Abstract
Nitrogen is an essential nutrient for plant growth. In the Rhizobium-legume symbiosis, root nodules are the sites of bacterial nitrogen fixation, in which atmospheric nitrogen is converted into a form that plants can utilize. While recent studies suggested an important role for the soybean (Glycine max) ecto-apyrase GS52 in rhizobial root hair infection and root nodule formation, precisely how this protein impacts the nodulation process remains undetermined. In this study, the biochemical characteristics of the GS52 enzyme were investigated. Computer modeling of the GS52 apyrase structure identified key amino acid residues important for catalytic activity, which were subsequently mutagenized. Although the GS52 enzyme exhibited broad substrate specificity, its activity on pyrimidine nucleotides and diphosphate nucleotides was significantly higher than on ATP. This result was corroborated by structural modeling of GS52, which predicted a low specificity for the adenine base within the substrate-binding pocket of the enzyme. The wild-type enzyme and its inactive mutant forms were expressed in soybean roots in order to evaluate the importance of GS52 enzymatic activity for nodulation. The results indicated a clear correlation between GS52 enzymatic activity and nodule number. Altogether, our study indicates that the catalytic activity of the GS52 apyrase, likely acting on extracellular nucleotides, is critical for rhizobial infection and nodulation.
Apyrases (EC 3.6.1.5) are divalent ion-dependent, trinucleotide and dinucleotide phosphatases ubiquitous in both eukaryotes and prokaryotes. Some of the diversity of apyrase function is attributable to the fact that the catalytic domains are located within cells or organelles, or outside of cells (Plesner, 1995). The latter apyrases, the so-called ecto-apyrases, are well known to modulate the concentration of extracellular nucleotides (Todorov et al., 1997; Marcus et al., 2003). Five characteristic apyrase conserved regions (ACRs) are found in members of the nucleoside triphosphate diphosphohydrolase (NTPDase)/apyrase family (Kirley et al., 2006). ACRs form the active site of apyrases, in which ACR1 and ACR4 correspond to the β- and γ-phosphate-binding domains of the actin/Hsp70/hexokinase superfamily (Hurley, 1996).
Nodulation is the result of a symbiosis between legume plants and soil bacteria called rhizobia (Oldroyd and Downie, 2008; Ferguson et al., 2010). Recently, phylogenetic analysis identified a legume-specific clade of apyrases (Cannon et al., 2003), some of which were shown to function in the nodulation process (Etzler et al., 1999; Day et al., 2000; Cohn et al., 2001; Govindarajulu et al., 2009). For example, the ecto-apyrase from Dolichos biflorus was reported to bind to lipochitooligosaccharide nodulation factors (Nod factors) derived from various rhizobia (Etzler et al., 1999). Nod factors are essential signals required for induction of the nodulation process (Oldroyd and Downie, 2008; Ferguson et al., 2010). Binding to the Nod factor increased the enzymatic activity of the D. biflorus ecto-apyrase, suggesting a possible signaling role for this enzyme (Etzler et al., 1999). Consistent with a role for ecto-apyrases in nodulation, the addition of antibodies directed against the ecto-apyrases of D. biflorus or soybean (Glycine max) was shown to inhibit nodulation (Etzler et al., 1999; Day et al., 2000).
The soybean ecto-apyrase GS52, originally identified in the wild soybean Glycine soja, is a member of the legume-specific clade and was shown to be mildly induced early during rhizobial infection (Day et al., 2000) but also during the later stages of nodule development (Brechenmacher et al., 2008; Govindarajulu et al., 2009). This enzyme was localized to the plant plasma membrane (Day et al., 2000). Strong ectopic expression of the GS52 gene in Lotus japonicus roots resulted in a marked increase in root hair infection as well as higher nodule numbers (McAlvin and Stacey, 2005). The stimulation of infection is especially interesting because it suggests a role for the enzyme very early in the symbiotic interaction. Conversely, RNA interference-mediated gene silencing of the GS52 gene significantly reduced nodule numbers in transgenic soybean roots (Govindarajulu et al., 2009). Collectively, the published data support a role for legume-specific ecto-apyrases in the nodulation process.
However, it is not clear whether the role mentioned above requires enzymatic activity, presumably acting to hydrolyze extracellular nucleotides. We initiated this study by first computationally modeling the GS52 protein structure, which led to the identification of key amino acid residues likely important for catalysis. These residues were subsequently mutated, and the enzymatic activity of these proteins was compared with the wild-type enzyme. Subsequent ectopic expression of both the mutant and wild-type enzymes in transgenic soybean and Medicago truncatula roots clearly showed that nodulation was dependent on the enzymatic activity.
RESULTS
Biochemical Characterization of Recombinant GS52 Apyrase
Previous publications did not describe the enzymatic properties of the GS52 apyrase. Therefore, the enzyme was expressed in Escherichia coli and purified (Supplemental Fig. S1), and its enzymatic properties were analyzed. The GS52 enzyme showed maximum activity at 37°C. Enzyme activity was relatively high over a broad range of pH values, with maximal activity observed at pH 6.5 to 7.0 using either ATP or ADP as substrate. This pH optimum matches that of the extracellular pH in the root hair zone of alfalfa (Medicago sativa), which was measured in the range of pH 6.4 to 6.7 (Felle et al., 1998). Similar measurements have not been done in soybean.
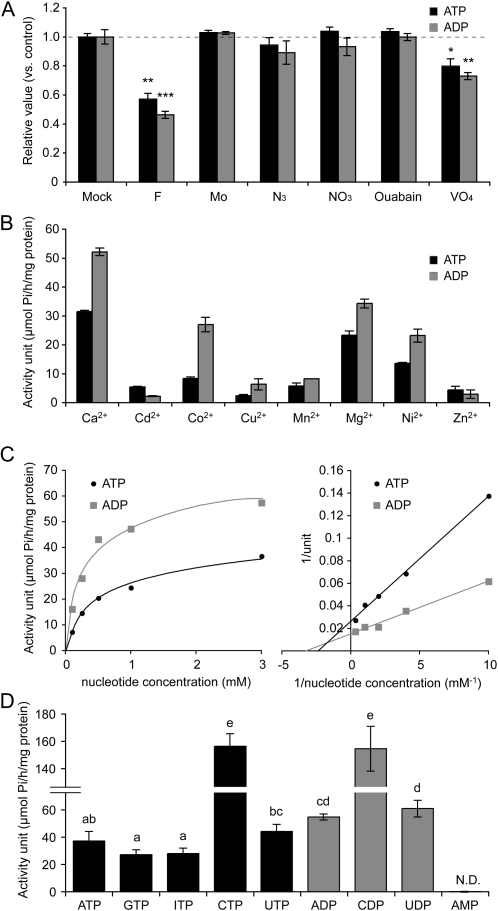
Apyrases are not sensitive to known inhibitors that act on various ATPases and phosphatases. Therefore, we measured the effects of the common inhibitors on the ATP/ADP hydrolysis activity of the GS52 apyrase. The enzyme was assayed in the presence of fluoride (NaF; an inhibitor of pyrophosphatase), molybdate (MoNa2O4; an inhibitor of acid phosphatase), azide (NaN3; an inhibitor of F-type ATPase), nitrate (NaNO3; an inhibitor of V-type ATPase), ouabain (an inhibitor of Na+-K+ ATPase), and vanadate (Na3VO4; an inhibitor of P-type ATPase). All of the inhibitors tested, except for fluoride and vanadate, had no influence on GS52 apyrase activity. Vanadate inhibited approximately 20% using either ATP or ADP as substrate. However, the concentration (1 mm) of vanadate used is rather high given that less than 500 μm vanadate is required for half-maximal inhibition of P-type ATPase activity (O’Neill and Spanswick, 1984). Indeed, 500 μm vanadate did not inhibit GS52 activity. Therefore, the effect is most likely due to nonspecific action as a general inhibitor against phosphatases. Fluoride inhibited GS52 activity approximately 50% at a concentration of 10 mm. This result is consistent with previous biochemical studies (Komoszyński, 1996), in which plant apyrases were specifically inhibited by fluoride based on comparative biochemical studies between plants and animals. Taken together, our results indicate that GS52 protein possesses typical plant apyrase properties and, furthermore, that the recombinant GS52 protein was enzymatically active. However, a plant apyrase inhibitor, NGXT191 (Windsor et al., 2003), previously shown to inhibit apyrases from Arabidopsis (Arabidopsis thaliana) and potato (Solanum tuberosum), had no inhibitory effect on GS52 apyrase activity at concentrations of 5 and 10 μg mL−1 (the relative activity values to mock treatment were 1.09 ± 0.05 and 1.15 ± 0.11 after incubation with NGXT at 5 and 10 μg mL−1, respectively). These results imply that the GS52 apyrase likely acts differently in comparison with the other plant apyrases, which are involved in plant cell growth (Steinebrunner et al., 2003; Wu et al., 2007).
