Abstract
Ralstonia solanacearum is the causal agent of bacterial wilt (BW), one of the most important bacterial diseases worldwide. We used cDNA microarray to survey the gene expression profile in transgenic tomato (Solanum lycopersicum) overexpressing Arabidopsis (Arabidopsis thaliana) CBF1 (AtCBF1), which confers tolerance to BW. The disease-resistant phenotype is correlated with constitutive expression of the Related-to-ABI3/VP1 (RAV) transcription factor, ethylene-responsive factor (ERF) family genes, and several pathogenesis-related (PR) genes. Using a transient assay system, we show that tomato RAV2 (SlRAV2) can transactivate the reporter gene driven by the SlERF5 promoter. Virus-induced gene silencing of SlERF5 and SlRAV2 in AtCBF1 transgenic and BW-resistant cultivar Hawaii 7996 plants gave rise to plants with enhanced susceptibility to BW. Constitutive overexpression of SlRAV2 in transgenic tomato plants induced the expression of SlERF5 and PR5 genes and increased BW tolerance, while knockdown of expression of SlRAV2 inhibited SlERF5 and PR5 gene expression under pathogen infection and significantly decreased BW tolerance. In addition, transgenic tomato overexpressing SlERF5 also accumulated higher levels of PR5 transcripts and displayed better tolerance to pathogen than wild-type plants. From these results, we conclude that SlERFs may act as intermediate transcription factors between AtCBF1 and PR genes via SlRAV in tomato, which results in enhanced tolerance to BW.
Tomato (Solanum lycopersicum) is the second most consumed vegetable worldwide. The productivity and quality of tomato fruits are often threatened by a broad range of plant diseases caused by fungi, bacteria, nematodes, and arthropods (Deslandes et al., 2002; Hemming et al., 2004). Ralstonia solanacearum is one of the most common soil-borne vascular diseases of the tomato crop; the resulting disease, bacterial wilt (BW), can be devastating and difficult to control by conventional approaches. Introgression of traits has played a pivotal role in developing BW-resistant varieties to reduce yield loss; however, only a few of the generated varieties show stable resistance because of the great diversity of pathogen strains (Hai et al., 2008). Genetic engineering is a promising alternative strategy to enhance plant disease resistance to a wide range of pathogens. The validity of this approach has been demonstrated in crops into which a wide array of plant disease resistance genes and pathogen virulence genes have been cloned. Although many genetic engineering programs in major tomato-growing areas worldwide focus on producing BW-tolerant varieties, the genetic network regulating plant tolerance to BW remains poorly understood. However, understanding plant defense mechanisms and responses to pathogens is critical to developing resistant tomato varieties (Robb et al., 2007).
Being sessile in nature, plants use a variety of strategies to protect themselves from pathogen infection. The protection is manifested by a single gene or a group of genes working in coordination to modulate specific defense responses via signal transduction cascades and transcriptional activation of many genes (Zhang et al., 2004a; Wang et al., 2005). The integrated defense systems are reflected in the expression of transcription factors and protein kinases as well as changes in cytosolic calcium fluxes, an increase in reactive oxygen species during the oxidative burst, and induction of hypersensitive cell death (the hypersensitive response; Gómez-Gómez, 2004; Ryan et al., 2007). The expression of various defense genes also leads to the production of defensive compounds, such as pathogenesis-related (PR) proteins and enzymes involved in the biosynthesis of protective secondary metabolites (Gu et al., 2002). Even though the functions of most PR gene products are unknown, some of these proteins, such as β-1,3-glucanase (PR2) and chitinase (PR3), are known to inhibit fungal growth, and thaumatin-like/osmotin (PR5) has been found to induce apoptosis (He et al., 2001; Gu et al., 2002).
Many PR genes induced during pathogen infection are up-regulated by one or more signaling molecules, such as salicylic acid (SA), ethylene, and jasmonic acid (JA; Koo et al., 2007). Recent evidence indicates that transcription factors play key roles in controlling the expression of PR genes; for instance, ethylene-responsive factor (ERF) proteins activate PR genes by binding to the GCC box (GCCGCC) of their promoters, thereby regulating the plant defense response to pathogen infection (Zhang et al., 2004a). Recently, AP2/EREBP (for apetala2/ethylene-responsive element-binding protein) proteins were shown to be integrators of biotic and abiotic stress responses through their interaction with cis-acting elements, the GCC box, and/or CRT/DRE (for C-repeat/dehydration response element; Park et al., 2001; Zhang et al., 2005). These proteins comprise unique transcription factors to the plant lineage and are classified into four subfamilies: AP2, DREB (for dehydration response element-binding protein), ERF, and RAV (for related to ABI3/VP1). The members of the ERF subfamily, which include tobacco (Nicotiana tabacum) ERF1 to -4, Arabidopsis (Arabidopsis thaliana) ERF1 to -5, ORA59, tomato Pti4 to -6, tomato ERF1 to -4, and tomato stress-responsive factor (TSRF1), have been identified as transcriptional activators that bind to the GCC box in response to biotic stresses (Gu et al., 2002; Chakravarthy et al., 2003; Zhang et al., 2007; Pré et al., 2008). Although AtERF4/7 also regulates genes by interacting with a GCC box, it is a transcriptional repressor and thus a negative regulator capable of modulating both biotic and abiotic stress responses (Yang et al., 2005). In addition, rice (Oryza sativa) TERF1, barley (Hordeum vulgare) HvRAF, and tomato TSRF1 are involved in the regulation of both biotic and abiotic stress tolerance (Jung et al., 2007; Gao et al., 2008). These findings strongly suggest that the induction of PR genes in plants is mediated by different ERF proteins and/or signaling molecules. By contrast, the regulation of PR genes by the subfamily members DREB and RAV in response to biotic stress remains unclear.
CBF/DREB1 (for CRT-binding factor or DRE-binding protein 1) genes, including CBF1 (DREB1B), CBF2 (DREB1C), and CBF3 (DREB1A), are located on Arabidopsis chromosome 4 (Gilmour et al., 1998). The CBF family can bind to CRT/DRE elements present in the promoters of cold-regulated (COR) genes, such as KIN1, COR15a, COR47, and RD29A, to induce these genes in response to low temperature and dehydration (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Sakuma et al., 2002). Moreover, overexpression of cDNA encoding CBF3 in transgenic Arabidopsis activated several stress-tolerance genes, thus enhancing the tolerance of plants to drought, freezing, and salt stresses (Liu et al., 1998; Gilmour et al., 2000; Sakuma et al., 2006). Previously, we have reported that transgenic tomato expressing Arabidopsis CBF1 (AtCBF1) cDNA is tolerant to various abiotic stresses such as chilling, oxidative stress, high salt, and water deficit (Hsieh et al., 2002a, 2002b; Lee et al., 2003). Here, we report that AtCBF1 transgenic tomato plants are tolerant to Ralstonia infection in greenhouse experiments and that AtCBF1 modulates the plant defense response against Ralstonia by repressing the proliferation of bacteria in vascular tissues. In addition, we have used cDNA microarray to identify downstream defense components that connect AtCBF1 with disease defense response. Our study provides new insights into signaling pathways and defines a possible mechanism of how AtCBF1 directly or indirectly regulates other AP2/EREBP transcription factors, thereby improving tolerance of tomato against Ralstonia.
RESULTS
Several Pathogenesis-Related Genes Are Activated in CBF1 Transgenic Plants
In previous studies, we have demonstrated that constitutive expression of AtCBF1 in tomato increased tolerance to chilling and water deficit (Hsieh et al., 2002a, 2002b). To identify the genes that were differentially expressed in AtCBF1 transgenic tomato plants, we now used subtractive hybridization and homemade microarray systems (Liu et al., 2006). Expression was increased by at least 2-fold for 25 genes in AtCBF1 transgenic plants compared with wild-type plants (Table I). Among those genes, the following were pathogenesis-related genes: PR3 (chitinase), PR5 (thaumatin-like protein), PR7 (endoproteinase), PR9 (peroxidase), and PR10 (RNase-like protein). Thus, heterologous expression of AtCBF1 appears to result in enhanced expression of several PR genes.
Table I. Putative target genes of heterologous AtCBF1 in transgenic tomato plants.
Sequences of cis-acting elements are as follows: CRT/DRE, CCGAC or RYCGAC (HvCBF); GCC, GCCGCC; RAV1A, CAACA; and RAV1B, CACCTG.
