Abstract
The micronutrient zinc is essential for all living organisms, but it is toxic at high concentrations. Here, to understand the effects of excess zinc on plant cells, we performed an iTRAQ (for isobaric tags for relative and absolute quantification)-based quantitative proteomics approach to analyze microsomal proteins from Arabidopsis (Arabidopsis thaliana) roots. Our approach was sensitive enough to identify 521 proteins, including several membrane proteins. Among them, IRT1, an iron and zinc transporter, and FRO2, a ferric-chelate reductase, increased greatly in response to excess zinc. The expression of these two genes has been previously reported to increase under iron-deficient conditions. Indeed, the concentration of iron was significantly decreased in roots and shoots under excess zinc. Also, seven subunits of the vacuolar H+-ATPase (V-ATPase), a proton pump on the tonoplast and endosome, were identified, and three of them decreased significantly in response to excess zinc. In addition, excess zinc in the wild type decreased V-ATPase activity and length of roots and cells to levels comparable to those of the untreated de-etiolated3-1 mutant, which bears a mutation in V-ATPase subunit C. Interestingly, excess zinc led to the formation of branched and abnormally shaped root hairs, a phenotype that correlates with decreased levels of proteins of several root hair-defective mutants. Our results point out mechanisms of growth defects caused by excess zinc in which cross talk between iron and zinc homeostasis and V-ATPase activity might play a central role.
Environmental pollution due to heavy metals represents a serious threat to living organisms. Zinc contamination in water and soil, for example, has become a serious environmental problem (Alkorta et al., 2004). Zinc concentration is rising through human activities such as mining, steel processing, and production of wastewater by industrial plants. Furthermore, abnormally high concentrations of zinc in plants cause secondary problems, because plants are at the bottom of food chains. On the other hand, zinc deficiency has several effects on human health, namely growth retardation and delayed sexual and bone maturation (Plum et al., 2010). Zinc deficiency in plants also reduces crop production (Hacisalihoglu and Kochian, 2003).
At relatively low concentrations, zinc is essential for plants as a cofactor of a large number of enzymes and proteins. However, excess zinc causes serious growth defects such as chlorosis and root growth inhibition (Marschner, 1995). It has been proposed that such growth defects might be a secondary effect of excess zinc caused by deficiency of other essential ions, such as iron or magnesium, which have ionic radii similar to zinc (Marschner, 1995). Because zinc can be easily substituted for these two metals in the active sites of enzymes or transporters, excess zinc is toxic by interfering with basic cellular functions (Marschner, 1995). For common bean (Phaseolus vulgaris), excess zinc has been reported to depress the activities of Rubisco and PSII, probably by competing with or replacing magnesium (Van Assche and Clijsters, 1986). Iron deficiency also leads to chlorosis due to reduction of the activity of enzymes that catalyze the biosynthesis of the plant chlorophyll precursor, protochlorophyllide (Spiller et al., 1982; Tottey et al., 2003). Information concerning the inhibitory effects of excess zinc on plant growth has been accumulated; however, it remains unclear how plants sense zinc and respond in order to cope with its excess.
The uptake of nutrients and ions and their repartitioning within the different intracellular compartments largely rely on proton gradients generated across the membranes by several proton pumps (Martinoia et al., 2007). For example, under iron deficiency, plant cells can take up iron via a three-step mechanism. First, iron mobility is enhanced by a proton gradient that is created by the plasma membrane H+-ATPases. Santi and Schmidt (2009) recently reported that transcripts of plasma membrane H+-ATPases encoding the genes AHA2 and AHA7 increased in Arabidopsis (Arabidopsis thaliana) roots under iron deficiency. Second, iron is converted from Fe3+ to Fe2+ by FRO2, ferric-chelate reductase (Yi and Guerinot, 1996; Robinson et al., 1999). Third, Fe2+ is taken up by IRT1, an iron and zinc transporter (Connolly et al., 2002; Vert et al., 2002).
On the other hand, as a strategy to avoid the toxicity of several ions, plant cells take advantage of such proton gradients to sequester the ions inside vacuoles (Dietz et al., 2001). During this process, the vacuolar H+-ATPase (V-ATPase), which is a heteromultimeric proton pump, together with H+-pyrophosphatase (V-PPase), plays a central role in vacuole acidification, energizing the active transport of ions across the tonoplast and coordinately regulating turgor pressure and cell elongation (Maeshima, 2000; Sze et al., 2002). Several zinc transporters and zinc-chelating proteins and peptides are involved in zinc homeostasis in plants (Krämer, 2010). Among them, tonoplast Zn2+/H+ antiporters MTP1 and MTP3 transport excess zinc from the cytosol to the vacuoles (Kobae et al., 2004; Desbrosses-Fonrouge et al., 2005; Arrivault et al., 2006; Gustin et al., 2009). In fact, more recently, it has been reported that the mtp1 mutant shows severe inhibition of root growth in the presence of excess zinc (Kawachi et al., 2009). Moreover, dysfunction of V-ATPase in the de-etiolated3-1 (det3-1) mutant, which has a mutation in the V-ATPase subunit C-encoding gene, leads to defects in cell elongation that result in plant dwarfism (Schumacher et al., 1999). Furthermore, the V-ATPase plays an important role in the trans-Golgi network (TGN) for cell expansion, which is involved in the synthesis and trafficking of cell wall components, such as cellulose (Brüx et al., 2008).
To analyze the molecular mechanism to cope with excess Zn, we have identified and reported on zinc-responsive proteins in the roots of Arabidopsis using a highly improved method of two-dimensional electrophoresis (Fukao et al., 2009). However, our previous approach failed to identify membrane proteins, including transporters and other proteins known to be responsive to and fundamentally important under excess zinc. Thus, accurate large-scale identification of response proteins under excess zinc is urgently needed to understand in detail the molecular and physiological effects of excess zinc on plant cells. In this study, to elucidate the effects of excess zinc on the protein levels in Arabidopsis in detail, we adopted an iTRAQ (for isobaric tags for relative and absolute quantification)-based quantitative proteomics approach. Our data revealed that the levels of 157 proteins increased more than 1.2-fold and 127 decreased to less than 0.83-fold in response to excess zinc in the wild type. Here, many membrane proteins were identified and quantified. In fact, we found that IRT1 and FRO2 levels particularly increased under excess zinc. In addition, the levels of several subunits of the V-ATPase decreased in response to excess zinc. This was further confirmed by the zinc-sensitive phenotype of the det3-1 mutant. Finally, we found that root hair morphology was abnormal in roots grown on excess zinc and that this phenotype was correlated with decreased amounts of proteins that have been previously reported in root hair-defective mutants.
In this study, we show that the combined analyses of proteomic approaches and use of mutant plants clarified zinc-induced molecular phenomena in plants.
RESULTS
iTRAQ Analysis of Zinc-Responsive Microsomal Proteins in the Wild Type
We recently reported that both shoot and root growth of Arabidopsis is significantly inhibited by the addition of 300 μm ZnSO4 in the growth medium (Fukao et al., 2009). In this study, microsomal proteins were prepared from roots grown for 10 d on Murashige and Skoog (MS) medium already containing 30 μm ZnSO4 or MS medium exogenously supplemented with 300 μm ZnSO4 (containing totally 330 μm ZnSO4; hereafter referred to as 300-Zn). The purity of the microsomal fraction was evaluated by immunoblot analysis (Supplemental Fig. S1). In order to identify zinc-responsive proteins, microsomal fractions from the wild type were analyzed by iTRAQ combined with highly sensitive and high-resolution liquid chromatography-tandem mass spectrometry. We summarized results using less than 5% false discovery rate (FDR) in each iTRAQ analysis (FDR = 4.06, 3.13, or 3.62; Elias and Gygi, 2007). In total, our approach allowed the identification and quantification of 521 proteins (Supplemental Table S1). Among the proteins identified, 27, 40, and 90 proteins reproducibly increased by more than 2.0-, 1.5-, and 1.2-fold, respectively (Supplemental Fig. S2). On the other hand, 12, 43, and 72 proteins decreased to less than 0.50-, 0.67-, and 0.83-fold, respectively (Supplemental Fig. S2). Particularly, zinc transporters or membrane proteins previously shown to be implicated in zinc transport are summarized in Supplemental Table S2 according to previous reports (Hall and Williams, 2003; Palmgren et al., 2008). Interestingly, two zinc transporters, IRT1 (AT4G19690) and MTP3 (AT3G58810), were highly increased, by 10.7- and 4.7-fold, respectively (Table I; Supplemental Table S2). While IRT1 is localized on the plasma membrane and actively imports both zinc and iron under normal growth conditions (Korshunova et al., 1999; Vert et al., 2002), MTP3 is localized on the tonoplast, where it transports zinc into the vacuoles (Arrivault et al., 2006).
Table I. Proteins with increased levels in response to zinc in the wild type.
