Abstract
The likelihood of duplicate gene retention following polyploidy varies by functional properties (e.g. gene ontologies or protein family domains), but little is known about the effects of whole-genome duplication on gene networks related by a common physiological process. Here, we examined the effects of both polyploid and nonpolyploid duplications on genes encoding the major functional groups of photosynthesis (photosystem I, photosystem II, the light-harvesting complex, and the Calvin cycle) in the cultivated soybean (Glycine max), which has experienced two rounds of whole-genome duplication. Photosystem gene families exhibit retention patterns consistent with dosage sensitivity (preferential retention of polyploid duplicates and elimination of nonpolyploid duplicates), whereas Calvin cycle and light-harvesting complex gene families do not. We observed similar patterns in barrel medic (Medicago truncatula), which shared the older genome duplication with soybean but has evolved independently for approximately 50 million years, and in Arabidopsis (Arabidopsis thaliana), which experienced two nested polyploidy events independent from the legume duplications. In both soybean and Arabidopsis, Calvin cycle gene duplicates exhibit a greater capacity for functional differentiation than do duplicates within the photosystems, which likely explains the greater retention of ancient, nonpolyploid duplicates and larger average gene family size for the Calvin cycle relative to the photosystems.
Polyploidy (whole-genome duplication [WGD]) has played an important role in the evolutionary history of angiosperms and has even been suggested to underlie their origin and radiation (De Bodt et al., 2005) as well as increasing the likelihood of surviving the Cretaceous-Tertiary extinction (Fawcett et al., 2009). It has been estimated that 15% of angiosperm speciation events involved polyploidy (Wood et al., 2009), and based solely on chromosome numbers, 30% or more of flowering plants are polyploid (Soltis et al., 2009). Synteny data from sequenced genomes provide evidence for a hexaploidy event in the common ancestor of the two largest clades of eudicots (Tang et al., 2008), and chromosomal diploids such as Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and poplar (Populus trichocarpa) show evidence of additional, subsequent polyploid duplications (Bowers et al., 2003; Sterck et al., 2005; Zhang et al., 2005; Tuskan et al., 2006). Thus, the true percentage of flowering plant taxa that are paleopolyploids is certainly higher, and it is clear from these species and from other, less fully characterized taxa (Blanc and Wolfe, 2004a; Schlueter et al., 2004; Pfeil et al., 2005; Cui et al., 2006; Schranz and Mitchell-Olds, 2006; Town et al., 2006; Barker et al., 2008) that flowering plant genomes comprise nested sets of duplications.
Much effort has been made to identify emergent effects of polyploidy: the universal “rules” by which polyploidy functions (Doyle et al., 2008). In Arabidopsis, genomic studies have begun to elucidate distinct patterns of gene retention and loss, correlating with functional classification, for both polyploid and nonpolyploid (NP) duplications (Blanc and Wolfe, 2004b; Seoighe and Gehring, 2004; Maere et al., 2005; Freeling and Thomas, 2006). Transcription factors and kinases, for example, exhibit high retention rates following polyploidy and low retention rates following NP duplication (Blanc and Wolfe, 2004b; Maere et al., 2005). Conversely, several Gene Ontology (GO) categories, including DNA metabolism, show the opposite pattern. Although taxonomic sampling is limited to date, in many cases these patterns appear to hold across a range of species. Grouping genes by protein family domains, Paterson et al. (2006) observed similar patterns across Arabidopsis, rice, yeast, and pufferfish. Barker et al. (2008) found consistent patterns of retention and loss by GO class across all tribes of Compositae, which have evolved separately for more than 30 million years following a shared paleopolyploid duplication. It is noteworthy, however, that these patterns differ substantially from those observed in Arabidopsis, suggesting that, at least in some cases, such patterns are lineage specific (Barker et al., 2008).
Despite the considerable progress that has been made in elucidating patterns of duplicate gene retention following polyploidy, as well as mechanisms driving these patterns, little is yet known of the effect of polyploidy on the gene networks that underlie key physiological or developmental processes. The behavior of genes has been studied in the context of individual gene families (Adams and Wendel, 2005), protein domains (Paterson et al., 2006), GO categories (Blanc and Wolfe, 2004b; Seoighe and Gehring, 2004; Maere et al., 2005; Freeling and Thomas, 2006), and coexpressed networks (Blanc and Wolfe, 2004b) but not in the framework of a physiological process.
The objective of this study was to characterize the effects of polyploidy, as well as NP duplications, on the network of functionally interrelated genes underlying photosynthesis, a key determinant of the ecological success and economic utility of plants. Photosynthesis is a prime example of how polyploids can differ phenotypically from their diploid progenitors. Polyploids consistently exhibit larger mesophyll cells with more chloroplasts and greater photosynthetic capacities per cell than their diploid progenitors (for review, see Warner and Edwards, 1993). The causes of these differences at the level of underlying genes are unknown.
The legume genus Glycine, which includes the cultivated soybean (Glycine max), has a history of recurring polyploidy. In addition to two paleopolyploidy events in the lineage leading to soybean (Fig. 1), the wild, perennial relatives of soybean underwent a burst of genome duplications within the last 100,000 years involving various combinations of extant diploid genomes (Doyle et al., 2004). Thus, Glycine is an attractive system for studying patterns of genome evolution following polyploidy, particularly in light of the fact that the soybean genome sequence was recently completed (Schmutz et al., 2010). Here, we utilized the genomic resources of soybean to investigate how two nested rounds of WGD, as well as NP duplications, have shaped the structure of photosynthetic gene families. Because the legume genus Medicago shared the oldest polyploidy event with Glycine (Fig. 1; Pfeil et al., 2005), we performed similar analyses on the model species barrel medic (Medicago truncatula) in order to determine the effects of a common polyploidy event in independently evolving lineages. Finally, we extended these analyses to the more distantly related eudicot model species Arabidopsis, which has also experienced two well-characterized paleopolyploid events (Fig. 1; Blanc et al., 2003; Bowers et al., 2003; Thomas et al., 2006), in order to look for patterns emerging from independent sets of nested genome duplications. Thus, in total, we have analyzed photosynthetic gene family evolution across three plant species and four genome duplication events.
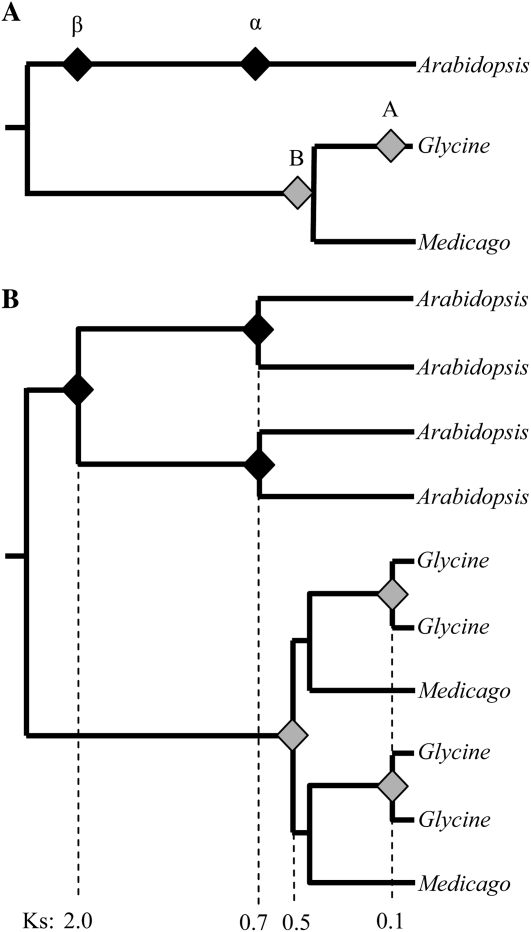
Figure 1.
Estimated timing of genome duplication events in Arabidopsis, Glycine, and Medicago. A, A simplified species tree showing the relative maximum ages of genome duplication events (designated by diamonds) as estimated by homeolog divergences (Ks). For duplication events in Arabidopsis, we follow the naming convention of Bowers et al. (2003), in which the most recent WGD is designated α and the older event is designated β. The duplication events in Glycine are designated A and B to highlight the fact that they are distinct from (and more recent than) the Arabidopsis WGDs. Because Medicago shared the B duplication event with Glycine, we refer to this duplication as B in Medicago as well, even though this is the most recent WGD in the Medicago lineage. B, A gene tree showing the expected topology if all homeologs have been retained in all three species.
RESULTS
Proteins of the Calvin cycle (CC), PSII, and PSI are encoded by 11, nine, and nine distinct nuclear gene families, respectively, whereas light-harvesting complex (LHC) genes cluster into 12 distinct but distantly related nuclear gene families (Das, 2004). Additional CC, PSII, and PSI proteins are encoded by the chloroplast, but because the focus of this study was on duplication events within the nuclear genome, plastid-encoded genes were not analyzed here. In all three species, CC gene families were the largest on average, and PSI gene families were the smallest (Table I). Glycine has experienced the most recent polyploid duplication of the three genera examined, and average photosynthetic gene family size was about twice as large in soybean as in barrel medic or Arabidopsis (Table I).
