Chloroplasts and their nonphotosynthetic relatives in the plastid organelle family evolved from a cyanobacterial endosymbiont (for review, see Timmis et al., 2004). The subsequent coevolution of the chloroplast and nuclear genomes produced an organelle that is eubacterial at its core but with extensive chloroplast-specific embellishments. Of the thousands of genes in the cyanobacterial ancestor, only approximately 100 are retained in chloroplast genomes. These genes fall into three categories: those encoding (1) components of the chloroplast gene expression machinery (RNA polymerase, ribosomal proteins, tRNAs, and rRNAs), (2) subunits of photosynthetic enzymes (Rubisco, PSII, the cytochrome b6f complex, PSI, the ATP synthase, and the NADH dehydrogenase), and (3) proteins involved in other metabolic processes (e.g. ClpP, AccD, Ycf1, and Ycf2). The chloroplast proteome has a complexity of several thousand proteins and is dominated by nuclear gene products that are synthesized in the cytosol and imported into the organelle. Many of these are encoded by genes of cyanobacterial ancestry that were transferred to the nucleus and that have retained their ancestral functions. As a result, the chloroplast gene expression and photosynthesis machineries consist of proteins that are derived from two physically separate genetic systems. Detailed knowledge of chloroplast gene expression and the nucleus-encoded proteins that influence it are prerequisites for understanding nuclear-organellar cross talk and chloroplast evolution, and will aid in optimizing transgene expression in the plastid compartment.
The use of genetic and biochemical approaches, together with the ability to manipulate the chloroplast genome in several species, have brought most aspects of chloroplast gene expression out of the “black box” and into the realm of concrete, mechanistic hypotheses. The intent of this contribution is to highlight new perspectives that have resulted from recent observations and instances in which current data warrant the revision of previous paradigms. For more comprehensive information, I refer the reader to recent reviews of chloroplast RNA metabolism (Stern et al., 2010), transcription (Liere and Börner, 2007; Lerbs-Mache, 2010), and translation (Peled-Zehavi and Danon, 2007). Mechanisms of chloroplast gene expression have been studied primarily in land plants and in the green alga Chlamydomonas reinhardtii. Here, I emphasize findings with land plants, as detailed reviews of chloroplast gene expression in Chlamydomonas have been published in a recent volume (Stern and Harris, 2009).
THE CORE GENE EXPRESSION MACHINERIES IN CHLOROPLASTS RETAIN STRONG RESEMBLANCE TO THOSE IN BACTERIA BUT HAVE ORGANELLE-SPECIFIC EMBELLISHMENTS
The bacterial ancestry of chloroplasts is readily apparent in the organization of chloroplast genomes and in the machineries for chloroplast transcription, translation, and RNA turnover. Polycistronic transcription units that resemble bacterial operons predominate in land plant chloroplasts (Bock, 2007). Chloroplast ribosomes are similar in protein content and antibiotic sensitivities to bacterial ribosomes (Peled-Zehavi and Danon, 2007). A bacterial-type RNA polymerase contributes to chloroplast transcription (Liere and Börner, 2007; Lerbs-Mache, 2010), and chloroplast RNA turnover employs ribonucleases that are derived from those in bacteria (Stern et al., 2010). Superimposed on this bacterial infrastructure are features that were acquired only after the chloroplast became incorporated into a eukaryotic cell. Examples include a plethora of introns, a phage-type RNA polymerase, the modification of mRNA sequences by RNA editing, and the processing of polycistronic primary transcripts to generate complex transcript populations.
CHLOROPLAST GENE EXPRESSION EMPLOYS UNUSUAL RNA-BINDING PROTEINS THAT EMERGED IN THE CONTEXT OF NUCLEAR-ORGANELLAR COEVOLUTION
The analysis of nonphotosynthetic mutants in maize (Zea mays), Arabidopsis (Arabidopsis thaliana), and Chlamydomonas has revealed numerous nucleus-encoded RNA-binding proteins that participate in the expression of chloroplast genes. Two major themes emerged from this large body of work: (1) the repertoire of nucleus-encoded chloroplast RNA-binding proteins is remarkably complex given the small coding capacity of the chloroplast genome; and (2) most such proteins belong to protein families that function almost exclusively in organellar gene expression. Both of these points are exemplified by the pentatricopeptide repeat (PPR) family, whose members are defined by degenerate 35-amino acid helical repeats (for review, see Schmitz-Linneweber and Small, 2008). PPR proteins are not represented in bacteria, and PPR proteins in eukaryotes function almost exclusively in organellar gene expression. The PPR family consists of over 400 members in angiosperms, approximately half of which are predicted to localize to chloroplasts and half to mitochondria. Several other predicted helical repeat protein classes have also been implicated in chloroplast RNA metabolism (Stern et al., 2010); it is likely that these share mechanistic similarities with PPR proteins, so these and PPR proteins are referred to together below as “PPR-like” proteins.
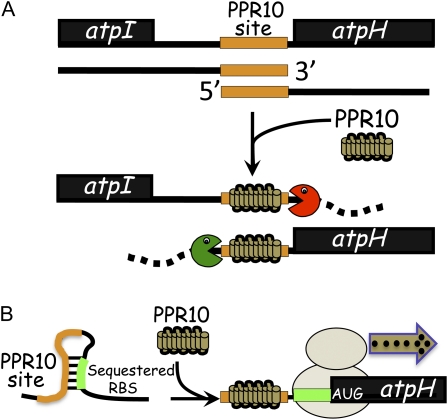
Current data support the view that PPR tracts are sequence-specific RNA-binding motifs that bind single-stranded RNA along a surface formed by stacked, helical repeating units (Schmitz-Linneweber and Small, 2008; Williams-Carrier et al., 2008; Prikryl et al., 2011). Genetic data have implicated PPR proteins in many aspects of organellar RNA metabolism, and it is often suggested that they mediate their multifarious effects by recruiting different effector proteins to specific RNA sites. Indeed, there is good evidence that an appended domain found in a subset of PPR proteins functions in this manner during the process of plant organellar RNA editing (see below). However, recent data also support an alternative view: that many of the functions attributed to PPR proteins may result directly from the unusual nature of the PPR-RNA interface, which sequesters an extended RNA segment such that it cannot interact with other proteins or RNAs (Prikryl et al., 2011; Fig. 1).
Figure 1.
Sequestration of a segment of single-stranded RNA by a long PPR tract may account for many functions attributed to PPR proteins. This is exemplified by PPR10, which stabilizes specific processed RNA termini (A) and enhances translation (B) by sequestering the same RNA segment (Pfalz et al., 2009; Prikryl et al., 2011). Analogous interactions could influence RNA processing, stability, and translation in other ways. A, Site-specific barrier activity of PPR10 defines processed RNA termini. The processed RNA termini derived from polycistronic transcripts spanning atpI and atpH are shown at the top. PPR10 binds this intergenic region and promotes the accumulation of processed RNAs by blocking exoribonucleases intruding from either direction. The processed atpI and atpH transcripts derive from different precursor molecules. B, Site-specific RNA-remodeling activity of PPR10 enhances translation. The atpH ribosome-binding site (RBS) is sequestered in a duplex with a portion of the PPR10-binding site. PPR10 binding frees the atpH ribosome-binding site for translation (Pfalz et al., 2009; Prikryl et al., 2011).
