Abstract
Tobacco (Nicotiana tabacum) plants synthesize nicotine and related pyridine-type alkaloids, such as anatabine, in their roots and accumulate them in their aerial parts as chemical defenses against herbivores. Herbivory-induced jasmonate signaling activates structural genes for nicotine biosynthesis and transport by way of the NICOTINE (NIC) regulatory loci. The biosynthesis of tobacco alkaloids involves the condensation of an unidentified nicotinic acid-derived metabolite with the N-methylpyrrolinium cation or with itself, but the exact enzymatic reactions and enzymes involved remain unclear. Here, we report that jasmonate-inducible tobacco genes encoding flavin-containing oxidases of the berberine bridge enzyme family (BBLs) are expressed in the roots and regulated by the NIC loci. When expression of the BBL genes was suppressed in tobacco hairy roots or in tobacco plants, nicotine production was highly reduced, with a gradual accumulation of a novel nicotine metabolite, dihydromethanicotine. In the jasmonate-elicited cultured tobacco cells, suppression of BBL expression efficiently inhibited the formation of anatabine and other pyridine alkaloids. Subcellular fractionation and localization of green fluorescent protein-tagged BBLs showed that BBLs are localized in the vacuoles. These results indicate that BBLs are involved in a late oxidation step subsequent to the pyridine ring condensation reaction in the biosynthesis of tobacco alkaloids.
Tobacco (Nicotiana tabacum) and other Nicotiana species synthesize nicotine to mount defensive responses to insect herbivores. Because the activation of acetylcholine receptors is inherently toxic to all heterotrophs with neuromuscular junctions, nicotine is thought to be a broadly effective plant defense metabolite (Baldwin, 2001). A reduction in the amount of nicotine in the tobacco leaf, due to either natural variation (Jackson et al., 2002) or genetic manipulation (Steppuhn et al., 2004), results in more pronounced damage by insect herbivores. Tobacco plants sense browsing insects on their leaves and increase the de novo synthesis of nicotine by utilizing the general jasmonate signaling pathway and nicotine-specific regulatory components (Shoji and Hashimoto, 2011). The highly bioactive Ile conjugate of jasmonate is perceived by a complex of CORONATINE INSENSITIVE1 and the transcriptional repressor JAZ proteins (Sheard et al., 2010), resulting in JAZ degradation and a subsequent release of transcription repression (Chini et al., 2007). Tobacco orthologs of these signaling proteins are utilized in tobacco to regulate jasmonate-inducible nicotine biosynthesis (Shoji et al., 2008). As reported recently (Shoji et al., 2010), several tobacco transcription factors, including ERF189, of an ethylene response factor (ERF) subfamily are encoded in clustered genes at the nicotine regulatory locus NICOTINE2 (NIC2) and bind to the GCC box element in the promoter of the putrescine N-methyltransferease gene (PMT; Hibi et al., 1994). ERF189 and related tobacco transcription factors are shown to directly and specifically activate all known structural genes in the nicotine pathway (Shoji et al., 2010).
The early enzymatic steps of nicotine biosynthesis are well established (Supplemental Fig. S1). The pyrrolidine ring of nicotine is derived from putrescine, via consecutive reactions involving the enzymes PMT and N-methylputrescine oxidase (Heim et al., 2007; Katoh et al., 2007), which together produce the N-methylpyrrolinium cation, whereas the pyridine ring uses enzymes involved in the early steps of NAD biosynthesis, such as Asp oxidase, quinolinic acid synthase, and quinolinic acid phosphoribosyl transferase (Sinclair et al., 2000; Katoh et al., 2006). The genes for these five enzymes are coordinately regulated in tobacco roots (Shoji et al., 2010). When isotopically labeled nicotinic acids were fed to tobacco plants, the isotope labels were incorporated into the pyridine ring of nicotine (Dawson et al., 1956). Interestingly, the tritium label at C-6 of nicotinic acid was specifically lost in isolated nicotine (Dawson et al., 1960; Leete and Liu, 1973), suggesting that the C-6 position is first oxidized and subsequently reduced during the incorporation of nicotinic acid into nicotine. The putative oxidoreductases involved in the activation of nicotinic acid and the mechanisms behind the nicotine-forming condensation reactions between a nicotinic acid-derived intermediate and the N-methylpyrrolinium cation are yet to be clarified. A pinoresinol-lariciresinol reductase/isoflavone reductase/phenylcoumaran benzylic ether reductase family oxidoreductase, A622, is required for the biosynthesis of tobacco alkaloids, possibly in a step to produce a nicotinic acid-derived precursor, but the exact enzymatic reaction catalyzed by A622 is not known (Deboer et al., 2009; Kajikawa et al., 2009).
Besides nicotine, other pyridine alkaloids also accumulate at substantial levels in tobacco roots, in elicited cultured tobacco cells, and in wild Nicotiana species (Supplemental Fig. S1; Saito et al., 1985; Shoji and Hashimoto, 2011). Anatabine is synthesized by the dimerization of a metabolite of nicotinic acid (Leete and Slattery, 1976), whereas the piperidine ring of anabasine is derived from Lys by way of cadaverine (Watson et al., 1990). Anatalline consists of two pyridyl rings and a central piperidine ring (Häkkinen et al., 2004). The biosynthesis of tobacco alkaloids consisting of more than two six-membered heterocyclic rings may involve the same ring condensation reactions as predicted for the production of nicotine. Indeed, suppression of A622, an enzyme proposed to form a coupling-competent nicotinic acid intermediate, in tobacco roots and cultured tobacco cells inhibited the formation of not only nicotine but also other pyridine alkaloids (Kajikawa et al., 2009). Nornicotine is formed from nicotine by cytochrome P450 monooxygenases of the CYP82E subfamily in tobacco leaf tissues (Siminszky et al., 2005; Xu et al., 2007; Lewis et al., 2010), but the pathway of nornicotine biosynthesis in other cell types is not established.
In this study, we identified vacuole-localized tobacco flavoproteins, berberine bridge enzyme-like proteins (BBLs), that were required for the synthesis of nicotine, anatabine, anabasine, and anatalline. The accumulation of a novel pyridine alkaloid in BBL-suppressed tobacco root tissues suggests that BBLs are involved in a late, possibly the final, step of tobacco alkaloid biosynthesis. The dimerization of a pyridine ring with an unsaturated pyrrolidine or piperidine ring appears to involve multiple oxidoreductases of different families.
RESULTS
Isolation of NIC-Regulated BBL Genes from Tobacco
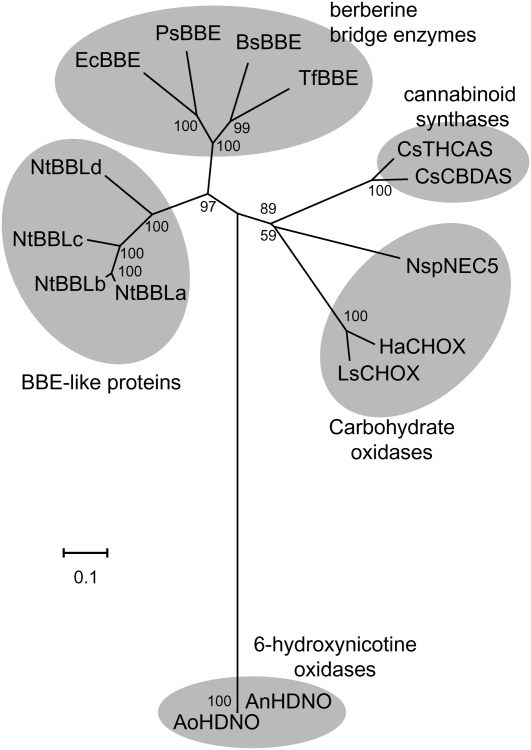
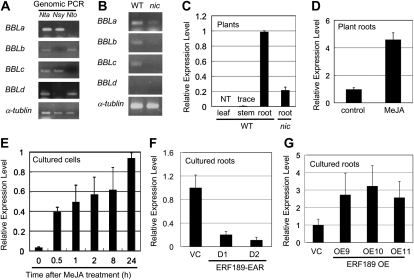
To identify novel genes that are controlled by the NIC regulatory loci, we compared comprehensive cDNA expression profiles of wild-type tobacco roots and nic1nic2 double mutant roots obtained using tobacco oligonucleotide microarrays (Katoh et al., 2007; Shoji et al., 2010). One tobacco gene (BBLa) encoding a BBL was identified as a putative target of the NIC regulatory loci. After searching the tobacco genome database (http://solgenomics.net/) and extensive PCR screening and sequencing, we recovered four tobacco BBL cDNAs encoding full-length proteins (BBLa, BBLb, BBLc, and BBLd). BBLb has been reported as a jasmonate-inducible gene in cultured tobacco BY-2 cells (Goossens et al., 2003). BBLa is 94%, 83%, and 63% identical in amino acid sequence to BBLb, BBLc, and BBLd, respectively (Supplemental Fig. S2). These tobacco BBLs constitute a distinct clade in the FAD-containing oxidoreductase family that includes berberine bridge enzymes (BBEs), carbohydrate oxidases, cannabinoid synthases, and 6-hydroxynicotine oxidases (Fig. 1). Tobacco BBL genes contained no introns. Genomic PCR using primers specific to each BBL gene amplified BBLa and BBLc from the genomic DNA of N. tabacum and Nicotiana sylvestris and BBLb and BBLd from that of N. tabacum and Nicotiana tomentosiformis (Fig. 2A). Therefore, the diploid progenitors of N. tabacum possess two related BBL genes: BBLa and BBLc probably originate from N. sylvestris, whereas BBLb and BBLd may be derived from N. tomentosiformis.