Table I. Photosynthetic gene families by functional group, and their sizes in soybean, barrel medic, and Arabidopsis.
| Functional Group | Protein | Gene Family Size | ||
| Soybean | Barrel Medic | Arabidopsis | ||
| CC | RbcS | 10 | 6 | 4 |
| FBPase | 10 | 3 | 3 | |
| TPI | 6 | 2 | 2 | |
| PGK | 4 | 2 | 3 | |
| GAPDH | 13 | 5 | 7 | |
| FBA | 14 | 4 | 8 | |
| TKL | 12 | 2 | 2 | |
| RPE | 4 | 3 | 3 | |
| PRI | 9 | 3 | 4 | |
| PRK | 2 | 1 | 1 | |
| SBPase | 3 | 1 | 1 | |
| Total/average | 87/7.9 | 32/2.9 | 38/3.5 | |
| PSII | PsbO | 4 | 2 | 3 |
| PsbP | 4 | 2 | 2 | |
| PsbQ | 3 | 1 | 2 | |
| PsbR | 2 | 1 | 1 | |
| PsbS | 3 | 1 | 1 | |
| PsbTn | 4 | 3 | 2 | |
| PsbW | 4 | 2 | 1 | |
| PsbX | 5 | 4 | 1 | |
| PsbY | 4 | 3 | 1 | |
| Total/average | 33/3.7 | 19/2.1 | 14/1.6 | |
| PSI | PsaD | 2 | 1 | 2 |
| PsaE | 4 | 2 | 2 | |
| PsaF | 1 | 1 | 1 | |
| PsaG | 2 | 1 | 1 | |
| PsaH | 4 | 1 | 2 | |
| PsaK | 2 | 1 | 1 | |
| PsaL | 2 | 2 | 1 | |
| PsaN | 2 | 1 | 1 | |
| PsaO | 2 | 1 | 1 | |
| Total/average | 21/2.3 | 11/1.2 | 12/1.3 | |
| LHC | LhcA1 | 2 | 1 | 1 |
| LhcA2 | 4 | 1 | 2 | |
| LhcA3 | 2 | 1 | 1 | |
| LhcA4 | 2 | 1 | 1 | |
| LhcA5 | 2 | 1 | 1 | |
| LhcA6 | 2 | 1 | 1 | |
| LhcB1 | 8 | 6 | 5 | |
| LhcB2 | 2 | 1 | 3 | |
| LhcB3 | 2 | 1 | 1 | |
| LhcB4 | 4 | 2 | 3 | |
| LhcB5 | 4 | 1 | 2 | |
| LhcB6 | 4 | 1 | 1 | |
| Total/average | 38/3.2 | 18/1.5 | 22/1.8 | |
| Combined | Total/average | 181/4.4 | 81/2.0 | 86/2.1 |
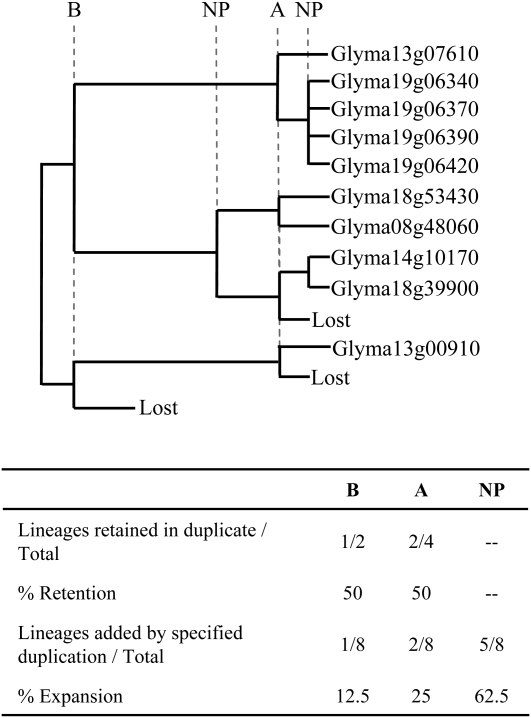
We quantified the contributions of the various duplication events to each gene family using two parameters: percentage retention and percentage expansion (Fig. 2). Percentage retention measures the percentage of genes duplicated by polyploidy that have survived in duplicate, and percentage expansion measures the relative contributions of polyploid versus NP duplications to current gene family size (for details, see “Materials and Methods”). Figure 2 illustrates these calculations using the Rubisco small subunit (RbcS) gene family in soybean. Gene trees and corresponding estimates of retention and expansion for each gene family and species are available in Supplemental File S1.
Figure 2.
An example of percentage retention and percentage expansion calculations, using the RbcS gene family in soybean. The RbcS protein is encoded by 10 genes in soybean, dispersed on five different chromosomes (8, 13, 14, 18, and 19; Schmutz et al., 2010). Gene pairs identified as homeologs from either the A or B polyploidy event reside in or near syntenic blocks and have the expected number of synonymous substitutions per synonymous site (Ks) for that duplication (for details, see “Materials and Methods”). For example, Glyma13g07610 and the tandem duplicates on chromosome 19 reside in a syntenic block spanning 50 genes on chromosome 13 and 77 genes on chromosome 19, with a mean Ks = 0.18. All of the 10 RbcS gene family members descended from one of two pre-B ancestors, such that the gene family has expanded by eight gene lineages. One of the two gene lineages added by the B duplication was subsequently lost, whereas two of four gene lineages added by A were subsequently lost. In addition to the two polyploid duplications, one NP duplication took place between B and A, and four NP duplications took place between A and the present. The four tandemly duplicated genes on chromosome 19 are the result of three recent and nearly simultaneous NP duplications.
Photosynthetic Homeolog Retention in Soybean Is High Compared with the Genome-Wide Average and Differs by Functional Group
Figure 3 summarizes retention of polyploid duplicates by photosynthetic gene family and species. Across all photosynthetic gene families in soybean, 78.7% (70 of 89) of pre-A gene lineages have retained duplicates from the A (most recent) polyploidy event, and 84.9% (152 of 179) of photosynthetic genes present today have a homeolog from the A duplication. In contrast, based on duplicate retention within internal synteny blocks, Schmutz et al. (2010) estimated that 43.4% of genes have retained homeologs from the A duplication genome wide. Thus, photosynthetic gene families exhibit a significantly higher rate of retention from the A duplication (χ21 = 124.3, P < 1.0 × 10−8) than the genome-wide average (Fig. 4A). Using a modified approach incorporating gene phylogenies (see “Materials and Methods”), we obtained a similar but slightly higher estimate of genome-wide retention (52.2%). Even compared with this upper estimate, photosynthetic gene families exhibit a significantly higher rate of retention from the A duplication (χ21 = 76.3, P < 1.0 × 10−8) than the genome-wide average.
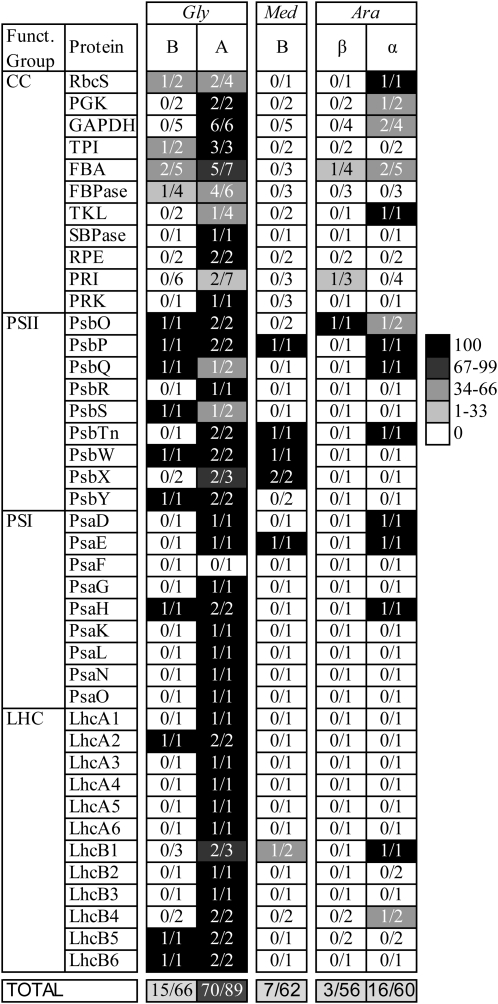
Figure 3.
Percentage retention of homeologs, given by gene family and species, for the CC, PSII and PSI, and LHC. Shading indicates percentage retention, and values indicate the number of gene lineages retained in duplicate over the number of gene lineages initially duplicated by the specified polyploidy event. Gly, soybean; Med, barrel medic; Ara, Arabidopsis.
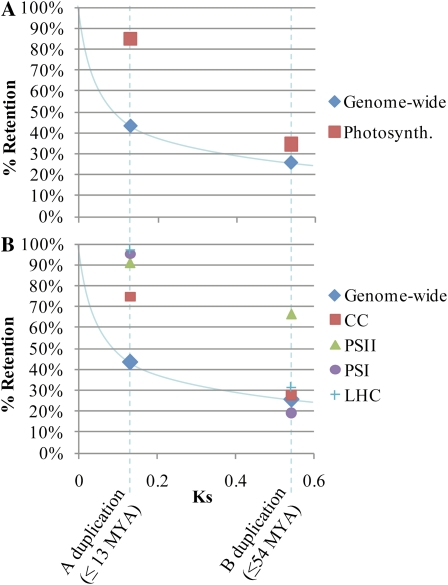
Figure 4.
Observed retention rates for photosynthetic genes following polyploidy in soybean compared with synteny-based estimates of genome-wide retention (Schmutz et al., 2010). A, Percentage retention of photosynthetic genes versus genome-wide retention. B, Percentage retention of photosynthetic genes, showing each functional group separately, versus genome-wide retention. The curved blue line in both panels shows the inferred genome-wide homeolog decay curve. MYA, Million years ago; Photosynth., all photosynthetic genes combined. [See online article for color version of this figure.]