| Clone Name | Accession No. | Description | Unigene No. | Corresponding Arabidopsis Gene | Ratioa | SGN Database: Tomato WGS Scaffolds (2.30) | cis-Acting Elements within the 2-kb Promoter | |||
| CRT/DRE | GCC | RAV1A | RAV1B | |||||||
| cLEY14E7 | BE449751 | Protein phosphatase 2C-like protein | SGN-U573715 | At2g25070 | 10.41 ± 2.41 | No hit found | –b | – | – | – |
| C6SR473 | CK574973 | Cys protease (PR7) | SGN-U580215 | At4g32940 | 6.49 ± 0.51 | SL2.30sc04948 | 1 | 0 | 3 | 0 |
| cLEX4M16 | AW219536 | Peroxidase (PR9) | SGN-U581155 | At5g05340 | 6.22 ± 3.96 | SL2.30sc03665 | 0 | 0 | 5 | 1 |
| LEC5R05G01 | CK468708 | PR10 protein | SGN-U578441 | At1g24020 | 5.90 ± 1.49 | SL2.30sc04828 | 0 | 0 | 6 | 0 |
| cLEX11D13 | AW621284 | Zinc transporter protein ZIP1 | SGN-U583586 | At1g05300 | 5.87 ± 1.78 | SL2.30sc03685 | 0 | 0 | 3 | 0 |
| cLEX8A20 | AW220124 | Dehydrin homolog C17 | SGN-U581375 | At1g20450 | 4.60 ± 1.55 | SL2.30sc04135 | 5 | 0 | 1 | 1 |
| LEEC101F05 | CK725213 | Acidic endochitinase precursor (PR3) | SGN-U566861 | At5g24090 | 3.80 ± 1.71 | SL2.30sc03902 | 1 | 0 | 3 | 0 |
| SF471 | CK574994 | Acidic 26-kD endochitinase precursor (PR3) | SGN-U581507 | At3g12500 | 3.65 ± 0.05 | SL2.30sc03665 | 1 | 0 | 4 | 0 |
| cLEX12C2 | AW621528 | Syntaxin-related protein Nt-syr1 | SGN-U584182 | At3g11820 | 3.53 ± 0.54 | SL2.30sc04133 | 1 | 0 | 5 | 0 |
| cLEW12A21 | BF096513 | Catalase isozyme 1 | SGN-U578839 | At4g35090 | 3.48 ± 0.03 | SL2.30sc05380 | 1 | 0 | 6 | 0 |
| cLEW19G18 | BF097084 | PTEN-like protein | SGN-U566184 | At3g19420 | 3.43 ± 0.00 | SL2.30sc03665 (gap in promoter) | – | – | – | – |
| cLEW27E20 | BF098457 | Senescence-associated protein-related | SGN-U578016 | At5g20700 | 3.21 ± 0.12 | SL2.30sc03731 | 1 | 0 | 5 | 0 |
| cLEX5K5 | AW219630 | DnaJ-like heat shock protein | SGN-U589575 | At4g13830 | 3.17 ± 1.20 | SL2.30sc03902 | 2 | 0 | 1 | 0 |
| cLEW8A6 | AW980043 | 1-Acylglycerol-3-phosphate acyltransferase | SGN-U575300 | At1g51260 | 3.00 ± 0.67 | SL2.30sc03876 | 0 | 0 | 6 | 0 |
| cLEX2M12 | AW219010 | Ubiquitin family protein | SGN-U567499 | At2g30100 | 2.71 ± 0.26 | SL2.30sc04474 | 1 | 0 | 3 | 0 |
| cLEX2M14 | AW219011 | Eukaryotic translation initiation factor 5A-1 | SGN-U578904 | At1g13950 | 2.65 ± 0.62 | SL2.30sc03701 | 0 | 0 | 3 | 0 |
| SF146 | CK574987 | Gly-rich protein | SGN-U313109 | At2g05440 | 2.59 ± 0.55 | SL2.30sc03852 (gap, 313 bp checked) | 1 | 0 | 1 | 0 |
| cLEW22K5 | BF097441 | Microsomal signal peptidase 25-kD subunit | SGN-U577878 | At2g39960 | 2.58 ± 0.18 | SL2.30sc06557 (gap, 1,534 bp checked) | 0 | 0 | 2 | 0 |
| Rs-Ck-1-G3 | CK715671 | Microsomal ω-6-desaturase | SGN-U574778 | At3g12120 | 2.52 ± 0.60 | SL2.30sc04057 | 2 | 0 | 3 | 0 |
| LERCD04N18 | CK715495 | Unknown protein | SGN-U582639 | At3g03870 | 2.52 ± 0.20 | SL2.30sc04199 | 0 | 1 | 6 | 0 |
| cLEW19L9 | BF097167 | COPl homolog | SGN-U579490 | At2g32950 | 2.21 ± 0.24 | SL2.30sc04607 | 4 | 0 | 7 | 0 |
| LEECI01D07 | CK720580 | Formate dehydrogenase | SGN-U579280 | At5g14780 | 2.19 ± 0.33 | SL2.30sc03665 | 1 | 0 | 4 | 0 |
| LERCD04N21 | CK715497 | Cys protease (PR7) | SGN-U578421 | At1g47128 | 2.16 ± 0.47 | SL2.30sc05611 | 1 | 0 | 3 | 0 |
| SF847 | AY257487 | PR-5 | SGN-U578836 | At4g11650 | 2.11 ± 0.39 | SL2.30sc03923 | 1 | 2 | 4 | 1 |
| cLEW26O13 | BF098337 | DnaJ-like heat shock protein | SGN-U579998 | At3g44110 | 2.02 ± 0.01 | SL2.30sc03604 | 0 | 0 | 8 | 0 |
| SF547 | CK664757 | β-1,3-Glucanase (PR2) | SGN-U581016 | At3g57270 | 1.29 ± 0.11 | SL2.30sc05010 | 0 | 1 | 4 | 1 |
Ratio = (fluorescence intensity of each cDNA for transgenic plants/fluorescence intensity of each cDNA for wild-type plants) ÷ (fluorescence intensity of ubiquitin for transgenic plants/fluorescence intensity of ubiquitin for wild-type). Each value is the mean ± sd of three independent experiments.
En dash (–), promoter sequence in SGN database is unavailable.
Progression of BW Is Delayed in Transgenic AtCBF1 Plants by Systemic Suppression of Bacterial Multiplication
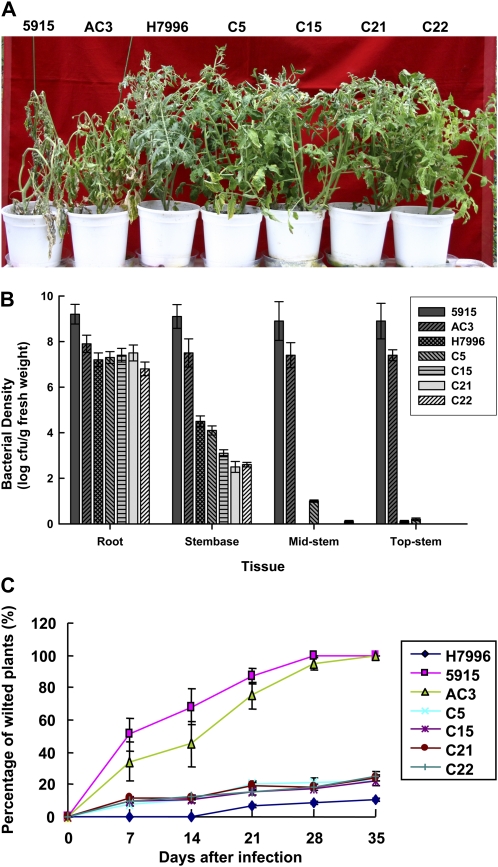
We hypothesized that up-regulation of PR genes in AtCBF1 transgenic tomato may enhance its resistance to Ralstonia infection. We observed transgenic plants in the greenhouse to discover which plants were more tolerant to pathogen attack. Ralstonia inoculation assay was performed to examine whether overexpression of AtCBF1 in tomato can enhance pathogen resistance. Tomato natural cv Hawaii 7996 (H7996) has displayed stable resistance against various R. solanacearum strains (Grimault et al., 1995). In this study, we used tomato cv H7996 and the background of the AtCBF1 transgenic plant 5915 as BW-resistant and -susceptible control, respectively. Four AtCBF1 constitutive overexpression lines (C5, C15, C21, and C22) with high expression but low insertion (one to two copies) of transgene were selected for further investigation. In parallel, we created transgenic tomato plants with the AtCBF1 gene driven by the abscisic acid (ABA)-inducible ABRC1 promoter (line AC3) for pathogen infection (Lee et al., 2003). Similar to the BW-resistant tomato cv H7996, the transgenic lines (C5, C15, C21, and C22) did not show any signs of wilting at 7 d post inoculation (dpi) with Ralstonia (Fig. 1A). Wild-type (5915) and AC3 plants without ABA treatment were severely wilted at 7 dpi (Fig. 1A). Upon ABA treatment, AC3 plants exhibited enhanced resistance to Ralstonia infection (data not shown). To further investigate the nature of the enhanced BW resistance seen in the transgenic lines, we monitored the in planta multiplication of Ralstonia after inoculation. The bacterial titers in various tissues of susceptible control plants (5915 and AC3) reached a very high level (≥107 colony-forming units [cfu] g−1 fresh tissue) at 7 dpi (Fig. 1B). By contrast, the internal bacterial titers in the transgenic lines and H7996 were much lower than those in 5915 and AC3 except in roots. In addition, the pattern and level of bacterial growth suppression in AtCBF1 transgenic lines was similar to that in H7996, the BW-resistant variety, with gradually declining levels of bacteria from the roots to the top stems (Fig. 1B).
Figure 1.
AtCBF1 transgenic plants exhibit enhanced resistance to Ralstonia. A, Test plants were inoculated with Ralstonia and then kept at 28°C with a photoperiod of 16 h. The photograph was taken at 7 dpi. The test plants comprised wild-type plants (susceptible variety 5915, the genetic background of transformants), a BW-resistant control variety (H7996), a control transgenic line (AC3; AtCBF1 driven by an ABA-inducible ABRC1 promoter), and T2 transgenic plants continuously expressing AtCBF1 (C5, C15, C21, and C22). B, Ralstonia multiplication in transgenic tomato plants was systemically suppressed. The bacterial titer inside the test plants was measured in different tissues at 7 dpi. The data are means of three independent measurements. C, Disease progression of BW was delayed in transgenic tomato plants. The response of plants subjected to BW bioassays was scored as the percentage of wilted plants over time.
To reveal the correlation between disease resistance and AtCBF1 expression, disease progression in H7996 and in transgenic lines exhibiting high levels of BW resistance were compared with that in wild-type and AC3 plants. Less than 20% of the transgenic and H7996 plants wilted during the test period (Fig. 1C), and AtCBF1 transgenic lines exhibited a disease incidence nearly equivalent to that of H7996 over the test period. By contrast, nearly 50% of the wild-type (5915) plants and 40% of AC3 plants wilted on 7 dpi, and all had withered on 35 dpi. Thus, we conclude that constitutive expression of AtCBF1 in transgenic tomato enhances BW resistance by systemic suppression of internal bacterial multiplication via the activation of PR proteins.