Microsomal proteins from the wild type grown on MS or 300-Zn medium were labeled with iTRAQ-114 and -115 reagent, respectively, and those from the det3-1 mutant grown on MS or 300-Zn medium were labeled with iTRAQ-116 and -117 reagent, respectively. Proteins that increased more than 2.0-fold were selected and ordered based on fold change values in wild type 300-Zn/wild type MS. Data are means ± sd of three independent experiments. * P < 0.05, ** P < 0.001, significantly different from the wild type or the det3-1 mutant on MS medium.
| Arabidopsis Genome Initiative Code | Protein | Peptidea | Coverageb | Ratioc (Wild Type 300-Zn/Wild Type MS) | Ratioc (det3-1 MS/Wild Type MS) | Ratioc (det3-1 300-Zn/det3-1 MS) |
| AT4G10480 | Nascent polypeptide-associated complex α-chain protein, putative/α-NAC, putative | 3.3 | 23.9 | 12.111 ± 3.947* | 0.689 ± 0.243 | 6.328 ± 3.262* |
| AT3G12390 | Nascent polypeptide-associated complex α-chain protein, putative/α-NAC, putative | 3.0 | 20.5 | 12.051 ± 3.382* | 0.957 ± 0.205 | 5.102 ± 3.486 |
| AT3G49470 | NACA2 (nascent polypeptide-associated complex subunit α-like protein 2) | 1.0 | 6.0 | 11.726 ± 7.023 | 0.720 ± 0.010** | 5.409 ± 1.887* |
| AT4G19690 | IRT1 (iron-regulated transporter 1); cadmium, iron, manganese, and zinc transmembrane transporter | 3.7 | 19.3 | 10.742 ± 5.370* | 0.664 ± 0.130* | 15.066 ± 8.097* |
| AT1G01580 | FRO2 (ferric reduction oxidase 2);ferric-chelate reductase | 12.0 | 18.0 | 8.449 ± 1.787** | 0.738 ± 0.085* | 5.764 ± 0.762** |
| AT1G09560 | GLP5 (germin-like protein 5); manganese ion binding/metal ion binding/nutrient reservoir | 4.0 | 15.7 | 5.026 ± 1.528* | 0.723 ± 0.133* | 3.940 ± 1.076* |
| AT3G58810 | MTP3/MTPA2; efflux transmembrane transporter/zinc ion transmembrane transporter | 4.3 | 8.7 | 4.708 ± 0.207** | 0.649 ± 0.052** | 4.658 ± 1.791* |
| AT1G17880 | Nascent polypeptide-associated complex domain-containing protein/BTF3b-like transcription factor, putative | 6.0 | 37.6 | 4.375 ± 1.211* | 0.875 ± 0.037* | 2.650 ± 1.013 |
| AT1G20200 | EMB2719 (embryo defective 2719) | 5.0 | 12.6 | 3.787 ± 1.055* | 0.893 ± 0.127 | 2.739 ± 2.509 |
| AT1G78000 | SULTR1;2 (sulfate transporter 1;2); sulfate transmembrane transporter | 5.7 | 9.5 | 3.736 ± 0.678** | 0.718 ± 0.048** | 2.357 ± 0.383* |
| AT1G73230 | Nascent polypeptide-associated complex domain-containing protein | 7.0 | 37.8 | 3.652 ± 0.649** | 0.862 ± 0.014** | 2.483 ± 0.874* |
| AT4G16190 | Cys proteinase, putative | 3.0 | 12.2 | 3.054 ± 0.374** | 0.614 ± 0.082** | 4.550 ± 0.083** |
| AT3G51800 | ATG2 (G2p-related protein);metalloexopeptidase | 8.3 | 22.8 | 2.820 ± 0.465* | 0.988 ± 0.112 | 2.278 ± 0.630* |
| AT1G78850 | Curculin-like (Man-binding) lectin family protein | 5.3 | 15.1 | 2.737 ± 0.536* | 1.014 ± 0.132 | 2.104 ± 0.774* |
| AT4G39090 | RD19 (responsive to dehydration 19); Cys-type peptidase | 5.3 | 17.0 | 2.719 ± 0.270** | 0.601 ± 0.076** | 4.205 ± 0.352** |
| AT3G53890 | 40S ribosomal protein S21 (RPS21B) | 1.7 | 17.5 | 2.661 ± 0.796* | 0.994 ± 0.118 | 1.690 ± 0.565 |
| AT3G10090 | 40S ribosomal protein S28 (RPS28A) | 3.0 | 39.1 | 2.531 ± 0.236** | 0.728 ± 0.010** | 2.015 ± 0.454* |
| AT5G64140 | RPS28 (ribosomal protein S28); structural constituent of ribosome | 3.0 | 39.1 | 2.521 ± 0.171** | 0.711 ± 0.025** | 2.038 ± 0.434* |
| AT2G07727 | Cytochrome b (MTCYB, COB, CYTB) | 1.0 | 3.3 | 2.492 ± 0.380* | 1.041 ± 0.019* | 1.093 ± 0.142 |
| AT3G30390 | Amino acid transporter family protein | 2.7 | 6.1 | 2.359 ± 0.100** | 0.789 ± 0.108* | 1.978 ± 0.393* |
| AT2G27730 | Identical to uncharacterized mitochondrial protein; similar to GB:CAN62125.1 and GB:CAO48731.1 | 1.7 | 19.8 | 2.351 ± 0.566* | 0.984 ± 0.056 | 1.159 ± 0.235 |
| AT4G32470 | Ubiquinol-cytochrome c reductase complex 14-kD protein, putative | 2.3 | 21.3 | 2.279 ± 0.547* | 0.897 ± 0.087 | 1.021 ± 0.163 |
| AT5G19820 | EMB2734 (embryo defective 2734); lyase | 2.7 | 3.0 | 2.247 ± 0.365* | 0.389 ± 0.044** | 3.975 ± 1.456* |
| AT1G48630 | Guanine nucleotide-binding family protein/activated protein kinase C receptor, putative/RACK, putative | 7.3 | 20.4 | 2.199 ± 0.401* | 0.357 ± 0.030** | 3.774 ± 1.809 |
| AT1G18080 | ATARCA (Arabidopsis thaliana homolog of tobacco ArcA); nucleotide binding | 14.0 | 38.3 | 2.123 ± 0.408* | 0.372 ± 0.023** | 3.714 ± 1.685 |
| AT1G51980 | Mitochondrial processing peptidase α-subunit, putative | 4.3 | 12.0 | 2.081 ± 0.383* | 0.878 ± 0.186 | 1.141 ± 0.082* |
| AT1G26630 | FBR12 (fumonisin B1-resistant 12); translation initiation factor | 6.3 | 38.8 | 2.076 ± 0.145** | 0.808 ± 0.025** | 1.464 ± 0.226* |
Peptide indicates average number of assigned peptides.
Coverage indicates average percentage of assigned peptides to the predicted protein.
The values were calculated as the ratio of 115 (wild type 300-Zn) to 114 (wild type MS) label, 116 (det3-1 MS) to 114 (wild type MS) label, or 117 (det3-1 300-Zn) to 116 (det3-1 MS) label.
According to the ATTEDII database, some nascent polypeptide-associated complex (NAC) domain-containing proteins are strongly related to ribosomal proteins at the transcription level (Obayashi and Kinoshita, 2010). In our study, AT4G10480, AT3G12390, AT3G49470, AT1G17880, and AT1G73230, which are NAC domain-containing proteins, were highly increased, by 12.1-, 12.1-, 11.7-, 4.4-, and 3.7-fold, respectively (Table I). The levels of these proteins might increase to maintain a low level of ribosomal proteins (Table II). Although it is not yet evident whether these proteins are membrane associated or not, their high response to zinc suggests an important role under excess zinc.
Table II. Proteins with decreased levels in response to zinc.
Microsomal proteins from the wild type grown on MS or 300-Zn medium were labeled with iTRAQ-114 and -115 reagent, respectively, and those from the det3-1 mutant grown on MS and 300-Zn medium were labeled with iTRAQ-116 and -117 reagent, respectively. Proteins that decreased less than 0.5-fold were selected and ordered based on fold change values in wild type 300-Zn/wild type MS. Data are means ± sd of three independent experiments. * P < 0.05, ** P < 0.001, significantly different from the wild type or the det3-1 mutant on MS medium.
| Arabidopsis Genome Initiative Code | Prot\in | Peptidea | Coverageb | Ratioc (Wild Type 300-Zn/Wild Type MS) | Ratioc (det3-1 MS/Wild Type MS) | Ratioc (det3-1 300-Zn/det3-1 MS) |
| AT1G10630 | ATARFA1F; GTP binding/phospholipase activator/protein binding | 6.0 | 33.3 | 0.496 ± 0.123** | 0.791 ± 0.024** | 0.595 ± 0.107** |
| AT3G60245 | 60S ribosomal protein L37a(RPL37aC) | 3.7 | 23.6 | 0.474 ± 0.239 | 1.022 ± 0.280 | 0.812 ± 0.177 |
| AT3G15950 | NAI2 (TSA1-LIKE); protein binding | 24.3 | 32 | 0.457 ± 0.018** | 1.207 ± 0.136 | 0.313 ± 0.107* |
| AT2G38380 | Peroxidase 22 (PER22, P22, PRXEA)/basic peroxidase E | 6.0 | 18.9 | 0.447 ± 0.071** | 1.107 ± 0.063* | 0.319 ± 0.161** |
| AT3G53480 | ATPDR9/PDR9 (pleiotropic drug resistance 9); ATPase, coupled to transmembrane movement of substances | 4.3 | 3.3 | 0.427 ± 0.211* | 0.746 ± 0.073* | 0.729 ± 0.371 |
| AT4G39200 | 40S ribosomal protein S25 (RPS25E) | 3.3 | 32.7 | 0.402 ± 0.246* | 0.703 ± 0.128* | 0.851 ± 0.366 |
| AT5G24780 | VSP1 (vegetative storage protein 1);acid phosphatase | 2.7 | 9.5 | 0.402 ± 0.136** | 0.549 ± 0.051** | 0.471 ± 0.097** |
| AT4G37410 | CYP81F4 (cytochrome P450, family 81, subfamily F, polypeptide 4); oxygen binding | 4.0 | 10 | 0.362 ± 0.157** | 0.582 ± 0.016** | 0.347 ± 0.131** |
| AT4G35790 | ATPLDDELTA (Arabidopsis thaliana phospholipase D δ); phospholipase D | 5.0 | 6.7 | 0.354 ± 0.101** | 1.001 ± 0.026 | 0.480 ± 0.137* |
| AT1G02920 | ATGSTF7 (glutathione S-transferase 11); glutathione transferase | 2.7 | 15.5 | 0.335 ± 0.140** | 0.605 ± 0.034* | 0.869 ± 0.179 |
| AT3G16780 | 60S ribosomal protein L19 (RPL19B) | 5.0 | 27 | 0.235 ± 0.100** | 1.134 ± 0.255 | 0.481 ± 0.285 |
| AT2G39310 | Jacalin-related lectin 22 (JAL22) | 4.3 | 11.9 | 0.217 ± 0.119** | 0.786 ± 0.059* | 0.259 ± 0.168** |
Peptide indicates average number of assigned peptides.