Although retention is high for photosynthetic gene families overall, retention rates following the A duplication differ significantly among the different functional groups (Fig. 4B). For the CC, 74.7% (65 of 87) of genes present today retain duplicates from the A polyploidy event. In contrast, within the three thylakoid membrane-associated protein complexes (PSII, PSI, and LHC), duplicate retention is much higher (90.9%, 95.2%, and 97.4%, respectively). Combined, retention for the three thylakoid-associated functional groups (94.7%) is significantly higher than for the CC (χ21 = 13.8, P = 2.0 × 10−4). However, despite lower retention than the thylakoid complexes, the CC still had higher retention of duplicates from the A polyploidy event than the upper estimate for the genome-wide average (χ21 = 17.6, P = 2.7 × 10−5).
In slight contrast to the significantly higher retention of A-homeologs than the genome-wide average, photosynthetic gene families overall have fractionated (returned to singleton status) at a rate more similar to the genome-wide level following the B duplication (Fig. 4A). For all photosynthetic gene families, 22.7% (15 of 66) of pre-B gene lineages have retained both duplicates from the B polyploidy event, and 34.6% (62 of 179) of photosynthetic genes present today retain homeologs from the B duplication. Based on internal synteny analysis, 25.9% of present-day genes genome wide retain duplicates from the B event (Schmutz et al., 2010). Thus, whereas retention of duplicate photosynthetic genes is double the genome-wide average following the A polyploidy event, retention is only 34% higher following the B polyploidy event. Nonetheless, even for the B duplication, retention in photosynthetic gene families is significantly higher than the genome-wide average (χ21 = 7.0, P = 0.008).
As with the A duplication, retention rates vary considerably by photosynthetic functional group following the B duplication (Fig. 4B). CC (27.6%, 24 of 87), PSI (19.0%, four of 21), and LHC (31.6%, 12 of 38) all exhibit equivalent retention rates to each other (χ22 = 1.1, P = 0.5851) and to the synteny-based estimate (25.9%) for the genome-wide average (χ21 < 1.0, P > 0.42). In contrast, duplicate retention in PSII is significantly higher (66.7%, 22 of 33) than in the other three functional groups (χ23 = 19.275, P = 2.4 × 10−4) and compared with the genome-wide average (χ21 = 28.4, P = 1.0 × 10−7).
The Contributions of Polyploid and NP Duplications to Gene Family Expansion Differ by Functional Group in Soybean
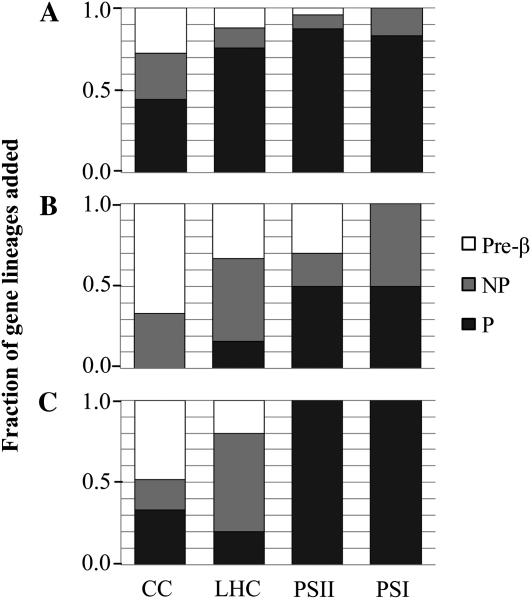
Supplemental Figure S1 shows the contributions of polyploidy and NP duplications to gene family expansion for each photosynthetic gene family. In soybean, the fraction of gene families retaining NP duplicates differs significantly among the four functional groups (P = 0.02, Fisher’s exact test). NP duplications have made a larger contribution to the expansion of CC gene families than to the expansion of LHC, PSII, or PSI (Table II; Fig. 5A). Seven of 11 CC gene families (64%) have expanded via NP mechanisms, compared with only two of nine (22%), one of nine (11%), and one of 12 (8%) for PSII, PSI, and LHC, respectively (Supplemental Fig. S1). On average, NP duplications have contributed 27.2% of total gene family expansion in the CC compared with 7.4%, 8.3%, and 5.0% for PSII, PSI, and LHC, respectively. Thus, gene family expansion is strongly biased toward polyploid duplication in the thyakoid-associated complexes and more balanced between polyploid and NP duplications for the enzymes of the CC (Table II; Fig. 5A).
Table II. Average percentage retention and percentage expansion by photosynthetic functional group following duplication events in soybean, barrel medic, and Arabidopsis.
Retention and expansion values were obtained by averaging the values from each gene family within a functional group. n, Number of nuclear gene families associated with each functional group; –, percentage retention not calculated for NP duplicates because, unlike polyploid duplicates, no record remains for those that were lost.
| Species | Percentage Retention/Percentage Expansion | ||||
| CC (n =11) | PSII (n = 9) | PSI (n = 9) | LHC (n = 12) | Combined (n = 41) | |
| Soybean, B | 15.0/6.9 | 66.7/25.9 | 11.1/4.2 | 25.0/8.3 | 28.4/11.0 |
| Soybean, A | 76.5/65.8 | 85.2/66.7 | 88.9/87.5 | 97.2/86.7 | 87.9/76.7 |
| Soybean, NP | –/27.2 | –/7.4 | –/8.3 | –/5.0 | –/12.3 |
| Barrel medic, B | 0.0/0.0 | 44.4/70.0 | 11.1/50.0 | 4.2/25.0 | 13.4/47.3 |
| Barrel medic, NP | –/100.0 | –/30.0 | –/50.0 | –/75.0 | –/52.7 |
| Arabidopsis, β | 5.3/17.9 | 11.1/12.5 | 0.0/0.0 | 0.0/0.0 | 6.3/15.3 |
| Arabidopsis, α | 30.9/50.0 | 38.9/87.5 | 33.3/100.0 | 12.5/31.3 | 25.4/57.0 |
| Arabidopsis, NP | –/32.1 | –/0.0 | –/0.0 | –/68.7 | –/27.8 |
Figure 5.
Fraction of gene lineages added by duplication type (polyploid [P], NP, pre-B/β) for the four photosynthetic functional groups. A, Soybean. B, Barrel medic. C, Arabidopsis.
The CC has also retained more duplicates that predate the B polyploidy event than have the thylakoid-associated complexes (Fig. 5A; Supplemental Fig. S2). We utilized Phytozome gene clusters (http://www.phytozome.net) to test if any of these pre-B duplications were part of the ancient hexaploidy shared by all eudicots (Tang et al., 2008). Phytozome gene clusters reconstruct ancestral gene sets for key phylogenetic nodes, including “Rosid (pre-hexaploidy)” and “Rosid (post-hexaploidy)” (Schmutz et al., 2010). Soybean genes that cluster at the Rosid (pre-hexaploidy) node but not at more recent nodes [such as Rosid (post-hexaploidy), “Eurosid I,” or “Legume”) were likely derived from this paleohexaploidy. None of the 21 pre-B duplications in the CC fit these criteria (15 of the 21 clustered only at older nodes, and six clustered at the more recent Eurosid I node). Additionally, the pre-B duplication in the phosphoglycerate kinase (PGK) gene family was a tandem duplication (that was subsequently duplicated again by the A polyploidy event to yield two tandem pairs, Glyma08g17600/Glyma08g17610 and Glyma15g41540/Glyma15g41550), providing additional evidence against WGD in this case.
In contrast to the 21 pre-B duplications observed in the CC, only four pre-B duplications were detected across the three thylakoid-associated functional groups. Two of these (one in the PSII gene family, PsbX, and one in the LHC gene family, LhcB4) cluster at nodes more recent than Rosid (pre-hexaploidy), and the other two (both in the LhcB1 gene family) cluster at older nodes. Thus, we find no evidence that any of the pre-B duplications in photosynthetic gene families were the result of the paleohexaploidy; instead, they were most likely NP duplications. The greater number of retained pre-B duplicates in the CC compared with the thylakoid-associated complexes, therefore, is consistent with the greater number of post-B NP duplications in the CC.
In soybean, therefore, PSII exhibits high levels of retention following polyploid duplication and minimal contribution from NP duplication to gene family expansion. In contrast, the CC exhibits the opposite pattern, with comparatively low levels of polyploid duplicate retention and a greater contribution from NP duplication, including a large number of pre-B NP duplications, to gene family expansion. PSI and the LHC exhibit intermediate patterns, with high retention of duplicates from the A WGD, comparable to PSII, but low retention of duplicates from the B WGD, comparable to the CC. As with PSII, NP duplications have made little contribution to gene family expansion in PSI and the LHC.
Patterns of Retention and Expansion Observed in Soybean Are Repeated across Species
The B polyploidy event took place in the common ancestor of Glycine and Medicago shortly before the two lineages diverged (Pfeil et al., 2005; Fig. 1A). We examined retention and expansion of photosynthetic gene families in barrel medic to see if similar patterns developed following the same WGD in an independently evolving lineage. As with soybean, PSII exhibits the highest average rate of retention of B duplicates, with the other three functional groups exhibiting notably lower retention rates (Table II). Also as with soybean, CC gene families exhibit the highest contributions of NP duplications to gene family expansion in barrel medic as well as the greatest number of pre-B duplications (Table II; Fig. 5B). Thus, consistent with the patterns observed in soybean, gene family expansion is biased toward polyploid duplication in the two photosystems (and, to a lesser extent, the LHC) and toward NP duplications in the CC (Table II Fig. 5B) in barrel medic.