All NTPDase/apyrase family members require divalent metal ions as cofactors for enzymatic activity. To determine the cofactor preference for GS52, we evaluated the effects of a variety of divalent ions on apyrase activity. As shown in Figure 1B, Ca2+ was found to be the most effective, whereas addition of Mg2+ supported only moderate activity. The order of preference was as follows: Ca2+ > Mg2+ > Ni2+ > Co2+ ≈ Mn2+ ≈ Cd2+ > Zn2+ ≈ Cu2+ for ATPase activity; Ca2+ > Mg2+ > Ni2+ ≈ Co2+ > Mn2+ ≈ Cu2+ > Cd2+ ≈ Zn2+ for ADPase activity. The activities were completely inhibited by the addition of 1 mm EDTA (data not shown). Therefore, GS52 apyrase activity is dependent on divalent ions as cofactors, with a preference for Ca2+.
Figure 1.
Characterization of soybean GS52 apyrase enzyme activity. A, Effects of ATPase inhibitors on GS52 apyrase activity. Histograms show mean ± se as relative values to those of mock treatment controls. The concentrations of inhibitors used are as follows: 10 mm NaF (F), 1 mm NaMoO4 (Mo), 10 mm NaN3 (N3), 50 mm NaNO3 (NO3), 1 mm ouabain, and 1 mm Na3VO4 (VO4). Asterisks indicate statistically significant differences compared with the controls: * P < 0.05, ** 0.001 < P < 0.01, *** P < 0.001. B, Effects of divalent ion cofactors on GS52 apyrase activity. ATPase or ADPase activity was measured in the presence of 1 mm divalent cations (Ca2+, Cd2+, Co2+, Cu2+, Mn2+, Mg2+, Ni2+, and Zn2+). C, Linear Michaelis-Menten plot and Lineweaver-Burk plot for enzymatic turnover of ATP and ADP by GS52 apyrase. The nucleotide concentration ranged from 0.1 to 3 mm. D, Substrate specificity of GS52 against nucleoside triphosphates (black bars), nucleotide diphosphates (gray bars), or a nucleoside monophosphate, AMP. N.D., Not detected. Different letters indicate significantly different results at P < 0.05. All experiments were performed at least three times.
The ADPase activity of GS52 was consistently more than 1.5-fold higher than the ATPase activity (Fig. 1B) and also showed higher affinity for ADP (i.e. ATPase, Km = 424 ± 24 μm, Vmax = 38.2 ± 1.9 μmol h−1 mg−1, Kcat/Km = 1.28 × 103 m−1 s−1; ADPase, Km = 309 ± 25 μm, Vmax = 65.8 ± 6.9 μmol h−1 mg−1, Kcat/Km = 3.02 × 103 m−1 s−1; Fig. 1C). Indeed, testing of GS52 activity against a broader range of nucleotide substrates showed a consistent pattern of greater activity against diphosphate nucleotides in comparison with triphosphate nucleotides (Fig. 1D). The GS52 enzyme also demonstrated higher activity against pyrimidine nucleotides, especially the cytidine nucleotides CTP and CDP, with the following kinetics values: CTPase, Km = 522 ± 32 μm, Vmax = 144.9 ± 10.1 μmol h−1 mg−1, Kcat/Km = 3.94 × 103 m−1 s−1; CDPase, Km = 1,056 ± 96 μm, Vmax = 232.6 ± 12.8 μmol h−1 mg−1, Kcat/Km = 3.12 × 103 m−1 s−1 (data not shown). The specificity constant Kcat/Km for ADP, CTP, and CDP was more than 3-fold higher in comparison with ATP, indicating that the enzyme acts on pyrimidine nucleotides and diphosphate nucleotides with higher catalytic efficiency. Phosphate released from AMP by GS52 was undetectable, suggesting that the enzyme possesses no 5′-nucleotidase activity.