Coverage indicates average percentage of assigned peptides to the predicted protein.
The values were calculated as the ratio of 115 (wild type 300-Zn) to 114 (wild type MS) label, 116 (det3-1 MS) to 114 (wild type MS) label, or 117 (det3-1 300-Zn) to 116 (det3-1 MS) label.
We also found some proteins whose amounts significantly decreased in response to excess zinc. Among the proteins that decreased to less than 0.50-fold that of control, NAI2 (AT3G15950) and jacalin-related lectin 22 (JAL22; AT2G39310) decreased to 0.46- and 0.22-fold, respectively (Table II). NAI2 has been reported to be a key factor in endoplasmic reticulum (ER) body formation, and JAL22 is a candidate component of the PYK10 complex, which is found in the ER body (Nagano et al., 2008). PYK10 (AT3G09260; Matsushima et al., 2004), which is a major component in the ER body, PYK10-binding protein 1 (AT3G16420; Matsushima et al., 2004), and JAL33 and JAL35 (AT3G16450 and AT3G16470, respectively; Nagano et al., 2008) also decreased, to 0.83-, 0.85-, 0.55-, and 0.86-fold, respectively (Supplemental Table S1). Taken together, the ER body might be a target of damage due to excess zinc. BIP1 (AT5G28540) and BIP2 (AT5G42020), which are members of the HSP70 family localized to the ER, decreased to 0.75- and 0.74-fold, respectively (Supplemental Table S1). These results suggest that excess zinc might affect not only the ER body but also the entire ER.
Shoot Chlorosis Due to Excess Zinc Is Caused by Iron Deficiency
As described above, IRT1 levels were highly increased (Table I). In addition, FRO2 was also highly increased, 8.4-fold (Table I). IRT1 and FRO2 levels have been reported to increase under iron deficiency (Connolly et al., 2002). It has been suggested that plants suffer an iron deficiency under excess zinc by a yet-unclear mechanism. To check whether iron concentration was decreased in shoots and roots of the wild type grown on 300-Zn medium, the concentrations of 15 elements are quantified by inductively coupled plasma atomic emission spectroscopy; the results are summarized in Table III. Interestingly, the concentration of iron was significantly decreased in roots and in shoots under 300-Zn medium by 30% and 20%, respectively (Table III). It appears that excess zinc affects the concentration of iron in plant tissues to a different extent in roots and shoots. Therefore, we focused on the relationship between excess zinc and iron and shoot chlorosis.
Table III. Effect of excess zinc on the level of several chemical elements.
The concentrations of 15 chemical elements were determined in the shoots and roots of wild-type plants grown either on MS or 300-Zn medium. Data shown represent means ± sd of three independent experiments.
| Additional 300 μm ZnSO4 | Root |
Shoot |
|||
| — | + | — | + | ||
| Zinc | mg g−1 | 0.752 ± 0.087 | 10.5 ± 0.4 | 0.114 ± 0.074 | 0.928 ± 0.011 |
| Iron | mg g−1 | 2.47 ± 0.12 | 1.73 ± 0.11 | 0.128 ± 0.001 | 0.103 ± 0.004 |
| Calcium | mg g−1 | 1.46 ± 0.08 | 1.68 ± 0.08 | 4.66 ± 0.13 | 4.42 ± 0.05 |
| Potassium | mg g−1 | 19.7 ± 2.1 | 15.3 ± 0.4 | 43.3 ± 0.6 | 36.5 ± 1.0 |
| Magnesium | mg g−1 | 1.78 ± 0.12 | 1.83 ± 0.07 | 3.14 ± 0.01 | 2.77 ± 0.04 |
| Sodium | mg g−1 | 1.16 ± 0.11 | 0.898 ± 0.189 | 1.66 ± 0.03 | 1.51 ± 0.046 |
| Phosphorus | mg g−1 | 8.28 ± 0.45 | 9.33 ± 0.13 | 8.96 ± 0.19 | 7.17 ± 0.09 |
| Sulfur | mg g−1 | 6.07 ± 0.35 | 6.66 ± 0.11 | 6.74 ± 0.14 | 10.3 ± 0.1 |
| Aluminum | μg g−1 | 14.0 ± 2.7 | 21.0 ± 1.4 | 13.9 ± 2.0 | 9.62 ± 1.34 |
| Boron | μg g−1 | 17.6 ± 2.6 | 14.5 ± 6.5 | 32.7 ± 0.9 | 38.1 ± 2.5 |
| Copper | μg g−1 | 7.80 ± 0.94 | 15.0 ± 4.8 | 5.58 ± 0.34 | 13.8 ± 2.7 |
| Manganese | μg g−1 | 29.2 ± 0.24 | 33.9 ± 2.2 | 69.3 ± 0.8 | 67.3 ± 0.6 |
| Cadmium | μg g−1 | 0.240 ± 0.134 | 0.270 ± 0.113 | 0.063 ± 0.052 | 0.039 ± 0.001 |
| Cobalt | μg g−1 | 5.77 ± 0.42 | 3.40 ± 0.31 | 0.362 ± 0.052 | 0.112 ± 0.055 |
| Chromium | μg g−1 | 3.24 ± 1.03 | 1.98 ± 0.32 | 0.962 ± 0.029 | 1.00 ± 0.06 |
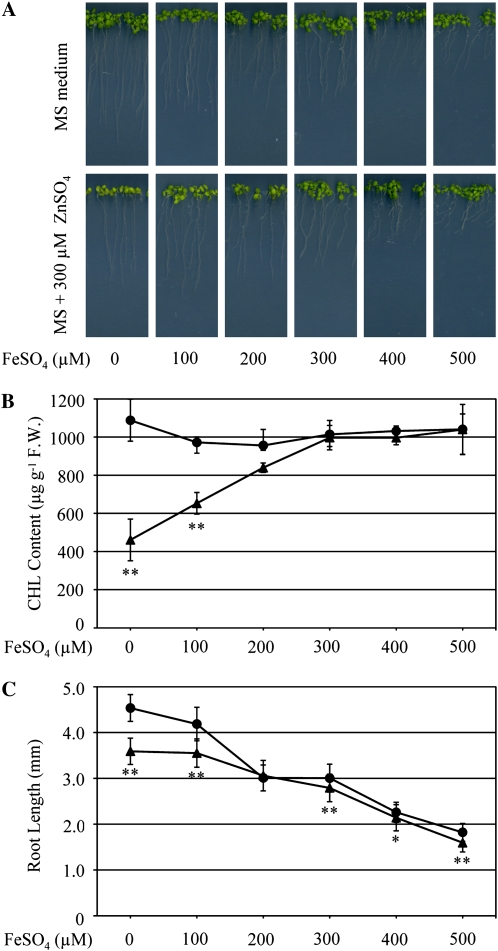
In order to test whether excess zinc causes shoot chlorosis through iron depletion, plants were grown on 300-Zn medium supplemented with various concentrations of FeSO4 (Fig. 1A). Chlorosis was evaluated by quantifying the chlorophyll content. Interestingly, the chlorophyll content in shoots recovered in a FeSO4 concentration-dependent manner (Fig. 1B). Shoot chlorosis fully recovered upon addition of 300 to 500 μm FeSO4 to 300-Zn medium. Together, these results strongly suggest that shoot chlorosis is caused by iron deficiency due to excess zinc. Consistently, root growth inhibition was alleviated upon addition of 200 μm FeSO4, and root lengths were similar in seedlings grown on MS and 300-Zn (Fig. 1C). Therefore, the reduction in root growth upon exposure to high zinc concentration is also attributable to zinc-induced Fe deficiency in these organs. Nonetheless, increased FeSO4 concentration (greater than 200 μm) in the growth medium under high-zinc conditions did not further restore root growth (Fig. 1C).
Figure 1.
Effects of exogenously supplied ZnSO4 and FeSO4 on chlorophyll content and root growth. A, Growth of wild-type seedlings on MS or 300-Zn medium supplemented with 100, 200, 300, 400, 500, or 0 μm FeSO4. B and C, Chlorophyll content (B) and root length (C) of 10-d-old seedlings grown on MS medium (black circles) and 300-Zn medium (black triangles) in A were measured. F.W., Fresh weight. Data are means ± sd of three independent experiments. Ten seedlings were grown in each experiment at different times. * P < 0.05, ** P < 0.001, significantly different from the data on MS medium.