Tolerance to Ralstonia Infection Is Not Affected by Exogenous GA3 Treatment in Transgenic Plants
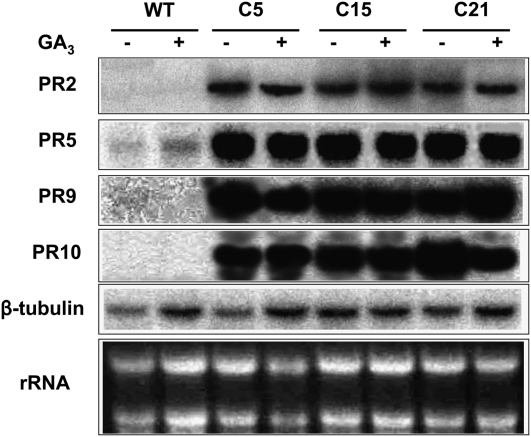
Constitutive expression of AtCBF1 in tomato resulted in a dwarf phenotype that was alleviated by the application of GA3 (Hsieh et al., 2002a, 2002b). To test whether GA3 is a common antagonist of the AtCBF network that not only restores the dwarf and late-flowering phenotype but also influences the expression of the PR genes, we examined the expression levels of PR genes in AtCBF1 transgenic lines (C5, C15, and C21) and wild-type plants in the presence and absence of GA3 treatment by northern-blot analyses (Fig. 2). The tested PR genes, such as PR2 (β-1,3-glucanase), PR5, PR9, and PR10, were up-regulated by 1.29-, 2.11-, 6.22-, and 5.90-fold, respectively, in AtCBF1 transgenic plants compared with wild-type tomato plants, as shown in Table I. Although the ratio of PR2 is only 1.29-fold, the mRNA density showed it significantly accumulated in AtCBF1 transgenic plants (Fig. 2). Northern-blot results showed that exogenous application of GA3 did not change the level of expression of PR genes in the transgenic plants. The transcripts of these genes were barely detectable in the wild-type plants with or without GA3. Thus, we conclude that the expression of PR genes in AtCBF1 transgenic plants results from the overexpression of AtCBF1 by a GA3-independent pathway.
Figure 2.
Northern-blot analyses of PR genes in transgenic tomato plants. Total RNA (10 μg) was extracted from wild-type plants (WT) and transgenic T1 plants overexpressing AtCBF1 (C5, C15, and C21). Probes used to hybridize total RNA were 32P-labeled PR2, PR5, PR9, PR10, and β-Tubulin cDNA fragments. Equal loading in each lane was verified by rRNA detection, which was carried out by ethidium bromide staining of the gel followed by visualization of bands under UV illumination. Experiments were performed in triplicate.
Wild-type and AtCBF1 transgenic tomato plants were infected with a high cell density of Ralstonia and allowed to grow under controlled conditions. In the absence of GA3 treatment, the fruit set, seed number, and fresh weight of the AtCBF1 transgenic lines were severely affected (Table II). However, the defects in fruit set, seed number, and fresh weight of AtCBF1 transgenic plants were partially to almost completely restored by GA3 treatment. After Ralstonia infection, all of the tested wild-type plants wilted too severely to reach the reproductive stage, while AtCBF1 transgenic plants survived and reproduced either with or without GA3 treatment. More importantly, not significant to little differences were found in the fruit yield between noninfected wild-type plants and Ralstonia-infected GA3-treated AtCBF1 transgenic lines (Table II; P > 0.05 in line C5 and C15, Student’s t test). These results strongly suggest that the protection of tomato plants from Ralstonia infection by overexpression of AtCBF1 is independent of the restoration of growth to normal levels by exogenous GA3.
Table II. The pathogen tolerance of transgenic tomato plants is not affected by exogenous GA3 treatment.
Data shown in each column, from top to bottom, are fruit number (FN) per plant, seed number (SN) per fruit, and fresh weight (FW; g) per plant. Each value is the mean ± sd (n = 5 individual plants). Wild-type and AtCBF1 transgenic plants were grown in pots with peat moss and watered every alternate day in a greenhouse with a 16/8-h photoperiod (daylight of about 120 μmol m−2 s−1, 26°C ± 2°C; night temperature of 22°C ± 2°C). For GA3 treatment, AtCBF1 transgenic and wild-type plants were sprayed with 5 mg L−1 GA3 three times per week (Hsieh et al., 2002a). One-month-old plants were inoculated with Ralstonia. Disease progression of BW was defined as wilted plant number divided by total plant number. Three months later, these plants were harvested, weighed for fresh weight, and calculated for fruit and seed numbers.
| Treatment | Wild Type | C5 | C15 | C21 | Wild Type + GA3 | C5 + GA3 | C15 + GA3 | C21 + GA3 |
| Control | ||||||||
| FN | 21.6 ± 4.1 | 6.0 ± 1.6 | 7.2 ± 1.6 | 1.6 ± 1.1 | 26.6 ± 4.1 | 24.8 ± 3.6 | 22.4 ± 3.2 | 17.4 ± 5.8 |
| SN | 48.7 ± 9.2 | 8.4 ± 2.7 | 6.8 ± 1.3 | 2.4 ± 0.9 | 43.7 ± 9.2 | 25.4 ± 3.0 | 22.6 ± 2.6 | 29.6 ± 14.8 |
| FW | 132.4 ± 7.1 | 80.6 ± 5.1 | 106.8 ± 9.2 | 85.0 ± 3.9 | 147.4 ± 7.1 | 133.4 ± 13.8 | 138.8 ± 13.6 | 127.6 ± 8.7 |
| Ralstonia | ||||||||
| FN | 0 ± 0 | 9.8 ± 1.4 | 10.8 ± 1.6 | 3.8 ± 1.0 | 0 ± 0 | 25.8 ± 3.7 | 20.6 ± 4.1 | 13.8 ± 2.6 |
| SN | 0 ± 0 | 8.3 ± 3.2 | 7.7 ± 1.5 | 3.8 ± 1.2 | 0 ± 0 | 14.6 ± 3.2 | 20.8 ± 7.2 | 14.3 ± 5.2 |
| FW | 12.3 ± 6.3 | 112.4 ± 14.8 | 119.0 ± 15.5 | 120.0 ± 11.5 | 13.6 ± 5.3 | 121.4 ± 4.0 | 118.2 ± 8.3 | 113.8 ± 7.2 |
AtCBF1 Binds to CRT/DRE But Not to the GCC Box
To study the DNA-binding activity of AtCBF1 to the GCC box, which is generally present in the promoter region of PR genes, we performed electrophoretic mobility shift assay (EMSA) experiments with a purified His-tagged AtCBF1 fusion protein. The results indicated that AtCBF1 recombinant protein binds the CRT/DRE sequence but not the GCC box and mutated CRT/DRE (Supplemental Fig. S1). Binding to this element was sequence specific, as the association was efficiently inhibited by a 10- to 100-fold excess of unlabeled competitive CRT/DRE fragment (Supplemental Fig. S1C). From these results, we conclude that AtCBF1 binds competitively to CRT/DRE but not to the GCC box in vitro.
Several AP2/EREBP Family Genes Are Up-Regulated in AtCBF1 Transgenic Tomato
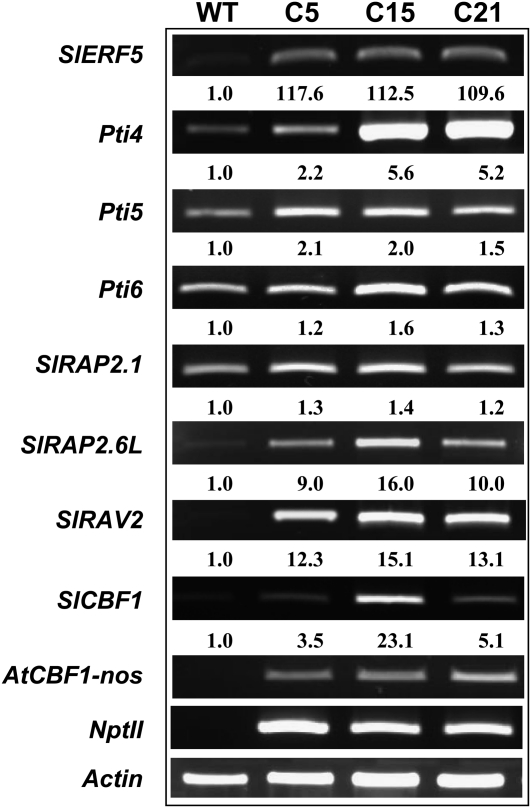
According to the obtained EMSA results, AtCBF1 specifically interacts with the CRT/DRE rather than the GCC box. Therefore, we hypothesized that AtCBF1 overexpression in tomato regulates PR genes through either an indirect pathway or an accessory protein. To identify potential intermediate modulators involved in the signaling cascade of AtCBF1 contributing to BW tolerance, we examined changes in mRNA level of several well-known GCC box-binding ERFs, such as Pti4, Pti5, and Pti6, in AtCBF1 transgenic tomato by using semiquantitative reverse transcription (RT)-PCR. In addition, we analyzed the expression patterns of newly identified ERFs and RAVs, such as SlERF5 and SlRAV2, and some tomato orthologs of putative Arabidopsis CBF1-regulated AP2/EREBP (Zhang et al., 2004b), such as SlRAP2.1 and SlRAP2.6-like genes, in AtCBF1 transgenic tomato. RAV transcription factors belong to a subfamily of the AP2/EREBP superfamily (Nakano et al., 2006). In Arabidopsis and in the rice genome, six members of the RAV family contain both AP2 and B3 domains (Nakano et al., 2006). However, the exact size of the tomato RAV family still remains unclear. Therefore, to determine the number of RAV genes that are expressed in AtCBF1 transgenic tomato, we performed RT-PCR with degenerate primers (Supplemental Table S1) designed from the B3 and AP2 domains of AtRAV2 (At1g68840). We identified two RAV genes, designated SlRAV1 and SlRAV2, that were expressed in AtCBF1 transgenic tomato plants. Afterward, the full-length RAV genes were obtained by using RACE or the genome walking method (for primers, see Supplemental Table S2). Among them, SlRAV2 was the major transcript up-regulated in AtCBF1 transgenic tomato plants. The mRNA transcripts of AP2/EREBP family genes (i.e. SlERF5, Pti4, Pti5, Pti6, SlRAP2.1, SlRAP2.6-like, SlRAV2, and SlCBF1) exhibited a moderate to strong increase in AtCBF1 transgenic tomato plants (C5, C15, and C21) as compared with the wild type (Fig. 3).