The lineage leading to Arabidopsis experienced two polyploidy events, designated β and α (Bowers et al., 2003; Fig. 1A), subsequent to the ancient hexaploidy at the base of the eudicots, and after divergence from the lineage (Eurosid I) that gave rise to legumes. Therefore, we examined duplicate retention and expansion of photosynthetic genes in Arabidopsis in order to see if the patterns observed in the two legume species also emerged following these completely independent (and older) duplications (Fig. 1).
Across all photosynthetic gene families in Arabidopsis, 26.7% (16 of 60) of pre-α gene lineages have retained duplicates from the α polyploidy event and 43.0% (37 of 86) of photosynthetic genes present today have a homeolog from the α duplication. After collapsing recent tandem duplicates into a single locus, as per Thomas et al. (2006), 40.5% (32 of 79) of photosynthetic genes present today retain homeologs from the α duplication. In contrast, across the whole genome, 28.5% (6,329 of 22,209) of genes retain α-homeologs (Thomas et al., 2006). Thus, as in soybean, photosynthetic genes in Arabidopsis have significantly higher retention of α duplicates than the genome-wide average (χ21 = 5.566, P = 0.018).
Also consistent with soybean, retention rates following the α duplication differed significantly among the different functional groups in Arabidopsis. Again, the CC has a retention rate (40.0%) approaching the genome-wide average (χ21 = 2.268, P = 0.132), whereas PSII (57.1%) is significantly higher (χ21 = 4.314, P = 0.038; Yate’s correction). PSI (50%) also exhibits higher retention than the genome-wide average, although due to the small numbers of genes involved, the difference is not statistically significant (χ21 = 1.768, P = 0.184). In contrast to soybean, the LHC has low retention of α-homeologs in Arabidopsis (22.2%), comparable to the genome-wide average (χ21 = 0.348, P = 0.555).
Again similar to the pattern observed in soybean, despite higher retention of duplicates following the α polyploidy event, photosynthetic genes in Arabidopsis exhibit retention rates following the β duplication that are lower than the genome-wide average. Across all photosynthetic gene families, only 10.1% (eight of 79) retain β-homeologs, compared with 21.4% (2,874 of 13,449) across the whole genome (χ21 = 5.922, P = 0.015; Bowers et al., 2003).
Consistent with the two legume lineages, PSII exhibits the highest retention rate from both polyploidy events in Arabidopsis (Table II). Also consistent with the legume lineages, the CC has the highest fraction of gene families (four of 11) that have expanded via NP duplication and higher percentage expansion via NP duplications than either photosystem (Table II; Supplemental Fig. S1). In addition, as in the legume species, CC gene families have retained more pre-β duplicates than any of the thylakoid-associated functional groups in Arabidopsis (Fig. 5C; Supplemental Fig. S2). Tang et al. (2008) generated gene clusters, including Arabidopsis genes, representing ancestral genes that were duplicated by the ancient hexaploidy event. We checked to see if any pre-β duplications collapse into these gene clusters, indicating that they were the result of the hexaploidy event. Of the 13 pre-β duplications within the CC, only one (in PGK) could be assigned to the hexaploidy. Thus, the majority of these pre-β duplications were likely also NP duplications. Of the two pre-β duplications in LHC gene families, one (LhcB4) was assigned to the hexaploidy. Neither photosystem retained pre-β duplications.
Thus, consistent patterns emerge across three species and two independent sets of WGDs. First, photosynthetic genes overall have higher retention of duplicates from the most recent polyploidy events than the genome-wide average, although CC genes exhibit retention rates comparable to the genome-wide average. Second, following older polyploidy events, photosynthetic genes overall exhibit fractionation comparable to or greater than the genome-wide average. Third, of the photosynthetic functional groups, PSII exhibits the highest retention of polyploid duplicates in the long term (Table II). Fourth, polyploid duplications have contributed more to gene family expansion in both photosystems than in the CC (Table II; Fig. 5). Fifth, the CC exhibits the highest level of gene family expansion via NP duplication (Fig. 5; Supplemental Fig. S2), including very old NP duplications that predate the B/β polyploidy events.
No Patterns of Duplicate Retention or Expansion Are Observed at the Gene Family Level
Despite consistent patterns at the level of photosynthetic functional groups, there is a striking absence of pattern in terms of homeolog retention at the level of individual gene families, whether looking across nested duplications within species, shared duplication events across species, or independent duplication events. For example, of the 23 gene families that have retained homeologs in Arabidopsis or soybean, only six have retained duplicates in both species (Fig. 3), and of the 15 photosynthetic gene families that have retained duplicates from polyploidy in Arabidopsis, only two have retained duplicates from both α and β (Fig. 3). Comparing percentage retention values across the 41 gene families, we observed negligible correlation (r ≤ 0.23) when comparing the B polyploidy event in soybean with either polyploidy event in Arabidopsis or when comparing the two nested duplications within Arabidopsis.
CC Duplicates Exhibit Greater Functional Divergence Than Duplicates in the Thylakoid-Associated Complexes
Consistent differences in retention and expansion at the level of the four photosynthetic functional groups suggest that different evolutionary forces are acting upon duplicates within each group. Because a common explanation for duplicate retention is functional differentiation, we looked for evidence of positive selection and/or expression divergence between duplicated photosynthetic genes.
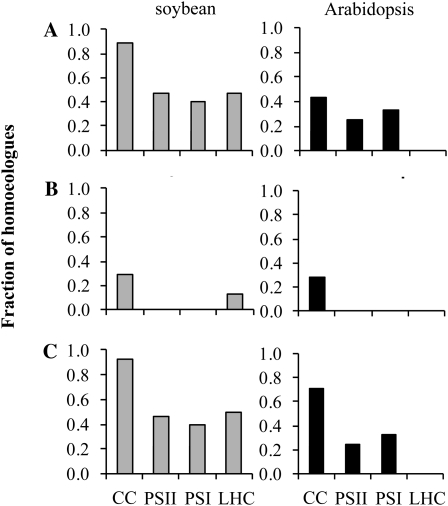
Global ω (synonymous substitutions per synonymous site [Ka/Ks]) was measured for gene pairs resulting from each duplication mechanism (Supplemental File S2). All photosynthetic gene family members in all four classes appeared to be under purifying selection in all three taxa (ω < 1; Supplemental Table S1). We then looked for local signatures of positive selection within sliding windows of both sequence and spatial domains (windows of either 30 adjacent codons in the primary sequence or windows of amino acid residues contained within 10-Å spheres in the folded protein) for duplicates from the most recent (A/α) polyploidy events in soybean and Arabidopsis. Using these more sensitive approaches, we found evidence for positive selection (ω > 1.2) within local domains for several gene families (Supplemental File S2). In soybean, the majority of A-homeolog pairs of CC genes (25 of 28) show evidence of positive selection, including duplicates from every gene family except phosphoribulokinase (PRK), whereas fewer than half of photosystem or LHC homeologs exhibit signatures of positive selection (Fig. 6A; Supplemental Table S1).
Figure 6.
Fractions of homeolog pairs from the most recent polyploidy event (α or A) in soybean and Arabidopsis exhibiting evidence for functional divergence. A, Fractions of homeologous duplicates exhibiting signatures of positive selection (sliding-window ω). B, Fractions of homeologous duplicates exhibiting divergence in expression profiles (r < 0.5). C, Fractions of homeologous duplicates exhibiting positive selection, expression divergence, or both.
In Arabidopsis, three of seven α-homeolog pairs of CC genes exhibit signatures of positive selection (RbcS, PGK, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH); Supplemental File S2). In each of the two photosystems, positive selection was detected for one of three homeolog pairs (PsaH from PSI and PsbQ from PSII). Only two α-homeolog pairs remain for LHC genes, and no evidence of positive selection was detected for either.
We explored the expression profiles of duplicated photosynthetic genes using data from several RNA-Seq experiments in soybean (Bolon et al., 2010, Libault et al., 2010; Supplemental File S3). CC duplicates exhibited lower average correlation coefficients than photosystem or LHC duplicates, regardless of duplication mechanism (Supplemental Table S2). For example, all duplicate pairs from the A polyploidy event maintain highly correlated expression profiles in PSI (r ≥ 0.85) and PSII (r ≥ 0.94). LHC gene pairs from the A duplication exhibited a somewhat greater diversity in expression profiles, with 15 of 17 A duplicates highly correlated (r ≥ 0.80), but two pairs (from LhcB1 and LhcB6) showed evidence of expression divergence (r < 0.3). In contrast, for 28 CC gene pairs from the A duplication for which both copies are expressed, eight exhibited divergent expression profiles (r < 0.55), including negative values for two pairs (from triose phosphate isomerase [TPI] and ribose 5-phosphate isomerase [PRI]; Fig. 6B; Supplemental File S3).