Structural Analysis of the GS52 Protein Model
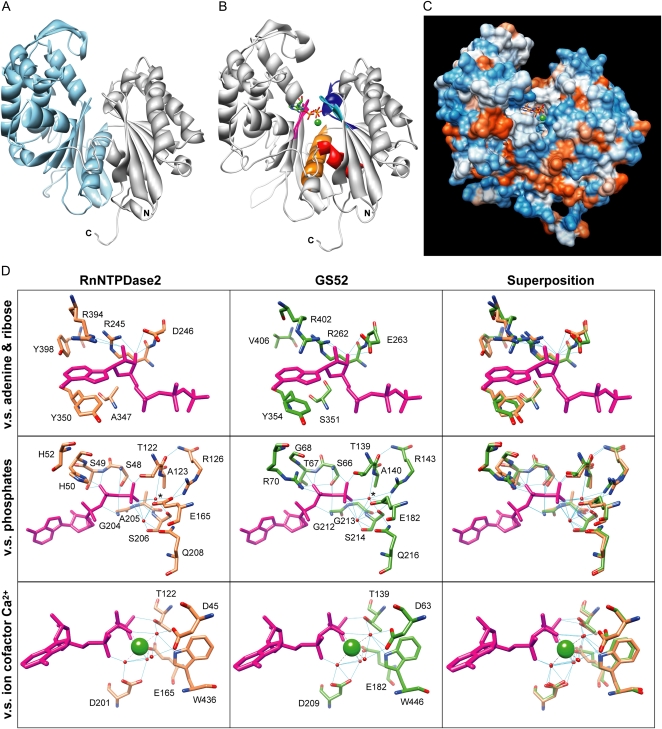
A putative three-dimensional (3-D) structure model for wild-type GS52 protein was created to understand the structural features of the protein. Searching in the Protein Data Bank (PDB), seven candidates (PDB codes 3cj1, 3aap, 1t6c, 3cer, 3mdq, 1u6z, and 3hi0) were found with low E-values (threshold < 0.001) against the GS52 protein sequence. In particular, two templates, 3cj1 (Rattus norvegicus NTPDase2; RnNTPDase2) and 3aap (Legionella pneumophila NTPDase; Lp1NTPDase), for which crystal structure coordinates were published recently (Zebisch and Sträter, 2008; Vivian et al., 2010), were selected for molecular modeling of the apo-form GS52 protein due to their high sequence identity (more than 25%), lowest E-values (0), and high probability of true positive hits (100%). The pairwise alignments showed that 394 and 350 amino acid residues of GS52 matched/aligned with that of templates 3cj1 and 3aap, respectively (data not shown). The corresponding ranges of aligned residues in the target and templates are as follows: 53 to 468 in GS52 versus 30 to 456 in 3cj1; 57 to 457 in GS52 versus 3 to 352 in 3aap. The predicted secondary structures were entirely conserved especially with 3cj1 in the ACRs (Supplemental Fig. S2A) and also with 3aap (data not shown). In order to construct the structural model for the GS52, we applied a multiple sequence alignment method (Martí-Renom et al., 2000) to align the GS52 and template sequences and modified misaligned regions in the alignment. Finally, homology modeling of the tertiary structure of the GS52 (residues 53–467), based on the crystal structures of 3cj1 and 3aap, was performed (Fig. 2A). The model was optimized by several steps of energy minimization of side chains and loop regions. The quality score of the models predicted by the modelEvaluator tool was 0.849, indicating that the model is close to the native structure (=1.00) and highly reliable. The final model of GS52 was superimposed onto the 3-D structure of the template 3cj1 for further verification (Supplemental Fig. S2B). Despite low sequence identity between GS52 and 3cj1, their overall folds were largely conserved. The root mean square differences in 389 atom pairs and in overall alignment between GS52 and 3cj1 were 0.514 and 0.564 Å, respectively. In a similar procedure of the apo-form modeling, the docking form model of GS52, which is a tertiary complex with the nonhydrolyzable ATP analog AMPPNP and cofactor Ca2+, was generated from the docking form of RnNTPDase2 and Lp1NTPDase (PDB codes 3cja and 3aar, respectively; Fig. 2B; Supplemental Fig. S2C). In this case, the coordinates of the substrate complex were taken from that of RnNTPDase2 (3cja). The quality score was 0.851 for the docking form. The structural difference between GS52 and 3cja was 0.466 and 0.499 Å in root mean square difference in 389 atom pairs and overall alignment, respectively. The structural homology and the sequence conservation of ACRs suggest that GS52 possesses the same scaffold as RnNTPDase2, while subtle differences in sequence and structure mold the enzymatic activity.
Figure 2.
Molecular modeling of the GS52 protein structure. A, The 3-D model for the apo form of GS52 apyrase. Residues 53 to 468 of GS52 are visualized with domain I (53–178 and 435–468) and domain II (179–434), which are colored in gray and blue-white. The models are oriented so that the membrane would be located below the protein. B, Docking form model with a substrate (shown in stick form) and a cofactor ion, Ca2+ (shown as a ball). The ACR1 to -5 domains are highlighted in the colors cyan, blue, orange, magenta, and red, respectively. C, Molecular surface colored by electrostatic surface potential. A substrate and Ca2+ are shown as sticks and a ball, respectively. D, Comparison of substrate-binding site between GS52 (middle panels) and RnNTPDase2 (left panels). The C-α coordinates of the residues that are involved in substrate binding are shown with a substrate (purple), water molecules (red balls), and a cofactor ion (a green ball). Superposed residues between Lp1NTPDase2 (orange) and GS52 (green) are shown in the right panels. Blue lines represent the hydrogen bonds. The putative nucleophilic water is labeled with an asterisk (middle row).
For more detailed analysis of the GS52 protein model, the substrate-binding site on the model was compared with that of RnNTPDase2. The binding site for the substrate and divalent ion, which is composed of 20 amino acid residues (Fig. 2D), is located in the cleft between the two domains (Fig. 2, A and B) and is quite accessible from the surface (Fig. 2C). Most of the coordinate or usage of residues was entirely conserved between GS52 and RnNTPDase2, although there were a few differences (Fig. 2D). For example, we found several differences in residue usage in the binding site. In the hydrophobic adenine-binding pocket (which is formed by R245, A347, Y350, R394, and Y398 on RnNTPDase2), V406 on GS52 (a corresponding residue to Y398) likely does not participate in the pocket due to its shorter side chain (Fig. 2D, top row). The distance from adenine ring C2 to the methyl group of the side chain on V406 of GS52 is 8.98 Å, whereas the distance to the hydroxyl group on the aromatic ring side chain of Y398 is 3.72 Å. At position A347 of RnNTPDase2, polar hydrophilic residue S351 was used in the pocket on GS52 (Fig. 2D). These residues used in the GS52 enzyme likely reduce the hydrophobicity of the pocket and the specificity of the enzyme toward the adenine base. This result supports our biochemical results, in which GS52 showed broader substrate specificity (Fig. 1D) and a relatively higher Km value (Fig. 1C) compared with the apyrases of Arabidopsis (e.g. the Km value of AtAPY1/AtAPY2 for ATPase activity is more than 10 times lower than that of GS52; Steinebrunner et al., 2000). Interestingly, these residues found in the binding site for the nucleotide base are highly conserved among the legume-specific apyrases (Supplemental Fig. S3) and may be a defining feature of these enzymes.
At the corresponding positions of H50 on RnNTPDase2 that interact with the β-phosphate of the substrate, G68 was positioned on GS52 (Fig. 2D, middle row). Its coordinate in the model does not appear to interact with the substrate’s phosphate tail. Instead, we found that R70, which has a flexible side chain, is likely involved in binding of the phosphate tail because the side chain of this residue points toward the interdomain cleft and has a predicted hydrogen bond with the β-phosphate. The coordinate of the side chain of R70 approximately corresponds to H50 of RnNTPDase2 (see the middle superimposed picture in Fig. 2D). Interestingly, an R70 side chain rotamer can interact with the α-phosphate of the substrate through a hydrogen bond (data not shown). This result is supported by the fact that a Lys of the corresponding position on actins interacts with the α-phosphate (Vorobiev et al., 2003). T67 on GS52 is also different from the corresponding residue S49 on RnNTPDase2 in the binding site for the phosphate tail. It is conceivable that these substitutions might be directly related to the increased specificity of GS52 for nucleoside diphosphates. Taken together, these results suggest that the use of different amino acid residues at several positions in the binding pocket of GS52 likely defines the enzymatic characteristics described in Figure 1D.