Iron Supply Is Sufficient for Chlorosis Recovery in irt1 Mutants
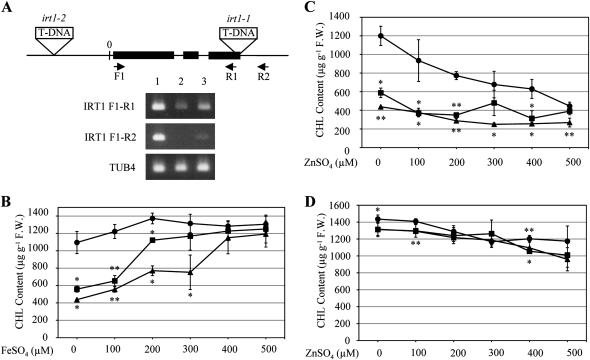
As previously mentioned, the protein level of the iron and zinc transporter IRT1 increased greatly in response to excess zinc (Table I). To gain further insight into the role of IRT1 under excess zinc, two IRT1 T-DNA insertion mutant lines, irt1-1 (SALK_054554C) and irt1-2 (SALK_024525), were characterized. First, semiquantitative reverse transcription-PCR analysis of the IRT1 gene revealed that IRT1 transcripts in irt1-1 were not detectable and that IRT1 transcripts in the irt1-2 mutant decreased, although shorter transcripts were present in both lines (Fig. 2A), suggesting that itr1-1 might be a null allele and irt1-2 a weak mutant of IRT1.
Figure 2.
Effects of exogenously supplied ZnSO4 and FeSO4 on chlorophyll content in irt1 mutants. A, IRT1 and TUB4 transcripts were amplified by PCR from the wild type (lane 1), irt1-1 (lane 2), and irt1-2 (lane 3). IRT1 transcript was amplified using primers F1 and R1 for 28 cycles of PCR and with primers F1 and R2 for 35 cycles of PCR. TUB4 transcript was amplified by 28 cycles of PCR. B and C, Chlorophyll content of 10-d-old seedlings of the wild type (black circles), irt1-1 (black triangles), and irt1-2 (black squares) grown on MS medium supplemented with 100, 200, 300, 400, 500, or 0 μm FeSO4 (B) or ZnSO4 (C). D, The effect of increasing zinc concentration on chlorophyll content in the wild type (black circles), irt1-1 (black triangles), and irt1-2 (black squares) grown on MS medium with 400 μm FeSO4. F.W., Fresh weight. Data are means ± sd of three independent experiments. Ten seedlings were grown in each experiment at different times. * P < 0.05, ** P < 0.001, significantly different from the wild type. Asterisks above and below the symbols show P values of irt1-1 and irt1-2, respectively.
It has been reported that the chlorophyll content in the irt1 mutant is extremely low even under normal growth conditions and that the content is recovered to wild-type levels by supplying iron in the growth medium (Vert et al., 2002). Therefore, the itr1-1 and irt1-2 mutants were grown on MS medium or on MS medium supplemented with FeSO4 or ZnSO4 (Fig. 2, B and C). Under these conditions, the chlorophyll content in irt1 mutants was lower than that of the wild type, even under normal growth conditions. Furthermore, the chlorophyll content dramatically recovered after supplying iron but not zinc (Fig. 2, B and C). Interestingly, we found that a concentration of 400 μm FeSO4 was enough for both irt1 mutant alleles to recover chlorophyll content to wild-type levels (Fig. 2B). In irt1 mutants, iron might be taken up by other iron transporters or might enter cells passively, because recovery of the chlorophyll content upon addition of iron was slower in irt1-1 than in irt1-2 (Fig. 2B). While root growth of irt1 mutants was more sensitive to excess zinc than the wild type, they were similarly affected under excess iron (Supplemental Fig. S3, A and B). Therefore, the zinc-induced increase in IRT1 protein levels is likely involved in the protection of plants from zinc toxicity.
Next, to further examine the cross talk between zinc and iron, irt1 mutants were grown on MS medium containing 400 μm FeSO4 and simultaneously supplemented with various concentrations of ZnSO4 (Fig. 2D). Under these growth conditions, the chlorophyll content was not affected significantly even by applying the high zinc concentration of 500 μm (Fig. 2D). Taken together, these results clearly demonstrated that chlorosis did not occur when iron was available at a sufficient concentration, even in the presence of excess zinc. On the other hand, it is noteworthy that the root length of the wild type and irt1 mutants was comparable under the above growth conditions (Supplemental Fig. S3C).
The Two Proton Pumps, V-ATPase and V-PPase, Are Inversely Affected by Excess Zinc in the Wild Type
Next, we focused on the V-ATPase subunits in the iTRAQ results, because the V-ATPase has a pivotal role in promoting turgor pressure and cell elongation (Schumacher et al., 1999; Krebs et al., 2010). Interestingly, seven subunits of the V-ATPase were identified, and the levels of three of them decreased in response to excess zinc, to less than 0.83-fold (Table IV). The amounts of subunits A, C, and E1 decreased to less than 0.83-fold, although the amounts of subunits D, G, H, and a3 were either slightly decreased or unaffected (Table IV).
Table IV. Effect of excess zinc on protein levels of V-ATPase subunits and V-PPase.
Microsomal proteins from the wild type grown on MS or 300-Zn medium were labeled with iTRAQ-114 and -115 reagent, respectively, and those from the det3-1 mutant grown on MS or 300-Zn medium were labeled with iTRAQ-116 and -117 reagent, respectively. Subunits of V-ATPase complex and V-PPase were selected and alphabetically ordered. Boldface entries indicate the V-ATPase subunits that were significantly decreased in both the wild type and the det3-1 mutant. Data are means ± sd of three independent experiments. N.D. indicates that a peptide and/or iTRAQ reporter ions were not detected. * P < 0.05, ** P < 0.001, significantly different from the wild type or the det3-1 mutant on MS medium.
| Arabidopsis Genome Initiative Code | Protein | Peptidea | Coverageb | Ratioc (Wild Type 300-Zn/Wild Type MS) | Ratioc (det3-1 MS/Wild Type MS) | Ratioc (det3-1 300-Zn/det3-1 MS) |
| AT1G78900 | VHA A | 25.0 | 47.2 | 0.809 ± 0.076* | 0.570 ± 0.036** | 0.758 ± 0.148 |
| AT1G76030 | VHA B1 | N.D. | N.D. | N.D. | ||
| AT4G38510 | VHA B2 | N.D. | N.D. | N.D. | ||
| AT1G20260 | VHA B3 | N.D. | N.D. | N.D. | ||
| AT1G12840 | VHA C | 7.3 | 21.3 | 0.795 ± 0.196 | 0.397 ± 0.054** | 0.791 ± 0.125* |
| AT3G58730 | VHA D | 8.3 | 30.4 | 0.893 ± 0.097 | 0.604 ± 0.048** | 0.832 ± 0.249 |
| AT4G11150 | VHA E1 | 12.7 | 48.0 | 0.832 ± 0.044* | 0.568 ± 0.045** | 0.766 ± 0.143 |
| AT3G08560 | VHA E2 | N.D. | N.D. | N.D. | ||
| AT1G64200 | VHA E3 | N.D. | N.D. | N.D. | ||
| AT4G02620 | VHA F | N.D. | N.D. | N.D. | ||
| AT3G01390 | VHA G1 | 5.3 | 59.1 | 0.983 ± 0.076 | 0.589 ± 0.060** | 0.847 ± 0.019** |
| AT4G23710 | VHA G2 | N.D. | N.D. | N.D. | ||
| AT4G25950 | VHA G3 | N.D. | N.D. | N.D. | ||
| AT3G42050 | VHA H | 8.3 | 25.6 | 0.835 ± 0.324 | 0.696 ± 0.065** | 0.859 ± 0.103 |
| AT2G28520 | VHA a1 | N.D. | N.D. | N.D. | ||
| AT2G21410 | VHA a2 | N.D. | N.D. | N.D. | ||
| AT4G39080 | VHA a3 | 7.0 | 9.5 | 1.074 ± 0.144 | 0.739 ± 0.063** | 1.044 ± 0.274 |
| AT4G34720 | VHA c1 | N.D. | N.D. | N.D. | ||
| AT1G19910 | VHA c2 | 2.0 | 15.2 | 1.274 ± 0.119* | 0.835 ± 0.046* | 1.384 ± 0.558 |
| AT4G39080 | VHA c3 | N.D. | N.D. | N.D. | ||
| AT1G75630 | VHA c4 | N.D. | N.D. | N.D. | ||
| AT2G16510 | VHA c5 | N.D. | N.D. | N.D. | ||
| AT4G32530 | VHA c”1 | N.D. | N.D. | N.D. | ||
| AT2G25610 | VHA c”2 | N.D. | N.D. | N.D. | ||
| AT3G28710 | VHA d1 | N.D. | N.D. | N.D. | ||
| AT3G28715 | VHA d2 | N.D. | N.D. | N.D. | ||
| AT5G55290 | VHA e1 | N.D. | N.D. | N.D. | ||
| AT4G26710 | VHA e2 | N.D. | N.D. | N.D. | ||
| AT1G15690 | AVP1 | 12.0 | 14.3 | 1.450 ± 0.172* | 1.331 ± 0.113* | 1.062 ± 0.256 |
Peptide indicates average number of assigned peptides.
Coverage indicates average percentage of assigned peptides to the predicted protein.
The values were calculated as the ratio of 115 (wild type 300-Zn) to 114 (wild type MS) label, 116 (det3-1 MS) to 114 (wild type MS) label, or 117 (det3-1 300-Zn) to 116 (det3-1 MS) label.