Figure 3.
RT-PCR analysis of SlERF genes in AtCBF1 transgenic tomatoes. Total RNA was isolated from AtCBF1 transgenic tomatoes (C5, C15, and C21) and wild-type (WT) plants and reverse transcribed into cDNA as the templates for RT-PCR. Tomato SlCBF1, SlERF5, Pti4, Pti5, Pti6, SlRAP2.1, SlRAP2.6-like (SlRAP2.6L), SlRAV2, Actin1, AtCBF1 transgene (AtCBF1-nos), and NptII transcripts were amplified by RT-PCR and analyzed on a gel stained with ethidium bromide.
SlERF5 and Pti6 Interact with the GCC Box
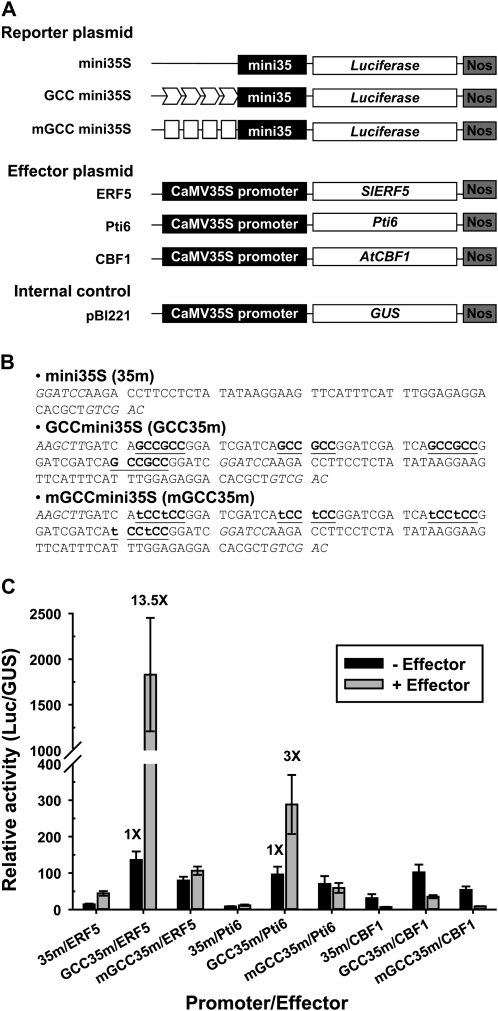
To investigate whether SlERF5 binds the GCC box, an element present in the promoters of PR genes, and directly regulates the expression of PR genes, we performed transactivation assays with Arabidopsis mesophyll protoplasts. We constructed a series of reporter plasmids with a firefly luciferase (Luc) reporter gene driven by a cauliflower mosaic virus 35S (CaMV35S) minimal promoter (mini35S), four GCC box repeats with a mini35S promoter (GCCmini35S), four mutant GCC box repeats with mini35S (mGCCmini35S), and effector plasmids with either AtCBF1 or SlERF5 cDNA, or Pti6 (positive control; Gu et al., 2002) driven by the CaMV35S promoter (Fig. 4A). The pBI221 plasmid containing the GUS gene driven by the CaMV35S promoter was used as an internal control. Plasmids were cotransfected into protoplasts and incubated for 20 h, and soluble proteins were extracted to determine transactivation of the reporter gene (Luc/GUS relative activity). At coexpression of 35S:Pti6 or 35S:SlERF5 with GCCmini35S, transactivation of the reporter gene was increased 3- to 13-fold compared with mGCCmini35S or mini35S, respectively (Fig. 4C). However, cotransfection of 35S:AtCBF1 with mGCCmini35S decreased the transactivation of the reporter gene to the basal level, in agreement with the EMSA results. These results indicated that SlERF5 and Pti6 but not AtCBF1 function as activators of GCC box-mediated transcription.
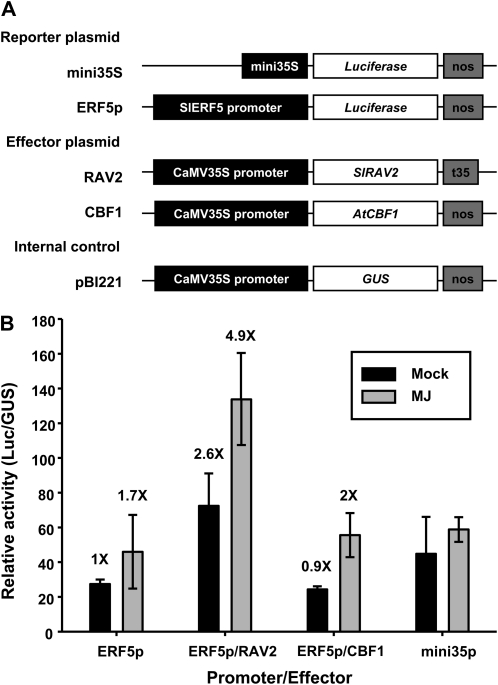
Figure 4.
SlERFs activate the reporter genes driven by the CaMV35S minimal promoter containing vicinal GCC boxes. A, Schematic diagrams of the reporter, effector, and internal control plasmids used in the transient transactivation assay in Arabidopsis leaf protoplasts. The reporter plasmids contain a repeat of four GCC or mGCC boxes fused to the CaMV35S minimal promoter and the firefly luciferase gene Luc. In the effector plasmids, Arabidopsis CBF1, tomato Pti6, and SlERF5 genes were under the control of a CaMV35S promoter. Nos denotes the terminator of nopaline synthase. The pBI221 vector contains a CaMV35S promoter driving GUS as the internal control. B, DNA sequences of the promoter region in the reporter plasmids. Sequences shown in boldface and underlined mark the wild-type and mutant GCC boxes, respectively. C, Transactivation of the Luc reporter gene by AtCBF1, Pti6, and SlERF5 in Arabidopsis protoplasts. Different effectors were cotransfected with the reporter and internal control plasmid (pBI221). The data represent means of three independent transient transformations. Error bars indicate sd. Transient transformations without the effector plasmid were used as a control.
SlRAV2 Interacts with a Promoter of SlERF5
SlERF5 and SlRAV2 contain one AP2 domain and belong to the ERF and RAV subfamily of AP2/EREBP proteins, respectively. Presumably, SlERF5 and SlRAV2, like other well-identified AP2/EREBPs, act as transcription factors to regulate gene expression in the nucleus. To verify this assumption, full-length SlERF5 and SlRAV2 coding regions were fused with yellow fluorescent protein (YFP) under the control of the 35S promoter and transiently expressed in Arabidopsis protoplasts. Indeed, we found that SlERF5 and SlRAV2 are localized in the nucleus (Supplemental Fig. S3).
The promoter sequences of SlERF5 and Pti6 were identified via the genome walking method and submitted to GenBank (accession nos. EU164418 and EU164419, respectively). Several RAV1A elements (CAACA) are present, but neither sequences for CRT/DRE nor a GCC box could be seen in the promoter regions of SlERF5 and Pti6 (Supplemental Fig. S2). To verify whether SlRAV proteins play the part of transacting factors binding to the SlERF5 promoter, we performed in vivo transactivation assays with a reporter plasmid carrying the Luc reporter gene driven by the SlERF5 promoter (776 bp). As a control, Luc driven by the CaMV35S minimal promoter (mini35S) was employed. The effector plasmids were coding sequence of SlRAV2 or AtCBF1 driven by the CaMV35S promoter (Fig. 5A). Methyl jasmonate (MJ), which acts as a global regulator of defense responses (Reymond and Farmer, 1998), was applied to mimic the pathogen or elicitor treatment. Coexpression of the SlERF5 promoter (ERF5p) with 35S:SlRAV2 resulted in an induction of transactivation of the reporter gene 2.6 times higher than the control; this induction even increased further to a level of 4.9 times that of the control (ERF5p reporter only) in the presence of MJ (Fig. 5B). By contrast, cotransfection of the SlERF5 promoter with 35S:AtCBF1 reduced the transactivation of the reporter gene to the basal level, with no effect by MJ on transactivation of the reporter gene. These results indicated that the SlERF5 promoter interacts with SlRAV2 but not with AtCBF1 and that MJ enhances the transactivation of SlERF5 and SlRAV2. Therefore, SlRAV2 and SlERF5/Pti6 may be intermediate transcription factors acting between AtCBF1 and PR genes. Taken together, we hypothesize that overexpression of AtCBF1 regulates some RAV genes to adjust ERF genes that further modulate the expression of PR genes in transgenic tomato, thus enhancing tolerance to Ralstonia infection.
Figure 5.
SlRAV2 interacts with the promoter of SlERF5. A, Schematic diagrams of the reporter, effector, and internal control plasmids used in the transient transactivation assay in Arabidopsis leaf protoplasts. The reporter plasmid contains the CaMV35S minimal promoter and the SlERF5 promoter sequence (776 bp) fused to the firefly luciferase gene Luc. In the effector plasmids, SlRAV2 and Arabidopsis CBF1 genes were driven under the control of the CaMV35S promoter. Nos and t35 denote the terminators of nopaline synthase and CaMV35S, respectively. The pBI221 vector contains a CaMV35S promoter driving GUS as the internal control. B, Transactivation of the Luc reporter gene by SlRAV2 and AtCBF1 in Arabidopsis protoplasts. Different effectors were cotransfected with the reporter and internal control plasmid (pBI221). Mock, Methanol; MJ, 30 μm MJ. The data represent means of three independent transient transformations. Error bars indicate sd. Transient transformation without the effector plasmid (ERF5p or mini35p) was used as a control.