We also explored the expression profiles of duplicated photosynthetic genes in Arabidopsis using public microarray data. Similar to soybean, duplicate genes from both photosystems exhibit highly correlated expression profiles (r > 0.9) regardless of duplication type (Supplemental Table S2; Supplemental File S3). The only exceptions were a pair of α-homeologs for PsbP (r = 0.55) and a pair of β-homeologs for PsbO (r = 0.08). In both cases, one of the two copies was effectively silent in all tissues and conditions examined (www.genevestigator.org; data not shown). Also similar to soybean, LHC duplicates exhibit a slightly greater diversity of expression profiles. Pre-β duplicates of LhcB4 and LhcB5 genes exhibit lower levels of coexpression (r = 0.68 and 0.52, respectively), but the three more recent duplicates for which expression profiles can be discriminated are highly coexpressed (ravg = 0.93, rmin = 0.90). The pre-β LhcB5 duplication involves a gene (AT1G76570) that appears to encode a structurally distinct LHC protein, with four transmembrane domains instead of the canonical three (making it more like the four-transmembrane protein, PsbS; Klimmek et al., 2006). This gene and one of the genes involved in the pre-β LhcB4 duplication exhibit expression profiles more like PsbS. Due to these structural and expression differences, Klimmek et al. (2006) proposed to reclassify these genes into distinct families (LhcB7 and LhcB8).
In contrast to the uniformly high level of coexpression within the two photosystems, Arabidopsis CC duplicates, as in soybean, exhibit considerable variation in degree of coexpression, with Pearson correlation coefficients ranging from −0.67 to 0.98 (Supplemental File S3). On average, expression profiles are less correlated for CC duplicates than for duplicates of photosystem or LHC genes, regardless of the mechanism of duplication (Supplemental Table S2).
Extending the analysis of coexpression beyond duplicates within gene families, all genes encoding subunits of PSI are highly coregulated (for all pairwise comparisons of PSI genes, ravg = 0.96 and rmin = 0.94; Supplemental Fig. S3). Excluding the silent PSII homeologs, all genes encoding subunits of PSII are also highly coregulated (ravg = 0.95, rmin = 0.83). LHC genes exhibit a greater diversity of expression profiles (ravg = 0.84, rmin = 0.51), but most of this diversity results from the expression profiles of the unusual LhcB5 (AT1G76570) and, to a lesser extent, LhcB4 (AT2G40100) genes. Otherwise, LHC genes are coexpressed, although not to the same extent as are photosystem genes (ravg = 0.91, rmin = 0.71). In contrast, CC genes show a greater variety of expression patterns (ravg = 0.16, rmin = −0.80; Supplemental Fig. S3). CC genes cluster into two general expression profiles, but even within these two clusters, there is a greater range of correlation coefficients than is found within PSI, PSII, or the LHCs.
Combining data on selection and expression, greater than 70% of all α/A-homeologs of CC genes exhibit some evidence for functional divergence (positive selection, expression divergence, or both), compared with 50% or less for PSI, PSII, and LHC, in both soybean and Arabidopsis (Fig. 6C).
DISCUSSION
Distinct patterns of duplicate retention and gene family expansion emerge when photosynthetic gene families are considered in their functional contexts. In particular, core PSII gene families exhibit relatively high levels of homeolog retention, and both photosystems exhibit low levels of NP duplicate retention in comparison with the CC in all three species.
This reciprocal pattern of duplicate retention suggests that different evolutionary forces are acting upon duplicates from the different photosynthetic functional groups. What might be the driving forces for these differences in duplicate retention? The pattern observed for PSII of high retention rates following genome duplications and low retention rates following single-gene duplications is the hallmark of “balanced gene drive” (Freeling and Thomas, 2006). According to the “balance hypothesis” (Papp et al., 2003), genes whose products function in multisubunit complexes or signaling cascades will tend to be dosage sensitive because changes in the stoichiometry of individual subunits lead to improper assembly and/or function of the complex, with deleterious consequences for the individual (Papp et al., 2003; Birchler et al., 2007; Birchler and Veitia, 2010; Innan and Kondrashov, 2010). In support of this hypothesis, Papp et al. (2003) showed in yeast that genes causing haploinsufficiency are more than twice as likely to function in complexes than are genes that do not. Similarly, the class of yeast genes that are lethal when overexpressed is significantly enriched for genes whose products function in complexes.
Such dosage sensitivity leads to distinct predictions about the retention of genes duplicated by small-scale processes versus those duplicated by polyploidy. Small-scale duplications that affect some but not all genes encoding subunits of a protein complex will be deleterious and should be actively eliminated from the genome by purifying selection to maintain gene balance. In contrast, WGDs affect all subunits of dosage-sensitive complexes (and are, therefore, referred to as “balanced” duplications; Papp et al., 2003). Consequently, dosage-sensitive genes duplicated by polyploidy should be maintained following polyploidy, again by purifying selection for gene balance. Numerous studies have demonstrated that polyploid genomes are enriched for genes whose products function in protein-protein complexes (e.g. ribosomal proteins, proteasomal proteins, and transcription factors; Blanc and Wolfe, 2004b; Seoighe and Gehring, 2004; Maere et al., 2005; Freeling and Thomas, 2006; Paterson et al., 2006).
The PSII complex is a large, multisubunit protein complex with a fixed subunit stoichiometry (Minagawa and Takahashi, 2004), and incompletely assembled or misassembled PSII complexes not only impair photosynthetic electron transport but also sensitize the plant to photooxidative damage (Baena-González and Aro, 2002; Hwang et al., 2008). We suggest that these properties make PSII genes dosage sensitive. Our observation that among the four photosynthetic functional groups, PSII exhibits the highest retention rates following polyploidy, and among the lowest retention rates for NP duplications, is consistent with this hypothesis (see below for a discussion of PSI).
In contrast, NP duplications are observed more frequently in CC gene families than in either photosystem in all three species, suggesting that single-gene duplications are less likely to be deleterious in the context of the CC. These observations are consistent with previous studies demonstrating that enzymes in general tend to be dosage insensitive (Kondrashov and Koonin, 2004).
Several CC duplicates have been retained for long periods of time. Based on the Ks distributions of duplicated genes in a variety of species, the fate of the vast majority of duplicated genes is nonfunctionalization within a few million years (Lynch and Conery, 2000). If these gene families are not dosage sensitive, why have so many duplicates persisted for so long? One possibility is that there may in fact be a selective advantage to increased dosage. Two CC enzymes (Rbcs and sedoheptulose-1,7-bisphosphatase [SBPase]) are thought to be rate limiting or near rate limiting in carbon fixation (Harrison et al., 1996; Sun et al., 2003), and overexpression of SBPase increases photosynthetic rates in tobacco (Nicotiana tabacum; Miyagawa et al., 2001; Lefebvre et al., 2005). Notably, though, SBPase is single copy in Arabidopsis and barrel medic, so clearly in these taxa at least there has not been strong selection for increased dosage of this gene family. The Rbcs gene family, in contrast, has expanded via both polyploidy and small-scale duplications in Arabidopsis and soybean. We did not find evidence of retention of duplicates from polyploidy in barrel medic, but its Rbcs gene family has expanded via recent single-gene duplications. Thus, it may be advantageous to increase gene dosage for Rbcs and possibly for other CC enzymes.
Alternatively, CC duplicates may have been retained because they evolved to serve new roles in the plant. Presumably due to genetic redundancy, many duplicate gene pairs experience a period of relaxed selective constraint (Lynch and Conery, 2000), which could facilitate subfunctionalization (partitioning of ancestral functions between paralogues), neofunctionalization (the acquisition of new functions by one or both paralogues), or escape from adaptive conflict (improvement of ancestral functions that were constrained when carried out by a single, ancestral gene; Des Marais and Rausher, 2008, Innan and Kondrashov, 2010). Subfunctionalized genes are retained because both copies are required to carry out the full suite of ancestral functions. Neofunctionalized genes and genes that have undergone escape from adaptive conflict are retained if the novel or improved functions confer a selective advantage for the host.
We looked for evidence of functional differentiation in the form of positive selection and/or divergence in expression profiles. Of the four photosynthetic functional groups, we found that CC A/α duplicates are the most likely to contain regions under positive selection and/or to exhibit divergence in expression profiles in both Arabidopsis and soybean. Thus, in general, it appears that photosystem genes are under strong purifying selection and are constrained to a narrow range of correlated expression profiles, whereas CC gene families are more likely to exhibit functional divergence.
Eight of 11 CC gene families encode enzymes that function in other pathways (either glycolysis or the oxidative pentose phosphate pathway [OPPP]), and these alternative pathways may provide “functional sinks,” or avenues for subfunctionalization or neofunctionalization that facilitate the retention of duplicated genes. Both the glycolytic and OPPP pathways are at least partially duplicated and spatially separated in plants, with distinct enzyme complements functioning in the plastid and cytosol in each pathway (Tobin and Bowsher, 2005). Within these different compartments, the two pathways exhibit multiple levels of regulation, and the amounts and activities of the various enzymes change with tissue type and developmental stage (Tobin and Bowsher, 2005). Thus, it seems plausible that greater opportunity exists for subfunctionalization and/or neofunctionalization among duplicates of genes encoding enzymes that function in glycolysis or OPPP in addition to the CC compared with enzymes restricted to the CC alone. Consistent with this hypothesis, the two smallest CC gene families in Arabidopsis, soybean, and barrel medic (SBPase and PRK) function exclusively in photosynthetic carbon fixation, whereas the two largest CC gene families (GAPDH and fructose-bisphosphate aldolase [FBA]) also participate in glycolysis.