Three additional classes of organellar RNA-binding proteins illustrate a similar theme, albeit on a smaller scale. The CRM (Barkan et al., 2007), PORR (Kroeger et al., 2009), and APO (Amann et al., 2004; Watkins et al., 2011) domains are represented in small plant-specific gene families. All members of these families are predicted to localize to chloroplasts or mitochondria, and all that have been studied bind RNA and promote intron splicing in plant organelles. That PPR, CRM, PORR, and APO proteins are dedicated to promoting organelle-specific steps in RNA metabolism implies a coevolutionary process through which these protein families were spawned in concert with the molecular processes they engender.
CHLOROPLAST GENE EXPRESSION IS REGULATED AT MANY STEPS
The balance of chloroplast-encoded proteins changes in response to developmental and environmental inputs. The developmental component is particularly important in multicellular plants, where members of the plastid organelle family adopt different forms in different cell types. For example, the amyloplasts in potato (Solanum tuberosum) tubers and the chromoplasts in tomato (Solanum lycopersicum) fruit lack chloroplast gene products involved in photosynthesis but maintain the expression of chloroplast genes involved in other aspects of cellular metabolism (Kahlau and Bock, 2008; Valkov et al., 2009). The effects of light and hormones are superimposed on developmental programs to further influence the synthesis of chloroplast gene products.
A key difference between chloroplasts and bacteria concerns the point at which gene expression is controlled: whereas transcription initiation is the most common point of regulation in bacteria, in chloroplasts, the regulation of posttranscriptional steps features prominently (Eberhard et al., 2002). Rapid progress in the identification of nuclear genes required for various steps in chloroplast gene expression has provided many candidates for regulatory factors. Examples of gene regulation and potential regulatory mechanisms are discussed in the context of each step of gene expression below.
The precise regulation of gene expression may be less important in land plant chloroplasts than in bacteria, where the efficient use of resources is likely to have a stronger impact on organismal fitness. In fact, the stoichiometric accumulation of subunits within each photosynthetic enzyme complex is mediated, in part, by proteolysis of unassembled subunits (Adam, 2007). The relative contributions of coordinated protein synthesis versus posttranslational proteolysis has been most thoroughly explored in Chlamydomonas, where a set of mechanisms known as control by epistasy of synthesis (CES) coordinate the synthesis of subunits of each photosynthetic enzyme complex via negative feedback loops that are triggered by specific unassembled subunits (Choquet and Wollman, 2009). The degree to which CES-type mechanisms exist in land plants is unclear. Several unassembled proteins that trigger CES in Chlamydomonas have been shown not to do so in land plants (McCormac and Barkan, 1999; Monde et al., 2000). On the other hand, a CES-like mechanism coordinates the synthesis of the nucleus- and plastid-encoded subunits of Rubisco in tobacco (Nicotiana tabacum; Rodermel et al., 1996; Wostrikoff and Stern, 2007). An alternative strategy for coordinating the expression of multiple chloroplast genes could employ nucleus-encoded proteins that promote the expression of subsets of chloroplast genes (e.g. RNA-binding proteins or σ-factors), as many such proteins have been described (Schmitz-Linneweber et al., 2005; Pfalz et al., 2009; Tillich et al., 2009; Lerbs-Mache, 2010).
CHLOROPLAST TRANSCRIPTION
Large-scale changes in chloroplast transcription occur in response to light, hormonal, and developmental signals (for review, see Liere and Börner, 2007). In barley (Hordeum vulgare), for example, chloroplast transcription peaks early during the differentiation of leaf cells (Baumgartner et al., 1989), and cytokinin and light act synergistically to stimulate the transcription of a subset of chloroplast genes in mature chloroplasts (Zubo et al., 2008). The psbD promoter is activated by blue light (Gamble and Mullet, 1989), and the relative rates of psbA and psaAB transcription change in response to different wavelengths of light, serving to optimize the ratio of PSII to PSI (Pfannschmidt et al., 1999).
Progress in elucidating the chloroplast transcription machinery has provided a basis for addressing the mechanisms underlying the control of chloroplast transcription (for review, see Liere and Börner, 2007). In addition to a bacterial-type, multisubunit plastid-encoded polymerase (PEP), chloroplasts in angiosperms and in the moss Physcomitrella harbor one or two nucleus-encoded RNA polymerases (NEPs). NEPs are single-subunit polymerases related to those in T7-type bacteriophage and in mitochondria. Initial data suggested a division of labor in which NEP transcribes “housekeeping” genes (e.g. genes for tRNAs, rRNAs, ribosomal proteins, PEP, ClpP, and AccD) and PEP transcribes genes involved in photosynthesis (Hajdukiewicz et al., 1997). The organization of chloroplast genes into two such regulons was proposed to comprise a developmental cascade, in which the activation of NEP early in chloroplast development supports the accumulation of chloroplast ribosomes and ultimately of PEP, which then transcribes photosynthetic genes at later stages (Mullet, 1993). This model in its simplest form has not stood the test of time. It is now clear that most chloroplast genes can be transcribed by either NEP or PEP, albeit from distinct promoters (for review, see Liere and Börner, 2007). Nonetheless, the absence of PEP does shift the balance of transcripts toward those involved in gene expression, and NEP-mediated transcription does predominate early in chloroplast development. Furthermore, neither NEP nor PEP is sufficient for the biogenesis of photosynthetically competent chloroplasts (Allison et al., 1996; Swiatecka-Hagenbruch et al., 2008), indicating that some chloroplast genes require one or the other polymerase for adequate expression.
PEP promoters are characterized by consensus sequences that resemble promoters recognized by σ-70 in Escherichia coli. Indeed, PEP promoters are recognized by nucleus-encoded proteins that are related to σ-70 (Lerbs-Mache, 2010; Schweer et al., 2010a). Chloroplast σ-factors are encoded by a small gene family in land plants. The presence of multiple σ-factors offers the potential to differentially regulate chloroplast gene subsets at the transcriptional level. In fact, there is evidence that different members of the σ-family are differentially regulated and target different chloroplast promoters. Nonetheless, the scope of this type of regulation seems to be limited, as reverse-genetic analyses in Arabidopsis demonstrate considerable functional redundancy among the different sigmas.
Evidence for nonredundant functions of each chloroplast σ-factor in Arabidopsis is summarized by Lerbs-Mache (2010). A compelling example involves SIG5. SIG5 is necessary for the use of the blue light-inducible psbD promoter, and SIG5 transcript levels are induced in response to blue light. Thus, the blue light regulation of SIG5 transcription may be sufficient to account for blue light induction of psbD. Another example involves the regulation of σ-factor activity by phosphorylation. SIG1 and SIG6 in Arabidopsis are substrates for phosphorylation in vivo (Shimizu et al., 2010) and in vitro (Schweer et al., 2010b), respectively. SIG1 phosphorylation changes in response to the oxidation state of the plastoquinone pool, and mutation of the phosphorylated amino acids alters the ratio of psbA to psaAB transcription (Shimizu et al., 2010). Several different protein kinases have been proposed to contribute to the regulation of σ factor activity (Ogrzewalla et al., 2002; Puthiyaveetil et al., 2008), but the signal transduction pathways that connect incident light to changes in chloroplast transcription are largely uncharacterized.