Virus-Induced Gene Silencing of SlERF5 and SlRAV2 Attenuates the Defense against BW in Tomato
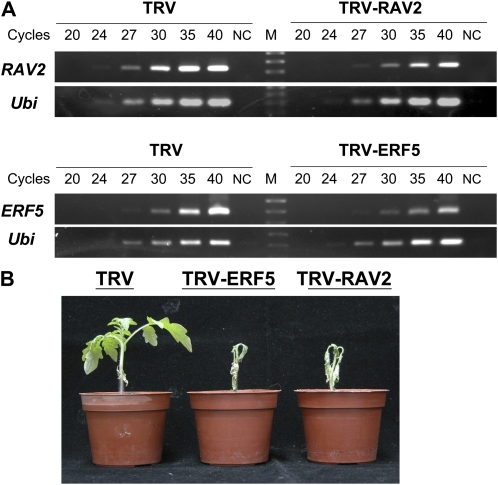
The tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) approach is an efficient silencing system to study the function of candidate genes responsible for certain disease resistance and their signaling pathways (Brigneti et al., 2004; Chen et al., 2009). To find out whether SlERF5 and SlRAV2 are involved in the BW-defense mechanism in both natural cv H7996 and AtCBF1 transgenic tomato plants, we performed experiments silencing these AP2/EREBP transcription factors. A mixture of Agrobacterium tumefaciens cultures containing TRV1 and TRV2-X (X = partial cDNA sequence of SlERF5 or SlRAV2; for primers, see Supplemental Table S2) T-DNA constructs was infiltrated into cotyledons of 10-d-old AtCBF1 transgenic seedlings as well as into cotyledons of the BW-resistant H7996 and the BW-susceptible varieties 5915 and L390 as controls. Fifteen days post agroinfiltration, total RNA was prepared from leaves and used for semiquantitative RT-PCR analyses of silenced genes, SlERF5 and SlRAV2. In TRV-ERF5- and TRV-RAV2-infected plants, the transcripts of SlERF5 and SlRAV2 were reduced compared with the TRV-only infected control (Fig. 6A). The Ubiquitin RNA served as an internal control for RNA quality. The Ubiquitin transcript levels were similar in silenced plants, TRV-ERF5 and TRV-RAV2, and TRV-only infected plants (Fig. 6A).
Figure 6.
Silencing of SlERF5 and SlRAV2 using TRV-based vector. BW-resistant tomato variety H7996 and AtCBF1 transgenic plants (CBF1) were infected with mixtures of Agrobacterium transformed with pTRV1 and pTRV2 (TRV) or pTRV2 carrying SlERF5 (TRV2-ERF5) or SlRAV2 (TRV2-RAV2) fragments. A, Semiquantitative RT-PCR analysis showing the effect of VIGS on tomato ERF5 and RAV2. For each sample, six amplification products (following 20, 24, 27, 30, 35, and 40 cycles of PCR) were analyzed. Ubiquitin product (Ubi) was used as a reference. Lane NC represents the negative control, in which the RT reaction mix without reverse transcriptase was used as a template. Lane M represents a DNA marker. B, TRV-alone, TRV-ERF5-, and TRV-RAV2-infected H7996 plants were treated with Ralstonia for 2 weeks.
Ten days post agroinfiltration, the plants were challenged with Ralstonia. Both visual symptom development and internal bacterial density in both the stem base and stem were determined at 5 dpi. All of the TRV-only infected tomato plants showed resistance to Ralstonia infection. AtCBF1 transgenic or H7996 tomato plants preinfected with TRV-ERF5 or TRV-RAV2, respectively, displayed a severe wilt phenotype after inoculation with Ralstonia (Fig. 6B). Furthermore, we carried out a bacterial titer assay for gene-silenced plants at 5 dpi (Table III). Here, tomato cv 5915, the background of AtCBF1 transgenic plants, and the susceptible cv L390 were used as the control to confirm the success of pathogen infection. These cultivars displayed a severe wilt phenotype with a very high bacteria level (mean value was greater than 109 cfu g−1 fresh weight at both stem bases and midstems; Table III). The stem base and midstem of SlERF5- and SlRAV2-silenced plants exhibited relatively higher levels of bacterial density compared with TRV-only control plants. These results indicated that silencing SlERF5 and SlRAV2 had indeed decreased the resistance of tomato to BW.
Table III. Assessment of Ralstonia density in silenced tomato plants.
AtCBF1 transgenic plants (CBF1OX) and BW-resistant tomato variety H7996 were infected with mixtures of Agrobacterium transformed with pTRV1 and pTRV2 (TRV-only control) or pTRV2 carrying SlRAV2 (SlRAV2) or SlERF5 (SlERF5) fragments. BW-susceptible tomato varieties L390, 5915 (the background of CBF1OX), and TRV-infected CBF1OX and H7996 plants were treated with Ralstonia. The bacterial titer inside the test plants was measured in stem bases and midstems at 5 dpi. The number of total assayed plants and positively detected plants (+) are indicated. Each value is the mean ± sd. Pairwise comparisons were made between wild-type plants (or TRV infected in H7996) and silenced plants with Student’s t test (a P < 0.01, b P < 0.05).
| Tomato Plants | Silenced Gene | Sample No. | Stem Bases | Midstems | ||
| + | Log | + | Log | |||
| cfu g−1 plant tissue | cfu g−1 plant tissue | |||||
| 5915 | ||||||
| Wild type | – | 30 | 30 | 9.1 ± 0.3 | 30 | 7.9 ± 0.9 |
| CBF1OX | TRV | 36 | 20 | 4.6 ± 0.6b | 10 | 3.0 ± 0.5b |
| CBF1OX | SlRAV2 | 35 | 27 | 8.7 ± 1.0 | 19 | 7.5 ± 0.7 |
| CBF1OX | SlERF5 | 36 | 25 | 8.0 ± 0.8a | 22 | 6.0 ± 1.2b |
| H7996 | ||||||
| Wild type | TRV | 36 | 22 | 3.6 ± 0.5 | 6 | 1.3 ± 0.4 |
| Wild type | SlRAV2 | 35 | 25 | 5.8 ± 0.6a | 18 | 4.3 ± 0.7a |
| Wild type | SlERF5 | 29 | 18 | 3.9 ± 0.7 | 11 | 2.4 ± 0.7b |
| L390 | ||||||
| Wild type | – | 6 | 6 | 10.7 ± 0.2 | 6 | 10.1 ± 0.4 |
Generation and Characterization of SlRAV2RNAi Knockdown as Well as 35S:SlRAV2 and 35S:SlERF5 Transgenic Tomato Plants
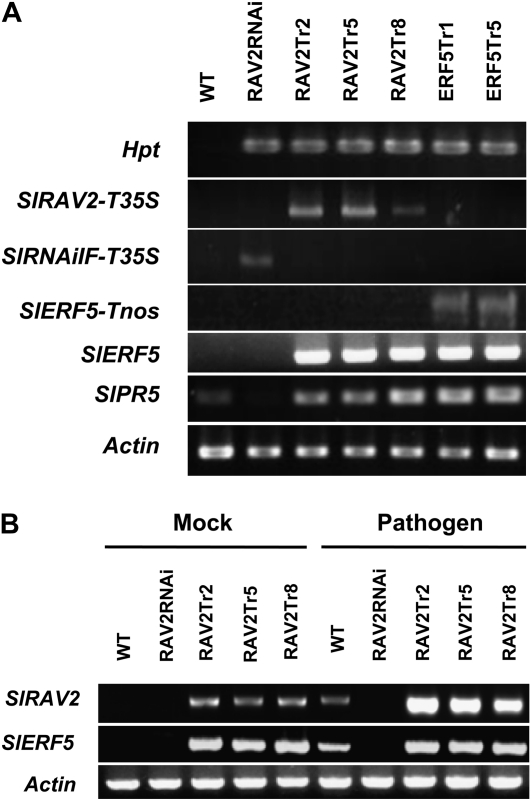
To further investigate the functions of SlRAV2 and SlERF5 in pathogen resistance, transgenic tomato plants with knockdown expression of SlRAV2 or overexpression of SlRAV2 or SlERF5 were generated. Transgenic plants with lower insertion (one to two copies) of transgene were selected for further study. Semiquantitative RT-PCR was performed to analyze the mRNA levels in these transgenic plants, including independent lines of SlRAV2 knockdown (RAV2RNAi), 35S:SlRAV2 (RAV2Tr2, -5, and -8), and 35S:SlERF5 (ERF5Tr1 and -Tr5) transformants. The mRNA levels of the hygromycin phosphotransferase gene (Hpt) and Actin were used as transgenic and internal controls, respectively. The foreign transcripts of SlRAV2 and SlERF5 transgenes with a 35S or nos terminator were expressed only in transgenic plants (Fig. 7A). In addition, SlERF5 and its downstream gene, SlPR5, were not only abundantly expressed in SlERF5 transgenic plants but also highly accumulated in SlRAV2 transgenic plants (Fig. 7A).
Figure 7.
Analysis of 35S:SlRAV2, SlRAV2RNAi, and 35S:SlERF5 transgenic tomato lines. A, Endogenous and transgenic mRNA transcript levels of SlRAV2, SlERF5, and SlPR5 genes in SlERF5 overexpression (ERF5Tr1 and ERF5Tr5), SlRAV2 overexpression (RAV2Tr2, RAV2Tr5, and RAV2Tr8), and SlRAV2 knockdown (RAV2RNAi) tomato plants. B, Expression of SlRAV2 and SlERF5 in wild-type (WT), RAV2Tr, and RAV2RNAi tomato lines under Ralstonia infection for 12 h. mRNA levels of the indicated genes in pathogen-treated (pathogen) and nontreated (mock) plants were determined by semiquantitative RT-PCR. SlRAV2-T35S and SlERF5-Tnos show transgenic expression amplified by the specific forward primers (SlRAV2-F and SlERF5-F) and the terminator reverse primers (35T-R and nos3′R; Supplemental Table S1). SlRNAiIF-T35S show the pRAV2RNAi fragment amplified by the 35T-R reverse primer and a forward primer (RNAiI-F) located in the intron of the RNAi vector pH7GWIWG2. Hpt and Actin expression levels were analyzed as a transgenic and a quantification control, respectively.