To further support our findings on zinc-responsive proteins in the wild type, we performed iTRAQ analysis of microsomal fractions from the det3-1 mutant. Among the 519 proteins identified in the det3-1 mutant, 22, four, and 39 proteins reproducibly increased by more than 2.0-, 1.5-, and 1.2-fold, respectively (Supplemental Fig. S2; Supplemental Table S3). On the other hand, 15, 92, and 154 proteins were decreased to less than 0.50-, 0.67-, and 0.83-fold, respectively (Supplemental Fig. S2; Supplemental Table S3). Consistently, the number and fold change of zinc highly responsive proteins (greater than 2.0-fold or less than 0.5-fold) were similar in the wild type and the det3-1 mutant (Tables I and II). Together, these results indicate that the responsiveness to excess zinc is basically not affected in det3-1.
To gain further insights into the inhibitory effect of excess zinc, we compared the results between proteins in the wild type grown on 300-Zn medium and untreated det3-1 mutant. Among 157 proteins (increased more than 1.2-fold on 300-Zn in the wild type), only 11 proteins were similarly increased in the det3-1 mutant (Supplemental Table S4), whereas 98 proteins were unaffected (1.2- to 0.83-fold) and 48 proteins were conversely decreased less than 0.83-fold (Supplemental Table S1). Basically, there were no proteins commonly increased more than 2.0-fold in both genotypes. On the other hand, among of 127 proteins (decreased less than 0.83-fold by 300-Zn in the wild type), 29 proteins were also decreased less than 0.83-fold in det3-1 (Supplemental Table S4). Eighty-seven proteins were not affected (1.2- to 0.83-fold), and only 11 proteins were conversely increased more than 1.2-fold (Supplemental Table S1). A close examination of the data revealed that three nucleosome assembly proteins, namely NAP1;1, NAP1;2, and NAP1;3, were collectively decreased in both the wild type and untreated det3-1 mutant (Supplemental Table S4). These results indicate that there is no strong correlation between proteins affected by excess zinc in the wild type and those in untreated det3-1 mutant.
The transcript levels of the V-ATPase C subunit in the det3-1 mutant are approximately reduced to half that of the wild type (Schumacher et al., 1999). Consistently, we found that the protein amount of the V-ATPase C subunit in det3-1 was approximately half that of the wild type (det3-1 MS/wild type MS ratio in Table IV), which is in agreement with the transcript levels published earlier (Schumacher et al., 1999). Moreover, the amounts of six other V-ATPase subunits were also decreased in the det3-1 mutant (det3-1 MS/wild type MS ratio in Table IV). The decrease of protein levels of V-ATPase subunits, such as A, C, and E1, also occurred under excess zinc in the wild type (wild type 300-Zn/wild type MS ratio in Table IV). Interestingly, while the amount of AVP1 (V-PPase), which is another proton pump on the tonoplast, increased in both the wild type on 300-Zn medium and untreated det3-1 mutant, it was unaffected in det3-1 on 300-Zn medium. Although this may suggest a compensatory role of V-PPase under certain circumstances, it does not seem to be a general mechanism in response to excess zinc (Table IV).
Root Growth of the det3-1 Mutant Is Severely Reduced by Excess Zinc
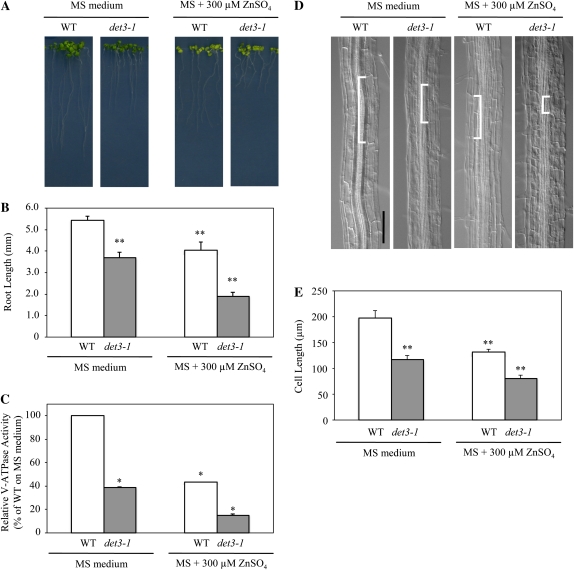
In order to examine the physiological role of the V-ATPase complex under excess zinc, the det3-1 mutant was grown on either MS or 300-Zn medium. The root length of the det3-1 mutant was reduced significantly compared with the wild type, even on the standard MS medium (Fig. 3). In addition, the root growth of the det3-1 mutant was more severely inhibited on 300-Zn medium than the wild type (Fig. 3, A and B). Considering these results, the V-ATPase activity in the wild type and the det3-1 mutant was directly measured on root microsomal fractions extracted from plants grown on either MS or 300-Zn medium. The V-ATPase activity in the det3-1 mutant grown on MS medium was approximately half that in the wild type, which is consistent with previous reports (Fig. 3C; Schumacher et al., 1999; Krebs et al., 2010). Interestingly, the V-ATPase activity in the wild type grown on 300-Zn medium was comparable to that of the untreated det3-1 mutant, while that of the det3-1 mutant grown on 300-Zn medium was one-fourth that of the wild type (Fig. 3C). These findings show a clear correlation between the reduction in V-ATPase subunit content, V-ATPase activity, and root length under conditions of excess zinc.
Figure 3.
Effects of excess zinc on phenotype of the det3-1 mutant. A and B, Wild-type (WT) and det3-1 mutant seedling growth (A) and root length (B) were measured. The sd values give an estimate of the variation in mean number of more than 10 plants. C, V-ATPase activity of microsomes from roots. Data are means ± sd of four independent experiments. About 1,200 seedlings were grown on 12 plates in each experiment at different times. D, Morphology of root cells in the wild type and the det3-1 mutant. White brackets show one cell in each root. Bar = 100 μm. E, Cell length in roots was measured at the cell maturation zone. The sd values give an estimate of the variation in mean cell length of more than 80 cells for each condition. About 200 seedlings were grown on two plates in parallel experiments. * P < 0.05, ** P < 0.001, significantly different from the wild type on MS medium.
Finally, to clarify whether inhibition of root growth by excess zinc is due to inhibition of cell elongation or a defect in cell cycling, cell lengths in roots were measured at the maturation zone. The cell length of untreated det3-1 mutants and the wild type grown on 300-Zn medium was similar and about half that of the untreated wild type (Fig. 3, D and E). These results indicate that root growth inhibition by excess zinc is due to inhibition of cell elongation rather than cell proliferation. In the det3-1 mutant, it has been reported that cell elongation in the hypocotyls, petioles, and inflorescence stems was more affected than that in roots (Schumacher et al., 1999). Consistently, under our growth conditions, cell elongation in det3-1 was also significantly affected in organs such as petioles and stems, as described previously (data not shown). Additionally, in the det3-1 mutant, we observed approximately 30% and 40% reduction of root length and cell length, respectively, upon growth on MS medium (Fig. 3, D and E).
The Inhibitory Effect of Excess Zinc on Cell Elongation Is Reversible
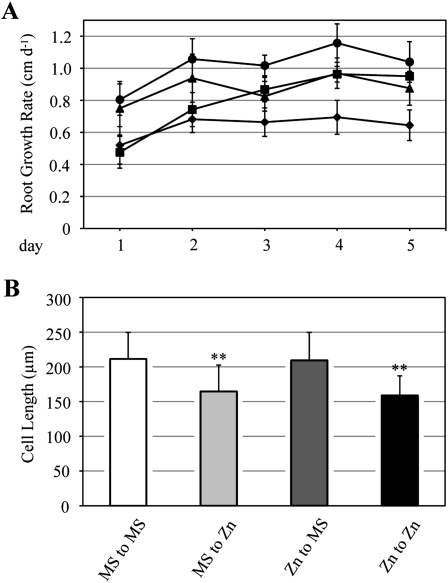
To examine whether the inhibitory effects of excess zinc on cell elongation are reversible on removing zinc, wild-type seedlings grown on either MS or 300-Zn medium for 10 d were transplanted to MS or 300-Zn medium. Then, the length of each root was scored every day for 5 d (Fig. 4A). Roots transplanted from MS to fresh MS medium were able to elongate about 1 cm per day. However, the growth rate of roots transplanted from MS to 300-Zn medium was inhibited, while the growth rate of roots transplanted from 300-Zn to MS medium was gradually restored with time (Fig. 4A).
Figure 4.
Reversibility of root growth inhibition due to excess zinc. A, Wild-type seedlings were grown on MS medium or 300-Zn medium for 10 d. Then, seedlings on MS medium were transplanted to MS medium (black circles) or 300-Zn medium (black triangles). Seedlings on 300-Zn medium were transplanted to MS medium (black squares) or 300-Zn medium (black diamonds). Length change of each root was measured every day for 5 d. The sd values give an estimate of the variation in mean number of more than 15 plants. Ten seedlings were grown at different times in two independent experiments. B, Cell length 5 d after transplantation was measured in the cell maturation zone. The sd values give an estimate of the variation in mean cell length of more than 80 cells for each condition. About 200 seedlings were grown on two plates in parallel. ** P < 0.001, significantly different from “MS to MS.”
Next, cell length was determined in the root maturation zone 5 d after transplantation, in a zone where cells were supposed to be actively dividing at the time of transplantation. We found that the length of cells in roots transplanted from 300-Zn to MS medium was comparable to that of roots transplanted from MS to MS medium (Fig. 4B). However, cell length of roots transplanted from MS medium to 300-Zn was inhibited to the same extent as that of roots transplanted from 300-Zn to 300-Zn. Altogether, these results clearly showed that cell elongation suppression by excess zinc could be reversed upon transfer to regular growth conditions.