Nonetheless, in soybean, A-homeologs of plastid-targeted and cytosolic CC genes exhibit comparable propensities for expression divergence (25% and 33%, respectively), and both are more likely to exhibit divergent expression patterns than are genes from either photosystem (Supplemental File S3). A-homeologs of both plastid-targeted and cytosolic CC genes are also more likely to have experienced positive selection than genes from either photosystem. So, although dual-function CC gene families may have additional avenues for functional divergence than their single-function counterparts, all CC gene families appear more able to diverge functionally than the gene families of the thylakoid-associated complexes (PSI, PSII, and LHC). This, in combination with the dosage insensitivity of enzymes, could explain the relatively greater retention of NP duplicates in the CC than in the thylakoid complexes.
The fact that PSII exhibits consistently higher retention of homeologs than the CC, despite no obvious differentiation in function between duplicates, further supports the hypothesis that these duplicated genes were simply locked in place by dosage constraints. This is not to say, however, that our analyses prove an absence of functional differentiation among the duplicates of PSII genes. Obviously, an overall positive correlation of expression between two genes does not preclude the possibility that the two copies have subfunctionalized or neofunctionalized at some finer scale. Indeed, careful molecular analyses of several PSII gene families have revealed differences in function. For example, using Arabidopsis T-DNA knockouts, Lundin et al. (2007) demonstrated that one copy of PsbO is more efficient than the other at supporting the oxygen-evolving capacity of PSII under photoinhibitory conditions, whereas the second copy regulates turnover of the D1 protein during the damage-repair cycle. Using transcriptional reporter gene fusions, Sawchuk et al. (2008) showed that LhcB2.1 (AT2G05100) is expressed at the onset of subepidermal leaf tissue development, whereas LhcB2.3 (AT3G27690) is not expressed until late in mesophyll differentiation. It is not unlikely that other PSII gene duplications have led to functional specialization as well. Our data indicate, however, that the realm of possibilities is narrower for gene families that function strictly in photosynthesis than for CC gene families, whose products participate in multiple pathways.
If gene balance requirements explain the relatively high retention rates of homeologs in PSII, why are duplicates retained for some PSII gene families but not others? Perhaps only a subset of the protein-protein interactions within the greater PSII complex are dosage sensitive. However, if this were the case, then we would expect to see homeologs from the same gene families retained across nested duplications, and across all three species, yet we do not. Furthermore, in Arabidopsis, one β-homeolog from PsbO and one α-homeolog of PsbP are silent, or nearly so. Obviously, these genes are not contributing to the balance of gene products (proteins).
Previous studies that supported the balance hypothesis have demonstrated a greater propensity for “connected” genes to be retained following polyploidy (Papp et al., 2003), but this is not to say that all such genes are retained. Similarly, not all unbalanced changes in dosage involving connected genes are deleterious. In yeast, 37% of the genes with minimal fitness deficiency as heterozygous knockouts are involved in protein complexes (Papp et al., 2003). This highlights a key challenge associated with the balance hypothesis: determining what precisely makes a gene dosage sensitive. Participation in a multiprotein complex alone is only weakly predictive. Recent studies at the protein level suggest that dosage sensitivity correlates with topological position within a protein complex (Veitia, 2005) and with the degree of protein “underwrapping” (the degree to which protein structural integrity is dependent on its interactive context; Liang et al., 2008), but the molecular basis for dosage sensitivity remains poorly understood.
For those genes that do indeed have dosage-related effects on fitness, dosage balance requirements are likely to be circumvented over time via other mechanisms (Aury et al., 2006; Ha et al., 2009). For example, changes in cis-regulatory sequences or abundance of trans-acting factors (including microRNAs; Birchler and Veitia, 2010) might eventually allow for balance in gene products to be achieved without maintaining balance in gene copy number. Alternatively, selection on individual members of a protein complex might cause a “ripple effect of adaptation” through the rest of the complex (Rodriguez et al., 2007), thereby altering balance constraints (Birchler and Veitia, 2010).
PSI exhibits similarly low levels of NP duplicate retention as PSII, consistent with dosage sensitivity, yet it also exhibits relatively low homeolog retention rates, comparable to the CC. However, with the exception of PsaF, genes encoding PSI subunits exhibit 100% retention from the A polyploidy event in soybean (compared with 76.5% for the CC), suggesting that there is some delay in homeolog loss within PSI. The PSI complex, therefore, may be sufficiently dosage sensitive for NP duplications to be selected against but less sensitive than PSII, due, for example, to a lower level of protein underwrapping (Liang et al., 2008). We searched the Liang et al. (2008) Arabidopsis data set for underwrapping estimates of photosystem proteins, but only one PSI protein (PsaE) was included. PsaE was estimated to have a relatively low degree of protein underwrapping (31.2%). It would be interesting to calculate underwrapping for all photosystem subunits to see if, indeed, PSII proteins exhibit greater underwrapping than PSI proteins.
Alternatively, changes in gene dosage may be more readily adjusted for in PSI at the level of expression or posttranslational modification, allowing for a quicker decay of homeologs despite initial dosage sensitivity. The PSI complex has fewer subunits than PSII (approximately 15 versus 25). Having to coordinate among fewer loci could allow the process of decoupling protein abundance from gene copy number to proceed more quickly.
Additionally, nearly two-thirds of the PSII subunits are encoded by the plastid (14 chloroplast-encoded subunits versus nine that are nucleus encoded) compared with only one-third for PSI (five chloroplast versus nine nuclear). Chloroplast number per cell increases with nuclear genome doubling (Warner and Edwards, 1993), which might serve initially to maintain dosage balance between nucleus-encoded gene products and chloroplast-encoded gene products. Coordination of nucleus- and plastid-encoded subunit stoichiometry, therefore, might represent a more significant challenge in PSII than in PSI, thereby driving longer term retention of balanced duplicates in PSII than in PSI.
In contrast, Duarte et al. (2010) demonstrated that nuclear genes encoding organelle-localized proteins tend to be maintained as singletons across a range of plant taxa (shared single-copy nuclear genes) and postulated that this was the result of selection to maintain dosage balance between nucleus-encoded and organelle-encoded subunits of signaling networks or protein complexes in the organelle (Duarte et al., 2010; Edger and Pires, 2009). Under this hypothesis, any duplication (polyploid or NP) affecting nuclear genes involved in such complexes would be unbalanced, because their interacting partners encoded by the organelle would not also be duplicated. The low level of duplicate retention (polyploid or NP) in PSI appears to be consistent with this hypothesis, but the elevated level of polyploid duplicate retention we observed in PSII does not. In the end, coordination of gene dosage between nuclear and organelle genomes remains poorly understood (Duarte et al., 2010; Edger and Pires, 2009) and will require further investigation.
Like the CC, LHC gene families tend to exhibit lower retention of homeologs and greater retention of NP duplicates than the photosystems. This suggests that the LHC is not dosage sensitive. Unlike the CC, however, we find relatively little evidence for functional differentiation among LHC duplicates, even those that have been retained since before the β/B polyploidy events. It is possible that these gene families are dosage sensitive, but only weakly so, or, as we speculate for PSI, that they are able to rapidly “correct” for dosage imbalances at the level of expression. Thus, selection could be too weak or too short-lived to result in elevated levels of homeolog retention or to stringently eliminate unbalanced NP duplicates. All LHC proteins are encoded by the nucleus, so if coordination of nucleus- and plastid-encoded subunit stoichiometry prolongs gene balance constraints, LHC homeologs would be expected to decay more rapidly than PSII homeologs.
Alternatively, the LHC may be dosage insensitive, and duplicate retention could be driven by functional differentiation that our analyses failed to detect. In Arabidopsis, most of the NP duplicates in the LHC resulted from recent duplications, and Affymetrix microarray probes do not discriminate among LHC paralogues. Thus, we were unable to compare the expression profiles of the recent tandem duplicates for LhcA2, LhcB1, or LhcB2. Using transcriptional reporter fusions, Sawchuk et al. (2008) have shown differences in expression domains across tissue types and developmental stages for duplicated Lhc genes.
Intriguingly, the LhcB1 gene family has experienced independent tandem duplications in all three lineages analyzed here: LhcB2 has undergone recent tandem duplication in Arabidopsis, and each of the major Lhc genes (LhcB1 to -3) has undergone recent tandem duplications in tomato (Solanum lycopersicum; Cannon et al., 2004). The major Lhc proteins are the most abundant proteins in the light-harvesting complex, and LhcB1 and LhcB2 play important roles in balancing excitation pressure between the two photosystems via state transitions (Tikkanen et al., 2006). The high rate of tandem duplication in the major Lhc gene families has been suggested to facilitate the tuning of light harvesting to different light conditions (Cannon et al., 2004). The major Lhc proteins are only peripherally associated with the photosystem protein complexes. The less intimate association with the other subunits of the photosystems, compared with the minor Lhc proteins, might reduce dosage balance constraints (Veitia, 2005), consistent with the higher rate of NP duplication in the major Lhc observed in several species.
CONCLUSION
In conclusion, we suggest that the photosystem protein complexes are dosage sensitive, which leads to retention of polyploid duplicates (as evidenced by very high retention rates following the most recent WGD in soybean) as well as active elimination of NP duplicates, via purifying selection, to maintain gene balance. Over time, balance of gene products is increasingly achieved via the regulation of expression and becomes decoupled from gene dosage. This relaxes selection on gene copy number. Because the photosystems are highly functionally constrained, there are few opportunities for subfunctionalization or neofunctionalization, and most of the “extra” gene copies then begin to decay. We see remnants of this process in silenced PsbO and PsbP homeologs in Arabidopsis. Consequently, homeolog retention rates begin to approach genome-wide levels for older polyploidy events. The CC, in contrast, is not dosage sensitive. Thus, redundant gene copies are neither actively eliminated nor maintained by selection, regardless of duplication mechanism (polyploidy or small-scale processes). These genes follow typical decay curves (Lynch and Conery, 2000; Blanc and Wolfe, 2004a; Schlueter et al., 2004; Maere et al., 2005), with most eventually being nonfunctionalized. However, in part because CC genes participate in multiple biochemical pathways, opportunities for functional differentiation (and long-term retention) are greater than for PSII or PSI. This, in turn, is manifested in older and larger gene families.