Figure 1.
Phylogenetic tree of BBE-like proteins. Shown is an unrooted neighbor-joining phylogenetic tree of tobacco BBE-like proteins (NtBBLs) and of related proteins belonging to four subgroups: BBEs, 6-hydroxynicotine oxidases, cannabinoid synthases, and carbohydrate oxidases. The tree was generated using MEGA4 software (Tamura et al., 2007) with the neighbor-joining algorithm. Bootstrap values (1,000 replicates) are indicated at branch nodes, and the scale bar indicates the number of amino acid substitutions per site. Species are as follows: NtBBLa (N. tabacum), NtBBLb (N. tabacum), NtBBLc (N. tabacum), NtBBLd (N. tabacum), EcBBE (Eschscholzia californica), BsBBE (Berberis stolonifera), PsBBE (Papaver somniferum), TfBBE (Thalictrum flavum), AoHDNO (Arthrobacter oxidans), AnHDNO (Arthrobacter nicotinovorans), CsCBDAS (C. sativa), CsTHCAS (C. sativa), HaCHOX (Helianthus annuus), LsCHOX (Lactuca sativa), and NspNEC5 (Nicotiana langsdorffii × Nicotiana sanderae).
Figure 2.
Expression profiles of tobacco BBL genes. A and B, PCR primers specific to each BBL member were used, and the tobacco α-Tubulin gene was amplified as a control. A, Genomic PCR analysis of N. tabacum (Nta) and its probable progenitors, N. sylvestris (Nsy) and N. tomentosiformis (Nto). B, Transcript levels of each BBL gene were assessed by RT-PCR in the roots of wild-type (WT) and nic1nic2 mutant (nic) tobacco plants. C to G, Quantitative RT-PCR analysis of BBL genes using BBL-consensus PCR primers. Transcript levels are shown as relative values. C, Organ-specific expression pattern in the wild-type plant and expression level in the nic root. NT, Not detectable. D, Treatment of tobacco roots with 100 μm MeJA for 24 h. E, Treatment of cultured tobacco cells with 50 μm MeJA for the periods indicated. F, BBL transcript levels in cultured tobacco roots of the VC line and two transgenic lines expressing a dominant-negative ERF189 form (ERF189-EAR, D1 and D1; Shoji et al., 2010). G, BBL transcript levels in cultured tobacco roots of the VC line and three transgenic lines overexpressing ERF189 (OE9, OE10, and OE11; Shoji et al., 2010).
Expression profiles of BBLs were analyzed by reverse transcription (RT)-PCR. By using specific primers to amplify each BBL gene, we showed that BBLa, BBLb, and BBLc are expressed in wild-type tobacco roots, but BBLd mostly is not (Fig. 2B). Levels of BBLa to -c expression were markedly low in the nic1nic2 mutant roots, indicating that they were positively regulated by the NIC loci. Quantitative RT-PCR using a common primer set that amplified all BBL genes was next conducted to examine tissue-specific and jasmonate-inducible expression patterns. BBLs were expressed strongly in the root but at very low or negligible levels in the stem and leaf of wild-type tobacco plants (Fig. 2C). In the nic1nic2 mutant roots, the abundance of the BBL transcripts was reduced by 80% (Fig. 2C). The application of 50 μm methyl jasmonate (MeJA) increased BBL expression 4- to 5-fold in the wild-type tobacco roots (Fig. 2D) and strongly induced BBL expression from an initially very low level within 30 min in cultured tobacco BY-2 cells (Fig. 2E). When the expression of the NIC2-locus ERF genes, which specifically activated all known structural genes of nicotine biosynthesis (Shoji et al., 2010), was suppressed by the constitutive expression of a dominant repressive form of ERF189 in two transgenic tobacco root lines (D1 and D2; Shoji et al., 2010), expression of the BBL genes was effectively reduced by more than 80% (Fig. 2F). Furthermore, overexpression of ERF189 in three transgenic tobacco root lines (OE9, OE10, and OE11; Shoji et al., 2010) resulted in 2- to 3-fold increases in the levels of the BBL transcripts (Fig. 2G). These expression patterns of BBLs are highly similar to those of structural genes involved in the biosynthesis and transport of nicotine (Hibi et al., 1994; Reed and Jelesko, 2004; Cane et al., 2005; Katoh et al., 2007; Kajikawa et al., 2009; Shoji et al., 2009, 2010).
Constitutive BBL Knockdown in Tobacco Hairy Roots
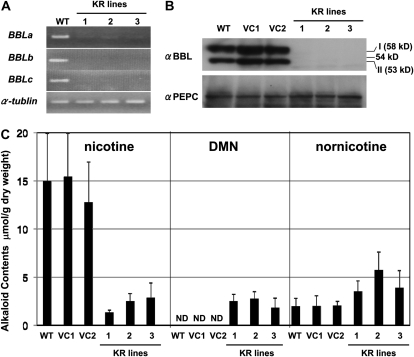
To investigate their possible roles in the biosynthesis of tobacco alkaloids, we first suppressed the expression of BBL genes constitutively with RNA interference (RNAi) in tobacco hairy roots, using the cauliflower mosaic virus 35S promoter. Three independent transgenic lines (KR1, KR2, and KR3) were obtained in which the transcript levels of BBLa, BBLb, and BBLc were highly reduced (Fig. 3A). When the accumulation of BBL proteins was analyzed by immunoblotting using the BBL-specific antiserum, the wild-type and two vector control (VC) hairy root lines showed signals for BBLs of 58 kD (isoform I) and 53 kD (isoform II), whereas the three KR lines did not (Fig. 3B). Since the predicted full-length forms of BBLa, BBLb, and BBLc are 62, 63, and 62 kD, respectively, BBLs are processed in tobacco cells to yield two smaller protein species. Suppression of BBL expression did not affect the growth of the hairy roots.
Figure 3.
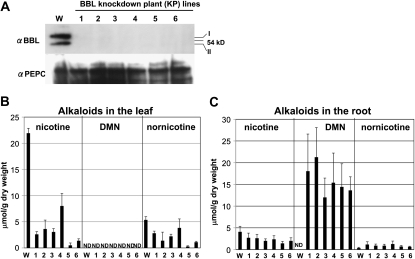
Down-regulation of BBL genes in transgenic tobacco roots. Tobacco hairy roots of the wild type (WT), two vector-transformed control lines (VC1 and VC2), and three RNAi lines (KR1, KR2, and KR3) were analyzed. A, RT-PCR analysis of BBLa, BBLb, and BBLc as well as the control α-Tubulin gene. B, Immunoblot analysis using the antisera against BBL and PEPC (loading control). BBLs existed in two forms with different molecular masses (I, 60 kD; II, 53 kD). C, Alkaloid contents of hairy roots. Data indicate mean values ± sd from three biological replicates. ND, Not detected.
