Abstract
The seed is a key evolutionary adaptation of land plants that facilitates dispersal and allows for germination when the environmental conditions are adequate. Mature seeds are dormant and desiccated, with accumulated storage products that are to be used by the seedling after germination. These properties are imposed on the developing embryo by a maturation program, which operates during the later part of embryogenesis. A number of “master regulators” (the “LEC genes”) required for the induction of the maturation program have been described, but it is not known what prevents this program from being expressed during early embryogenesis. Here, we report that Arabidopsis (Arabidopsis thaliana) embryos mutant for strong alleles of DICER-LIKE1, the enzyme responsible for the biosynthesis of microRNAs (miRNAs), mature earlier than their wild-type counterparts. This heterochronic phenotype indicates that miRNAs are key regulators of the timing of the maturation program. We demonstrate that miRNAs operate in part by repressing the master regulators LEAFY COTYLEDON2 and FUSCA3 and identify the trihelix transcription factors ARABIDOPSIS 6B-INTERACTING PROTEIN1-LIKE1 (ASIL1) and ASIL2 and the histone deacetylase HDA6/SIL1 as components that act downstream of miRNAs to repress the maturation program early in embryogenesis. Both ASIL1 and HDA6/SIL1 are known to act to prevent the expression of embryonic maturation genes after germination, but to our knowledge, this is the first time they have been shown to have a role during embryogenesis. Our data point to a common negative regulatory module of maturation during early embryogenesis and seedling development.
One of the reasons for the evolutionary success of seed plants is their ability to generate a resistant structure, the seed, which facilitates dispersal and reinitiates development only in the appropriate environmental conditions. In Arabidopsis (Arabidopsis thaliana), wild-type embryos follow a predictable pattern of cell divisions, going through a series of stages named after the shape of the embryo: preglobular, globular, transition, heart, torpedo, bent green cotyledon, and mature (Jürgens and Mayer, 1994). These stages encompass two major phases of development. The first part of embryogenesis, until the heart stage, is devoted to patterning, setting up the embryonic axes, meristems, and tissue types (Jenik et al., 2007). The heart-to-late-heart stage transition marks the onset of embryonic maturation, first evidenced by the appearance of chlorophyll autofluorescence in the epidermis of the hypocotyl, signaling the beginning of proplastid maturation to chloroplasts (Mansfield and Briarty, 1991). The embryos turn green in color and start accumulating storage products at the early torpedo stage. The photosynthetic activity of the embryonic chloroplasts may contribute to the biosynthesis of storage lipids (Ruuska et al., 2004). In Arabidopsis, the storage products include seed storage proteins and storage lipids (very-long-chain fatty acids, polyunsaturated fatty acids, and triacylglycerols; Baud et al., 2008). Once the embryos fill the seed, they acquire desiccation tolerance, desiccate, and enter dormancy (Vicente-Carbajosa and Carbonero, 2005; Baud et al., 2008).
Because the maturation program directs seed dormancy, it needs to be carefully timed, ensuring it starts midembryogenesis and is repressed after germination. A complex network is involved in timing maturation during embryo development, including positive regulators of maturation during midembryogenesis and negative regulators after germination. The existence of repressors of maturation in early embryogenesis has been postulated, but none have been identified so far (for review, see Baud et al., 2008; Santos-Mendoza et al., 2008).