Various other proteins have been detected in chloroplasts that bind DNA or that are associated with the chloroplast “transcriptionally active chromosome” (for review, see Liere and Börner, 2007; Lerbs-Mache, 2010). Mutations in several of the corresponding genes compromise chloroplast gene expression (Pfalz et al., 2006). However, more detailed analyses will be required to determine which of these proteins are true transcription factors and which function in other DNA-associated processes or in posttranscriptional processes that are coupled to transcription.
The termination of transcription in chloroplasts has received relatively little attention. Many 3′ RNA termini are not products of transcription termination but rather result from the processing of longer transcripts (for review, see Stern et al., 2010). However, PEP, NEP, and T7 RNA polymerases terminate in vitro at strong intrinsic bacterial terminators, which consist of GC-rich RNA hairpins followed by several contiguous uridines (Chen and Orozco, 1988; Jeng et al., 1990; Kühn et al., 2007). Thus, it seems likely that NEP and PEP respond to sequences with these features in vivo. As only a handful of 3′ regions have been assayed for transcription termination activity, more comprehensive assays may yet uncover efficient transcription terminators in chloroplasts.
RNA SPLICING
A set of approximately 20 introns (one group I and approximately 19 group II introns) was acquired early during the evolution of land plants and is shared by most land plants today (Turmel et al., 2006). Algal chloroplast introns were acquired independently of those in land plants, so the chloroplast intron content in Chlamydomonas is entirely different. All chloroplast introns were derived from “self-splicing” group I or group II ribozymes. However, self-splicing has not been reported for any introns in land plant chloroplasts, and it is now abundantly clear that their splicing is protein dependent.
Group I and group II introns have distinct structures and splicing mechanisms. Both intron types are found in bacteria, albeit in small numbers, where they splice with the aid of conserved intron-encoded proteins called maturases (Pyle and Lambowitz, 2006). Chloroplast introns, however, are degenerate and require interactions with “host”-encoded proteins of diverse evolutionary origin. A parallel to the intron fragmentation that is believed to underlie the evolution of nuclear spliceosomal snRNAs can be seen in the transspliced group II introns in chloroplasts, two of which are found in Chlamydomonas and one in land plants.
Approximately 14 proteins in land plants and three in Chlamydomonas have been identified that are necessary to splice chloroplast group II introns and that associate with intron RNAs in vivo (de Longevialle et al., 2010; Stern et al., 2010). All but one of these proteins are encoded by the nuclear genome, the exception being the maturase encoded in the trnK intron in land plants. It is anticipated that many such proteins promote splicing by guiding intron folding into a catalytically active structure; biochemical data support this view for CRS1 (Ostersetzer et al., 2005), which is required for the splicing of the chloroplast atpF intron in angiosperms. Concepts to emerge from the characterization of chloroplast group II intron ribonucleoprotein particles (RNPs) are summarized below.
Chloroplast Group II Intron RNPs Are Complex
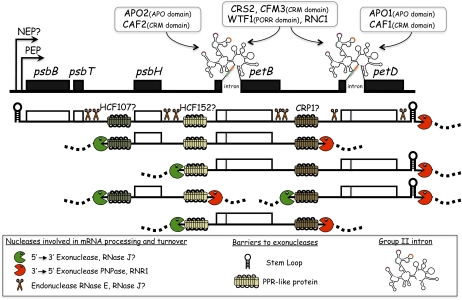
Studies of group I and group II intron splicing in bacteria and yeast revealed simple RNPs involving one or two proteins and an intron that is inherently self-splicing (Pyle and Lambowitz, 2006). By contrast, chloroplast group II intron RNPs are more protein than RNA. For example, six different proteins have been shown to associate with, and to be required for the splicing of, the maize petD intron (Fig. 2). This scenario is typical of the other group II introns in land plant chloroplasts.
Figure 2.
Model for expression of the chloroplast psbB gene cluster. The psbB gene cluster spans five genes and generates approximately 20 processed transcripts via intercistronic processing and the splicing of group II introns in the petB and petD genes (Barkan, 1988; Westhoff and Herrmann, 1988). The diagram shows a subset of the processed transcripts. Splicing factors for the petB and petD introns are indicated above, with their organelle-specific RNA-binding domains specified in parentheses (for review, see de Longevialle et al., 2010; Watkins et al., 2011). The position of a bound PPR-like protein defines the positions of processed RNA termini in each intercistronic region by blocking exoribonucleases; genetic data implicate HCF107, HCF152, and CRP1 in the indicated events (Barkan et al., 1994; Felder et al., 2001; Meierhoff et al., 2003), but these interactions have not been confirmed biochemically. The endonucleases RNAse E and RNAse J are posited to cleave rather nonspecifically at unstructured AU-rich regions to provide exonuclease access to internal RNA regions. A generic version of this model was presented previously, based on the effects of PPR10 on two other transcription units (Pfalz et al., 2009).
There Is No Chloroplast “Spliceosome”
A spliceosome of uniform composition catalyzes the splicing of most nuclear pre-mRNA introns. By contrast, different chloroplast splicing factors act combinatorially to promote the splicing of different intron subsets. Furthermore, the proteins that participate in chloroplast splicing are unrelated to those that participate in nuclear splicing.
Invention of Novel RNA-Binding Domains, and Cooption of Preexisting RNA-Binding Domains for Plant Organellar Splicing
The CRM, PORR, and APO domains were initially recognized to be RNA-binding domains through the analysis of chloroplast RNA splicing, and all analyzed members of these protein families participate in intron splicing in organelles. Thus, the abundance of group II introns in plant organelles may have been the driving force for the evolution of these protein families. On the other hand, several chloroplast splicing factors maintain a strong resemblance to bacterial proteins. Examples include maize CRS2, which is related to bacterial peptidyl tRNA hydrolase, and Chlamydomonas Raa2, which is related to bacterial pseudouridine synthase. Minor evolutionary tinkering was sufficient to impart novel activities upon these ancient proteins.
Splicing as a Regulatory Step?
The regulation of splicing could, in principle, serve to adjust the balance of gene expression in chloroplasts. As for other steps in chloroplast gene expression, it is not known to what extent splicing efficiency limits the final output of gene product. However, weak mutant alleles of several maize chloroplast splicing factors condition a distinct mutant phenotype (Watkins et al., 2007), indicating that these proteins and the splicing events they promote are limiting for gene expression. The degree to which these and other splicing factors perform a regulatory role remains to be determined.
RNA EDITING
Another distinguishing feature of gene expression in land plant chloroplasts is the posttranscriptional modification of mRNA sequences by RNA editing (for review, see Chateigner-Boutin and Small, 2010). mRNA editing has not been observed in bacteria or in algal organelles. However, this property is shared with plant mitochondria, and indeed, RNA editing in plant mitochondria and chloroplasts is believed to have a common evolutionary origin. In angiosperms, chloroplast mRNA editing is limited to the change of specific cytidine residues to uridine and occurs at approximately 40 positions. This is a rapidly evolving feature, as more than half of the edited sites differ between monocot and dicot plants. Chloroplast genomes that “lack” a particular edited site generally encode a uridine at the corresponding position. Most editing events in chloroplasts are important for gene function: some create start codons, and some modify the coding sequence such that a deleterious amino acid is changed to a conserved and functional one.