The fact that individual photosynthetic gene families do not exhibit consistent patterns of retention or loss across the three species examined here, or across nested polyploidy events within Arabidopsis (Figs. 3 and 5), might seem to argue against the conclusion that higher level functional groups are shaped by specific evolutionary forces. However, random mutational processes are likely to be driving both retention of dosage-insensitive CC gene duplicates (by facilitating functional divergence) and loss of dosage-sensitive PSII duplicates (by decoupling gene dosage from the amount of gene product). Thus, dosage sensitivity could produce a high overall rate of retention of polyploid duplicates in PSII, for example, despite individual gene families escaping this selective pressure by mutations that break the linkage between gene dosage and the abundance of gene product. Because these mutations are random, different gene families fractionate in different species or following different polyploidy events in the same species.
The abundance of genomics studies observing trends in retention might give an exaggerated sense of consistency in terms of how particular genes respond to duplication. At least in the case of photosynthetic genes, such patterns dissolve when looking at the level of individual gene families, serving as a reminder that genome-level patterns are tendencies and not absolutes. Additionally, most studies looking at the behavior of broad functional classes of genes are restricted to a few species. Barker et al. (2008) found very different patterns of retention following polyploidy in the Compositae than have been observed in Arabidopsis, suggesting that duplicate gene evolution following polyploidy may follow family-specific trajectories. Additional studies like ours will help to reveal the extent to which “omics”-level patterns carry through to individual gene families and to what extent patterns observed in one species can be extended to other species.
This study differs from previous genomics-level studies of polyploidy in that it investigates gene families in their specific physiological contexts. The reciprocal pattern of duplicate retention observed here, between the photosystems on one hand and the CC and LHC on the other, would not be detected when grouping genes by the functional categories used in these earlier studies, such as protein domains (Paterson et al., 2006) or Gene Ontology (GO) categories (Blanc and Wolfe, 2004b; Maere et al., 2005). First, the enzymes in a biochemical pathway (e.g. the CC) or the subunits of a protein complex (e.g. PSII) are not generally characterized by common protein domains. Second, there are sufficient inconsistencies in GO annotations to preclude effective analysis of biochemical pathways and/or protein complexes via gene ontologies. For example, although there is a GO cellular component category for PSII (GO:000953), this GO term has not been assigned to the Arabidopsis genes encoding three of the nine PSII subunits (PsbS, PsbW, and PsbX). Similarly, there is a GO biological process term for “carbon fixation” (GO:0015977), but gene families encoding four of 11 CC enzymes are not associated with this GO term. Thus, additional studies guided specifically by physiological or biochemical context will provide a valuable complement to existing studies using more generically assigned functional classifications.
MATERIALS AND METHODS
Tentative Consensus-Based Analyses
Protein sequences were obtained from The Arabidopsis Information Resource Web site (http://www.arabidopsis.org) for all Arabidopsis (Arabidopsis thaliana) genes involved directly in the CC, PSI and PSII, and LHC. Arabidopsis protein sequences were used to query the soybean (Glycine max; release 12.0) and the barrel medic (Medicago truncatula; release 8.0) gene indices maintained by the Dana Farber Cancer Institute (http://compbio.dfci.harvard.edu/tgi/plant.html) using TBLASTN. Sequences for all tentative consensus (TC) BLAST hits and corresponding Arabidopsis coding sequences were translated to protein sequence and aligned using ClustalW, with default parameters, in BioEdit. Alignments were adjusted by eye as necessary. Singleton EST BLAST hits were excluded from analysis due to the frequency of errors in single EST sequences.
Gene phylogenies were constructed from the aligned sequences using maximum parsimony, as implemented in PAUP 4.0 (Swofford, 2003). For alignments with fewer than 12 genes (including Arabidopsis, soybean, and barrel medic), a full branch-and-bound search was performed. For alignments with 12 or more genes, a heuristic search was performed, with 1,000 random addition sequence replicates, using Tree-Bisection-Reconnection branch swapping. To estimate divergences among soybean and/or barrel medic genes, the number of synonymous substitutions per synonymous site (Ks) was calculated for each paralogous gene pair by the method of Yang and Neilsen (2000), as implemented in PAML (Yang, 1997), from the sequence alignments used to construct gene phylogenies. Ks values for pairs of Arabidopsis genes were taken from Blanc and Wolfe (2004a) or calculated as with soybean and barrel medic. Ks values were averaged for nodes joining more than two genes.
Duplications in soybean and barrel medic were categorized as resulting from polyploidy or NP duplication based on Ks values and gene tree topology as follows. The approximately 50-million-year duplication event (hereafter referred to as B) occurred in the common ancestor of Medicago and Glycine (Pfeil et al., 2005; Fig. 1). The median Ks value for this duplication event is 0.54 (0.40–0.72 ± 1 sd; Schlueter et al., 2004). The median Ks value for the Glycine-specific, approximately 10-million-year duplication (hereafter referred to as A) is 0.13 (0.08–0.19; Egan and Doyle, 2010; Schmutz et al., 2010, J.A. Schlueter, unpublished data). A Ks value within either of these ranges for a pair of soybean genes or within the older range for a pair of barrel medic genes was taken as evidence of duplication by polyploidy (homeology). Because the B duplication was shared by Glycine and Medicago, a barrel medic sequence is expected to be sister to each soybean lineage descended from this duplication. Because the A polyploidy was Glycine specific, no barrel medic sequence should nest within soybean lineages resulting from this duplication. Gene phylogenies with this expected topology were considered further evidence for homeology. Duplicate sequences were identified as homeologs if they were supported by both Ks and gene tree topology. Due to the frequency of gene losses, duplicates in barrel medic or soybean were also considered homeologs if supported by Ks even if gene losses in the other species had to be inferred. Duplicates were also considered homeologs if Ks was outside of the range for that polyploidy event, but within 0.1 (B) or 0.02 (A) of the confidence interval for the polyploidy, and rejecting the event increased the number of losses inferred.
Genomic Synteny-Based Analyses
Arabidopsis protein sequences for photosynthetic genes were used to query the soybean genome sequence (Glyma1 assembly; http://www.phytozome.net/soybean.php) and release 2.0 of the M. truncatula genome sequence (http://www.medicago.org/genome/downloads/Mt2) using TBLASTN. TC sequences identified through the TC-based analyses were also used to search the respective genome sequences using BLASTN. The coding sequences of Arabidopsis and all soybean loci showing significant BLAST scores (<1e-5) were aligned using ClustalW in BioEdit with default parameters. Ks and ω (Ka/Ks) were calculated by the method of Yang and Neilson (2000), as implemented in PAML (Yang, 1997). Gene trees were constructed following the same methods used with the TC sequences.
In the absence of subsequent rearrangements, genes duplicated by polyploidy should reside in syntenic blocks (Zhang et al., 2002; Blanc et al., 2003; Bowers et al., 2003). We determined whether soybean photosynthetic genes reside in or near (within 500 genes of) internal synteny blocks, as identified by Schmutz et al. (2010). Pairs of gene family members residing within syntenic blocks were designated homeologs. Ks estimates were used to determine from which polyploidy event (B or A) homeologs were derived. For gene pairs close to, but not within, syntenic blocks, we manually searched for evidence of local synteny in a region of approximately 200 kb centered on each gene using the Phytozome soybean genome browser (http://www.phytozome.net/cgi-bin/gbrowse/soybean/). Gene pairs within 500 genes of a synteny block that showed evidence for local synteny (at least three additional homeologous gene pairs within 200 kb) were also designated homeologs. For genes not residing in or near syntenic blocks, we concluded that their homeologs have been lost. Duplicate gene pairs that were not assigned to the B or A polyploidy events were assigned to one of three NP bins: pre-B, B-A, or A-present, based on Ks and gene tree topology.
Genome-wide estimates of homeolog retention were obtained from Schmutz et al. (2010) or by using a modified approach as follows. Gene families, blocks, and synonymous distances were identified as by Schmutz et al. (2010). Using the gene family clusters and Clustal multiple sequence alignments, gene trees were constructed with RAxML. An in-house Python script sequentially broke down each RAxML tree file into pairwise relationships and assigned pairwise Ks values to the nodes separating those pairs. In instances where more than one pair of genes characterized a node, we calculated a mean Ks value and assigned that mean to the node. All Ks values between 0.06 and 0.39 were assigned to the A duplication. To estimate genome-wide retention for the A duplication, the number of genes associated with this Ks range was divided by the total number of genes in syntenic blocks.
