3/AD-13 were transformed into yeast strain AH109 simultaneously according to the protocol. The transformants cotransformed with plasmids encoding OsMADS6 and OsMADS13 were used as a positive control (Favaro et al., 2002), and the transformants containing plasmids pGADT7 and pGBKT7 were used as a negative control. The interaction was judged by the growth condition on selective mediums (−Trp/−Leu/−His/+3-AT) according to the protocol from the company.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of osmads13 mutants and abnormal ovule development of osmads13-3.
Supplemental Figure S2. Schematic representation of dl-sup mutants and the phenotype of dl-sup6.
Supplemental Figure S3. OsMADS3 does not interact with OsMADS13 in yeast cells.
Supplemental Table S1. Primers used in this research.
Acknowledgments
We acknowledge Prof. Martin M. Kater for providing the allele of osmads13-1 and Prof. Hiro-Yuki Hirano for the weak allele of dl-2. We thank the anonymous reviewers for comments on the manuscript, Dr. Hao Yu and Dr. Ning Jiang for editing the manuscript, and Z.J. Luo and M.J. Chen for genetic mutant screening.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. (2000) Molecular and genetic analyses of the silky1 gene reveal conservation floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJ, van Tunen AJ. (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7: 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theissen G. (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater MM, Colombo L. (2007) Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chu H, Yuan Z, Zhang DB. (2006a) Isolation and genetic analysis for rice mutants treated with 60 Co γ-ray. J Xiamen Univ (Nat Sci) 45: 82–85 [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH. (2006b) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Chu HW, Qian Q, Liang WQ, Yin CS, Tan HX, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al. (2006) The Floral Organ Number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S. (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Clifford H. (1987) Spikelet and Floral Morphology. Smithsonian Institution Press, Washington, DC [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Colombo L, Battaglia R, Kater MM. (2008) Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci 13: 444–450 [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, van Went J, Dons HJ, Angenent GC, van Tunen AJ. (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z. (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX. (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. (1999) PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J 18: 4023–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Fan J, Li W, Dong X, Guo W, Shu H. (2007) Ectopic expression of a hyacinth AGL6 homolog caused earlier flowering and homeotic conversion in Arabidopsis. Sci China C Life Sci 50: 676–689 [DOI] [PubMed] [Google Scholar]
- Favaro R, Immink RG, Ferioli V, Bernasconi B, Byzova M, Angenent GC, Kater M, Colombo L. (2002) Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol Genet Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L. (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S, Immink RG, Angenent GC. (2004) Conservation and diversity in flower land. Curr Opin Plant Biol 7: 84–91 [DOI] [PubMed] [Google Scholar]
- Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM. (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XC, Liang WQ, Yin CS, Ji SM, Wang H, Su X, Guo CC, Kong HZ, Xue HW, Zhang DB. (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Hsu HF, Huang CH, Chou LT, Yang CH. (2003) Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 44: 783–794 [DOI] [PubMed] [Google Scholar]
- Hu LF, Liang WQ, Yin CS, Cui X, Zong J, Wang X, Hu JP, Zhang DB. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y. (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54: 147–156 [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. (2009) SEPALLATA3: the 'glue' for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al. (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L. (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. (2004) Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K. (2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Lee DY, Lee J, Moon S, Park SY, An G. (2007) The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49: 64–78 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Son JS, Nam J, Jeong DH, Lee K, Jang S, Yoo J, Lee J, Lee DY, et al. (2003) Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol 44: 1403–1411 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Jia RD, Yin CS, Zong J, Kong HZ, Zhang DB. (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder H, Rudall P. (2005) Evolutionary history of Poales. Annu Rev Ecol Evol Syst 36: 107–124 [Google Scholar]
- Liu C, Thong Z, Yu H. (2009) Coming into bloom: the specification of floral meristems. Development 136: 3379–3391 [DOI] [PubMed] [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pè M, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L. (1999) OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev Genet 25: 237–244 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. (1996) Diversification of C-function activity in maize flower development. Science 274: 1537–1540 [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W. (2006) The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol Biol Evol 23: 2492–2504 [DOI] [PubMed] [Google Scholar]
- Nardmann J, Zimmermann R, Durantini D, Kranz E, Werr W. (2007) WOX gene phylogeny in Poaceae: a comparative approach addressing leaf and embryo development. Mol Biol Evol 24: 2474–2484 [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF. (2001a) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF. (2001b) Conversion of leaves into petals in Arabidopsis. Curr Biol 11: 182–184 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. (2006) Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Morel P, Negrutiu I, Trehin C. (2009) Time to stop: flower meristem termination. Plant Physiol 150: 1764–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. (1995) Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheimer R, Kellogg EA. (2009) Evolution of AGL6-like MADS box genes in grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell 21: 2591–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Fischer RL. (1993) The ovule and the embryo sac. Plant Cell 5: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema AS, Vandenbussche M, Koes R, Heijmans K, Gerats T. (2010) Variations on a theme: changes in the floral ABCs in angiosperms. Semin Cell Dev Biol 21: 100–107 [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Zethof J, Gerats T, Vandenbussche M. (2009) The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J 60: 1–9 [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Stuppy W, Jennifer C, Kellogg EA, Briggs BG. (2005) Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am J Bot 92: 1432–1443 [DOI] [PubMed] [Google Scholar]
- Seok HY, Park HY, Park JI, Lee YM, Lee SY, An G, Moon YH. (2010) Rice ternary MADS protein complexes containing class B MADS heterodimer. Biochem Biophys Res Commun 401: 598–604 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Drews GN, Meyerowitz EM. (1998) Non-autonomy of AGAMOUS function in flower development: use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development 125: 4303–4312 [DOI] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T. (2009) A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev 23: 1791–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H. (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42: 115–149 [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S. (2009) bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BE, Hake S. (2009) Translational biology: from Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene T, Vekemans D, Becker A, Melzer S, Geuten K. (2010) Expression divergence of the AGL6 MADS domain transcription factor lineage after a core eudicot duplication suggests functional diversification. BMC Plant Biol 10: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue YB, Cheng ZK. (2010a) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Melzer R, Theissen G. (2010b) Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets.’. Plant J 64: 177–190 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Zanis MJ, Kellogg EA, Schmidt RJ. (2007) Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc Natl Acad Sci USA 104: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Nagato Y, Kurata N, Nonomura KI. (2011) Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev Biol 351: 208–216 [DOI] [PubMed] [Google Scholar]
- Yamaki S, Satoh H, Nagato Y. (2005) Gypsy embryo specifies ovule curvature by regulating ovule/integument development in rice. Planta 222: 408–417 [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB. (2009) RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong HZ, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H. (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack JH, Arrington JM, Hu Y, Landherr LL, dePamphilis CW, Becker A, Theissen G, Ma H. (2006) Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol Dev 8: 30–45 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA. (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 2342–2353 [Google Scholar]
- Zhang X, Zong J, Liu J, Yin JY, Zhang DB. (2010) Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J Integr Plant Biol 52: 1016–1026 [DOI] [PubMed] [Google Scholar]