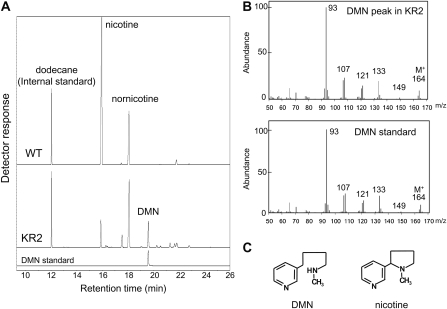
Tobacco alkaloids consisted mostly of nicotine, in addition to a smaller amount of nornicotine, in the wild-type and VC hairy roots (Fig. 3C). Nicotine levels were much lower in the KR lines (1–3 μmol g−1 dry weight) than the wild-type and VC lines (13–16 μmol g−1 dry weight). In contrast, nornicotine levels were slightly higher in the KR lines (4–6 μmol g−1 dry weight) than the wild-type and VC lines (2 μmol g−1 dry weight). Interestingly, an unknown compound was detected in the KR lines at a retention time of 19.7 min in the gas-liquid chromatograms (Fig. 4A). This novel metabolite was identified as dihydrometanicotine (DMN; Fig. 4C) by comparing its retention time in the gas-liquid chromatograms (Fig. 4A), its mass spectra (Fig. 4B), and its mobility shift in the thin-layer chromatograms (Fig. 5C), with the authentic compound. DMN has been isolated as a catabolite of nicotine in mammals (De Clercq and Truhaut, 1962; Neurath et al., 1966) but had not been found in plants. DMN was only detectable in the KR lines, where it accumulated at levels comparable to nicotine (2–3 μmol g−1 dry weight; Fig. 3C).
Figure 4.
Identification of DMN in the BBL-suppressed tobacco roots. A, Gas-liquid chromatograms of alkaloid fractions from the cell extracts of the wild type (WT) and the KR2 line, along with the DMN standard. DMN (retention time, 19.7 min) accumulated in the KR2 roots but not in wild-type roots. B, Mass fragment patterns of the DMN peak in the KR2 roots and of the authentic DMN. C, Chemical structures of DMN and nicotine.
Figure 5.
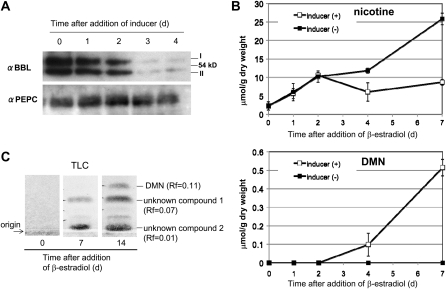
Time courses of inducible BBL suppression and alkaloid accumulation in transgenic tobacco roots. A, Immunoblot analysis of BBL and PEPC proteins in the inducible BBL RNAi root line (XN1) after the roots were treated with β-estradiol for the period indicated. B, Accumulation of nicotine and DMN in XN1 roots cultured in the absence or presence of β-estradiol. Data indicate mean values ± sd from three biological replicates. C, Metabolite analysis by thin-layer chromatography (TLC). Nitrogen-containing compounds were detected using Dragendorff’s reagent. At least two unidentified compounds (RF values of 0.01 and 0.07), in addition to DMN (RF of 0.11), were detectable after the inducer treatment.
Inducible BBL Knockdown in Tobacco Hairy Roots
To examine the time courses of the reduction in nicotine and the accumulation of DMN after suppression of the BBL genes, we used an inducible expression system (the XVE-β-estradiol system; Zuo et al., 2000) to drive RNAi-based gene suppression. Two independent tobacco hairy root lines (iKR1 and iKR2) were obtained in which expression of the BBL genes was efficiently suppressed after the addition of the inducer β-estradiol to the culture medium. In the iKR1 line, the levels of the BBL proteins began to decrease after 2 d of treatment and were very low by day 3 (Fig. 5A). We did not observe inhibitory effects of the inducer on root growth in the iKR lines or on the accumulation of tobacco alkaloids in wild-type and vector control roots (Kajikawa et al., 2009). When the iKR line was treated with the inducer, the formation of nicotine continued for 2 d and then stopped, whereas the untreated iKR roots continued to synthesize nicotine (Fig. 5B). DMN became detectable after 2 d of treatment and continued to increase thereafter. It should be noted, however, that the accumulation of DMN was very small compared with the suppressed level of nicotine synthesis. For example, at day 7, the production of as much as 17 μmol g−1 dry weight of nicotine was estimated to be inhibited by the suppression of BBLs, but DMN merely accumulated to a level of 0.5 μmol g−1 dry weight. Similar results were obtained in another inducible BBL-knockdown line, iKR2 (Supplemental Fig. S3). When the alkaloid extracts from iKR hairy roots were analyzed by thin-layer chromatography, two metabolites with RF values of 0.01 and 0.07, in addition to DMN (RF of 0.11), were detected using Dragendorff’s reagent after the suppression of BBLs (Fig. 5C). Although the chemical structures of these metabolites were not determined, their reactivity toward Dragendorff’s reagent suggests that they contain nitrogen. These two compounds also accumulated in the BBL-suppressed lines KR1 to -3 but not in the untreated iKR lines, VC lines, or wild-type tobacco roots (data not shown). Thus, at least three metabolites, including DMN, accumulate in tobacco cells as a consequence of the BBL suppression.
BBL Knockdown in Transgenic Tobacco Plants
To evaluate the consequences of the BBL suppression in tobacco plants, we constitutively suppressed the expression of the BBL genes by RNAi in transgenic tobacco plants. Six independent transgenic lines (KP1, KP2, KP3, KP4, KP5, and KP6) showed little accumulation of BBL proteins in the immunoblot analysis (Fig. 6A). Suppression of BBLs did not affect the growth or development of the KP tobacco plants. We analyzed tobacco alkaloids in the leaves and roots of 1-month-old plants. Nicotine was the predominant alkaloid in both parts in wild-type plants (Fig. 6, B and C). In the leaves, the amounts of nicotine and nornicotine (μmol g−1 dry weight) were much lower in the KP plants (1–8 and 0.3–3 μmol g−1 dry weight, respectively) than wild-type plants (22 and 5 μmol g−1 dry weight, respectively). We did not detect DMN in the leaves of either wild-type or KP plants. In the roots, the nicotine content was considerably lower in the KP plants (1–3 μmol g−1 dry weight) than wild-type plants (4 μmol g−1 dry weight), whereas the nornicotine content was somewhat higher in the KP plants (0.6–1 μmol g−1 dry weight) than in the wild-type plants (0.4 μmol g−1 dry weight). Interestingly, DMN was the most abundant alkaloid in the roots of the KP plants, reaching 12 to 21 μmol g−1 dry weight, but was absent in the wild-type roots (Fig. 6C). DMN may accumulate in the root tissue, possibly because of its inability to be transported to the aerial tissues via the xylem.
Figure 6.
BBL down-regulation in tobacco plants. Tobacco plants of the wild type (W) and six independent BBL RNAi lines (KP1–KP6) were grown for 1 month after germination and analyzed for gene expression and alkaloids. A, Immunoblot analysis of BBL and PEPC (control) proteins in the root. B and C, Levels of nicotine, DMN, and nornicotine in the fourth newest leaf (B) and the root (C). Data indicate mean values ± sd from three replicates. ND, Not detected.
BBL Knockdown in Cultured Tobacco Cells
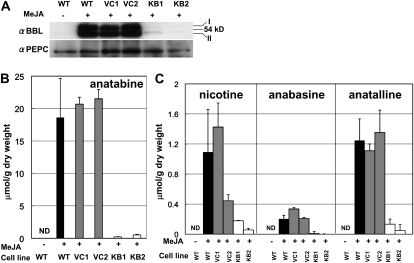
Next, we examined whether BBLs are required for the synthesis of anatabine, anabasine, and anatalline, which consist entirely of the pyridine moiety. Cultured tobacco BY-2 cells mainly produce these pyridine-type alkaloids upon elicitation by MeJA, since they do not synthesize the N-methylpyrrolinium cation, due to very inefficient expression of the N-methylputrescine oxidase genes (Shoji and Hashimoto, 2008). The metabolic impact of BBL suppression was thus examined in the MeJA-elicited BY-2 cells. Two control cell lines (VC1 and VC2) were transformed with an empty vector, and two KB cell lines (KB1 and KB2) were transformed with the BBL RNAi vector. An immunoblot analysis showed that 50 μm MeJA induced the expression of BBL proteins in the wild-type and VC cell lines but not in the KB line cells (Fig. 7A). Anatabine was the major alkaloid in the MeJA-elicited wild-type and VC cells; nicotine and anatalline were the next most abundant alkaloids, and anabasine was a minor alkaloid (Fig. 7B). The amount of anatabine in the two KB cell lines was 1% to 3% of that in the wild-type and VC cell lines. Other tobacco alkaloids (nicotine, anatalline, and anabasine) also accumulated at highly reduced levels in the KB cell lines compared with the wild-type and VC cell lines (Fig. 7C). DMN and other novel metabolites were not detected in the MeJA-elicited KB cell lines when analyzed by gas-liquid chromatography.