Root Hair Development Is Altered by Excess Zinc
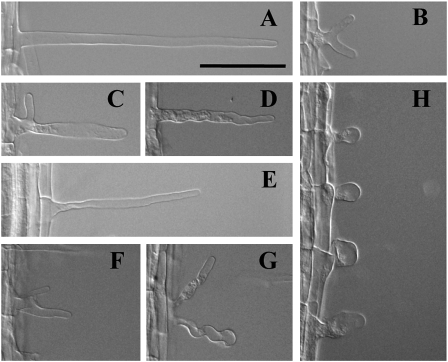
During our microscopic analysis of root cell length, we noticed that the majority of root hairs had an abnormal shape when plants were grown on 300-Zn medium. Root hairs in roots grown on 300-Zn medium were either branched at the proximal region (Fig. 5, B, C, F, and G), wavy (Fig. 5D), or rounded (Fig. 5H). This root hair morphology is reminiscent of previously reported root hair-defective mutants. For example, the rhd mutant series was isolated based on abnormal root hair morphology (Schiefelbein and Somerville, 1990). While proper initiation of root hairs is disrupted in the rhd1 mutant, normal hair elongation is affected in rhd2, rhd3, and rhd4 mutants. In addition, rhd2, shv1, shv2, and shv3 mutants failed to develop beyond the transition to tip growth, whereas bst1, cen1, cen2, cen3, rhd3, and scn1 mutants failed to control the shape of the root hair during tip growth (Parker et al., 2000). Although the above mutants were isolated by a forward genetics approach, the csld mutant series, which turned out to show abnormal root hair development, was isolated by a reverse genetics approach to investigate the functions of the cellulose synthase superfamily (Bernal et al., 2008). Also, root hairs treated with taxol, a microtubule-stabilizing drug, or oryzalin, a microtubule-depolymerizing drug, showed abnormal morphology (Bibikova et al., 1999). It is worth mentioning that abnormally shaped root hairs were found both in wild-type and det3-1 mutant roots grown on 300-Zn medium but never in roots grown on MS medium. Thus, the defect in V-ATPase activity in det3-1 mutants is not the cause of abnormal root hairs; rather, excess zinc is the primary cause of root hair growth defects, regardless of plant genotype.
Figure 5.
Abnormal root hair morphology on excess zinc. Root hairs of the wild type and the det3-1 mutant grown on MS or 300-Zn medium for 10 d are shown. A to D, Root hairs of the wild type on MS medium (A) and on 300-Zn medium (B–D). E to H, Root hairs of the det3-1 mutant on MS medium (E) and on 300-Zn medium (F–H). Bar = 100 μm.
Based on these findings, we reexamined our iTRAQ data and found that the amounts of several proteins that are involved in root hair development were indeed decreased in the wild type and det3-1 mutants grown on 300-Zn medium (Table V). For example, RHD3 and MRH5/SHV3 decreased to 0.59- and 0.64-fold in the wild type on 300-Zn medium and to 0.63- and 0.68-fold in the untreated det3-1 mutant, respectively. Also, TUB2 decreased to 0.66- and 0.54-fold in the wild type on 300-Zn medium and the untreated det3-1 mutant, respectively. On the other hand, TUA4 was not affected by zinc in the wild type. Interestingly, actin isoforms ACT7 and ACT8 were not affected by excess zinc, although the growth rate of root hairs is reported to be inhibited by the actin antagonist latrunculin B (Bibikova et al., 1999). Tubulins and actins might not be the direct cause of abnormal root hair observed in 300-Zn medium, because they were decreased even in untreated det3-1 mutant (det3-1 MS/wild type MS ratio in Table V).
Table V. Zinc-responsive proteins related to root hair morphology.
Microsomal proteins from the wild type grown on MS or 300-Zn medium were labeled with iTRAQ-114 and -115 reagent, respectively, and those from the det3-1 mutant grown on MS or 300-Zn medium were labeled with iTRAQ-116 and -117 reagent, respectively. Proteins related to root hair morphology were selected and ordered based on fold change values in wild type 300-Zn/wild type MS. Data are means ± sd of three independent experiments. * P < 0.05, ** P < 0.001, significantly different from the wild type or the det3-1 mutant on MS medium.
| Arabidopsis Genome Initiative Code | Protein | Peptidea | Coverageb | Ratioc (Wild Type 300-Zn/Wild Type MS) | Ratioc (det3-1 MS/Wild Type MS) | Ratioc (det3-1 300-Zn/det3-1 MS) |
| AT3G13870 | RHD3 (root hair defective 3) | 6.0 | 10.4 | 0.588 ± 0.183* | 0.906 ± 0.120 | 0.632 ± 0.109* |
| AT4G26690 | MRH5/SHV3(morphogenesis of root hair 5) | 5.0 | 8.7 | 0.639 ± 0.291 | 1.042 ± 0.224 | 0.678 ± 0.255 |
| AT5G62690 | TUB2 (tubulin β-2) | 6.0 | 17.6 | 0.660 ± 0.220 | 0.714 ± 0.078* | 0.543 ± 0.022** |
| AT1G04820 | TUA4 (tubulin α-4 chain) | 8.7 | 21.2 | 0.903 ± 0.039* | 0.668 ± 0.039** | 0.674 ± 0.142* |
| AT5G09810 | ACT7 (actin 7) | 10.0 | 29.3 | 1.077 ± 0.064 | 0.836 ± 0.051* | 0.767 ± 0.101* |
| AT1G49240 | ACT8 (actin 8) | 12.3 | 37.1 | 1.099 ± 0.076 | 0.836 ± 0.060* | 0.775 ± 0.107* |
Peptide indicates average number of assigned peptides.
Coverage indicates average percentage of assigned peptides to the predicted protein.
The values were calculated as the ratio of 115 (wild type 300-Zn) to 114 (wild type MS) label, 116 (det3-1 MS) to 114 (wild type MS) label, or 117 (det3-1 300-Zn) to 116 (det3-1 MS) label.
DISCUSSION
iTRAQ analysis has been developed to identify marker proteins for disease and to investigate the effects of drug treatment for clinical application (Latterich et al., 2008; Zamò and Cecconi, 2010). This technique is now widely applied to plants to understand changes in protein profiles during growth or due to environmental stimuli and stress (Lücker et al., 2009; Schneider et al., 2009; Pang et al., 2010). However, few studies have directly evaluated the physiological importance of proteins identified by iTRAQ analysis. Here, we investigated proteomic changes in Arabidopsis roots under excess zinc. Then, we subsequently evaluated the physiological roles of some key genes by analyzing in detail their corresponding mutants. We successfully identified key proteins that led us to explain unambiguously the phenotypes caused by excess zinc, such as chlorosis, root growth inhibition, and abnormal morphology of root hairs.
It has been known for decades that excess zinc leads to shoot chlorosis (Fig. 1; Marschner, 1995). However, proteins involved in this process have remained elusive. In this study, the iron concentration was significantly decreased in shoot tissues, which explained the increased amounts of IRT1 and FRO2. Actually, the ferric chelate reductase activity was increased in plants treated with 100 μm ZnSO4 for 4 d (Becher et al., 2004). Furthermore, it has been shown that IRT1 transports zinc in addition to iron by directly measuring zinc uptake activity and by complementation experiments in yeast (Eide et al., 1996; Korshunova et al., 1999). The ability to take up iron and zinc by IRT1 is inhibited by the addition of excess zinc and iron, respectively (Rogers et al., 2000). The competitive inhibition of iron uptake by excess zinc should be alleviated, since chlorosis was recovered when iron was supplied with zinc (Fig. 1). It is worth noting that in Arabidopsis roots, although MTP3 transcript levels increased upon growth on medium containing 500 μm ZnSO4 (Arrivault et al., 2006), the mRNA and protein of the IRT1 gene were not detectable when treated with similar ZnSO4 concentrations (Connolly et al., 2002). For the latter report, seedlings were grown for 2 weeks on Gamborg’s B5 medium (containing 7 μm ZnSO4; Gamborg et al., 1968) and transferred subsequently and grown for 3 d on iron-deficient medium supplied with 500 μm ZnSO4. However, in our study, seeds were germinated and then seedlings were continuously grown on MS medium supplied with 300 μm ZnSO4. Thus, the response of IRT1 to excess zinc might be different from our findings for the above reasons.
Excess Zinc Reduced Root Growth by Inhibiting Cell Expansion
Our iTRAQ analysis revealed that levels of three out of seven subunits of the V-ATPase were significantly decreased in both the wild type and the det3-1 mutant in response to excess zinc (Table IV). In addition, the V-ATPase activity in the wild type under excess zinc decreased to the same level as that of the untreated det3-1 mutant (Fig. 3). The VHA-a subunit has three isoforms in Arabidopsis (Sze et al., 2002). VHA-a2 and VHA-a3 are found in the tonoplast, whereas VHA-a1 is localized in the TGN (Dettmer et al., 2006; Brüx et al., 2008). Krebs et al. (2010) showed that zinc tolerance in a vha-a2 vha-a3 double mutant is reduced. Therefore, zinc might be less sequestrated into vacuoles under excess zinc because of V-ATPase dysfunction on the tonoplast. Actually, Dietz et al. (2001) reported that barley (Hordeum vulgare) root growth was significantly impaired by cadmium, which is a zinc analog, when simultaneously treated with bafilomycin A1, which is an inhibitor of V-ATPase activity (Ratajczak, 2000).