Comparison of Structure and Enzymatic Activity between Wild-Type and Mutant GS52 Enzymes
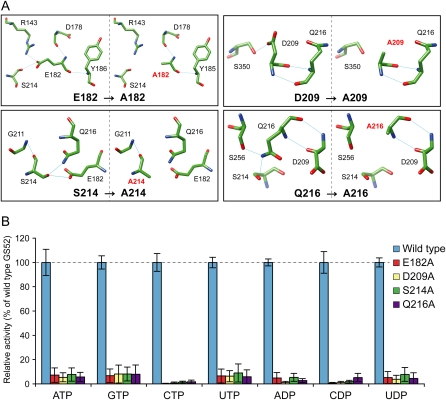
Using PCR-based site-directed mutagenesis, four mutant forms of the GS52 apyrase were constructed in which Ala residues were substituted for E182, D209, S214, or Q216. E182 is positioned within an α-helix in ACR3, and the three other residues, D209, S214, and Q216, are located in the two β-strand regions that are separated by a coil in ACR4 (Supplemental Fig. S2A). Similar mutations in mammalian ecto-NTPDases were previously shown to completely inactivate enzyme activity (Smith and Kirley, 1999; Drosopoulos et al., 2000; Yang et al., 2001; Kirley et al., 2006). The critical nature of these residues for enzyme catalysis is also supported by the GS52 molecular model. For example, while several GS52 residues appear to position the nucleophilic water molecule (which is critical to initiate nucleotide hydrolysis for direct in-line nucleophilic attack on the terminal phosphate of the substrate), the water molecule forms direct hydrogen bonds with the side chain of E182 and S214 and water-mediated hydrogen bonds with the side chain of Q216 and the backbone carbonyl group of S214 (Fig. 2D). The phosphate moieties of the substrate form direct hydrogen bonds with the backbone amino group of S214 as well as water-mediated hydrogen bonds with the backbone carbonyl group of S214 and with the side chains of E182, D209, S214, and Q216 (Fig. 2D). The cofactor ion is also bound to the side chains of E182 and D209 via water-mediated hydrogen bonds (Fig. 2D). Substrate binding and cofactor coordination by these residues suggest that mutations of these residues are likely to disrupt enzymatic activity.
Prior to in vitro and in vivo experiments using the GS52 mutant constructs, the mutational effects were computationally evaluated also on the protein structure (Fig. 3A). In addition to losing interaction with a nucleophilic water molecule and phosphate groups of the substrate, all mutants that we generated (E182A, D209A, S214A, and Q216A) are predicted to change the number of hydrogen bonds between the mutated residue and the other residues (Fig. 3A). For example, the D209A mutant protein lost the hydrogen bonds across D209 with S350. Similarly, mutation in E182, S214, and Q216 removed hydrogen bonds with R143/S214, E182/S216, and S214/S256, respectively. These results suggest that the mutant proteins are likely to exhibit low structural stability at the active site due to the loss of hydrogen bonds at each mutated residue (Fig. 3A). Mutational analysis by the MUpro program, a prediction tool of protein stability changes for single-site mutations (Cheng et al., 2006), predicted that the mutations would decrease the structural stability by the following confidence scores: −0.570 for E182A, −0.565 for D209A, −0.551 for S214A, and −0.573 for Q216A. Indeed, a delocalized conformation change in mutants of human NTPDase3, which has mutations corresponding to E182A and Q216A of GS52, has been reported (Yang et al., 2001). These decreased structure stabilities are expected to contribute to the reduction of protein accessibility or affinity for the substrate in addition to loss of direct interaction with the substrate.
Figure 3.
Comparison of core site structure and enzymatic activity between wild-type and mutant GS52 proteins. A, Structural comparison of the mutated residue and its surrounding residues between the wild type and four mutant proteins. Blue lines represent hydrogen bonds. Note that changes were detected in the number of hydrogen bonds in the mutated proteins as compared with that of the wild type. B, The enzymatic activities of the bacterially expressed recombinant proteins of wild-type and mutant GS52s were assayed as in Figure 1. Results are expressed as activity percentage relative to wild-type GS52. Values represent means ± se from at least three independent experiments.
The in silico predictions of mutant GS52 activity were validated by enzyme assays using recombinant proteins expressed in E. coli (Fig. 3B; Supplemental Fig. S1B). These assays demonstrated very low (less than 10% of the wild type) or undetectable activity in all mutants.
GS52 Enzymatic Activity Is Required for Enhanced Nodulation in Transgenic Roots
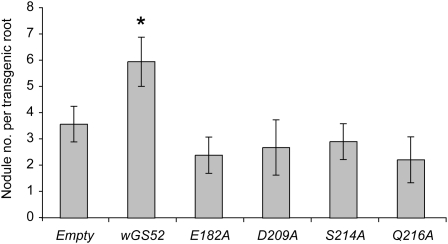
Consistent with a previous study in which the GS52 gene was ectopically expressed in L. japonicus roots (McAlvin and Stacey, 2005), transgenic roots of either M. truncatula or soybean expressing the wild-type GS52 gene from the strong figwort mosaic virus promoter produced significantly higher (greater than 1.5-fold) nodule numbers compared with roots transformed with the empty vector (Fig. 4; Supplemental Fig. S4). In contrast, none of the roots expressing the mutant GS52 genes demonstrated a higher number of nodules (Fig. 4; Supplemental Fig. S4). These results indicate that GS52 enzymatic activity is crucial for the enhancement of nodulation seen in the transgenic roots.
Figure 4.
Nodule formation on transgenic soybean roots expressing wild-type or mutant GS52 proteins. The wild-type GS52 gene (wGS52) or mutated versions (E182A, D209A, S214A, and Q216A) were expressed in soybean roots by hairy root transformation. The histogram shows mean values with se of nodule numbers in transgenic roots (n ≥ 15) 6 weeks after inoculation with B. japonicum. The asterisk indicates a statistically significant difference compared with the empty vector control (P < 0.05). Data are representative of three independent experiments.
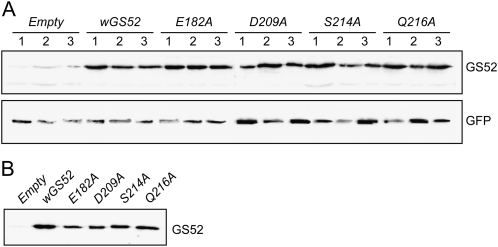
Changes in the GS52 protein sequence could affect in planta protein stability, resulting in reduced enzyme levels or targeting of the protein to the membrane. As shown in Figure 5A, each transgenic protein was expressed strongly and equally in all transgenic root tissues based on western blot using an anti-GS52 antibody, whereas only the native level of GS52 protein was seen in roots transformed with the empty vector. Similar results were obtained when microsomal fractions were assayed (Fig. 5B). Therefore, these control experiments suggest that the mutant proteins were expressed at levels comparable to the wild type and targeted appropriately to the microsomal membrane fraction, probably to the plasma membrane. Gene expression was also analyzed by quantitative real-time reverse transcription (RT)-PCR in the transgenic roots expressing both wild-type GS52 and the mutant proteins. In all cases, mRNA levels were high and comparable (Fig. 6). Therefore, the data suggest that the inability of the mutant proteins to increase nodulation in transgenic roots is due to the loss of enzymatic activity and not due to differences in gene or protein expression.