The chemistry of the editing reaction is believed to involve cytidine deamination (for review, see Chateigner-Boutin and Small, 2010). Thus, the editing machinery has been anticipated to consist of two types of component: (1) “specificity factors” that target a nucleotide for editing, and (2) a deaminase enzyme. Elucidation of the specificity factors began with the discovery that the PPR protein CRR4 is required to edit a nucleotide in the ndhD mRNA (Kotera et al., 2005). CRR4 binds with specificity to a short RNA harboring the edited site (Okuda et al., 2006). The position of the CRR4 binding site correlates well with in vivo and in vitro analyses of cis-elements required for editing: a region of approximately 30 nucleotides is sufficient to specify most edited sites, with the edited nucleotide found near the 3′ end of this region.
CRR4 belongs to a subfamily of PPR proteins that include appended motifs at their C terminus called E and DYW (Lurin et al., 2004). Following the discovery of CRR4, genetic analyses identified many more RNA-editing factors in chloroplasts and also in plant mitochondria. Almost all of these are either PPR-E or PPR-E-DYW proteins. These results strongly implicate both the E and DYW motifs in the editing process. Mutagenesis experiments have confirmed the “specificity” function to reside in the PPR tract and have shown the E domain to be essential for editing (Okuda et al., 2007, 2009). The DYW motif, however, can be deleted from several editing factors without loss of activity (Okuda et al., 2009). It has been proposed that the E-DYW appendage recruits the editing enzyme and/or itself has enzymatic activity (Salone et al., 2007; Hammani et al., 2009; Okuda et al., 2009).
These results paint a picture in which the subset of PPR proteins harboring E or E-DYW extensions are the primary specificity factors for RNA editing in chloroplasts; the PPR tract binds approximately 20 nucleotides upstream of the targeted C residue, and the E-DYW motif promotes editing in an as yet undetermined manner. Some such proteins specify editing at a single site, but many target multiple sites and are proposed to recognize a degenerate sequence (Heller et al., 2008; Hammani et al., 2009). Thus, the approximately 40 PPR proteins of this type in land plant chloroplasts are more than sufficient to account for known editing events. Members of the abundant chloroplast ribonucleoprotein (cpRNP) family of RNA-binding proteins have also been shown to affect several editing sites in vitro (Hirose and Sugiura, 2001) and in vivo (Tillich et al., 2009). cpRNPs bind RNA rather nonspecifically, so the mechanism by which they influence RNA editing is likely to differ from that of PPR editing factors.
An open question concerns the role of RNA editing in chloroplast gene regulation. The results of a comprehensive analysis (Peeters and Hanson, 2002) suggested that the low editing efficiencies in nongreen maize plastids are unlikely to be a limiting factor in gene expression. Another question concerns the evolutionary forces that produced plant organellar RNA editing. An attractive model coined “genomic debugging” (Maier et al., 2008) posits that the ease of evolving new specificity factors for RNA editing (i.e. PPR-E class proteins) allowed the fixation of otherwise deleterious mutations in the chloroplast genome.
mRNA STABILIZATION AND DECAY
The stabilities of chloroplast mRNAs vary considerably and can change in response to light and during leaf development (Klaff and Gruissem, 1991; Baumgartner et al., 1993; Kim et al., 1993). The results summarized below support the view that the stabilizing influence of chloroplast RNA-binding proteins, especially PPR-like proteins, is layered upon an RNA turnover machinery borrowed from bacteria, to determine RNA half-life in chloroplasts.
Chloroplast mRNAs typically survive for many hours (Klaff and Gruissem, 1991; Kim et al., 1993) and are much more stable than are typical mRNAs in bacteria. Nonetheless, the chloroplast ribonucleases that are implicated most strongly in RNA decay are closely related to those in bacteria (Stern et al., 2010). The most thoroughly studied pathway for RNA decay in chloroplasts involves the 3′→5′ exonuclease polynucleotide phosphorylase (for review, see Schuster and Stern, 2009). As in bacteria, chloroplast polynucleotide phosphorylase activity is stimulated by 3′ polyadenylation of its RNA substrate, and it is blocked by stable 3′ RNA structures. Recent results show that chloroplasts also have a protein-based mechanism for stabilizing 3′ termini that has no apparent analog in bacteria: a bound PPR protein can block 3′ exonucleases in vivo and in vitro (Hattori and Sugita, 2009; Pfalz et al., 2009; Prikryl et al., 2011). There is strong genetic evidence for a 5′→3′ exonuclease activity in chloroplasts whose activity can likewise be blocked by a stable RNA structure or bound PPR-like protein (for review, see Stern et al., 2010). The 5′→3′ exonuclease activity has not been identified, but it is likely to reside in chloroplast RNAse J, whose ortholog in bacteria has both endonuclease and 5′→3′ exonuclease activity (Condon, 2007).
The rate-limiting step in RNA decay in chloroplasts, as in bacteria, is endonucleolytic cleavage, which generates products that are accessible to exonucleases by removing protective features at the RNA termini (for review, see Stern et al., 2010). However, the identities of the relevant endonucleases remain a mystery. A chloroplast protein called CSP41 exhibits endoribonuclease activity in vitro, but compelling evidence that CSP41 influences RNA decay in vivo has not emerged. RNAse E and RNAse J perform this function in bacteria, cleaving at unstructured AU-rich sequences (Condon, 2007). E. coli lacks RNAse J, whereas Bacillus subtilis lacks RNAse E, but cyanobacteria and higher plant chloroplasts harbor both enzymes. Arabidopsis mutants lacking chloroplast RNAse E do not exhibit a global increase in mRNA levels (Walter et al., 2010), indicating that RNAse E cannot be the sole activity that initiates chloroplast mRNA decay. Given the activities observed for chloroplast RNAse E in vitro (Schein et al., 2008) and the fact that bacterial RNAse E and RNAse J have similar endonuclease activities, it may be that RNAses E and J in chloroplasts act redundantly to initiate mRNA decay.
The parameters that determine the rate of the initiating endonucleolytic cleavages for chloroplast RNA decay are not known. These are likely to include the sequence and structure of the mRNA, its extent of ribosome association, and the presence of other proteins (particularly PPR-like proteins) that mask or expose potential RNase cleavage sites. In any case, it can be anticipated that regulation of the ribonucleases and stabilizing proteins underlies the regulation of chloroplast RNA stability, and that the details of these mechanisms will be forthcoming in the near future.
INTERCISTRONIC AND 5′ mRNA PROCESSING
A particularly striking feature of gene expression in land plant chloroplasts is the complexity of the RNA populations arising from most genes. This phenomenon is exemplified by the psbB gene cluster (Barkan, 1988; Westhoff and Herrmann, 1988; Fig. 2). Multiple mRNA isoforms arise from the processing of polycistronic transcripts between coding regions (“intercistronic processing”), in the 5′ untranslated region (UTR; “5′ processing”), and by the removal of introns. The mechanisms and functional significance of these events in chloroplasts have been long-standing questions. Recent findings have begun to clarify these issues and have challenged models that have prevailed ever since these phenomena were recognized more than 20 years ago.