What is the cause for the cell expansion defect in the det3-1 mutant? Here, we have to consider that the reduction of growth due to excess zinc might be primarily caused by defects in the TGN, because the amount of VHA-a3 was not affected by excess zinc in this study. Unfortunately, although VHA-a1 was not detected in the iTRAQ analysis, VHA-A and VHA-E1 decreased in response to excess zinc (Table IV). The morphology of Golgi stacks and Golgi-derived vesicles was severely changed in a vha-A mutant (Dettmer et al., 2005). Also, the organization of Golgi stacks was abnormal in a vha-E1 mutant (Strompen et al., 2005). Furthermore, it has been demonstrated that Arabidopsis VHA-E isoforms are functionally able to complement a vma4 zinc-sensitive mutant, which bears a mutation in the single yeast subunit E isoform (Dettmer et al., 2010). Interestingly, a vma4 mutant grown on medium supplemented with 5 mm ZnCl2 was complemented by expression of VHA-E1 and VHA-E3 but not VHA-E2. If we assume that a reduced V-ATPase activity in the TGN can alone cause an inhibition of cell expansion (Brüx et al., 2008), it is plausible to consider that the severe phenotype observed in the det3-1 mutant under excess zinc is essentially caused by a defect in TGN acidification, leading to a cell wall defect. Therefore, the reduced V-ATPase activity observed in this study appears to be a symptom of zinc toxicity that disrupts TGN functionality.
Aldolase, which catalyzes an aldol cleavage of Fru 1,6-bisP to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate, interacts with the V-ATPase B subunit (Barkla et al., 2009). It has been proposed that aldolase regulates cell elongation mediated by V-ATPase, because root growth was inhibited in aldolase-antisense transgenic rice (Oryza sativa; Konishi et al., 2005). In our iTRAQ analysis, Fru-bisP aldolase (AT3G52930) was decreased in both the wild type under excess zinc and untreated det3-1 mutants to 0.79-fold (Supplemental Table S1). The decrease in aldolase might explain the inhibition of root growth under excess zinc. Both root and cell lengths in the wild type grown on excess zinc were comparable to those of untreated det3-1 mutant roots (Figs. 2–4). Therefore, excess zinc should affect not only the amount of V-ATPase subunits but also aldolase, and thus cell elongation.
Finally, this study demonstrated that growth defects caused by excess zinc were quickly restored by removing zinc, which is also consistent with a previous report (Fig. 4; Kawachi et al., 2009). These observations support plant cells growing slowly under ATP-limiting conditions and reestablishing the growth rate when the conditions are suitable. This might be a strategy for plants to survive under undesirable conditions.
The Formation of Abnormal Root Hairs under Excess Zinc
Root hairs are affected by changes in nutrient status, because they are the major site for the uptake of water and nutrients (Gilroy and Jones, 2000). It has been reported that changes in ion concentrations affect root hair morphology. For example, manganese deficiency leads to abnormal root hairs in Arabidopsis, and the concentration of manganese, iron, and zinc is reduced in manganese-deficient conditions (Yang et al., 2008). The abnormal root hairs formed due to excess zinc might be caused by other factors, because the concentrations of manganese and zinc increased under excess zinc (Table III). In our study, the amount of RHD3 was reduced by up to 60% that of the wild type and det3-1 under 300-Zn medium (wild type 300-Zn/wild type MS ratio and det3-1 300-Zn/det3-1 MS ratio in Table V). RHD3 is unique among the four RHD genes, because mutations in this gene affect both root and root hair morphology. The rhd3 mutant not only has shorter root hairs but also root hairs with a wavy appearance that are occasionally more highly branched and slightly shorter than the wild type. Finally, RHD3 might contribute to a root growth defect under excess zinc, because RHD3 encodes a protein with GTP-binding motifs that may be required during vacuole enlargement (Wang et al., 1997). Finally, among the six proteins identified, only RHD3 and MRH5/SHV3 were specifically decrease upon exposure to excess zinc. Thus, the disrupted root hair morphology in the wild type and the det3-1 mutant is probably caused by the decreased amounts of these two proteins.
CONCLUSION
In this study, to better understand zinc tolerance mechanisms, we used the iTRAQ method. Arabidopsis grown on excess zinc showed three major visible phenotypes, namely chlorosis, root growth inhibition, and abnormal morphology of root hairs. The iTRAQ data identified several zinc-responsive proteins and also clarified that excess zinc-induced reductions in V-ATPase protein level and activity are the cause of zinc toxicity symptoms, such as reduced root growth and phenocopying aspects of the det3-1 mutant. Therefore, the iTRAQ method is useful both for the investigation of proteins responsive to several environmental conditions and as an approach to discover the molecular network responsible for mutant phenotypes at the protein level. iTRAQ analysis has recently started to be used in plant science. This technique will be more widely available and useful in the near future and will, we believe, contribute to solving various puzzling biological processes.
MATERIALS AND METHODS
Sample Preparation
Arabidopsis (Arabidopsis thaliana) ecotype Columbia or the det3-1 mutant was germinated on sterile plates of MS medium containing 2.3 mm MES-KOH, pH 5.7, 1.0% (w/v) Suc, and 1.5% agar or on MS medium supplemented with 300 μm ZnSO4 (Murashige and Skoog, 1962). About 100 seeds per genotype and per sample were sown on plates (10 × 14 × 1.5 cm) that contained 50 mL of MS medium. In total, 1,200 Arabidopsis seedlings were vertically grown for 10 d at 22°C under 16-h-light/8-h-dark conditions. Roots (approximately 0.4 g fresh weight) were harvested after 3 h from the beginning of the light period and homogenized with buffer A (50 mm HEPES-KOH, pH 7.5, 5 mm EDTA, 400 mm Suc, and protease inhibitor cocktail). The homogenates were centrifuged at 1,000g at 4°C for 20 min, and the supernatants were centrifuged at 8,000g at 4°C for 20 min. The supernatants were centrifuged at 100,000g at 4°C for 60 min to prepare the microsomal fraction. The pellets were dissolved in iTRAQ buffer (Applied Biosystems). The protein concentration was determined by the Bradford method (Bio-Rad). Three replicates were prepared from three independent experiments of samples grown at different time.
Peptide Labeling with iTRAQ Reagents
Each 20 μL of microsomal protein fraction (2.5 mg mL−1) was reduced by tris-(2-carboxyethyl) phosphine at 60°C for 60 min and then alkalized by methyl methanethiosulfonate at room temperature for 10 min. Samples were digested by 10 μL of trypsin (1 mg mL−1) at 37°C for 16 h The peptides from the wild type grown on MS medium, the wild type grown on 300-Zn medium, det3-1 mutant grown on MS medium, and det3-1 mutant grown on 300-Zn medium were labeled with iTRAQ-114, -115, -116, and -117 reagents, respectively, at room temperature for 60 min. Mixed peptides were manually separated by 25, 50, 75, 100, 200, 350, and 1,000 mm NaCl using strong cation exchange (Applied Biosystems) and then desalted on Sep-Pak C18 cartridges (Waters). The labeled peptides were concentrated by a vacuum concentrator.
Mass Spectrometric Analysis
iTRAQ analysis was performed by using an LTQ-Orbitrap XL-HTC-PAL-Paradigm MS4 system. The iTRAQ-labeled peptides were loaded on the column (75 μm internal diameter, 15 cm; L-Column; CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics). Buffers were 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in water (A) and 0.1% (v/v) acetic acid and 90% (v/v) acetonitrile in water (B). A linear gradient from 5% to 45% B for 70 min was applied, and peptides eluted from the column were introduced directly into an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) with a flow rate of 200 nL min–1 and a spray voltage of 2.0 kV. The range of the mass spectrometric scan was mass-to-charge ratio 450 to 1,500, and the top three peaks were subjected to tandem mass spectrometry analysis. The obtained spectra were compared against data in The Arabidopsis Information Resource 8 (http://www.arabidopsis.org/) using the MASCOT server (version 2.2; Matrix Science) with the following search parameters: threshold set-off at 0.05 in the ion-score cutoff; protein identification cutoff set to two assigned spectra per predicted protein; peptide tolerance at 10 ppm; tandem mass spectrometry tolerance at ±0.2 D; peptide charge of 2+ or 3+; trypsin as the enzyme and allowing up to one missed cleavage; iTRAQ label and methyl methanethiosulfonate on Cys as a fixed modification; and oxidation on Met as a variable modification. iTRAQ data for three biological replicates were analyzed by MASCOT, then only data with FDR of less than 5% were used for subsequent data analysis. Only proteins that were identified in all three independent experiments were considered. Statistical analysis was conducted using Student’s t test.
T-DNA Insertion Mutants
T-DNA insertion mutants SALK_054554C (irt1-1) and SALK_024525 (irt1-2) were obtained from the Arabidopsis Biological Resource Center at Ohio State University.
RNA Extraction and Reverse Transcription-PCR
Total RNA was extracted from 10-d-old roots using TRIzol reagent (Invitrogen). Reverse transcription was performed using a kit (ReverTra Ace; Toyobo) with an oligo(dT)12-18 primer. IRT1 (At4g19690) and TUBULIN4 (TUB4; At5g44340) were amplified by PCR using the following primers: 5′-ATGGCTTCAAATTCAGCACTTCTCATG-3′ (sense; F1) and 5′-GAGCTGTGCATTTGACGAATCATCTTC-3′ (antisense; R1) or 5′-TGATTTTATCAACAGAACCCGGTTAGAGAAC-3′ (antisense; R2) for IRT1 and 5′-GAGGGAGCCATTGACAACATCTT-3′ (sense) and 5′-GCGAACAGTTCACAGCTATGTTCA-3′ (antisense) for TUB4. IRT1 was amplified in 28 cycles using F1 and R1 and in 35 cycles using F1 and R2. TUB4 was amplified in 28 cycles.