Figure 5.
Expression levels of GS52 proteins in transgenic soybean roots expressing wild-type and mutant GS52 genes. GS52 expression levels of wild-type GS52-expressing and mutant GS52-expressing transgenic roots were assessed by western-blot analysis using an anti-GS52 polyclonal antibody. A, Ten micrograms of crude protein preparation (S10 fraction) from each of three individual transgenic roots were loaded. The transgene marker, GFP, was used as an internal control. B, One microgram of membrane-derived microsomal protein (P100 fraction) was loaded.
Figure 6.
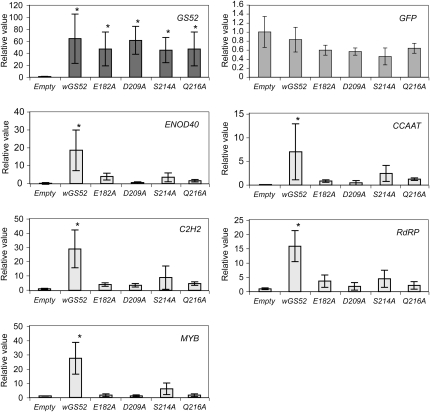
Gene expression levels of nodulation marker genes in transgenic soybean roots expressing wild-type and mutant GS52s. Expression levels of marker genes expressed during nodule organogenesis were measured by quantitative real-time RT-PCR. Data were normalized by the reference gene Cons6 (see “Materials and Methods”) and converted into a value relative to that of the empty vector control. The following genes were tested: ENOD40 (early nodulin 40), C2H2 zinc finger (Glyma13g40240), MYB transcription factor (Glyma17g07330), CCAAT box (Glyma10g10240), and RdRP (RNA-dependent RNA polymerase; Glyma01g01210). Expression levels of GS52 and GFP are shown as controls.
In addition to the production of a visible nodule structure, nodulation is also accompanied by the strong expression of specific plant genes (Libault et al., 2009). As shown in Figure 6, a number of genes known to be induced during nodulation (i.e. ENOD40, C2H2 zinc finger protein gene, CCAAT-binding transcription factor gene, MYB transcription factor gene, and RdRP) exhibited markedly higher expression levels in transgenic roots expressing the wild-type GS52 gene. In contrast, these genes were expressed in roots expressing the mutant GS52 proteins at levels similar to those transformed with the empty control vector (Fig. 6). These results indicate that the increase in expression of nodulation-specific genes is due to the stronger nodulation response exhibited in those plants transformed with the wild-type GS52 gene.
DISCUSSION
In this study, we addressed the question of whether the enzymatic activity of GS52 is essential for nodulation. The hypothesis was substantiated by mutation studies and biochemical analyses. Our results provide crucial information to help define the specific function of the GS52 apyrase in the nodulation process.
It is worth noting that the GS52 enzyme expressed and purified from E. coli lacked the transmembrane domain and would be expected to also lack the side chain glycosylation known to occur in planta. It was reported previously that the bacterial recombinant ecto-apyrases of pea (Pisum sativum) and Arabidopsis exhibited enzymatic activity (Steinebrunner et al., 2000; Kawahara et al., 2003). Therefore, the data suggest that neither the transmembrane domain nor glycosylation is required for enzymatic activity. The same is the case in animals, where bacterially expressed NTPDase1, -2, and -3 exhibited similar biochemical properties to those isolated directly from animal tissue (Zebisch and Sträter, 2007).
The recent crystallization of two ecto-NTPDases, RnNTPDase2 and Lp1NTPDase (Zebisch and Sträter, 2008; Vivian et al., 2010), provided excellent structural templates for homology modeling of the GS52 protein structure. The availability of these more recent structures allowed more detailed and reliable modeling relative to the former modeling of the potato apyrase (Kozakiewicz et al., 2008), which was constructed based on distant bacterial apyrase proteins (PDB code 1t6c/1t6d). The high-quality, 3-D model of GS52 apyrase provided predictions of the critical amino acid found within the active site of the protein (Fig. 2D). Perhaps most interesting is the finding that the legume-specific clade of ecto-apyrase, which includes GS52, has conserved amino acid residues in the binding site of the nucleotide base (Supplemental Fig. S3). This defines the basis of the hydrophobic interaction between the adenine base and the binding pocket on GS52 and implies the low base specificity of the enzyme, which may enable the utilization of both purine and pyrimidine nucleotides. These structural predictions were validated by the analysis of GS52 enzyme activity (i.e. GS52 exhibited broader specificity for all types of nucleoside triphosphates and diphosphates; Fig. 1D). Specifically, GS52 showed higher activity toward diphosphate nucleotides (approximately 1.5 times higher than for ATP) and the highest activity for CTP and CDP (approximately four times higher than for ATP). These characteristics are found in other legume-specific apyrases. For example, Etzler et al. (1999) reported that the D. biflorus ecto-apyrase has broad specificity for nucleoside triphosphates and diphosphates (including GTP, CTP, UTP, and ADP in addition to ATP). Likewise, the pea and cowpea (Vigna unguiculata) apyrases, PnAPY1 and VsNTPDase1, which are also members of the legume-specific apyrase family (Cannon et al., 2003), exhibited similar broad substrate specificity as GS52 (Kiba et al., 2006; Takahashi et al., 2006). More interestingly, the amino acid residues comprising the hydrophobic pocket for the adenine ring are grouped differently in legume-specific and nonspecific apyrases, whereas the other residues involved in the substrate binding are conserved among all plant apyrases (Supplemental Fig. S3). The other plant apyrases harbor stronger hydrophobic residues in their adenine-binding pocket in comparison with the legume-specific apyrases. Further structure-function studies of the GS52 enzyme (e.g. comprehensive mutational analysis in the substrate-binding pocket) are required to disclose the unique catalytic characteristics of the legume-specific apyrases.
The biochemical features lead to the hypothesis that nucleotides other than ATP (e.g. cytidine nucleotides or nucleoside diphosphates) may be important for the legume-Rhizobium symbiosis. This hypothesis is consistent with the report that exogenous addition of ADP, but not ATP, rescued nodulation on RNA interference GS52 soybean roots as well as stimulating nodulation when added to wild-type roots (Govindarajulu et al., 2009). As discussed in this paper, the data suggest that GS52 apyrase activity serves to maintain an optimal extracellular concentration of nucleotides (presumably ADP) that is crucial for the rhizobial infection. The release of ATP from M. truncatula root hairs was visualized using a reporter protein composed of a cellulose-binding domain fused to the ATP-requiring luciferase enzyme (Kim et al., 2006). Given that nucleotides other than ATP are detectable by the enzyme (Manandhar and Van Dyke, 1974), it is possible that other nucleotides are released from the tips of legume root hair cells.