It had been widely assumed that intercistronic mRNA processing in chloroplasts results from site-specific endonucleolytic cleavages that simultaneously generate adjacent processed 5′ and 3′ termini. This hypothesis was based on low-resolution mapping data that placed processed 5′ and 3′ ends near one another in various intergenic regions. The first hint that this view may be incorrect came with the mapping of the processed RNA termini between the petB and petD open reading frames (ORFs) in maize (Barkan et al., 1994): the 5′ end of the processed RNA from the downstream ORF (petD) maps approximately 30 nucleotides upstream of the 3′ end of the processed RNA from the upstream ORF (petB), proving that this pair of processed termini do not result from a single endonucleolytic cleavage event (Fig. 2).
Recent data show that this spatial relationship between processed 5′ and 3′ termini arising from the same intergenic region is common and provide strong evidence for a mechanism of intercistronic processing that does not involve site-specific endonucleolytic cleavage. During a study of the maize protein PPR10 (Pfalz et al., 2009), processed termini in the atpI-atpH, psaJ-rpl33, and psbH-petB intergenic regions were mapped precisely. In each case, the processed RNAs overlap by approximately 25 nucleotides, as had been shown previously for the petB-petD region (Figs. 1 and 2). By contrast, we are aware of only one instance in which processed 5′ and 3′ termini within an intergenic region have been shown unambiguously not to overlap in this manner, and even these termini appear to arise from independent processing events (Hashimoto et al., 2003).
There is strong evidence that the processed termini in the atpI-atpH and psaJ-rpl33 intergenic regions arise in the following manner (Pfalz et al., 2009; Prikryl et al., 2011). PPR10 binds to these two intergenic RNAs at sites that have similar sequences, and blocks the progress of exoribonucleases approaching from either the 5′ or 3′ direction (Fig. 1). This results in the accumulation of processed RNAs whose 5′ or 3′ terminus is defined by the upstream or downstream edge, respectively, of bound PPR10. This model was validated by experiments showing that recombinant PPR10 is sufficient to block both 5′ and 3′ exoribonucleases in vitro (Prikryl et al., 2011). Furthermore, PPR10 in conjunction with a generic 5′ exonuclease is sufficient to generate a 5′ terminus that corresponds precisely to the PPR10-dependent processed end found in vivo, proving that no additional proteins are required. This mechanism is likely to be the rule rather than the exception, as genetic data link three other PPR proteins (CRP1, HCF152, and PPR38) to the accumulation of processed transcripts with 5′ or 3′ ends mapping in three other intergenic regions (Barkan et al., 1994; Meierhoff et al., 2003; Hattori and Sugita, 2009; Fig. 2). Thus, “processed” mRNAs might be more accurately described as metastable degradation intermediates resulting from a site-specific blockade to the exonucleases involved in bulk RNA decay. An alternative mechanism for intercistronic processing has also been proposed, involving a PPR protein harboring an appendage with RNA cleavage activity (Okuda et al., 2009). The site-specific barrier and site-specific cleavage mechanisms are not mutually exclusive, although current data suggest the barrier mechanism to be the predominant one.
This scenario for intercistronic RNA processing requires a means for exonucleases to bypass stabilizing elements at transcript termini. We proposed a model involving the same housekeeping endonucleases that trigger mRNA decay (Pfalz et al., 2009; Fig. 2). This model is supported by the phenotype of Arabidopsis mutants lacking chloroplast RNAse E, which have defects in the processing of several polycistronic transcripts (Walter et al., 2010). However, many processing events were not disrupted in these mutants, indicating that other endonucleases (perhaps RNAse J) contribute as well.
Genetic data implicate PPR-like proteins not only in intercistronic RNA processing but also in the stabilization of specific processed 5′ ends in the chloroplasts of land plants and Chlamydomonas (Stern et al., 2010). 5′ processing and intercistronic processing had appeared to be distinct processes, but it now appears that they involve the same basic mechanism: the stalling of exonucleases at specific sites by a bound PPR-like protein. 5′ processing results from site-specific protection from a 5′→3′ exonuclease (likely RNAse J) near the transcription start site, whereas intercistronic processing involves protection from exonucleases intruding from either direction. Taken together, the results to date suggest that the machineries for 5′ and intercistronic mRNA processing in chloroplasts arose by the superposition of newly evolved PPR-like proteins upon a mechanism for RNA turnover that was borrowed from bacteria.
Chloroplasts contain abundant cpRNP proteins that are related to nuclear hnRNPs and that have also been implicated in mRNA stabilization. cpRNPs can stabilize RNAs in chloroplast extracts (Schuster and Gruissem, 1991; Nakamura et al., 2001), and a role for a cpRNP in the stabilization of several chloroplast mRNAs was confirmed in an in vivo analysis (Tillich et al., 2009). That being said, genetic data point to PPR-like proteins as the primary protein class involved in chloroplast RNA stabilization (Stern et al., 2010), suggesting that the long RNA-protein interface presented by a long PPR tract (Prikryl et al., 2011) provides a particularly effective barrier to nucleases.
CHLOROPLAST TRANSLATION
Several observations have highlighted translation as an important control point in chloroplast gene expression (Peled-Zehavi and Danon, 2007). (1) The translation of some chloroplast mRNAs is rapidly induced by light. (2) Translation rate has been shown to be a rate-limiting step in the expression of many chloroplast genes in Chlamydomonas (Eberhard et al., 2002) and can be regulated by the assembly status of the multimeric complexes harboring plastid gene products (Choquet and Wollman, 2009). (3) Genetic screens have identified numerous nucleus-encoded proteins that are required for the translation of specific chloroplast RNAs, demonstrating a large investment of the host genome in promoting chloroplast gene expression at the translational level. Adding to the intrigue are hints that chloroplast translation may involve mechanisms that are distinct from those in bacteria. Shine-Dalgarno elements are not evident in many chloroplast mRNAs, leading to speculation about novel ribosome recruitment mechanisms. Furthermore, whereas translational modulators in chloroplasts are consistently activators of chloroplast translation, translational regulation in bacteria is generally mediated by negative regulators.
Chloroplast ribosomes are formed from rRNAs and proteins that retain a strong resemblance to those in bacteria, and they function in conjunction with bacterial-type initiation and elongation factors (Peled-Zehavi and Danon, 2007). With this as backdrop, the discussion below starts from the parsimonious viewpoint that translation and its regulation in chloroplasts and bacteria involve similar mechanisms. Features of chloroplast translation that seem at odds with this perspective are evaluated in this evolutionary context.