We used RT-PCR to examine changes in mRNA levels of SlRAV2 and SlERF5 in pathogen-infected RAV2Tr and RAV2RNAi transgenic tomato. The transcription of SlRAV2 and SlERF5 was up-regulated by Ralstonia infection in wild-type plants (Fig. 7B). The level of SlERF5 and SlRAV2 mRNA transcripts was high in RAV2Tr lines as compared with the wild type under normal conditions but absent in the RAV2RNAi line even after treatment with the pathogen. Taken together, our results support the notion that SlRAV2 may be a key factor regulating SlERF5 gene expression. Hence, the SlRAV2 and SlERF5 transgenic plants were further evaluated for resistance to pathogen infection.
Constitutive Expression of SlERF5 and SlRAV2 in Tomato Confers Tolerance, While Knockdown of SlRAV2 Expression Causes Hypersensitivity to BW
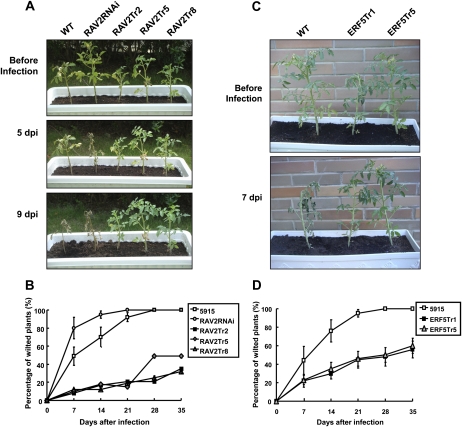
SlRAV2- and SlERF5-overexpressing transgenic tomato plants exhibited a slightly dwarf phenotype (Fig. 8, A and C) and generated less fruit and seeds under normal conditions, while the knockdown expression of SlRAV2 in tomato promoted plant growth and development (Fig. 8A, top panel). However, how SlRAV2 and SlERF5 participate in tomato growth and development remains to be further investigated. SlRAV2 and SlERF5 transgenic tomato plants were then subjected to Ralstonia challenge to verify their functions in the defense mechanism. The RAV2RNAi knockdown line already presented a severely wilted phenotype at 5 dpi, while the wild type wilted at 7 to 9 dpi (Fig. 8, A and B). On the other hand, all of the transgenic plants overexpressing either SlRAV2 or SlERF5 exhibited more resistance to BW (Fig. 8). When plants were inoculated with Ralstonia, both transgenic and wild-type plants showed reduction in PSII efficiency and chlorophyll content (Supplemental Fig. S4). The reduction in maximum photochemical efficiency of PSII in the dark-adapted state was on average 75% in wild-type (5915) plants, whereas transgenic lines showed reductions of 88% for RAV2RNAi, 19% for RAV2Tr, and 37% for ERF5Tr lines. Similarly, the chlorophyll content remained higher in RAV2Tr and ERF5Tr transgenic plants in comparison with the RAV2RNAi knockdown lines and wild-type plants after pathogen infection. The differences in PSII efficiency and chlorophyll content between wild-type plants and transgenic RAV2Tr and ERF5Tr tomato under pathogen treatment were statistically significant (P < 0.01, Student’s t test). Overall, these findings indicated that SlERF5 and SlRAV2 play crucial roles in the basal defense of tomato plants against BW and that SlRAV2 may be a key regulator involved in plant defense.
Figure 8.
Comparison of 35S:SlRAV2, SlRAV2RNAi, and 35S:SlERF5 transgenic lines with the wild type under pathogen infection. A, Wild-type plants (cv 5915; WT), T2 SlRAV2 knockdown (SlRAV2RNAi), and overexpression lines (RAV2Tr2, RAV2Tr5, and RAV2Tr8) were inoculated with Ralstonia. The photographs were taken at 0, 5, and 9 dpi. B, The percentage of wilted plants was calculated at 7, 14, 21, 28, and 35 dpi. C, Wild-type plants (5915) and SlERF5 T2 overexpression lines (ERF5Tr1 and ERF5Tr5) were inoculated with Ralstonia and then kept at 28°C with a photoperiod of 16 h. The photographs were taken at 0 and 7 dpi. D, The percentage of wilted plants was measured. Each value represents the mean ± sd (n = 10 individual plants) of three independent experiments.
DISCUSSION
CBF genes have been considered “master switches” that increase freezing tolerance in Arabidopsis plants via the activation of COR genes (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). Its tomato orthologs CBF1 and CBF2 are up-regulated by chilling and drought stress but not by other types of stress, such as high salinity or ABA treatment (Zhang et al., 2004b). In a BW-susceptible tomato variety, 5915, expression of SlCBF1, but not SlCBF2 and SlCBF3, was up-regulated by pathogen infection (Supplemental Fig. S5). However, it still remained unclear whether CBF regulons directly participate in the biotic stress response. In this study, we showed that overexpression of AtCBF1 in tomato leads to the constitutive accumulation of several PR genes (Table I; Fig. 2) and further enhanced tolerance to BW by suppressing the proliferation of Ralstonia. Furthermore, the degree and nature of enhanced resistance to BW observed in most of the tested transgenic lines was similar to that in H7996, a natural BW-resistant tomato cultivar (Fig. 1; Table III). Previously, BW resistance in H7996 was found to be related to suppressed internal pathogen multiplication rather than to the efficiency of root invasion or upward movement (Wang et al., 2000).
Accumulating evidence suggests that different ERF transcription factors induce a diverse set of PR genes under biotic and abiotic stresses (Park et al., 2001; Zhang et al., 2005, 2007). In agreement with our observations, overexpression of tomato Pti4 and Arabidopsis ERF1 in transgenic Arabidopsis plants led to the constitutive activation of several PR genes, resulting in enhanced tolerance against certain bacterial and fungal pathogens (Gu et al., 2002). Interaction of TSRF1 with the GCC box in the promoters of PR genes in response to Ralstonia infection was demonstrated in tobacco and tomato (Zhang et al., 2004a, 2007). Constitutive expression of tomato JERF3 in transgenic tobacco activated the expression of PR genes and resulted in enhanced salt tolerance (Wang et al., 2004). In addition, ectopic expression of the pepper (Capsicum annuum) pathogen-induced transcription factor CaRAV1 in transgenic Arabidopsis plants induced some PR genes and enhanced the resistance of plants against infection by Pseudomonas syringae pv tomato strain DC3000 (Sohn et al., 2006). Recently, Endres et al. (2010) reported that tobacco RAV2 is an important factor in the viral suppression of silencing and that the role of RAV2 is to divert host defenses toward responses that interfere with antiviral silencing.
Within the AP2/EREBP family, the AP2 subfamily members are involved in plant development, and some ERF subfamily members are likely involved in the responses to biotic and abiotic stresses (Sakuma et al., 2002; Nakano et al., 2006). The members of different subfamilies specifically bind to different cis-acting elements, such as the CRT/DRE, the GCC box, and/or the RAV1A/B elements (Sohn et al., 2006). With respect to the mechanism by which the expression of PR genes in AtCBF1 transgenic tomato plants is regulated, two hypotheses may be proposed. The first hypothesis is that ectopic overexpression of AtCBF1 directly activates PR gene expression. However, the results of EMSA and transactivation assays revealed that AtCBF1 did not interact with the GCC box (Fig. 4; Supplemental Fig. S1). Thus, this hypothesis might be excluded. CBF/DREB was found to bind to the common core region of CCGNC of CRT/DRE and the GCC box with different affinities in vitro (Sakuma et al., 2002). Therefore, we cannot entirely exclude the possibility that a fraction of the heterologous AtCBF1 protein overproduced in tomato plants might partially bind to the GCC box in the promoter region of PR genes.
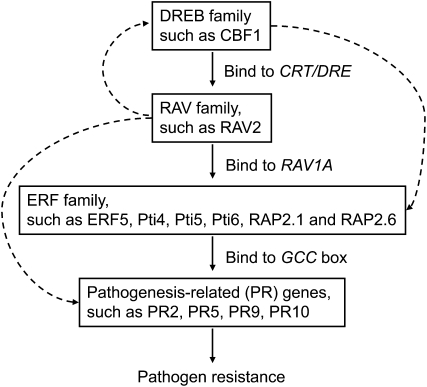
The second hypothesis is based on an indirect activation of PR genes. We hypothesized that AtCBF1 interacts with CRT/DRE elements in the SlRAV2 promoter, leading to up-regulated expression of SlRAV2; this in turn elevates the expression of other ERFs (e.g. SlERF5). Subsequently, these ERFs specifically interact with the GCC box in the promoters of PR genes, thus enhancing transgenic plant resistance to Ralstonia infection (Fig. 9). In Arabidopsis, the promoter regions of some AP2/ERF genes contain several CRT/DRE elements (Supplemental Table S3); among them, ERF1, ERF2, ERF4, RAP2.1, RAP2.6, and RAV1 were identified as cold-inducible downstream genes of the CBF/DREB transcriptional factor (Fowler and Thomashow, 2002; Sharabi-Schwager et al., 2010). In addition, there are two or three CRT/DRE elements present in the promoter region of rice RAV genes, such as Os01g04800 (−1,895 and −2,371 from ATG), Os05g47650 (−373 and −2,032), and Os07g17230 (−683, −1,188, and −2,256). Completion of the tomato genome sequencing project (the Sol Genomics Network) may reveal more information regarding whether a CBF1-binding site exists in the promoter region of tomato AP2/ERF transcription factors (Supplemental Table S4). We surveyed the cis-acting elements of the SlRAV2 promoter and found that there is one CRT/DRE and one CRT/DRE-like element presented (Supplemental Fig. S6). Transactivation assays with Arabidopsis mesophyll protoplasts proved that AtCBF1 can transactivate SlRAV2 gene expression (Supplemental Fig. S7). Furthermore, there are several RAV1A elements presented in the promoters of SlERF5 and Pti6 (Supplemental Fig. S2; Supplemental Table S4), and SlRAV2 can transactivate SlERF5 gene expression (Fig. 5). In addition, overexpression of SlERF5 increases PR5 gene expression, while overexpression of SlRAV2 enhances both the expression of SlERF5 and its downstream PR5 in tomato plants (Figs. 7 and 8). Mounting evidence suggests that overexpression of ERF genes activates the expression of some PR genes, which results in enhanced tolerance to biotic and abiotic stresses (Park et al., 2001; Wang et al., 2004; Zhang et al., 2005, 2007). The VIGS assay and pathogen challenge test in SlERF5 and SlRAV2 overexpression and SlRAV2 knockdown tomato plants performed in our study also verify that SlRAV2 and SlERF5 participate in the enhancement of BW tolerance (Figs. 6–8).