Previous mutational studies based on the biochemical analysis of mammalian NTPDases revealed that mutations at amino acid residues corresponding to E182, D209, S214, and Q216 on GS52 completely inactivated enzymatic function (E174 and S218 for NTPDase1; E182, D219, S224, and Q226 for NTPDase3; Kirley et al., 2006). These data were interpreted to indicate a loss of nucleotide binding due to a conformational change in these mutants. The recent crystal structure of RnNTPDase2 (Zebisch and Sträter, 2008) demonstrated that these residues (E165, D201, S206, and Q208 on NTPDase2) have critical interactions with the substrate complex (nucleotide, cofactor ion, and the nucleophilic water). In agreement with previous reports, computational modeling of the GS52 structure predicted that mutations in the corresponding residues in GS52 would cause a loss of catalytic activity due to loss of the interaction with the substrate complex as well as disruption of the structural stability of the enzyme protein (Fig. 3A). This was responsible for the loss of enzymatic activity in the GS52 mutants (Fig. 3B).
Consistent with data from L. japonicus (McAlvin and Stacey, 2005), M. truncatula or soybean roots transformed with wild-type GS52 formed significantly more nodules when inoculated with the compatible Rhizobium. In contrast, no increase in nodulation was found in roots expressing the four inactive apyrase proteins (Fig. 4; Supplemental Fig. S4). In all cases, the level of gene and protein expression was equal in these different plants, and the apyrase proteins were found in the membrane fraction (Fig. 5). Hence, the data clearly indicate that GS52 apyrase enzymatic activity is crucial for the stimulation of rhizobial infection and nodulation seen in the transgenic roots. Therefore, the data are consistent with a role for the GS52 ecto-apyrase in maintaining extracellular nucleotide levels to maximize the symbiotic interaction. The key rhizobial signal required for nodulation is the lipochitooligosaccharide Nod factor, which is essential for the induction of many of the early nodulation responses (Oldroyd and Downie, 2008; Hamel and Beaudoin, 2010). A key event triggered by Nod factor addition is calcium spiking in the root hair, whose frequency and amplitude are apparently a crucial, specific signal to initiate downstream intracellular signaling events (Oldroyd and Downie, 2004). Another early root hair-associated event in response to either Nod factor addition or rhizobial inoculation is the release of reactive oxygen species and nitric oxide (Gibson et al., 2008). The release of these compounds is also induced by exogenous addition of ATP (Tanaka et al., 2010a). In addition, calcium oscillations resembling, but not identical to, the calcium spiking seen in legume root hair cells upon Nod factor addition were seen in Arabidopsis roots upon the addition of nucleotides (Tanaka et al., 2010b). Therefore, at least superficially, some of the responses attributed to Nod factor addition in legumes are also features of the plant response to extracellular nucleotides. The addition of Nod factors to root hairs of M. truncatula triggers extracellular nucleotide release (Supplemental Fig. S5). Hence, it is tempting to speculate that Nod factor-triggered release of nucleotides may be causal for some of the early nodulation responses and that ecto-apyrases act to modulate the levels of extracellular nucleotides to maximize their effects. A key to testing this hypothesis will be to learn more about how plants recognize and respond to extracellular nucleotides and then to apply this knowledge to the Rhizobium-legume symbiosis.
MATERIALS AND METHODS
Molecular Modeling
For molecular modeling of the GS52 protein, the HHPred protein homology detection Web server (Söding et al., 2005) was used to detect available homologous template proteins in the PDB whose structures were characterized. The templates were selected based on their E-values (i.e. the expected probability of a hit being a random nonhomologous protein in the searched database). The homologous templates with very low E-values (e.g. as low as 0) were chosen as templates in order to build the tertiary structure models for the GS52 protein. The secondary structure of GS52 generated by the HHPred Web server was compared with the positions of α-helices and β-strands in the crystal structures of template proteins. An alignment of the target GS52 sequence to the template structures was constructed by the structure-sequence alignment module of the modeler program (Eswar et al., 2008). A model of GS52 was built from the alignment using the modeler program. The model was optimized by several cycles of energy minimization and careful manual curing of the coordinates of several amino acid residues in the 3-D structure of GS52 to improve the quality of the model. The absolute quality score of the model (i.e. the expected similarity between the model and the unknown native structure) in terms of the global distance test-total score (Zemla, 2003), a standard score between 0 and 1 used to quantify the structural similarity ranging from completely dissimilar to identical, was assessed by the modelEvaluator program (Wang et al., 2009). The models of GS52 shown in the figures were created using the Chimera 1.5 software (Pettersen et al., 2004). The mutation tool of the Chimera program allowed us to change amino acid side chains with a rotamer library.
Gene Constructs
PCR-based mutagenesis using a type IIs restriction enzyme was carried out according to a published protocol (Ko and Ma, 2005). To generate each mutant, the GS52 gene was amplified as two separate PCR fragments using designed primers (Supplemental Table S1), each of which was produced by either a primer pair that included wild-type GS52 (wGS52) forward primer and a mutagenic reverse primer or a pair that included wGS52 reverse primer and a mutagenic forward primer. The fragments were digested with the SapI (New England Biolabs), a type IIs restriction enzyme. The digested fragments were then ligated using T4 DNA ligase. The ligated fragments were purified and cloned into the EcoRV site on pBluescript II SK+. Each mutation was confirmed by sequencing.
To express a hexa-His-tag fusion protein of wild-type or mutant enzymes in Escherichia coli, the DNA fragment carrying the coding region (lacking the transmembrane region) was PCR amplified from the each clone in pBluescript II SK+ (see above) using primers 5′-CGCGGATCCGACCCAATATCATGATGGGAACATC-3′ and 5′-CGCGGATCCTTAAATAAAATACATTAATCGATC-3′ (BamHI sites are underlined). The PCR product was digested with BamHI and cloned into the pET15b (+) or pET22b (+) vector (Novagen). The constructs were again sequenced to confirm orientation and expected nucleotide substitution.
To generate constructs for Agrobacterium rhizogenes-mediated hairy root transformation, DNA fragments for full-length wGS52 or mutagenized GS52 genes (mGS52s) were cloned into the pCGT3305 vector (kindly provided by Dr. Christopher G. Taylor). Subsequently, the fragment containing the figwort mosaic virus promoter, wGS52 or mGS52, the hemagglutinin epitope tag sequence, and the NOS terminator was excised from the vector with AscI and AsiSI restriction enzymes (New England Biolabs) and cloned into the pAKK1467B transformation vector (Govindarajulu et al., 2008). This vector contains a GFP reporter driven by the constitutive superubiquitin promoter; thereby permitting the detection of transgenic roots by GFP expression.
GS52 Recombinant Protein Expression and Purification
Each recombinant plasmid was introduced into E. coli strain BL21 (DE3) pLysS or Rosetta (Novagen). Transformed strains were cultured at 37°C for 3 to 4 h (optical density at 600 nm = approximately 0.6) and further incubated at 20°C for 4 h after addition of isopropyl β-d-thiogalactopyranoside at 1 mm final concentration. Cells were disrupted through a precooled French press cell; the lysate was then separated into soluble and insoluble fractions by centrifugation. Expressed proteins were confirmed by 10% SDS-PAGE. The resulting protein was purified by immobilized metal affinity chromatography using Talon resin (Clontech), following the manufacturer’s manual.