Plastid-Specific Ribosomal Proteins
Chloroplast ribosomes include several “plastid-specific ribosomal proteins” (PSRPs; Yamaguchi and Subramanian, 2003; Beligni et al., 2004), which have been invoked as candidates for mediators of light-regulated chloroplast translation. However, a cryo-electron microscopy study of the spinach (Spinacia oleracea) chloroplast ribosome suggested instead that several PSRPs play structural roles, compensating for the loss of specific rRNA elements (Sharma et al., 2007). That study and a related one in Chlamydomonas (Manuell et al., 2007) did highlight variants of conserved ribosomal proteins as candidates for participation in chloroplast-specific mechanisms. For example, chloroplast-specific extensions on ribosomal protein S21 in spinach (Sharma et al., 2007) and ribosomal protein S2 in Chlamydomonas (Manuell et al., 2007) are positioned to contact the mRNA 5′ UTR during translation initiation.
PSRP1 has proven to be neither a ribosomal protein nor plastid specific (Sharma et al., 2010). In fact, PSRP1 and its bacterial orthologs inhibit translation by blocking tRNA binding sites. Furthermore, the gene encoding the cyanobacterial ortholog is strongly repressed after illumination, providing a molecular link between incident light and cyanobacterial translation. This observation led the authors to propose the intriguing possibility that PSRP1 abundance or activity may likewise be repressed in the light and that this might underlie the global enhancement of plastid translation after a shift from dark to light.
Mechanism of Start Codon Recognition
The most familiar mode of ribosome recruitment in bacteria involves the Shine-Dalgarno interaction: the pairing of the 3′ end of the 16S rRNA with complementary sequences upstream of the start codon. The consensus bacterial Shine-Dalgarno element has the sequence GGAGG and is centered approximately 10 nucleotides upstream from the start codon. Approximately one-third of chloroplast genes in land plants are preceded by predicted Shine-Dalgarno elements at the consensus location, and several of these have been confirmed to enhance translation (Peled-Zehavi and Danon, 2007). However, the majority of chloroplast genes lack properly positioned Shine-Dalgarno elements, leading to speculation about alternative modes for start codon selection. Two hypotheses are often suggested: that mRNA-specific translational activators can substitute for Shine-Dalgarno elements, and that plastid-specific ribosomal proteins play a role in Shine-Dalgarno-independent translation.
It is useful to consider what is known about bacterial translation to assess whether unique mechanisms need to be invoked to explain observations in chloroplasts. One important point is that Shine-Dalgarno elements are far from universal in bacterial genes (Nakagawa et al., 2010): for example, only approximately 39% of genes in cyanobacteria have apparent Shine-Dalgarno elements. Another lesson from bacteria is that 30S ribosomal subunits bind nonspecifically to single-stranded RNA and that a structure-free region spanning approximately 60 nucleotides centered on the start codon is important for optimal translation (de Smit and van Duin, 2003; Kudla et al., 2009). Current data support the notion that translation initiation in bacteria involves an unstructured RNA landing pad from which the 30S subunit can slide bidirectionally to access the start codon (de Smit and van Duin, 2003).
Can fundamentally similar mechanisms account for observations in chloroplasts? Perhaps so. A recent study showed that in both chloroplasts and bacteria, translation initiation regions lacking a Shine-Dalgarno sequence are less structured than are those harboring a Shine-Dalgarno element, suggesting that start codon accessibility is particularly critical in the absence of a Shine-Dalgarno interaction (L. Scharff and R. Bock, unpublished data). Furthermore, the chloroplast translational activators whose mechanisms are best understood seem to function by maintaining a structure-free zone for the ribosome (see below).
The possibility that 5′→3′ ribosome scanning contributes to start codon recognition in chloroplasts was raised in two studies, which reported preferential use of upstream start codons in reporter constructs (Hirose and Sugiura, 2004; Drechsel and Bock, 2010). It should be noted, however, that related observations have been made in E. coli (Adhin and van Duin, 1990), where ribosomes lack “specialized” structures and where polycistronic mRNAs are not generally processed prior to translation. Thus, these observations in chloroplasts may not reflect chloroplast-specific mechanisms but may instead be manifestations of differences in RNA structure resulting from differences in sequence, temperature, and intracellular milieu.
Nucleus-Encoded Translational Activators
Analyses of nonphotosynthetic mutants in plants and Chlamydomonas have revealed numerous nucleus-encoded proteins that influence chloroplast translation (for review, see Peled-Zehavi and Danon, 2007). These proteins are invariably activators, and they act specifically on one or several chloroplast mRNAs. Many of them are PPR-like proteins and, where tested, they act via the 5′ UTR of the target mRNAs. Furthermore, many proteins that activate translation also stabilize the same mRNA.
Two well-characterized examples in Chlamydomonas act on the petA (Boulouis et al., 2011) and psbD (Schwarz et al., 2007) mRNAs. In both cases, genetic data provide evidence that a PPR-like protein binds the mRNA 5′ end and stabilizes the RNA downstream. These proteins each interact with a second protein that binds the adjacent RNA segment and enhances the translation of the downstream ORF. PPR-like proteins also activate the translation of specific mRNAs in the chloroplasts of land plants via interaction with specific 5′ UTRs (Barkan et al., 1994; Sane et al., 2005; Schmitz-Linneweber et al., 2005; Pfalz et al., 2009).
Two general mechanisms for translational activation can be envisioned: the recruitment of ribosomes or translation factors, or the maintenance of an RNA structure (or lack of structure) that is attractive to ribosomes. Phylogenetic arguments and the available mechanistic data for chloroplast translational activators support the latter view. In vitro assays revealed the likely mechanism by which PPR10 enhances translation of the atpH ORF (Prikryl et al., 2011): when PPR10 binds to the atpH 5′ UTR, it remodels the RNA such that the atpH ribosome-binding region is freed from a secondary structure (Fig. 1B). Genetic data support an analogous mechanism for two translational activators in Chlamydomonas chloroplasts (Stampacchia et al., 1997; Schwarz et al., 2007). Detailed study of additional examples will be necessary to determine whether other types of activation mechanism are also at play.
The light regulation of psbA translation has attracted particular attention. The psbA gene encodes the D1 reaction center protein of PSII, which is subject to light-induced damage that necessitates new D1 synthesis for PSII repair. Light activates the initiation of psbA translation via the psbA 5′ UTR (for review, see Peled-Zehavi and Danon, 2007). Genetic screens identified the Arabidopsis protein HCF173 (Schult et al., 2007) and the Chlamydomonas protein Tba1 (Somanchi et al., 2005) as being required specifically for psbA translation. HCF173 and Tba1 are unrelated, they are not PPR-like proteins, and their mechanisms of action are unknown. A biochemical approach in Chlamydomonas led to a model for the regulation of psbA translation via a set of RNA-binding proteins whose activity is modulated by redox poise (for review, see Peled-Zehavi and Danon, 2007). However, there is no evidence for a related system in land plants, and recent reports are at odds with several aspects of that model (for review, see Zerges and Hauser, 2009).