Figure 7.
BBL down-regulation in cultured tobacco cells. Cultured BY-2 cells of the wild type (WT), two vector-transformed control cell lines (VC1 and VC2), and two BBL RNAi lines (KB1 and KB2) were treated with 50 μm MeJA for 48h (+), and their protein levels and alkaloid contents were analyzed. Wild-type BY-2 cells were also cultured in the absence of MeJA (−). A, Immunoblot analysis of BBL and PEPC (control) proteins. B, Anatabine content. C, Levels of nicotine, anabasine, and anatalline. Note that the scale is different from that in B. Data indicate mean values ± sd from three replicates. ND, Not detected.
Subcellular Localization of BBL Proteins
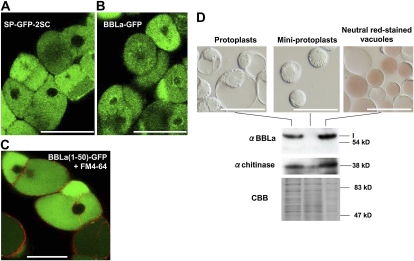
To examine the subcellular localization of the BBL proteins, we expressed a fusion protein comprising the full-length BBLa and GFP in cultured tobacco BY-2 cells and analyzed its distribution by confocal laser microscopy. The cultured cells contained large central vacuoles, which could be labeled by the vacuole-targeting marker SP-GFP-2SC (Mitsuhashi et al., 2000; Fig. 8A). BBLa-GFP was consistently observed in the central vacuoles (Fig. 8B). Accordingly, secretion signal peptides were predicted at the N termini of BBLa (21 amino acids), BBLb (22 amino acids), and BBLc (22 amino acids) by the motif prediction program SignalP (Emanuelsson et al., 2007; Supplemental Fig. S2). To test the functions of these N-terminal sequences, we fused the N-terminal 50 amino acids of BBLs to GFP and expressed BBL(1-50)-GFP proteins in cultured tobacco cells. BBLa(1-50)-GFP was transported to the vacuole, which was demarked by the FM4-64-labeled tonoplast (Fig. 8C). Similarly, BBLb(1-50)-GFP and BBLc(1-50)-GFP were also located in the central vacuoles (Supplemental Fig. S4). These results indicate that the N-terminal region of BBLs contains the vacuolar sorting determinants.
Figure 8.
Subcellular localization of BBLa protein in cultured tobacco cells. A, Full-length BBLa protein was fused at its C terminus to GFP and stably expressed in cultured BY-2 cells. Fluorescence was observed with a laser scanning confocal microscope. B, Confocal image of BY-2 cells expressing SP-GFP-2SC, which had been shown to be located in the vacuoles (Mitsuhashi et al., 2000). C, An N-terminal 50-amino acid fragment of BBLa was fused to GFP and stably expressed in the tobacco cells. The transformed tobacco cells were pulse labeled by the fluorescent dye FM4-64 and inspected 10 h later when the dye had been shown to primarily label the vacuolar membrane (Shoji et al., 2009). Bars = 50 μm. D, Immunoblots of subcellular fractions. Vacuoles and cytoplasm-rich miniprotoplasts were prepared from protoplasts of a BBLa-overexpressing tobacco cell line by Percoll gradient centrifugation. Vacuoles were stained with a 10 μg mL−1 neutral red solution for 10 min. Immunoblots probed with antisera against BBL and class I chitinase (vacuolar resident protein) are shown, together with a Coomassie Brilliant Blue (CBB)-stained gel as a loading control. Bars = 100 μm. [See online article for color version of this figure.]
To further support the vacuolar localization of BBL proteins, vacuole-rich vesicles and cytoplasm-rich miniprotoplasts were purified by Percoll gradient centrifugation of protoplasts prepared from the BBLa-expressing BY-2 cells (Hamada et al., 2004; Fig. 8D). Immunoblotting with an antiserum against an established vacuole-resident protein, class I chitinase (Matsuoka et al., 1995), confirmed the purity of the prepared fractions. A BBL of 58 kD (isoform I) was detected by the antiserum in the protoplasts and the vacuole-rich vesicles but not in the miniprotoplasts, indicating that BBLs are highly enriched in the vacuole-rich vesicles. The signal intensity of a smaller BBL, isoform II (53 kD), was below the detection limit.
Biochemical Properties of Recombinant BBL Proteins Produced in Yeast
To gain insights into the biochemical properties of BBLs, we expressed recombinant BBL proteins as secreted forms in the methyrotrophic yeast Pichia pastoris and purified them from the culture medium. The predicted N-terminal signal sequence of BBLs was substituted with the signal sequence of yeast α-factor, and the Strep tag was added to the C terminus of BBLs. Since we obtained the highest expression level for BBLa, compared with BBLb and BBLc, we purified the recombinant BBLa from the culture medium. The recombinant BBLa (95 kD) was much larger than the theoretical size of BBLa (60 kD), which lacked the predicted secretion signal sequence, and was found to be glycosylated, as revealed by periodic acid-Schiff (PAS) staining (Fig. 9A). When the purified BBLa protein was treated by an endoglycosidase, EndoHf, the deglycosylated form had a theoretical molecular mass of 60 kD and was insensitive to PAS staining (Fig. 9, A and B), indicating that the secreted recombinant BBLa protein was highly N-glycosylated by the endogenous glycosyltransferases of the yeast host. The two endogenous BBL isoforms (53 and 58 kD; lane 2 in Fig. 9B) found in the MeJA-treated tobacco BY-2 cells were smaller than the 60-kD deglycosylated BBLa expressed in yeast (lane 1 in Fig. 9B).
Figure 9.
Biochemical characterization of the recombinant BBLa protein produced in the Pichia cell culture. A, BBLa purified from the culture medium of P. pastoris was glycosylated. The purified protein was treated with (+) the endoglycosidase EndoHf and analyzed by SDS-PAGE. Separated proteins were stained with Coomassie Brilliant Blue (CBB) or the carbohydrate-staining PAS reagent after being transferred to a polyvinylidene difluoride membrane. Approximately 1 μg of the native protein and approximately 5 μg of the deglycosylated protein were loaded in the lanes. B, Immunoblot analysis of the deglycosylated recombinant BBLa protein produced from the Pichia culture (lane 1) and BBL proteins present in MeJA-treated wild-type BY-2 cells (lane 2). The antiserum against BBL was used for detection. C, Absorbance spectra of the deglycosylated recombinant BBLa protein (0.23 mg mL−1 in 10 mm sodium phosphate buffer, pH 7.0) and a standard solution of FAD. D, Fluorescence emission spectra of the deglycosylated recombinant BBLa protein before (control) and after the treatment with sodium dithionite. The protein samples (0.23 mg mL−1 in 10 mm sodium citrate buffer, pH 4.0) were irradiated at 450 nm.
The concentrated solution of the recombinant BBLa protein that had been purified from the yeast culture medium and subsequently deglycosylated was yellowish and showed absorbance maxima at 350 and 439 nm, which resembled the absorbance spectrum of free FAD (Fig. 9C). The fluorescence emission of the BBLa protein exhibited a maximum at 514 nm, and treatment of the enzyme solution with the flavin-reducing reagent sodium dithionite effectively quenched the fluorescence (Fig. 9D). These spectrophotometric properties suggest that the recombinant BBLa protein has a flavin molecule. All the known members of the BBE family are flavoproteins, containing an enzyme-bound FAD (Brandsch et al., 1987; Dittrich and Kutchan, 1991; Carter and Thornburg, 2004; Sirikantaramas et al., 2004). Indeed, a putative flavin-binding site is found in the BBL protein sequences (indicated by a dashed line in Supplemental Fig. S2).