Figure 9.
Proposed role of AP2/EREBP superfamily members in the plant defense pathway. The model illustrates the genetic interactions between AP2/EREBP transcription factors in the regulation of PR genes, which leads to enhanced tolerance to Ralstonia, in AtCBF1 transgenic tomato plants.
Many AP2/EREBP genes have been shown not only to be induced by pathogen infection but also to be regulated by stress-related plant hormones, such as ethylene, JA, and SA (Gutterson and Reuber, 2004). Chen et al. (2009) reported that mitogen-activated protein kinase-, JA/ethylene-, and SA-related defense signaling pathways are involved in the resistance in tomato to BW. Ectopic expression of CARAV1 in Arabidopsis strongly induced the expression of some PR genes regulated by the SA-dependent signaling pathway, such as PR1, PR2, and PR5 (Sohn et al., 2006). In this study, endogenous expression of SlCBF1, SlRAV2, and SlERF5 was induced by pathogen infection (Fig. 7; Supplemental Fig. S5), and SlPR5 transcripts accumulated to high levels in all of the AtCBF1, SlRAV2, and SlERF5 transgenic tomato plants (Figs. 2 and 7). Therefore, SA may play an important role as an intermediary in the defense mechanism between AtCBF1, SlRAV2, SlERF5, and the PR genes. In addition, SlERF5 was up-regulated by SlRAV2 and enhanced the level of induction by exogenous MJ in the transactivation assay (Fig. 5). As described by Chen et al. (2009), SA- and JA/ethylene-dependent pathways may interact synergistically, rather than antagonistically, in tomato defense mechanisms. JA may also play a regulatory role in the defense mechanism of the CBF-RAV-ERF-PR signaling cascade.
In summary, this study provides evidence that AtCBF1 is involved in the regulation of subsets of RAV family, ERF family, and PR genes that are related to the biotic stress response. Our observations indicate that the RAV2 transcription factor may comprise a key modulator in the plant defense signal pathway (Fig. 9). However, further studies are needed to understand in more detail the mechanism of the RAV2-mediated signaling cascade in plant defense. In addition to the AP2 domain, RAV transcription factors have another DNA-binding domain, the B3 domain, which can recognize the RAV1B element (CACCTG), as reported previously (Kagaya et al., 1999). Interestingly, we did not find a RAV1B element in the promoter region of tomato AP2/EREBP. The existence of novel RAV1B-like/B3-binding elements or the participation of posttranslational modifications and/or protein-protein interactions in the RAV-mediated defense mechanism need to be further investigated. AtCBF1 has been introduced into the tomato genome previously, resulting in transgenic plants that were tolerant to four different kinds of stress: chilling, oxidative stress, high salt, and water deficit (Hsieh et al., 2002a, 2002b; Lee et al., 2003). In this report, we observed that overexpression of either AtCBF1, SlERF5, or SlRAV2 in tomato plants conferred an enhancement of Ralstonia tolerance. These observations indicate that a targeted transgenic approach with a single transgene may be sufficient to enhance plant resistance to several environmental stresses, including abiotic and biotic stresses, and thus may be applied for crop improvement.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) cv CL5915-93D4-1-0-3 (5915) was provided by AVRDC-The World Vegetable Center, in Tainan, Taiwan, and was used as the background line for transformation. Before surface sterilization, seeds were soaked for 1 h at 32°C, treated with 1% (v/v) NaOCl for 10 min, washed several times with sterile water for 5 min, and then germinated on Murashige and Skoog basal medium at 26°C with a 16/8-h photoperiod at 120 μmol m−2 s−1.
Resistance Scoring of Transgenic Tomato Plants
Tomato plants were inoculated with Ralstonia solanacearum as described (Chen et al., 2009). The resistance of transgenic tomato plants to BW was evaluated as described previously (Lin et al., 2004; Chen et al., 2009). Ralstonia strain Pss4 (race 1, biovar 3; suspension A600 = 0.6, about 2 × 108 cfu mL−1) was used as the inoculum. Additionally, for Ralstonia colonization experiments, 10 plants were randomly harvested from each treatment at each sampling time. Three independent experiments were performed. Plants were uprooted, soil was washed off, and plants were soaked in 70% (v/v) ethanol for 3 to 5 min, rinsed twice in sterile water, and blotted to dryness on paper towels. For BW evaluation, tomato varieties H7996 and an ABA-inducible promoter driving AtCBF1 in a 5915 variety (AC3) line were used as resistant and susceptible controls, respectively (Wang et al., 2000). H7996, L390, and 5915 seeds were kindly provided by AVRDC.
Vector Construction and Plant Transformation
Construction of the binary vector carrying pCaMBIA2301/35S:AtCBF1 and Agrobacterium tumefaciens-mediated tomato transformation were carried out as described (Hsieh et al., 2002a, 2002b; Lee et al., 2003). For constitutive overexpression in tomato, constructs p35S:SlERF5 and p35S:SlRAV2 were prepared by inserting the SlERF5 and SlRAV2 coding sequences between the CaMV35S promoter and the nos or the 35S terminator in pCAMBIA1390/35S (Hsiao et al., 2007) and pH2GW7 (for primers, see Supplemental Table S2), respectively, both of which contain Hpt. For knockdown expression in tomato, the binary vector pSlRAV2RNAi was constructed by inserting a SlRAV2 N-terminal region (amino acids 27–65) into pH7GWIWG2, followed by transformation into tomato plants by the Agrobacterium-mediated transformation method.
Molecular Characterization of Transgenic Tomato Plants
Transgenic tomato plants were selected on 100 mg L−1 kanamycin (pCAMBIA2301/35S:AtCBF1) or 20 mg L−1 hygromycin (p35S:SlERF5, p35S:SlRAV2, and pSlRAV2RNAi). All transgenic plants were analyzed by Southern- and northern-blot hybridization or RT-PCR, as described previously (Hsieh et al., 2002a, 2002b). The following probes were used for northern-blot hybridization: tomato β-tubulin, PR2 (β-1,3-glucanase; accession no. CK664757), PR5-like (accession no. AY257487), PR9-like (peroxidase; accession no. AW219536), and PR10-like (RNase-like; accession no. CK468708). cDNA fragments were excised from pT7Blue (R) vector as probes and labeled with [α-32P]dCTP by the random primer method (Feinberg and Vogelstein, 1983).
Microarray Analysis
We previously constructed a tomato cDNA microarray comprising 12,448 cDNA clones derived from 5,600 tomato root EST clones and 15 libraries from stress-treated wild-type tomato plants. AtCBF1 transgenic tomato RNA and control plant RNA were probed. Probe labeling, hybridization, and scanning of the cDNA microarray were performed as described previously (Liu et al., 2006).
Determination of Chlorophyll Fluorescence Values and Chlorophyll Content
Chlorophyll fluorescence values were measured using a pulse-activated modulation fluorimeter (Walz). Chlorophyll content in leaves was determined by extraction with N,N-dimethylformamide as described (Moran and Porath, 1980). Absorption of the extracts was measured at 664 and 647 nm. Chlorophyll content was calculated with use of the following equation: total chlorophyll content = 7.04 A664 + 20.27 A647.
RT-PCR Analysis
Total RNA was isolated from leaves of wild-type and transgenic tomato plants by use of TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RT was conducted as described by the manufacturer (Promega). PCR involved gene-specific primers of SlCBF1 (AY497899), SlERF5 (AY559315), Pti4 (U89255), Pti5 (U89256), Pti6 (U89257), SlRAV2 (EU164417), SlRAP2.1 (AK246512), SlRAP2.6-like (EU164420), and SlActin1 (U60480; Supplemental Table S1). PCR was conducted in a final volume of 25 μL containing cDNA reverse transcribed from 30 ng of total RNA, 1× Taq buffer (Violet), 0.2 mm of each deoxyribonucleotide triphosphate, 2 units of Taq DNA polymerase (Violet), and 100 pmol of each primer (Supplemental Table S1). The following amplification program was used: one cycle of 95°C for 3 min; 25 to 30 cycles of 95°C for 25 s, 58°C for 30 s, and 72°C for 1 min; and then one cycle of 72°C for 7 min. The RT-PCR products were resolved on a 1% agarose gel and visualized by ethidium bromide staining.
Promoter Isolation
Genomic DNA was extracted from leaves of wild-type tomato plants (Murray and Thompson, 1980). Genome walking was performed as described by the manufacturer (BD GenomeWalker Universal Kit; Clontech). In addition to genome walking, inverse PCR was used to extend the SlERF5 (EU164418) and Pti6 (EU164419) promoter sequences and to obtain the full-length SlRAV2 (EU164417) gene by use of specific primers (Supplemental Table S2). Two micrograms of tomato genomic DNA was digested with HindIII and self-ligated as the template for inverse PCR. The following amplification program was used for the first PCR of genome walking and inverse PCR: one cycle of 95°C for 1 min; seven cycles of 94°C for 25 s and 72°C for 3 min; 32 cycles of 94°C for 25 s and 67°C for 3 min; and then one cycle of 67°C for 7 min. The program for the second PCR of genome walking and inverse PCR was as follows: one cycle of 95°C for 1 min; five cycles of 94°C for 25 s and 72°C for 3 min; 25 cycles of 94°C for 25 s and 67°C for 3 min; and then one cycle of 67°C for 7 min. The PCR products were ligated into the pGEMT Easy vector (Promega) for DNA sequencing.