Quantification of Chlorophyll
About 10 Arabidopsis seedlings per plate were grown on MS medium with the indicated zinc and/or iron concentrations for 10 d. Three replicates were prepared from three independent experiments at different times. Chlorophyll content was measured as described by Arnon (1949).
Determination of Metal Content in Arabidopsis Tissues
About 100 Arabidopsis seedlings per plate were grown on MS or 300-Zn medium for 10 d. Shoots and roots from about 1,500 Arabidopsis seedlings were harvested and dried at 80°C for 48 h. Three sets of samples were prepared. Dried tissues (20 mg) were digested with ultrapure HNO3 for 22 min at 130°C using Teflon vessels (ETHOS-1600). Then, the content of metallic elements was determined by inductively coupled plasma atomic emission spectroscopy (IRIS ICARP; Jarrel Ash Nippon). The analytical precision for elemental analysis was confirmed using the National Institute for Environmental Studies standard (nos. 1 and 9) to be better than 10% for all elements analyzed. Three replicates were prepared in parallel.
Measurement of V-ATPase Activity
V-ATPase activity was measured as NO3−-inhibited, Cl−-stimulated, and vanadate-insensitive ATPase activity according to Schumacher et al. (1999). Each 20-μL microsomal protein fraction (0.1 mg mL−1) was used for activity measurement. The composition of the measuring buffer was 25 mm Tricine-Tris (pH 8.0), 2 mm NaMoO4, 2 mm MgSO4, 1 mm NaN3, 0.002% Brij58, 0.5 mm dithiothreitol, 0.2 mm vanadate, and 1 mm ATP. NO3−-inhibited activity was measured in the presence of 50 mm KNO3, and Cl−-stimulated activity was measured in the presence of 50 mm KCl. The values were determined in three independent measurements, and the value for the wild type grown on MS medium was set to 100%. Four replicates were prepared from four independent experiments at different times.
Measurement of Root Cell Length
Roots grown for 10 d on MS or 300-Zn medium were fixed in formalin-acetic acid-alcohol and cleared using chloral solution as described by Tsuge et al. (1996). Whole roots were observed with a M165 FC stereoscopic microscope (Leica Microsystems) connected to a Leica DFC300 FX digital camera and a Leica DM2500 Nomarski differential interference contrast microscope connected to a Leica DFC310 FX digital camera. Root cell length was measured as described by Ferjani et al. (2007). The values give an estimate of the mean cell length of over 80 cells in each condition.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Purity check of the microsomal fraction.
Supplemental Figure S2. Numbers of zinc-responsive proteins in the wild type and the det3-1 mutant.
Supplemental Figure S3. Effect of exogenously supplied ZnSO4 and FeSO4 on root growth of irt1 mutants.
Supplemental Table S1. List of all proteins identified by iTRAQ analysis of zinc-treated wild-type and det3-1 mutant roots.
Supplemental Table S2. Effect of excess zinc on transporters and membrane proteins reported to be implicated in zinc transport.
Supplemental Table S3. List of all proteins identified by iTRAQ analysis of zinc-treated det3-1 mutant roots.
Supplemental Table S4. List of commonly increased or decreased proteins in zinc-treated wild-type and untreated det3-1 mutant roots.
Acknowledgments
We thank Dr. Gorou Horiguchi (Rikkyo University) for kindly providing the det3-1 mutant seeds.
References
- Alkorta I, Hernández-Allica J, Becerril JM, Amezaga I, Albizu I, Garbisu C. (2004) Recent findings on the phytoremediation of soils contaminated with environmentally toxic heavy metals and metalloids such as zinc, cadmium, lead, and arsenic. Rev Environ Sci Biotechnol 3: 71–90 [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Krämer U. (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Hernández-Coronado M, Pantoja O. (2009) Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21: 4044–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U. (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WGT. (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. (1999) Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J 17: 657–665 [DOI] [PubMed] [Google Scholar]
- Brüx A, Liu T-Y, Krebs M, Stierhof Y-D, Lohmann JU, Miersch O, Wasternack C, Schumacher K. (2008) Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses-Fonrouge A-G, Voigt K, Schröder A, Arrivault S, Thomine S, Krämer U. (2005) Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett 579: 4165–4174 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof Y-D, Schumacher K. (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Liu TY, Schumacher K. (2010) Functional analysis of Arabidopsis V-ATPase subunit VHA-E isoforms. Eur J Cell Biol 89: 152–156 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Schubert D, Calvo-Weimar O, Stierhof Y-D, Schmidt R, Schumacher K. (2005) Essential role of the V-ATPase in male gametophyte development. Plant J 41: 117–124 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Tavakoli N, Kluge C, Mimura T, Sharma SS, Harris GC, Chardonnens AN, Golldack D. (2001) Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J Exp Bot 52: 1969–1980 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Ferjani A, Fujiwara M, Nishimori Y, Ohtsu I. (2009) Identification of zinc-responsive proteins in the roots of Arabidopsis thaliana using a highly improved method of two-dimensional electrophoresis. Plant Cell Physiol 50: 2234–2239 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones DL. (2000) Through form to function: root hair development and nutrient uptake. Trends Plant Sci 5: 56–60 [DOI] [PubMed] [Google Scholar]
- Gustin JL, Loureiro ME, Kim D, Na G, Tikhonova M, Salt DE. (2009) MTP1-dependent Zn sequestration into shoot vacuoles suggests dual roles in Zn tolerance and accumulation in Zn-hyperaccumulating plants. Plant J 57: 1116–1127 [DOI] [PubMed] [Google Scholar]
- Hacisalihoglu G, Kochian LV. (2003) How do some plants tolerate low levels of soil zinc? Mechanisms of zinc efficiency in crop plants. New Phytol 159: 341–350 [DOI] [PubMed] [Google Scholar]
- Hall JL, Williams LE. (2003) Transition metal transporters in plants. J Exp Bot 54: 2601–2613 [DOI] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Mori H, Tomioka R, Lee Y, Maeshima M. (2009) A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol 50: 1156–1170 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M. (2004) Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol 45: 1749–1758 [DOI] [PubMed] [Google Scholar]
- Konishi H, Maeshima M, Komatsu S. (2005) Characterization of vacuolar membrane proteins changed in rice root treated with gibberellin. J Proteome Res 4: 1775–1780 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Krämer U. (2010) Metal hyperaccumulation in plants. Annu Rev Plant Biol 61: 517–534 [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KAS, Marten I, Stierhof Y-D, Hedrich R, Schumacher K. (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M, Abramovitz M, Leyland-Jones B. (2008) Proteomics: new technologies and clinical applications. Eur J Cancer 44: 2737–2741 [DOI] [PubMed] [Google Scholar]
- Lücker J, Laszczak M, Smith D, Lund ST. (2009) Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genomics 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M. (2000) Vacuolar H(+)-pyrophosphatase. Biochim Biophys Acta 1465: 37–51 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2 Academic Press, London [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. (2004) NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16: 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I. (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49: 969–980 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. (2010) Coexpression landscape in ATTED-II: usage of gene list and gene network for various types of pathways. J Plant Res 123: 311–319 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Clemens S, Williams LE, Krämer U, Borg S, Schjørring JK, Sanders D. (2008) Zinc biofortification of cereals: problems and solutions. Trends Plant Sci 13: 464–473 [DOI] [PubMed] [Google Scholar]
- Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X. (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9: 2584–2599 [DOI] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS. (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum LM, Rink L, Haase H. (2010) The essential toxin: impact of zinc on human health. Int J Environ Res Public Health 7: 1342–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak R. (2000) Structure, function and regulation of the plant vacuolar H(+)-translocating ATPase. Biochim Biophys Acta 1465: 17–36 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Eide DJ, Guerinot ML. (2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci USA 97: 12356–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Schellenberg M, Meyer S, Keller F, Gehrig P, Riedel K, Lee Y, Eberl L, Martinoia E. (2009) Quantitative detection of changes in the leaf-mesophyll tonoplast proteome in dependency of a cadmium exposure of barley (Hordeum vulgare L.) plants. Proteomics 9: 2668–2677 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller SC, Castelfranco AM, Castelfranco PA. (1982) Effects of iron and oxygen on chlorophyll biosynthesis. I. In vivo observations on iron and oxygen-deficient plants. Plant Physiol 69: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, Dettmer J, Stierhof Y-D, Schumacher K, Jürgens G, Mayer U. (2005) Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant J 41: 125–132 [DOI] [PubMed] [Google Scholar]
- Sze H, Schumacher K, Müller ML, Padmanaban S, Taiz L. (2002) A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H(+)-ATPase. Trends Plant Sci 7: 157–161 [DOI] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Van Assche F, Clijsters H. (1986) Inhibition of photosynthesis in Phaseolus vulgaris by treatment with toxic concentration of zinc: effect on ribulose-1,5-bisphosphate carboxylase/oxygenase. J Plant Physiol 125: 355–360 [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Yang TJW, Perry PJ, Ciani S, Pandian S, Schmidt W. (2008) Manganese deficiency alters the patterning and development of root hairs in Arabidopsis. J Exp Bot 59: 3453–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Zamò A, Cecconi D. (2010) Proteomic analysis of lymphoid and haematopoietic neoplasms: there’s more than biomarker discovery. J Proteomics 73: 508–520 [DOI] [PubMed] [Google Scholar]