When the recombinant BBLa protein and crude cell extracts from the tobacco root or elicitor-treated cultured tobacco cells were used in the in vitro enzyme assay, we did not detect oxidative conversion of DMN to nicotine or other alkaloids (data not shown). The exact enzymatic reaction catalyzed by BBLs needs to be studied further.
DISCUSSION
Here, we identified novel BBL genes required for the biosynthesis of tobacco alkaloids. Spectrophotometric analyses of a recombinant BBL protein that had been produced and purified from the culture medium of a yeast cell culture suggest that BBLs contain FAD. BBLs belong to an oxidoreductase subfamily that includes BBE (Fig. 1) within a larger vanillyl-alcohol oxidase flavoprotein superfamily (Leferink et al., 2008). The three-dimensional x-ray crystal structures of BBE, glucooligosaccharide oxidase, 6-hydroxy-d-nicotine oxidase, and aclacinomycin oxidoreductase showed that the flavoproteins of the BBE subfamily comprise two domains: a conserved FAD-binding domain and an α/β-domain with a seven-stranded, antiparallel β-sheet forming the less-conserved substrate-binding domain. BBE (Winkler et al., 2008), glucooligosaccharide oxidase (Huang et al., 2005), and aclacinomycin oxidoreductase (Alexeev et al., 2007) bicovalently attach FAD to the protein via two amino acid residues, His and Cys, whereas 6-hydroxy-d-nicotine oxidase covalently binds the flavin cofactor only via a His residue (Koetter and Schulz, 2005). In BBLs, the His residue, which forms the covalent linkage of FAD in these four enzymes, is conserved, but the Cys residue, which is used for the bicovalent linkage, is missing, except for BBLd, whose expression was not detected in the tobacco roots (Fig. 2B), as in the case of 6-hydroxy-d-nicotine oxidase (Supplemental Fig. S2). Besides these flavoproteins, for which crystal structures have been solved, many other BBE subfamily oxidoreductases catalyze the oxidation of a variety of metabolites, with the consumption of molecular oxygen and the production of hydrogen peroxide, and generally contain covalently tethered FAD (Leferink et al., 2008). Covalent flavinylation is thought to increase the redox potential of the cofactor and thus its oxidation power.
Metabolic functions of BBLs were analyzed by suppressing their expression in transgenic tobacco systems that normally synthesize pyridine-type alkaloids. Suppressed expression of BBL genes in tobacco hairy roots and tobacco plants effectively inhibited the formation of nicotine, whereas in jasmonate-elicited cultured tobacco cells, BBL suppression severely inhibited the formation of a major alkaloid, anatabine, as well as of minor tobacco alkaloids, nicotine, anabasine, and anatalline. These pyridine-type alkaloids are the products of condensation between a pyridine ring and another pyridine ring (anatabine, anabasine, and anatalline) or a pyrrolidine ring (nicotine). Therefore, BBLs are required for a presumed step in the activation of nicotinic acid or a condensation reaction yielding bicyclic pyridine alkaloids.
Expression profiles of BBL genes are also consistent with their involvement in the biosynthesis of tobacco alkaloids. Nicotine is formed almost exclusively in the roots, and its synthesis is enhanced by herbivory on leaves by way of the general jasmonate signaling pathway and the specific nicotine regulatory loci, NIC1 and NIC2. Tobacco BBL genes are specifically expressed in the roots, induced by jasmonate treatment, and positively regulated by the NIC loci. Dominant negative suppression and overexpression of a NIC2-locus ERF gene showed that the NIC2 locus regulates the expression of BBL genes. These expression patterns are shared by all known tobacco structural genes involved in nicotine biosynthesis (Shoji et al., 2010).
To narrow down the enzymatic step catalyzed by BBL oxidoreductases, it is informative to analyze metabolites that accumulate in otherwise alkaloid-synthesizing tobacco cells when the BBL reaction is inhibited. A novel alkaloid, DMN, began to accumulate as soon as the expression of BBLs was inducibly suppressed in tobacco hairy roots. DMN has been identified as a metabolite of nicotine in the rat (De Clercq and Truhaut, 1962) and was reported in cigarette smoke (Neurath et al., 1966) but has not been reported in tobacco plants, indicating that the BBL-catalyzed reaction normally proceeds very efficiently in planta. In the BBL-suppressed tobacco plants, DMN was retained in the roots, constituting the major alkaloid in this organ, but was totally absent in the leaves, suggesting that DMN is not transported from the roots to the aerial parts. The root-to-leaf transport system for tobacco alkaloids, possibly involving putative transporters, may not recognize DMN. Alternatively, DMN may not be exported from a subcellular compartment, possibly the vacuole, in the root cells for the long-distance transport. The chemical structure of DMN, in which the N-(methylamino)butyl moiety is attached to the C-3 position of the pyridine ring, shows that the condensation of the pyridine ring and the N-methylpyrroline ring has been completed before the BBL-catalyzed oxidation acts on an unknown reaction intermediate. Possibly, DMN may be an in vivo substrate of BBLs. Our inability to demonstrate the conversion of DMN to nicotine by a yeast-produced BBL protein might be caused by a lack of putative posttranslational processing of the nascent BBL polypeptide in the yeast. The two native BBL isoforms (I and II) detected in alkaloid-producing tobacco cells are considerably smaller in molecular mass than the yeast-produced deglycosylated BBL protein, indicative of posttlanslational processing of BBLs in tobacco cells, which might be important for their catalytic activation. Alternatively, DMN might be derived from an unstable BBL substrate. At least two metabolites accumulated, in addition to DMN, when the BBL reaction was inhibited in the tobacco cells that otherwise produced nicotine. These unidentified metabolites likely contain nitrogen, based on their reactivity toward Dragendorff’s reagent. The accumulation of multiple metabolites suggests that the intermediate accumulating immediately after the blockage of the BBL reaction is unstable and readily metabolized in tobacco cells. An analogous situation is found for the tobacco orphan reductase A622 (Kajikawa et al., 2009). When the expression of A622 was suppressed in alkaloid-producing tobacco cells, the formation of tobacco alkaloids was effectively inhibited, with a concomitant accumulation of nicotinic acid N-glucoside. The glucoside, however, is not a substrate for A622 but is thought to be a detoxification product of nicotinic acid. Although the results presented here strongly suggest that BBLs catalyze a late oxidation step in tobacco alkaloid biosynthesis, we cannot rule out the possibility that BBLs are accessory factors required for the full activities of biosynthetic enzymes.
BBLs are recruited to the vacuole when analyzed as GFP-fused constructs or by subcellular fractionation. The N-terminal 50 amino acid residues of BBLs are sufficient for targeting GFP to the vacuole, probably by way of the endoplasmic reticulum (ER), and contain predictable signal peptides. Interestingly, the characterized BBE family proteins of plant origin all contain N-terminal cleavable signal peptides (Dittrich and Kutchan, 1991; Bird and Facchini, 2001; Carter and Thornburg, 2004; Sirikantaramas et al., 2004; Taura et al., 2007b). The cannabidiolic acid synthase and Δ1-tetrahydrocannabinolic acid synthase of Cannabis sativa are probably glycosylated and recruited to the vacuole (Sirikantaramas et al., 2004; Taura et al., 2007b). The nectarin V protein of ornamental tobacco (Nicotiana langsdorffii × Nicotiana sanderae) possesses Glc oxidase activity and is secreted into the floral nectar (Carter and Thornburg, 2004). The BBE of Papaver somniferum is transported to the vacuole, although it is presumed to be active in the lumen of the ER or an ER-derived vesicle that is destined for the vacuole (Bird and Facchini, 2001). The subcellular localization of BBLs in the vacuole indicates that a late step of nicotine biosynthesis is catalyzed in this organelle or in the lumen of the ER or an ER-derived, vacuole-targeted vesicle. Determining the subcellular distribution of other enzymes involved in nicotine biosynthesis will clarify the compartmentation of the pathway. There may be unexpected transmembrane trafficking of intermediates.