Arabidopsis Protoplast Transient Expression and Reporter Gene Activity Assay
For the reporter gene constructs, the CaMV35S promoter in pJD301 was replaced by the 35S minimal promoter from −42 to +8 containing the TATA box. The GCC and mutant GCC box sequences (Fig. 4B) were multimerized four times and placed upstream of the 35S minimal promoter and the SlERF5 promoter (−776 to +23; Supplemental Fig. S2A) and fused to the Luc gene. For effector plasmids, the Luc gene in pJD301 was replaced by the coding regions of AtCBF1, SlERF5, Pti6, and SlRAV2. The pBI221 plasmid containing the GUS gene driven by the CaMV35S promoter was used as an internal control for transactivation assay.
Arabidopsis (Arabidopsis thaliana) protoplasts were isolated from 4-week-old leaves and transfected by a modified polyethylene glycol method as described (Abel and Theologis, 1994; Wu et al., 2009). Ten micrograms of reporter plasmid and 5 μg of effector plasmid or control plasmid (pUC18) were cotransfected into 4 × 104 protoplasts with 10 μg of internal control plasmid pBI221. The transfected cells were incubated for 20 h at 22°C under light, harvested by centrifugation at 100g for 2 min, and then lysed in lysis buffer (Promega). Luciferase activity was measured by use of a luciferase assay kit (Promega) according to the manufacturer’s instructions, and GUS activity was determined (Lu et al., 1998).
TRV-Based VIGS Assay
VIGS vectors (pTRV1 and pTRV2) and construction procedures for their derivatives have been described (Liu et al., 2002; Chen et al., 2009). SlRAV2 and SlERF5 cDNA fragments (301 and 318 bp, respectively) were obtained by PCR using specific primers (RAV2-VIGS-F/RAV2-VIGS-R and ERF5-VIGS-F/ERF5-VIGS-R; Supplemental Table S2) and recombined into pTRV2 to generate pTRV2-RAV2 and pTRV2-ERF5. For the VIGS assay, pTRV1 and pTRV2 and its derivatives (pTRV2-RAV2 and pTRV2-ERF5) were introduced into Agrobacterium strain GV3101 by electroporation. BW-resistant tomato variety H7996 and BW-susceptible variety L390 were grown in pots at 24°C in a growth chamber under a 16-h-light/8-h-dark cycle. The TRV inoculation procedure was performed as described (Dinesh-Kumar et al., 2003). The efficiency of VIGS in TRV-only, TRV-ERF5-, and TRV-RAV2-silenced tomato leaves on day 15 post agroinfiltration was examined by semiquantitative RT-PCR using specific primers (ERF5-RT-F/ERF5-RT-R, RAV2-RT-F/RAV2-RT-R, and UBI3-F/UBI3-R). On day 10 post agroinfiltration, TRV-, TRV-ERF-, and TRV-RAV2-infiltrated tomato, including H7996 and AtCBF1 transgenic plants, were inoculated with Ralstonia (2 × 108 cfu mL−1) by root drenching. Five days later, 1-cm sections from the midstem and stem base of these Ralstonia-inoculated plants were weighed and ground, and then the bacterial density was measured by direct plating. A lower inoculum dose (5 × 105 cfu mL−1) was used for the L390 susceptible control. For the assay of each gene, 10 to 12 plants were used in each experiment, and three independent experiments were performed. Pairwise comparisons were made between wild-type (or TRV-infected plant in H7996) and silenced plants with Student’s t test.
Gene accession numbers of all sequence data from this article can be found in “Materials and Methods” and Supplemental Tables S1 and S4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. EMSA characterization of the DNA-binding affinity of the recombinant AtCBF1 protein.
Supplemental Figure S2. Promoter sequences of SlERF5 and Pti6.
Supplemental Figure S3. Subcellular localization of SlERF5 (EYFP::SlERF5) and SlRAV2 (EYFP::SlRAV2) in Arabidopsis protoplasts.
Supplemental Figure S4. Photosynthesis efficiency and chlorophyll content of 35S:SlRAV2, SlRAV2RNAi, and 35S:SlERF5 transgenic lines under pathogen infection.
Supplemental Figure S5. Expression of tomato endogenous CBF genes under Ralstonia infection.
Supplemental Figure S6. Genome sequence of SlRAV2.
Supplemental Figure S7. CBF1 activates the reporter gene driven by the SlRAV2 promoter.
Supplemental Table S1. Oligonucleotides used for RT-PCR.
Supplemental Table S2. Oligonucleotides used for genome walking, inverse PCR, and vector construction.
Supplemental Table S3. Prediction of the CBF1-binding elements in the promoter region of the Arabidopsis AP2/ERF genes.
Supplemental Table S4. Prediction of the AP2/ERF-binding elements in the promoter region of tomato AP2/ERFs.
Acknowledgments
We thank AVRDC for technical assistance and providing biological materials. We thank Dr. Choun-Sea Lin (Agricultural Biotechnology Research Center, Academia Sinica [ABRC, AS]) and Ms. Fu-Hui Wu (ABRC, AS) for technical support in protoplast isolation and transfection as well as Ms. Chia-Hui Liao (Academia Sinica Biotechnology Center in Southern Taiwan) for tomato transformation. We thank Drs. Su-Chiung Fang (Academia Sinica Biotechnology Center in Southern Taiwan) and Heiko Kuhn (Editorial Office of ABRC, AS) for helpful discussions and manuscript editing.
References
- Abel S, Theologis A. (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5: 421–427 [DOI] [PubMed] [Google Scholar]
- Brigneti G, Martín-Hernández AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JD. (2004) Virus-induced gene silencing in Solanum species. Plant J 39: 264–272 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Lin YM, Chao TC, Wang JF, Liu AC, Ho FI, Cheng CP. (2009) Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol Plant 136: 324–335 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99: 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. (2003) Virus-induced gene silencing. Methods Mol Biol 236: 287–294 [DOI] [PubMed] [Google Scholar]
- Endres MW, Gregory BD, Gao Z, Foreman AW, Mlotshwa S, Ge X, Pruss GJ, Ecker JR, Bowman LH, Vance V. (2010) Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog 6: e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Zhang H, Tian Y, Li F, Zhang Z, Lu X, Chen X, Huang R. (2008) Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep 27: 1787–1795 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L. (2004) Plant perception systems for pathogen recognition and defence. Mol Immunol 41: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Grimault V, Prior P, Anaïs G. (1995) A monogenic dominant resistance of tomato to bacterial wilt in Hawaii 7996 is associated with plant colonization by Pseudomonas solanacearum. J Phytopathol 143: 349–352 [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB. (2002) Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14: 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hai TTH, Esch E, Wang J-F. (2008) Resistance to Taiwanese race 1 strains of Ralstonia solanacearum in wild tomato germplasm. Eur J Plant Pathol 122: 471–479 [Google Scholar]
- He P, Warren RF, Zhao T, Shan L, Zhu L, Tang X, Zhou JM. (2001) Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 14: 1453–1457 [DOI] [PubMed] [Google Scholar]
- Hemming MN, Basuki S, McGrath DJ, Carroll BJ, Jones DA. (2004) Fine mapping of the tomato I-3 gene for fusarium wilt resistance and elimination of a co-segregating resistance gene analogue as a candidate for I-3. Theor Appl Genet 109: 409–418 [DOI] [PubMed] [Google Scholar]
- Hsiao P, Sanjaya, Su RC, Teixeira da Silva JA, Chan MT. (2007) Plant native tryptophan synthase beta 1 gene is a non-antibiotic selection marker for plant transformation. Planta 225: 897–906 [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT. (2002a) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT. (2002b) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129: 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M. (2007) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225: 575–588 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Koo YJ, Kim MA, Kim EH, Song JT, Jung C, Moon JK, Kim JH, Seo HS, Song SI, Kim JK, et al. (2007) Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol Biol 64: 1–15 [DOI] [PubMed] [Google Scholar]
- Lee JT, Prasad V, Yang PT, Wu JF, Ho THD, Charng YY, Chan MT. (2003) Expression of Arabidopsis CBF1 regulated by an ABA/stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ 26: 1181–1190 [Google Scholar]
- Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, Black L, Green SK, Wang JF, Cheng CP. (2004) Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res 13: 567–581 [DOI] [PubMed] [Google Scholar]
- Liu N-Y, Ko S-S, Yeh K-C, Charng Y-Y. (2006) Isolation and characterization of tomato Hsa32 encoding a novel heat-shock protein. Plant Sci 170: 976–985 [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM. (1998) Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J Biol Chem 273: 10120–10131 [DOI] [PubMed] [Google Scholar]
- Moran R, Porath D. (1980) Chlorophyll determination in intact tissues using N,N-dimethylformamide. Plant Physiol 65: 478–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Robb J, Lee B, Nazar RN. (2007) Gene suppression in a tolerant tomato-vascular pathogen interaction. Planta 226: 299–309 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Huffaker A, Yamaguchi Y. (2007) New insights into innate immunity in Arabidopsis. Cell Microbiol 9: 1902–1908 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M, Lers A, Samach A, Guy CL, Porat R. (2010) Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J Exp Bot 61: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. (2006) Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61: 897–915 [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R. (2004) Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol 55: 183–192 [DOI] [PubMed] [Google Scholar]
- Wang JF, Olivier J, Thoquet P, Mangin B, Sauviac L, Grimsley NH. (2000) Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain-specific locus. Mol Plant Microbe Interact 13: 6–13 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li W, Chen J, Yang Y, Zhang Z, Zhang H, Wang XC, Huang R. (2007) Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol Biol 63: 63–71 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. (2004a) Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol 55: 825–834 [DOI] [PubMed] [Google Scholar]
- Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF. (2004b) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 39: 905–919 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang Z, Chen J, Chen Q, Wang XC, Huang R. (2005) Expressing TERF1 in tobacco enhances drought tolerance and abscisic acid sensitivity during seedling development. Planta 222: 494–501 [DOI] [PubMed] [Google Scholar]