Relationship between mRNA Processing and Translational Efficiency
The enhancement of translational efficiency is often invoked as the raison d’être for the pervasive intercistronic mRNA processing in chloroplasts. This is an appealing possibility, but the body of evidence to date does not provide strong evidence in favor of this view. First, many chloroplast genes are represented solely by polycistronic mRNAs. Second, for several genes that are represented by processed monocistronic mRNAs, processing has been shown not to be necessary for translation. This was demonstrated to be the case in vivo for maize petB and petD: when antibodies to PetB and PetD were used to immunoselect polysomes engaged in PetB and PetD synthesis, all transcripts containing spliced petB or petD sequences were recovered regardless of the upstream or downstream sequences (Barkan, 1988). A similar conclusion was drawn for atpH based on results from a tobacco chloroplast in vitro translation system (Yukawa et al., 2007). Furthermore, downstream ORFs in engineered polycistronic transcription units in tobacco chloroplasts can be translated efficiently without processing (Staub and Maliga, 1995). There is also evidence, however, that some mRNA processing events do enhance translational efficiency. A compelling example comes from an analysis of the psaC-ndhD transcription unit (Hirose and Sugiura, 1997): in the tobacco chloroplast in vitro translation system, the psaC and ndhD ORFs were translated more efficiently as monocistronic than as dicistronic RNAs. This was shown to be due to an inhibitory interaction between a sequence in the psaC coding region and its complement in the ndhD 5′ UTR.
A key argument that has been used to support the idea that intercistronic processing enhances translation derives from the genetic analysis of the PPR-like proteins CRP1, HCF107, and CRR2: in crp1, hcf107, and crr2 mutants, the loss of specific processed mRNAs correlates with reduced translational efficiencies (Barkan et al., 1994; Felder et al., 2001; Hashimoto et al., 2003). However, recent results with PPR10 warrant consideration of an alternative explanation for those correlations. The ppr10 mutant phenotype parallels those of crp1, hcf107, and crr2 in that specific processed atpH transcripts are absent and the atpH translation rate is also reduced. Both of those effects result from the stable association of PPR10 with the atpH 5′ UTR: bound PPR10 simultaneously blocks 5′→3′ RNA degradation and remodels the adjacent translation initiation region to expose the ribosome-binding site (Pfalz et al., 2009; Prikryl et al., 2011; Fig. 1). These findings undermine the notion that the loss of processed RNAs in crp1, hcf107, and crr2 mutants is the sole cause of reduced translational efficiency. Instead, it may be that it is the presence of CRP1, HCF107, and CRR2 proteins on their target 5′ UTRs that enhances translation, and that the reduced translation and loss of processed mRNAs in the mutants are independent effects of the absence of these proteins.
On balance, the current evidence argues against translational enhancement as a significant driving force for the evolution of chloroplast mRNA processing, although processing likely enhances translational efficiency in some cases. In fact, it can be anticipated that a newly acquired mRNA processing event will, over evolutionary time, become increasingly important for optimal translation due to relaxed constraints on flanking RNA sequences: If an ORF can be separated from cotranscribed sequences by processing, then it will be released from prior evolutionary constraints that would have limited inhibitory interactions with sequences found elsewhere on the RNA precursor. Although it is possible that chloroplast-specific mechanisms are at play during translation initiation and translational activation, perhaps it is too soon to let go of the conservative view that differences between chloroplasts and bacterial translation lie in the types of RNA-binding proteins that are available to modulate RNA structure and processing (e.g. PPR-like proteins) rather than in the translation process itself.
COUPLING OF CHLOROPLAST TRANSCRIPTION WITH DOWNSTREAM EVENTS
Bacterial transcription and translation are said to be “coupled” in that ribosomes initiate translation soon after the start codon exits RNA polymerase. The long lifetime of chloroplast mRNAs and the fact that they can be translated after separation from downstream RNA sequences imply that translation is not obligatorily coupled to transcription in chloroplasts. Nonetheless, translation in chloroplasts may generally initiate on nascent transcripts during the process of transcription. Chloroplast DNA is in contact with the stroma, which is the location of many RNA-binding proteins and ribosomes. Therefore, it can be expected that RNA-binding proteins and ribosomes begin to associate with nascent transcripts in a cotranscriptional manner. This view is supported by the fact that the chloroplast splicing factor APO1 (Watkins et al., 2011) colocalizes with the nucleoid (Amann et al., 2004). The degree to which RNA processing and translation are “cotranscriptional” versus “posttranscriptional” may simply reflect the underlying kinetics: events involving RNA-protein interactions that form more slowly will generally be posttranscriptional, whereas those involving interactions that form rapidly will generally be cotranscriptional.
There is evidence for the cotranslational insertion of some chloroplast-encoded proteins into the thylakoid membrane via a conserved “Sec” machinery (Zhang et al., 2001). In addition, translational pausing has been shown to accompany the integration of D1 into the membrane (Kim et al., 1991; Zhang et al., 2000). Whether this pausing is a consequence of the engagement of the nascent peptide with the membrane integration machinery or serves as a means to facilitate membrane integration remains to be resolved.
PERSPECTIVE
It is gratifying to look back upon the past decade and to recognize the remarkable progress that has been made in understanding the mechanisms of chloroplast gene expression. Some processes that had appeared to be complex now appear to have rather simple underlying mechanisms. For example, the complexity of intercistronic RNA processing now appears to distill down to a set of PPR-like proteins that bind specific RNA sites and block housekeeping exonucleases. Likewise, the process of RNA editing appears to involve a set of PPR proteins that target a multitude of sites but that recruit a shared enzymatic machinery. Other aspects of chloroplast gene expression that had been anticipated to involve simple mechanisms have turned out to be surprisingly complex. A prime example is chloroplast RNA splicing, which involves “self-splicing” RNAs whose splicing requires a multitude of different proteins. This complexity can also be seen in the interplay between NEP, PEP, and the various PEP-associated σ-factors that promote chloroplast transcription.
The evolution of chloroplast gene expression systems continues to present a set of intriguing puzzles. Proteins of bacterial ancestry serve in core gene expression processes (transcription, translation, and RNA turnover). Acquired aspects of chloroplast gene expression (RNA editing, protein-facilitated group II intron splicing, and intercistronic RNA processing) were imparted by the superposition of newly evolved proteins upon these ancient core machineries. The environment of plant organelles seems to have been particularly permissive for the evolution of novel RNA-binding motifs, including the multitude of diversified PPR proteins. In fact, it seems likely that the complex RNA metabolism and complex RNA-binding protein repertoires in plant organelles arose through an as yet mysterious coevolutionary process.
Another set of unanswered questions concern the regulation of chloroplast gene expression. There is no single step in gene expression that is the primary regulated step; rather, each step can contribute to different patterns of chloroplast gene expression under different conditions. Now that the nuts and bolts of chloroplast gene expression are understood in considerable detail, the field is poised to understand how chloroplast gene expression responds to light quality and quantity, stress, and developmental cues. A current challenge is to identify the subset of nucleus-encoded factors whose activity is limiting for gene expression and the signal transduction pathways that link these regulators to external signals.
Acknowledgments
I apologize to the authors of the many papers I am unable to cite due to space constraints. I am grateful to Kamel Hammani, Anastassia Khrouchtchova, Thomas Börner, Bill Zerges, and Kenny Watkins for useful discussions and for comments on the manuscript. I also thank Lars Scharff for discussions and for communicating unpublished data and Ros Williams-Carrier and Jana Prikryl for help in preparing the figures.