Refolding of GS52 proteins from inclusion bodies was performed according to the refolding cycloamylose method (Machida et al., 2000). Washed inclusion bodies were resuspended and denatured using guanidine hydrochloride. Four refolding reactions using different surfactant solutions were set up to determine which conditions allowed for successful refolding of the protein; the conditions tested were 0.05% (v/v) Tween 40, Tween 60, cetyltrimethylammonium bromide, or myristylsulfobetaine, all in 50 mm Tris buffer with 2 mm DL-cystine. After 1-h reactions, cycloamylose solution (Takara Bio) was added at a final concentration of 0.6%, and the preparations were incubated overnight. Solutions were centrifuged at 12,000g for 10 min, and supernatants were collected and tested using the apyrase assay described below. Based on these enzyme assays, we selected the protein sample from the refolding reaction with Tween 40.
Further purification was performed by gel filtration through Sephadex G-25 resin (GE Healthcare Life Sciences) and ultrafiltration with 30-kD cutoff Vivaspin columns (Sartorius). Protein content was determined according to the method of Bradford (1976) using a Bio-Rad Protein Assay Kit and bovine serum albumin as a standard.
Apyrase Assay
Activity assays, unless otherwise mentioned, were performed at 37°C for 30 min in 25 μL of a solution containing 40 mm succinate buffer or Tris-MES buffer (pH 6.5), 1 mm CaCl2, and 1 mm of the corresponding substrates. The reaction was initiated by addition of purified enzyme and incubated under the conditions indicated in each of the figure legends. Reactions were terminated by the addition of an equal volume of 10% (v/v) trichloroacetic acid and chilling on ice. Fifty microliters of ferrous sulfate-ammonium molybdate reagent (Taussky and Shorr, 1953) was added to each sample, and the A660 was measured with a Biotek Synergy HT spectrophotometer (BioTek). The absorbance data were calculated in comparison with that of phosphorus standards (Sigma) to determine the amount of released phosphate.
Hairy Root Transformation and Nodulation Assays
Hairy root transformation and nodulation assays were performed as described in our previous publication (Govindarajulu et al., 2008). Briefly, apical stem cuttings consisted of a shoot apex and a single fully expanded trifoliate leaf from 2-week-old soybean plants (Glycine max ‘Williams 82’) and were inserted into rock wool cubes (FibrGro), which were inoculated with A. rhizogenes strain K599 transformed with the apyrase GS52 constructs or empty vector (as a control). After 1 week, shoots with transgenic hairy roots were transferred to individual pots containing sterile vermiculite-perlite. After 2 weeks of plant growth, a bacterial suspension of Bradyrhizobium japonicum (USDA110) was used to inoculate each pot (10 mL of B. japonicum suspension with optical density at 600 nm = 0.8). Four weeks after inoculation, transgenic roots were isolated based on the detection of GFP fluorescence using a Leica MZFLIII stereomicroscope (Leica) outfitted with a 470-nm excitation/525-nm emission filter set (Chroma). The nodulation phenotype (i.e. nodule number) was analyzed only on these transgenic roots.
Hairy root transformation of Medicago truncatula was performed as described previously (Boisson-Dernier et al., 2001) with certain modifications. M. truncatula (cv Jemalong A17) seeds were sterilized and then germinated in the dark on wet filter paper on vertically aligned plates. Freshly germinated seedlings with radicles (greater than approximately 10 mm) were sectioned approximately 3 mm from the root tip with a sterile scalpel and then inoculated by coating the cut surface with A. rhizogenes transformed with the apyrase GS52 constructs or empty vector control. Inoculated seedlings were grown for 2 to 3 weeks under a 16-h light period at 25°C on B&D medium (Broughton and Dilworth, 1971) with 6% (w/v) Kalys agar (HP696-7470; Kalys) on a slanted 243- × 243- × 18-mm (Nunc) plastic plate. Seedlings with transgenic roots were selected by their GFP fluorescence using the above-mentioned stereomicroscope and then transferred to pots containing sterile vermiculite. Composite M. truncatula seedlings with transgenic roots were inoculated with Sinorhizobium meliloti strain ABS7M (10−6 bacteria mL−1). After 3 weeks of growth, plants were washed with water, and nodule numbers were scored only from roots showing a clear GFP signal.
Quantitative Real-Time RT-PCR Analysis
The synthesis of cDNA, PCR conditions, data analysis, and primer sequences were identical to those used in our previous publications (Govindarajulu et al., 2009; Libault et al., 2009). Primers for Cons6, Cons7, and Cons16 (Libault et al., 2008) were used to normalize gene expression levels.
Western-Blot Analysis
Extracted total protein or microsomal protein was subjected to SDS-PAGE and electrophoretically transferred to microporous polyvinylidene difluoride membranes. Transblots were blocked and subsequently incubated in primary antibody, rabbit anti-GS52 polyclonal antibody (Day et al., 2000) or rabbit anti-GFP polyclonal antibody (Invitrogen). Immunoreactive bands were visualized with horseradish peroxidase-conjugated secondary antibodies using Pierce ECL Substrate (Thermo Scientific). Signals on the immunoblots were collected and analyzed using a Fuji LAS3000 luminescent image system (FujiFilm).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bacterial expression of recombinant GS52 proteins.
Supplemental Figure S2. Comparison of secondary or tertiary structure between GS52 and RnNTPDase2.
Supplemental Figure S3. Partial alignment of GS52, RnNTPDase2, and other plant apyrases.
Supplemental Figure S4. Nodulation phenotype of M. truncatula transgenic roots expressing wild-type or mutant GS52 proteins.
Supplemental Figure S5. Exogenous addition of Nod factors increases the concentration of extracellular nucleotides at the tips of M. truncatula root hairs.
Supplemental Table S1. Primer sequences used for site-directed mutagenesis.
Acknowledgments
We are grateful to Dr. Christopher G. Taylor (Donald Danforth Plant Science Center) for providing the plasmid vectors and Dr. Stanley J. Roux (University of Texas) for providing NGXT191. Special thanks to Dr. Seth D. Findley (University of Missouri) for critical comments on the manuscript and Mr. Zheng Wang (University of Missouri) for technical support in using the modelEvaluator program.