For Arabidopsis, duplications resulting from the two most recent polyploidy events (designated β and α by Bowers et al., 2003; Fig. 1) were identified previously by Blanc et al. (2003), Bowers et al. (2003), and Thomas et al. (2006; α duplication only) using combinations of genomic synteny information, comparative phylogenetics, and estimates of sequence divergence (Ks). Lists of homeologs identified in these studies are available at http://wolfe.gen.tcd.ie/blanc/supp/functional.html (Blanc et al., 2003; Bowers et al., 2003) and http://genome.cshlp.org/content/16/7/934/suppl/DC1 (Thomas et al., 2006). We searched these lists in order to identify homeologs within the photosynthetic gene families investigated here. For all but three pairs of photosynthetic genes, the Blanc et al. (2003) and Bowers et al. (2003) data sets were consistent. The data sets differ for one pair each of PGK, PsbTn, and LhcB4, and these discrepancies were resolved as described in Supplemental Materials and Methods S1. As with soybean, gene pairs not identified as homeologs were assigned to one of three NP duplication bins (pre-β, β-α, or α-present) based on Ks and gene tree topology.
For each photosynthetic gene family, we quantified the contributions of the various duplication events to each gene family using two parameters: percentage retention and percentage expansion (Fig. 2). Percentage retention for a given WGD was calculated by dividing the number of gene lineages duplicated by the WGD that are retained in duplicate today by the total number of gene lineages duplicated by the WGD. Percentage expansion was calculated by dividing the number of gene lineages added by the given duplication event (β/B, α/A, or NP) by the total number of gene lineages added since immediately before the β/B event. Mean percentage retention and percentage expansion for each function group (CC, PSII, PSI, or LHC) were calculated by averaging the values obtained for each gene family assigned to that functional group. This method weights each gene family equally, regardless of size.
Overall percentage retention was also calculated for all photosynthetic genes combined, and for each functional group separately, in two ways. First, the number of lineages retaining duplicates from the specified duplication was divided by the total number of lineages present immediately prior to that duplication. Second, the number of genes present today that retain a homeolog from the specified duplication was divided by the total number of genes present today. Both methods differ from the mean percentage retention method in that they effectively weight large gene families more heavily than small gene families. Of the two methods to calculate overall percentage retention, the second method is comparable to the methods used by Schmutz et al. (2010) and was used for comparison with genome-wide retention estimates. This method yields higher estimates than the first because each retained homeolog pair counts as two, whereas singletons count only as one in both numerator and denominator.
Tests of Selection
Global Ka/Ks values (ω) were calculated for all pairwise combinations of gene family members in Arabidopsis and soybean using the method of Yang and Nielsen (2000), as implemented in PAML (Yang, 1997). Sliding-window Ka/Ks calculations were performed on homeolog pairs from the recent polyploidy events in Arabidopsis and soybean (α and A, respectively) using the Web tool Sliding Window Analysis of Ka and Ks (Liang et al., 2006). We only analyzed duplicates from the α and A duplications because these were the only duplication bins for which all functional groups (CC, PSII, PSI, and LHC) have retained duplicates in both species. For three-dimensional analyses, Protein Data Bank (PDB) files were obtained from the Research Collaboratory for Structural Bioinformatics PDB Web site (http://www.rcsb.org/pdb/). We used the default window sizes of 30 amino acids (one dimensional) and 10 Å (three dimensional). The PDB files used are given in Supplemental File S2.
Expression Analyses
Correlation coefficients for photosynthetic genes in soybean were calculated from 15 RNA-Seq experiments (cDNA libraries deep sequenced on the Illumina/Solexa platform; Bolon et al., 2010; Libault et al., 2010). Tissue sources were as follows: four developmental stages of seed (25–50 mg, 50–100 mg, 100–200 mg, and 200–300 mg) from one low-protein near-isogenic line and one high-protein near-isogenic line (Bolon et al., 2010), root tips from 3-d-old seedlings, roots from 18-d-old plants, root nodules collected 32 d after Bradyrhizobium japonicum inoculation, leaves from 18-d-old plants, apical meristems, open flowers, and 2- to 3-cm green seed pods (Libault et al., 2010).
For all photosynthetic genes in Arabidopsis, pairwise Pearson correlation coefficients (r) were calculated from publicly available microarray data using the Web tool CressExpress (http://www.cressexpress.org; Srinivasasainagendra et al., 2008), with default settings, and including all available tissue types and experiments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The contributions of polyploidy and NP duplications to gene family expansion for each photosynthetic gene family in soybean, barrel medic, and Arabidopsis.
Supplemental Figure S2. Total number of gene duplicates retained by duplication category and functional group in soybean, barrel medic, and Arabidopsis.
Supplemental Figure S3. Heat maps of expression correlation coefficients by functional group in Arabidopsis.
Supplemental Table S1. Sliding-window estimates of selection (ω) by functional group for the most recent polyploidy events (α and A) in soybean and Arabidopsis.
Supplemental Table S2. Average, minimum, and maximum levels of expression correlation between duplicate gene pairs by duplication type and functional group in soybean and Arabidopsis.
Supplemental File S1. Gene trees and corresponding estimates of retention and expansion by species for each photosynthetic gene family.
Supplemental File S2. Global and sliding-window estimates of selection by species and duplication category for each photosynthetic gene family.
Supplemental File S3. Pearson correlation coefficients of expression profiles by species for each photosynthetic gene family.
Supplemental Materials and Methods S1. A description of how discrepancies were resolved between different published lists of homeologs in Arabidopsis.
Acknowledgments
We thank Gary Stacey, Yung-Tsi Bolon, Bindu Joseph, Steven Cannon, and Michelle Graham for early access to their soybean RNA-Seq data sets. We also thank Steven Cannon for providing the raw data used in calculating genome-wide estimates of polyploid duplicate retention in soybean reported by Schmutz et al. (2010). We thank Tom Owens, Steven Cannon, and all members of the Doyle laboratory for helpful critiques of the manuscript.
References
- Adams KL, Wendel JF. (2005) Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171: 2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Aro EM. (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. (2008) Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol 25: 2445–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. (2007) Biological consequences of dosage dependent gene regulatory systems. Biochim Biophys Acta 1769: 422–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004a) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004b) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, et al. (2010) Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LY, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, et al. (2006) Widespread genome duplications throughout the history of flowering plants. Genome Res 16: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das VSR. (2004) Photosynthesis: Regulation under Varying Light Regimes. Science Publishers, Enfield, NH [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20: 591–597 [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. (2008) Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Rauscher JT, Brown AHD. (2004) Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biol J Linn Soc Lond 82: 583–597 [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires JC, Leebens-Mack J, dePamphilis CW. (2010) Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. (2009) Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res 17: 699–717 [DOI] [PubMed] [Google Scholar]
- Egan AN, Doyle J. (2010) A comparison of global, gene-specific, and relaxed clock methods in a comparative genomics framework: dating the polyploid history of soybean (Glycine max). Syst Biol 59: 534–547 [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Ha M, Kim ED, Chen ZJ. (2009) Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci USA 106: 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EP, Lloyd JC, Raines CA. (1996) The effect of reduced SBPase levels on leaf carbon metabolism. J Exp Bot 47: 1306 [Google Scholar]
- Hwang HJ, Nagarajan A, McLain A, Burnap RL. (2008) Assembly and disassembly of the photosystem II manganese cluster reversibly alters the coupling of the reaction center with the light-harvesting phycobilisome. Biochemistry 47: 9747–9755 [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S. (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. (2004) A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet 20: 287–290 [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Lawson T, Zakhleniuk OV, Lloyd JC, Raines CA, Fryer M. (2005) Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol 138: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Plazonic KR, Chen J, Li WH, Fernández A. (2008) Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genet 4: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Zhou W, Landweber LF. (2006) SWAKK: a Web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res 34: W382–W384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Lundin B, Hansson M, Schoefs B, Vener AV, Spetea C. (2007) The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J 49: 528–539 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J, Takahashi Y. (2004) Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth Res 82: 241–263 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S. (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotech 10: 965–969 [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. (2006) Many gene and domain families have convergent fates following independent WGD events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54: 441–454 [DOI] [PubMed] [Google Scholar]
- Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. (2007) Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc Natl Acad Sci USA 104: 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchuk MG, Donner TJ, Head P, Scarpella E. (2008) Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol 148: 1908–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. (2006) Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18: 1152–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Gehring C. (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, dePamphilis CW, Wall PK, Soltis PS. (2009) Polyploidy and angiosperm diversification. Am J Bot 96: 336–348 [DOI] [PubMed] [Google Scholar]
- Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. (2008) CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol 147: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouzé P, Van de Peer Y. (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167: 165–170 [DOI] [PubMed] [Google Scholar]
- Sun N, Ma L, Pan D, Zhao H, Deng XW. (2003) Evaluation of light regulatory potential of Calvin cycle steps based on large-scale gene expression profiling data. Plant Mol Biol 53: 467–478 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. (2008) Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Piippo M, Suorsa M, Sirpiö S, Mulo P, Vainonen J, Vener AV, Allahverdiyeva Y, Aro EM. (2006) State transitions revisited: a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol Biol 62: 779–793 [DOI] [PubMed] [Google Scholar]
- Tobin AK, Bowsher CG. (2005) Nitrogen and carbon metabolism in plastids: evolution, integration, and coordination with reactions in the cytosol. Adv Bot Res 42: 113–165 [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Veitia RA. (2005) Gene dosage balance: deletions, duplications and dominance. Trends Genet 21: 33–35 [DOI] [PubMed] [Google Scholar]
- Warner DA, Edwards GE. (1993) Effects of polyploidy on photosynthesis. Photosynth Res 35: 135–147 [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z, Neilsen R. (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17: 32–43 [DOI] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. (2002) Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol 19: 1464–1473 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu GH, Guo XY, Fan LJ. (2005) Two ancient rounds of polyploidy in rice genome. J Zhejiang Univ Sci B 6: 87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]