It should be noted that, in tobacco hairy roots, the nornicotine level did not decrease, or actually increased, after knockdown of the BBL genes (Fig. 3). Previously, we observed that levels of nicotine and nornicotine did not correlate in tobacco hairy roots when the regulatory NIC loci were mutated in combinations (Hibi et al., 1994). In the tobacco leaf, nornicotine is synthesized from nicotine in an oxidative demethylation reaction catalyzed by nicotine N-demethylases of the cytochrome P450 monooxygenase subfamily CYP82E (Siminszky et al., 2005). When three known nicotine N-demethylase genes were inactivated by mutations, the triple mutant tobacco plants still contained a small amount of nornicotine (Lewis et al., 2010). There might be an alternative pathway of nornicotine biosynthesis that does not utilize nicotine as the sole, direct intermediate. In addition to its preferred substrate N-methylputrescine, tobacco N-methylputrescine oxidase converts putrescine to Δ1-pyrroline (Heim et al., 2007; Katoh et al., 2007). If Δ1-pyrroline condenses with a derivative of nicotinic acid in tobacco plants, nornicotine could be generated by a route independent of nicotine. BBLs might be dispensable for such a nicotine-independent pathway of nornicotine formation.
MATERIALS AND METHODS
Plant Materials and Genetic Transformation
Binary vectors were introduced into Agrobacterium rhizogenes by electroporation. To generate transgenic hairy roots, leaf discs were prepared from 4- to 6-week-old tobacco (Nicotiana tabacum ‘Petit Havana SR1’) plants, were sterilized, and were inoculated with A. rhizogenes ATCC15834 as described by Kanegae et al. (1994). After selection and disinfection on solid Murashige and Skoog (MS) medium containing 250 mg L−1 cefotaxime and 15 mg L−1 hygromycin (for pXVE-BBLRNAi) or 50 mg L−1 kanamycin (for pHANNIBAL-BBLRNAi), hairy roots were subcultured in liquid MS medium with 3% Suc every 2 weeks. Transgenic tobacco plants were generated with Agrobacterium tumefaciens strain EHA105, as described by Horsch et al. (1985). Transgenic T0 and T1 plants were selected for resistance against 50 mg L−1 kanamycin, and the transgenic plants of the T1 generation were used in this study. Cultured tobacco BY-2 cells (cv Bright Yellow-2) were grown in liquid MS medium supplemented with 20 mg L−1 KH2PO4, 0.5 g L−1 MES, and 0.2 mg L−1 2,4-dichlorophenoxyacetic acid. Transgenic tobacco BY-2 cells were generated with A. tumefaciens strain EHA105, as described by An (1985). Four-day-old BY-2 cells were elicited with 50 μm MeJA in an auxin-free medium (Shoji et al., 2008), whereas 1-month-old tobacco plants (cv Burley 21) were treated with 100 μm MeJA for 1 d, and the root tissues were analyzed for up-regulated expression of nicotine biosynthesis-related genes (Shoji et al., 2002). Seeds of Nicotiana sylvestris and Nicotiana tomentosiformis were obtained from the Leaf Tobacco Research Center of Japan Tobacco, Inc.
Vector Construction
For constitutive expression, the coding region of BBLa cDNA was amplified by PCR and cloned into pGWB2 (Nakagawa et al., 2007). For subcellular localization assays, the coding region of BBLa cDNA excluding the stop codon and the partial fragments (+1–150, with the adenine in the translational start codon ATG as +1) of BBLa, BBLb, and BBLc cDNAs were amplified by PCR and cloned into pGWB5 (Nakagawa et al., 2007). For RNAi-mediated constitutive suppression, a partial fragment of BBLa cDNA (+1,312–1,654) was amplified by PCR, subcloned into pHANNIBAL (Wesley et al., 2001), and then transferred to the binary vector pBI121 (Clontech) to provide pHANNIBAL-BBLRNAi, whereas the inducible RNAi vector pXVE-BBLRNAi was constructed by transferring the RNAi cassette containing an inverted repeat of the partial BBLa fragment and the PDK intron from pHANNIBAL-BBLRNAi to a multicloning site downstream of the OLexA promoter in pER8 (Zuo et al., 2000).
Genomic PCR, RT-PCR, and Quantitative RT-PCR
Genomic DNA was isolated from root tissues of N. tabacum cv Petit Havana SR1, N. sylvestris, and N. tomentosiformis by using the PureLink Plant Total DNA Purification kit (Invitrogen). The following PCR primers were used to amplify genomic DNA (10 ng) with ExTaq DNA polymerase (TaKaRa Bio) for 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s: 5′-AAACTGCTACTGGAGCTGTTAC-3′ and 5′-TCTTCGCCCATGGCTTTTCGGTCT-3′ for BBLa, 5′-ACAAAGAATGATCAAAGTAG-3′ and 5′-TCTTCGCCCATGGCTTTTCGGTCT-3′ for BBLb, 5′-CTACTAGTGGAGCAGGAGAA-3′ and 5′-ACTCCGAATTTTCTGGACAG-3′ for BBLc, 5′-AAGGAATCATGCTGGTAATAG-3′ and 5′-TGCTGGCTCGGGAAATGGCA-3′ for BBLd, and 5′-AGTTGGAGGAGGTGATGATG-3′ and 5′-TATGTGGGTCGCTCAATGTC-3′ for tobacco α-tubulin genes.
Total RNA was isolated by using an RNeasy Plant Mini kit (Qiagen). cDNA was synthesized from 1 μg of total RNA by SuperScript II reverse transcriptase (Invitrogen) and amplified using ExTaq DNA polymerase (TaKaRa Bio) under the following PCR conditions: 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The BBL and α-tubulin cDNAs were amplified with the same primers used for the genomic PCR. The reactions for plant roots were repeated 24 cycles for BBLa and α-tubulin cDNAs and 30 cycles for BBLb, BBLc, and BBLd cDNAs. The number of reaction cycles for hairy roots was 22 for BBLa, BBLb, and BBLc cDNAs and 24 for α-tubulin cDNAs.
For quantitative RT-PCR, cDNA was amplified using the LC480 real-time thermal cycler system (Roche Applied Science) with SYBER Premix ExTaq (Perfect Real Time; TaKaRa Bio). The thermal program was 5 min at 95°C followed with 45 cycles of 10 s at 95°C, 10 s at 60°C, and 10 s at 72°C. The specificity of the reactions was confirmed by the machine’s standard melt curve method. The primers used were 5′-CTGCTGATAATGTCGTTGATGCTC-3′ and 5′-CACCTCTGATTGCCCAAAACAC-3′ for BBLs and 5′-AAGCCCATGGTTGTTGAGAC-3′ and 5′-GTCAACGTTCTTGATAACAC-3′ for the tobacco EF-1α gene as a control.
Production of BBL Antiserum
A partial cDNA fragment of BBLa (+1,006 to the stop codon) was amplified and cloned into pDONR221 (Invitrogen) by BP reaction and transferred into an Escherichia coli expression vector, pET-DEST42 (Invitrogen). The recombinant His-tagged BBLa protein was induced in the E. coli strain BL21 (DE3) (Invitrogen) by the addition of 1 mm isopropylthio-β-galactoside for 3 h at 37°C. The recombinant protein was extracted in denatured extraction buffer (8 m urea, 20 mm sodium phosphate, pH 7.4, 0.5 m NaCl, and 10 mm dithiothreitol) by sonication and purified by nickel-nitrilotriacetic acid agarose (Qiagen) according to the manufacturer’s instructions. Eluted fractions containing the recombinant protein were combined and dialyzed against the extraction buffer containing 4 m urea. The recombinant protein was further purified by gradient elution (from 0 to 1 M NaCl) from the Q-Sepharose Fast Flow column (GE Healthcare). BBLa antisera were produced in rabbits by Hokkaido System Science.