References
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, et al. (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact 21: 631–645 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, McCombie WR, Sato S, Tabata S, Denny R, Palmer L, Katari M, Young ND, Stacey G. (2003) Evolution and microsynteny of the apyrase gene family in three legume genomes. Mol Genet Genomics 270: 347–361 [DOI] [PubMed] [Google Scholar]
- Cheng J, Randall A, Baldi P. (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 62: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Cohn JR, Uhm T, Ramu S, Nam YW, Kim DJ, Penmetsa RV, Wood TC, Denny RL, Young ND, Cook DR, et al. (2001) Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol 125: 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G. (2000) Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant Microbe Interact 13: 1053–1070 [DOI] [PubMed] [Google Scholar]
- Drosopoulos JH, Broekman MJ, Islam N, Maliszewski CR, Gayle RB, III, Marcus AJ. (2000) Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39: requirement of glutamate 174 and serine 218 for enzyme activity and inhibition of platelet recruitment. Biochemistry 39: 6936–6943 [DOI] [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426: 145–159 [DOI] [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB. (1999) A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA 96: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M. (1998) The role of ion fluxes in Nod factor signalling in Medicago sativa. Plant J 13: 455–463 [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. (2008) Molecular determinants of a symbiotic chronic infection. Annu Rev Genet 42: 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Elmore JM, Fester T, Taylor CG. (2008) Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol Plant Microbe Interact 21: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Kim SY, Libault M, Berg RH, Tanaka K, Stacey G, Taylor CG. (2009) GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol 149: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Beaudoin N. (2010) Chitooligosaccharide sensing and downstream signaling: contrasted outcomes in pathogenic and beneficial plant-microbe interactions. Planta 232: 787–806 [DOI] [PubMed] [Google Scholar]
- Hurley JH. (1996) The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct 25: 137–162 [DOI] [PubMed] [Google Scholar]
- Kawahara T, Toyoda K, Kiba A, Miura A, Ohgawara T, Yamamoto M, Inagaki Y, Ichinose Y, Shiraishi T. (2003) Cloning and characterization of pea apyrases: involvement of PsAPY1 in response to signal molecules from the pea pathogen Mycosphaerella pinodes. J Gen Plant Pathol 69: 33–38 [Google Scholar]
- Kiba A, Toyoda K, Yoshioka K, Tsujimura K, Takahashi H, Ichinose Y, Takeda T, Kato T, Shiraishi T. (2006) A pea NTPase, PsAPY1, recognizes signal molecules from microorganisms. J Gen Plant Pathol 72: 238–246 [Google Scholar]
- Kim SY, Sivaguru M, Stacey G. (2006) Extracellular ATP in plants: visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirley TL, Crawford PA, Smith TM. (2006) The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses. Purinergic Signal 2: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JK, Ma J. (2005) A rapid and efficient PCR-based mutagenesis method applicable to cell physiology study. Am J Physiol Cell Physiol 288: C1273–C1278 [DOI] [PubMed] [Google Scholar]
- Komoszyński MA. (1996) Comparative studies on animal and plant apyrases (ATP diphosphohydrolase EC 3.6.1.5) with application of immunological techniques and various ATPase inhibitors. Comp Biochem Physiol B Biochem Mol Biol 113: 581–591 [DOI] [PubMed] [Google Scholar]
- Kozakiewicz A, Neumann P, Banach M, Komoszyński M, Wojtczak A. (2008) Modeling studies of potato nucleoside triphosphate diphosphohydrolase NTPDase1: an insight into the catalytic mechanism. Acta Biochim Pol 55: 141–150 [PubMed] [Google Scholar]
- Libault M, Joshi T, Takahashi K, Hurley-Sommer A, Puricelli K, Blake S, Finger RE, Taylor CG, Xu D, Nguyen HT, et al. (2009) Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol 151: 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1: 44–54 [Google Scholar]
- Machida S, Ogawa S, Xiaohua S, Takaha T, Fujii K, Hayashi K. (2000) Cycloamylose as an efficient artificial chaperone for protein refolding. FEBS Lett 486: 131–135 [DOI] [PubMed] [Google Scholar]
- Manandhar MSP, Van Dyke K. (1974) Delayed luminescence analysis (DLA) of purine and pyrimidine ribose and deoxyribose nucleotide triphosphates in picomole quantities. Microchem J 19: 42–51 [Google Scholar]
- Marcus AJ, Broekman MJ, Drosopoulos JH, Islam N, Pinsky DJ, Sesti C, Levi R. (2003) Metabolic control of excessive extracellular nucleotide accumulation by CD39/ecto-nucleotidase-1: implications for ischemic vascular diseases. J Pharmacol Exp Ther 305: 9–16 [DOI] [PubMed] [Google Scholar]
- Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29: 291–325 [DOI] [PubMed] [Google Scholar]
- McAlvin CB, Stacey G. (2005) Transgenic expression of the soybean apyrase in Lotus japonicus enhances nodulation. Plant Physiol 137: 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- O’Neill SD, Spanswick RM. (1984) Effects of vanadate on the plasma membrane ATPase of red beet and corn. Plant Physiol 75: 586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Plesner L. (1995) Ecto-ATPases: identities and functions. Int Rev Cytol 158: 141–214 [DOI] [PubMed] [Google Scholar]
- Smith TM, Kirley TL. (1999) Site-directed mutagenesis of a human brain ecto-apyrase: evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry 38: 321–328 [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. (2005) The HHPred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner I, Jeter C, Song C, Roux SJ. (2000) Molecular and biochemical comparison of two different apyrases from Arabidopsis thaliana. Plant Physiol Biochem 38: 913–922 [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ. (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Toyoda K, Hirakawa Y, Morishita K, Kato T, Inagaki Y, Ichinose Y, Shiraishi T. (2006) Localization and responsiveness of a cowpea apyrase VsNTPase1 to phytopathogenic microorganisms. J Gen Plant Pathol 72: 143–151 [Google Scholar]
- Tanaka K, Gilroy S, Jones AM, Stacey G. (2010a) Extracellular ATP signaling in plants. Trends Cell Biol 20: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Swanson SJ, Gilroy S, Stacey G. (2010b) Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol 154: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussky HH, Shorr E. (1953) A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem 202: 675–685 [PubMed] [Google Scholar]
- Todorov LD, Mihaylova-Todorova S, Westfall TD, Sneddon P, Kennedy C, Bjur RA, Westfall DP. (1997) Neuronal release of soluble nucleotidases and their role in neurotransmitter inactivation. Nature 387: 76–79 [DOI] [PubMed] [Google Scholar]
- Vivian JP, Riedmaier P, Ge H, Le Nours J, Sansom FM, Wilce MC, Byres E, Dias M, Schmidberger JW, Cowan PJ, et al. (2010) Crystal structure of a Legionella pneumophila ecto-triphosphate diphosphohydrolase, a structural and functional homolog of the eukaryotic NTPDases. Structure 18: 228–238 [DOI] [PubMed] [Google Scholar]
- Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC. (2003) The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA 100: 5760–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tegge AN, Cheng J. (2009) Evaluating the absolute quality of a single protein model using structural features and support vector machines. Proteins 75: 638–647 [DOI] [PubMed] [Google Scholar]
- Windsor B, Roux SJ, Lloyd A. (2003) Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana Nat Biotechnol 21: 428–433 [DOI] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ. (2007) Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol 144: 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Hicks-Berger CA, Smith TM, Kirley TL. (2001) Site-directed mutagenesis of human nucleoside triphosphate diphosphohydrolase 3: the importance of residues in the apyrase conserved regions. Biochemistry 40: 3943–3950 [DOI] [PubMed] [Google Scholar]
- Zebisch M, Sträter N. (2007) Characterization of rat NTPDase1, -2, and -3 ectodomains refolded from bacterial inclusion bodies. Biochemistry 46: 11945–11956 [DOI] [PubMed] [Google Scholar]
- Zebisch M, Sträter N. (2008) Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling. Proc Natl Acad Sci USA 105: 6882–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A. (2003) LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res 31: 3370–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]