References
- Adam Z. (2007) Protein stability and degradation in plastids. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Heidelberg, pp 315–338 [Google Scholar]
- Adhin MR, van Duin J. (1990) Scanning model for translational reinitiation in eubacteria. J Mol Biol 213: 811–818 [DOI] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J. (2004) ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP. (2007) The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 13: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Walker M, Nolasco M, Johnson D. (1994) A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J 13: 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. (1989) Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol 89: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. (1993) Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development: evidence for selective stabilization of psbA mRNA. Plant Physiol 101: 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Yamaguchi K, Mayfield SP. (2004) The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth Res 82: 315–325 [DOI] [PubMed] [Google Scholar]
- Bock R. (2007) Structure, function, and inheritance of plastid genomes. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Heidelberg, pp 29–63 [Google Scholar]
- Boulouis A, Raynaud C, Bujaldon S, Aznar A, Wollman F-A, Choquet Y. (2011) Critical role of MCA1 in the control of cytochrome f synthesis. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. (2010) Plant RNA editing. RNA Biol 7: 213–219 [DOI] [PubMed] [Google Scholar]
- Chen L-J, Orozco EM., Jr (1988) Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res 16: 8411–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wollman F. (2009) The CES process. Stern D, Harris E, , The Chlamydomonas Sourcebook: Organellar and Metabolic Processes. Academic Press, Oxford, pp 1027–1064 [Google Scholar]
- Condon C. (2007) Maturation and degradation of RNA in bacteria. Curr Opin Microbiol 10: 271–278 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C. (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3: 691–705 [DOI] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. (2003) Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol 331: 737–743 [DOI] [PubMed] [Google Scholar]
- Drechsel O, Bock R. (2011) Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res 39: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA. (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plücken H, Klaff P, Stein B, Bechtold N, Westhoff P. (2001) The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13: 2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble PE, Mullet JE. (1989) Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J 8: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Hattori M, Sugita M. (2009) A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J 276: 5860–5869 [DOI] [PubMed] [Google Scholar]
- Heller WP, Hayes ML, Hanson MR. (2008) Cross-competition in editing of chloroplast RNA transcripts in vitro implicates sharing of trans-factors between different C targets. J Biol Chem 283: 7314–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M. (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J 16: 6804–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M. (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J 20: 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M. (2004) Multiple elements required for translation of plastid atpB mRNA lacking the Shine-Dalgarno sequence. Nucleic Acids Res 32: 3503–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng ST, Gardner JF, Gumport RI. (1990) Transcription termination by bacteriophage T7 RNA polymerase at rho-independent terminators. J Biol Chem 265: 3823–3830 [PubMed] [Google Scholar]
- Kahlau S, Bock R. (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klein PG, Mullet JE. (1991) Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J Biol Chem 266: 14931–14938 [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22: 447–463 [DOI] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. (1991) Changes in chloroplast mRNA stability during leaf development. Plant Cell 3: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Bohne AV, Liere K, Weihe A, Börner T. (2007) Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell 19: 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2010) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T. (2007) Transcription and transcriptional regulation in chloroplasts. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Heidelberg, pp 121–174 [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier UG, Bozarth A, Funk HT, Zauner S, Rensing SA, Schmitz-Linneweber C, Börner T, Tillich M. (2008) Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell AL, Quispe J, Mayfield SP. (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol 5: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac DJ, Barkan A. (1999) A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11: 1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde RA, Zito F, Olive J, Wollman FA, Stern DB. (2000) Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6/f complex. Plant J 21: 61–72 [DOI] [PubMed] [Google Scholar]
- Mullet JE. (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103: 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Miura K, Gojobori T. (2010) Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci USA 107: 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Ohta M, Sugiura M, Sugita M. (2001) Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J Biol Chem 276: 147–152 [DOI] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. (2002) The plastid transcription kinase from mustard (Sinapis alba L.): a nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem 269: 3329–3337 [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104: 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. (2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem 281: 37661–37667 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17: 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters NM, Hanson MR. (2002) Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA 8: 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled-Zehavi H, Danon A. (2007) Translation and translational regulation in chloroplasts. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Heidelberg, pp 249–281 [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen J. (1999) Photosynthetic control of chloroplast gene expression. Nature 397: 625–628 [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A. (2011) Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF. (2008) The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci USA 105: 10061–10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A, Lambowitz A. (2006) Group II introns: ribozymes that splice RNA and invade DNA. Gesteland R, Cech T, Atkins J, , The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 469–506 [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L. (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C. (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett 581: 4132–4138 [DOI] [PubMed] [Google Scholar]
- Sane AP, Stein B, Westhoff P. (2005) The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J 42: 720–730 [DOI] [PubMed] [Google Scholar]
- Schein A, Sheffy-Levin S, Glaser F, Schuster G. (2008) The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P. (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G, Gruissem W. (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J 10: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G, Stern D. (2009) RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci 85: 393–422 [DOI] [PubMed] [Google Scholar]
- Schwarz C, Elles I, Kortmann J, Piotrowski M, Nickelsen J. (2007) Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G. (2010a) Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription: recent lessons from Arabidopsis thaliana. Eur J Cell Biol 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Link B, Link G. (2010b) AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J 62: 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Dönhöfer A, Barat C, Marquez V, Datta PP, Fucini P, Wilson DN, Agrawal RK. (2010) PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G). J Biol Chem 285: 4006–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK. (2007) Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA 104: 19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Kato H, Ogawa T, Kurachi A, Nakagawa Y, Kobayashi H. (2010) Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc Natl Acad Sci USA 107: 10760–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi A, Barnes D, Mayfield SP. (2005) A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J 42: 341–352 [DOI] [PubMed] [Google Scholar]
- Stampacchia O, Girard-Bascou J, Zanasco J-L, Zerges W, Bennoun P, Rochaix J-D. (1997) A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell 9: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. (1995) Expression of a chimeric uidA gene indicates that polycistronic mRNAs are efficiently translated in tobacco plastids. Plant J 7: 845–848 [DOI] [PubMed] [Google Scholar]
- Stern D, Harris E.editors (2009) The Chlamydomonas Sourcebook: Organellar and Metabolic Processes. Academic Press, Oxford [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M, Emanuel C, Hedtke B, Liere K, Börner T. (2008) Impaired function of the phage-type RNA polymerase RpoTp in transcription of chloroplast genes is compensated by a second phage-type RNA polymerase. Nucleic Acids Res 36: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C. (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc Natl Acad Sci USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. (2006) The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23: 1324–1338 [DOI] [PubMed] [Google Scholar]
- Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, Bock R, Cardi T. (2009) Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol 150: 2030–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. (2010) Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A. (2007) A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A. (2011) Arabidopsis APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P, Herrmann RG. (1988) Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur J Biochem 171: 551–564 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Kroeger T, Barkan A. (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D. (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR. (2003) Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur J Biochem 270: 190–205 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Kuroda H, Sugiura M. (2007) A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. Plant J 49: 367–376 [DOI] [PubMed] [Google Scholar]
- Zerges W, Hauser C. (2009) Protein synthesis in the chloroplast. Stern D, Harris E, , The Chlamydomonas Sourcebook: Organellar and Metabolic Processes. Academic Press, Oxford, pp 967–1026 [Google Scholar]
- Zhang L, Paakkarinen V, Suorsa M, Aro EM. (2001) A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J Biol Chem 276: 37809–37814 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM. (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]