Immunoblot Analysis
Root tissues and cultured cells were frozen in liquid nitrogen, homogenized by mortar and pestle, thawed, and then immediately mixed with CelLytic P Cell Lysis Reagent (Sigma). After centrifugation of the homogenate, 3 μg of soluble protein in the supernatant was separated by SDS-PAGE (10% T) and transferred onto a BioTrace polyvinylidene difluoride membrane (Pall) using a Transblot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). The membrane was blocked in 1× Tris-buffered saline (TBS) buffer containing 3% (w/v) bovine serum albumin for 1 h. Polyclonal anti-BBLa rabbit serum and polyclonal anti-phosphoenolpyruvate carboxylase (PEPC) rabbit serum (Chemicon) were diluted 1:1,000 in 1× TBS buffer containing 0.05% Triton X-100 and incubated with the membranes for 1 h. Similarly, polyclonal anti-class I chitinase rabbit serum was diluted 1:500 in 1× TBS buffer containing 0.05% Triton X-100 and incubated with the membranes for 1 h. After washing, horseradish peroxidase-conjugated anti-mouse IgG from sheep and horseradish peroxidase-conjugated anti-rabbit IgG from donkey (GE Healthcare) were used as secondary antibodies to detect target proteins with the ECL-Plus Western Blotting System (GE Healthcare).
Preparations of Vacuoles and Miniprotoplasts
Vacuoles and miniprotoplasts were prepared from tobacco protoplasts according to Hamada et al. (2004). Five-day-old tobacco BY-2 cells were incubated with an enzyme solution containing 2% Sumizyme C, 0.2% Sumizyme AP2 (both from Shin-Nihonkagaku Industries), and 0.45 m sorbitol, at 30°C (pH 5.5) for 2 h, gently homogenized with a Teflon homogenizer, and fractionated by density gradient centrifugation in a solution containing 37% Percoll, 6.5 mm HEPES-KOH (pH 7.3), 0.49 m Suc, 0.62 m sorbitol, and 0.04 m MgCl2, at 25,000g for 30 min. Cytoplasm-rich miniprotoplasts and vacuole-rich vesicles were separately collected from the corresponding fractions, washed twice with cold 0.6 m mannitol, and suspended in 10 volumes of an ice-cold extraction buffer containing 50 mm HEPES-KOH (pH 7.5), 5 mm EDTA, 0.25 m sorbitol, 2 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, complete protease inhibitor (Roche), and 1 mm dithiothreitol. The fractionated preparations were homogenized, centrifuged at 13,000g at 4°C for 30 min, and subjected to SDS-PAGE.
Confocal Microscopy
Tobacco BY-2 cells expressing GFP-fused proteins were examined with a C1-ECLIPSE E600 confocal laser scanning microscope (Nikon) equipped with an argon laser, the GFP(R)-BP filter, and the HQ-FITC-BP filter for GFP fluorescence or with a helium-neon laser and the G-2A filter for FM4-64 fluorescence. Tonoplasts of tobacco BY-2 cells were labeled with FM4-64 (Invitrogen) at 32 μm for 12 h.
Metabolite Analyses
Plant samples (50 mg dry weight) were lyophilized, homogenized, and soaked in 4 mL of 0.1 n sulfuric acid. The homogenate was sonicated for 15 min and centrifuged at 3,100 rpm for 15 min. The supernatant was neutralized by adding 0.4 mL of 25% ammonia water. The mixture (1 mL) was loaded onto an Extrelut-1 column (Merck) and eluted with 6 mL of chloroform. The eluent was dried at 37°C. The dry residues were dissolved in ethanol containing 0.1% dodecane as an internal standard and analyzed by gas-liquid chromatography (GC-2010; Shimadzu) or gas-liquid chromatography-mass spectrometry (Hewlett Packard 5890 SERIES II/JEOL MStation MS700 system; Agilent) with an Rtx-5 Amine capillary column (i.d. of 0.25 mm, film thickness of 0.50 μm, 30 m; Restek). Nicotine, nornicotine, anatabine, and anabasine were purchased from Wako or Sigma. The anatalline standard was a gift from Dr. K.-M. Oksman-Caldentey (Häkkinen et al., 2004). Dihydrometanicotine was purchased from Toronto Research Chemicals.
Thin-layer chromatography was used to evaluate alkaloid profiles. Alkaloid extracts (10 μL) prepared as above were spotted on Merck silica gel 60 F254 plates (20 × 20 cm, 0.2-mm layer) and developed with a solvent system comprising chloroform:methanol:25% ammonia water (85:15:2, v/v/v). Nitrogen-containing compounds were detected after spraying with Dragendorff’s reagent (Harborne, 1984).
Expression of BBLs in Yeast
A partial coding region of BBLa cDNA (+64 to the stop codon) excluding the putative signal peptide region was fused to the Strep tag II sequence (Schmidt and Skerra, 2007) at the N terminus by PCR and cloned into the PmlI site of pPICZαB (Invitrogen), which was then transformed to the Pichia pastoris SMD1168H strain (Invitrogen) by using the Pichia EasyComp kit (Invitrogen). Pichia cells grown in BMGY medium were collected and resuspended in 1,000 mL of modified BMMY medium containing 100 mm sodium citrate buffer (pH 5.5), 1% (v/v) methanol, 0.5% (w/v) casamino acid, and 10 mg L−1 riboflavin, as described by Taura et al. (2007a). The culture was shaken (90 rpm) at 20°C for 4 d with the addition of a 0.5% volume of methanol every 24 h. The culture was centrifuged, and the supernatant was concentrated by the addition of 70% (w/v) ammonium sulfate for 12 h. The precipitant was dialyzed against 20 mm sodium acetate buffer (pH 5.5). The EndoHf endoglycosidase (1,000 units; New England Biolabs), which had been fused to maltose-binding protein, was added and incubated at 37°C for 3 h. The EndoHf protein was removed by passing the samples through an amylose resin column (New England Biolabs), and the eluents were dialyzed against Strep-Tacin buffer W containing 100 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1 mm EDTA. Recombinant BBLa was purified with a Strep-Tacin Sepharose affinity column (IBA) according to the manufacturer’s instructions and dialyzed against 10 mm sodium phosphate buffer (pH 7.0) or 10 mm sodium citrate buffer (pH 4.0).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers AB604219 (NtBBLa), AM851017 (NtBBLb), AB604220 (NtBBLc), AB604221 (NtBBLd), P30986 (EcBBE), AF049347 (BsBBE), P93479 (PsBBE), AY610511 (TfBBE), P08159 (AoHDNO), AJ507836 (AnHDNO), AB292682 (CsCBDAS), AB057805 (CsTHCAS), AF472609 (HaCHOX), AF472608 (LsCHOX), and AF503442 (NspNEC5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biosynthesis of tobacco alkaloids.
Supplemental Figure S2. Alignment of deduced amino acid sequences of tobacco BBLs and related proteins.
Supplemental Figure S3. Time courses of inducible BBL suppression and alkaloid accumulation in the transgenic tobacco root line iRK2.
Supplemental Figure S4. Subcellular localization of BBLb and BBLc proteins in cultured tobacco cells.
Acknowledgments
We thank Ikuko Nishimura (Kyoto University), Tsuyoshi Nakagawa (Shimane University), Num Hai Chua (Rockefeller University), Peter Waterhouse (Commonwealth Scientific and Industrial Research Organization Plant Industry), and Ken Matsuoka (Kyushu University) for providing the SP-GFP-2SC-expressing tobacco BY-2 cells, pGWB2 and pGWB5 vectors, pER8 vector, pHANNIBAL vector, and class I chitinase antiserum, respectively. We are also grateful to Junko Tsukamoto (Nara Institute of Science and Technology) for gas chromatography-mass spectrometry.
References
- Alexeev I, Sultana A, Mäntsälä P, Niemi J, Schneider G. (2007) Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc Natl Acad Sci USA 104: 6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. (1985) High efficiency transformation of cultured tobacco cells. Plant Physiol 79: 568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127: 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Bird DA, Facchini PJ. (2001) Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213: 888–897 [DOI] [PubMed] [Google Scholar]
- Brandsch R, Hinkkanen AE, Mauch L, Nagursky H, Decker K. (1987) 6-Hydroxy-D-nicotine oxidase of Arthrobacter oxidans: gene structure of the flavoenzyme and its relationship to 6-hydroxy-L-nicotine oxidase. Eur J Biochem 167: 315–320 [DOI] [PubMed] [Google Scholar]
- Cane KA, Mayer M, Lidgett AJ, Michael A, Hamill JD. (2005) Molecular analysis of alkaloid metabolism in AABB v. aabb genotype Nicotiana tabacum in response to wounding of aerial tissues and methyl jasmonate treatment of cultured roots. Funct Plant Biol 32: 305–320 [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. (2004) Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol 134: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Dawson RF, Christman DR, Anderson RC, Solt ML, D’Adamo A, Weiss U. (1956) Biosynthesis of the pyridine ring of nicotine. J Am Chem Soc 78: 2645–2646 [Google Scholar]
- Dawson RF, Christman DR, D’Adamo A, Solt ML, Wolf AP. (1960) The biosynthesis of nicotine from isotropically labeled nicotinic acids. J Am Chem Soc 82: 2628–2633 [Google Scholar]
- Deboer KD, Lye JC, Aitken CD, Su AK, Hamill JD. (2009) The A622 gene in Nicotiana glauca (tree tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Mol Biol 69: 299–312 [DOI] [PubMed] [Google Scholar]
- De Clercq M, Truhaut R. (1962) Sur les mėcanismes d’action de la nicotine. Bull Soc Chim Biol (Paris) 44: 227–234 [PubMed] [Google Scholar]
- Dittrich H, Kutchan TM. (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA 88: 9969–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Goossens A, Häkkinen ST, Laakso I, Seppänen-Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Söderlund H, Zabeau M, et al. (2003) A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA 100: 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkkinen ST, Rischer H, Laakso I, Maaheimo H, Seppänen-Laakso T, Oksman-Caldentey K-M. (2004) Anatalline and other methyl jasmonate-inducible nicotine alkaloids from Nicotiana tabacum cv. By-2 cell cultures. Planta Med 70: 936–941 [DOI] [PubMed] [Google Scholar]
- Hamada T, Igarashi H, Itoh TJ, Shimmen T, Sonobe S. (2004) Characterization of a 200 kDa microtubule-associated protein of tobacco BY-2 cells, a member of the XMAP215/MOR1 family. Plant Cell Physiol 45: 1233–1242 [DOI] [PubMed] [Google Scholar]
- Harborne JB. (1984) Phytochemical Methods: A Guide to Modern Techniques of Plant Analysis, Ed 2. Chapman and Hall, London [Google Scholar]
- Heim WG, Sykes KA, Hildreth SB, Sun J, Lu RH, Jelesko JG. (2007) Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry 68: 454–463 [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y. (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eicholtz D, Rogers SG, Fraley RT. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Huang C-H, Lai W-L, Lee M-H, Chen C-J, Vasella A, Tsai Y-C, Liaw S-H. (2005) Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8α-N1-histidyl FAD. J Biol Chem 280: 38831–38838 [DOI] [PubMed] [Google Scholar]
- Jackson DM, Johnson AW, Stephenson MG. (2002) Survival and development of Heliothis virescens (Lepidoptera: Noctuidae) larvae on isogenic tobacco lines with different levels of alkaloids. J Econ Entomol 95: 1294–1302 [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Hirai N, Hashimoto T. (2009) A PIP-family protein is required for biosynthesis of tobacco alkaloids. Plant Mol Biol 69: 287–298 [DOI] [PubMed] [Google Scholar]
- Kanegae T, Kajiya H, Amano Y, Hashimoto T, Yamada Y. (1994) Species-dependent expression of the hyoscyamine 6β-hydroxylase gene in the pericycle. Plant Physiol 105: 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh A, Shoji T, Hashimoto T. (2007) Molecular cloning of N-methylputrescine oxidase from tobacco. Plant Cell Physiol 48: 550–554 [DOI] [PubMed] [Google Scholar]
- Katoh A, Uenohara K, Akita M, Hashimoto T. (2006) Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol 141: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetter JW, Schulz GE. (2005) Crystal structure of 6-hydroxy-D-nicotine oxidase from Arthrobacter nicotinovorans. J Mol Biol 352: 418–428 [DOI] [PubMed] [Google Scholar]
- Leete E, Liu Y. (1973) Metabolism of [2-3H]- and [6-3H]-nicotinic acid in intact Nicotiana tabacum plants. Phytochemistry 12: 593–596 [Google Scholar]
- Leete E, Slattery SA. (1976) Incorporation of [2-14C]- and [6-14C]nicotinic acid into the tobacco alkaloids: biosynthesis of anatabine and α,β-dipyridyl. J Am Chem Soc 98: 6326–6330 [DOI] [PubMed] [Google Scholar]
- Leferink NGH, Heuts DPHM, Fraaije MW, van Berkel WJH. (2008) The growing VAO flavoprotein family. Arch Biochem Biophys 474: 292–301 [DOI] [PubMed] [Google Scholar]
- Lewis RS, Bowen SW, Keogh MR, Dewey RE. (2010) Three nicotine demethylase genes mediate nornicotine biosynthesis in Nicotiana tabacum L.: functional characterization of the CYP82E10 gene. Phytochemistry 71: 1988–1998 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K. (1995) Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol 130: 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N, Shimada T, Mano S, Nishimura M, Hara-Nishimura I. (2000) Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Neurath G, Dünger M, Gewe J, Lüttich W, Wichern H. (1966) Unterschung der Flüchtigen Basen des Tabakrauches. Beitr Tabakforsch 3: 563–569 [Google Scholar]
- Reed DG, Jelesko JG. (2004) The A and B loci of Nicotiana tabacum have non-equivalent effects on the mRNA levels of four alkaloid biosynthesis genes. Plant Sci 167: 1123–1130 [Google Scholar]
- Saito K, Noma M, Kawashima N. (1985) The alkaloid contents of sixty Nicotiana species. Phytochemistry 24: 477–480 [Google Scholar]
- Schmidt TGM, Skerra A. (2007) The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2: 1528–1535 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. (2008) Why does anatabine, but not nicotine, accumulate in jasmonate-elicited cultured tobacco BY-2 cells? Plant Cell Physiol 49: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. (2011) Nicotine biosynthesis. Ashihara H , Plant Metabolism and Biotechnology. John Wiley and Sons (in press) [Google Scholar]
- Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, Hashimoto T. (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22: 3390–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Ogawa T, Hashimoto T. (2008) Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol 49: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Shoji T, Winz R, Iwase T, Nakajima K, Yamada Y, Hashimoto T. (2002) Expression patterns of two tobacco isoflavone reductase-like genes and their possible roles in secondary metabolism in tobacco. Plant Mol Biol 50: 427–440 [DOI] [PubMed] [Google Scholar]
- Siminszky B, Gavilano L, Bowen SW, Dewey RE. (2005) Conversion of nicotine to nornicotine in Nicotiana tabacum is mediated by CYP82E4, a cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 14919–14924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SJ, Murphy KJ, Birch CD, Hamill JD. (2000) Molecular characterization of quinolinate phosphoribosyltransferase (QPRtase) in Nicotiana. Plant Mol Biol 44: 603–617 [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F. (2004) The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J Biol Chem 279: 39767–39774 [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT. (2004) Nicotine’s defensive function in nature. PLoS Biol 2: E217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taura F, Dono E, Sirikantaramas S, Yoshimura K, Shoyama Y, Morimoto S. (2007a) Production of Δ(1)-tetrahydrocannabinolic acid by the biosynthetic enzyme secreted from transgenic Pichia pastoris. Biochem Biophys Res Commun 361: 675–680 [DOI] [PubMed] [Google Scholar]
- Taura F, Sirikantaramas S, Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S. (2007b) Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett 581: 2929–2934 [DOI] [PubMed] [Google Scholar]
- Watson AB, Brown AM, Colquhoun IJ, Walton NJ, Robins DJ. (1990) Biosynthesis of anabasine in transformed root cultures of Nicotiana species. J Chem Soc Perkin Trans 1990: 2607–2610 [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winkler A, Lyskowski A, Riedl S, Puhl M, Kutchan TM, Macheroux P, Gruber K. (2008) A concerted mechanism for berberine bridge enzyme. Nat Chem Biol 4: 739–741 [DOI] [PubMed] [Google Scholar]
- Xu D, Shen Y, Chappell J, Cui M, Nielsen M. (2007) Biochemical and molecular characterization of nicotine demethylase in tobacco. Physiol Plant 129: 307–319 [Google Scholar]
- Zuo J, Niu Q-W, Chua NH. (2000) Technical advance. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]