The central positive regulators of the seed maturation program are the “LEC genes” (the B3 domain transcription factors LEAFY-COTYLEDON2 [LEC2] and FUSCA3 [FUS3] and the B subunits of the NF-Y family of trimeric transcription factors LEC1 [also called NF-YB9] and LEC1-LIKE [L1L, NF-YB6]) and the B3 domain transcription factor ABA INSENSITIVE3 (ABI3). Single loss-of-function mutations in all of these genes result in deficiencies in the accumulation of storage products, desiccation intolerance, and/or a heterochronic change of cotyledons into true leaves. These factors regulate each other at the transcriptional level in embryos, and their interactions vary depending on tissue type. The hormones abscisic acid (ABA) and GA interact with this network of transcription factors. High ratios of ABA to GA promote maturation via ABI3 and ABI5 (Santos-Mendoza et al., 2008). In vitro studies have suggested that LEC1 and L1L interact with NY-YC1, -2, and -6 to up-regulate the induction of storage product genes by ABA (Yamamoto et al., 2009). Several lines of evidence have identified other positive regulators, acting either at the same level or downstream of the LEC genes, by binding to the promoters of genes encoding storage proteins and other genes involved in seed maturation: bZIP transcription factors that cooperate with the NF-Y complexes (ABI5, bZIP10, bZIP25, bZIP53, and bZIP67; Bensmihen et al., 2002; Alonso et al., 2009; Yamamoto et al., 2009); MYB transcription factors (AtMYB115 and AtMYB118; Zhang et al., 2009; Wang et al., 2009); and MADS box transcription factors (AGAMOUS-LIKE15 [AGL15] and AGL18; Zheng et al., 2009).
A number of factors are known to prevent the expression of embryonic traits after seed germination (for review, see Zhang and Ogas, 2009). This negative regulation involves transcriptional mechanisms via the B3 domain proteins VP1/ABI3-LIKE or HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE (VAL1/HSI2) and VAL2/HSL1 and the trihelix protein ARABIDOPSIS 6B-INTERACTING PROTEIN1-LIKE1 (ASIL1). Epigenetic factors also repress the maturation program in seedlings, including histone deacetylases (HDA6/SIL1 and HDA19), Polycomb group proteins (SWINGER, CURLY LEAF, and MEDEA), and chromatin remodelers (BRAHMA, AtSWI3c, BSH, and PICKLE). These transcriptional and epigenetic regulators appear to act both directly and indirectly (by modulating the actions of GA) to prevent the expression of the LEC genes and of the genes encoding storage products. It is not known whether any of these regulators or other factors are responsible for the repression of the maturation early in embryogenesis.
MicroRNAs (miRNAs) are 21-nucleotide single-stranded RNA molecules that act by binding complementary target mRNAs to promote their cleavage or interfere with translation (for review, see Voinnet, 2009). miRNAs are generated by cleavage of a precursor miRNA by a complex that includes the RNase III DICER-LIKE1 (DCL1), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the C2H2-zinc finger protein SERRATE (SE). The resulting miRNAs are then methylated by HUA-ENHANCER1 (HEN1) and incorporated into an RNA-induced silencing complex, which leads the miRNA to its target and effects either cleavage or repression of translation. ARGONAUTE proteins such as AGO1 and AGO10/ZWILLE/PINHEAD are central components of miRNA RNA-induced silencing complexes. Null alleles of dcl1 are embryonic lethal, but embryos mutant for other elements of the miRNA pathway have either nonoverlapping phenotypes or no observable phenotype (Lynn et al., 1999; Lu and Fedoroff, 2000; Chen et al., 2002; Grigg et al., 2005; Ronemus et al., 2006; Kurihara et al., 2009). Null alleles of dcl1 do accumulate storage products (Schwartz et al., 1994), but other than interactions of miR159 with the ABA pathway during germination (Reyes and Chua, 2007), no connections have been reported between the miRNA pathway and seed maturation.
Here, we report that miRNAs are key repressors of the maturation program during embryogenesis. We demonstrate that embryos mutant for strong dcl1 alleles are heterochronic, maturing much earlier than normal. This heterochronic phenotype requires the action of the LEC genes LEC2 and FUS3 downstream of DCL1. We also present the first evidence, to our knowledge, for the repression of maturation during early embryogenesis by ASIL1, ASIL2, and HDA6/SIL1. These genes also act downstream of the miRNA pathway. Our data suggest that one or more miRNA targets sit at the top of the regulatory cascade, controlling both activators and repressors of the embryonic maturation program.
RESULTS
Precocious Maturation in dcl1 Embryos
During an ethyl methanesulfonate screen for embryo-defective mutants (Jurkuta et al., 2009), we isolated a mutant with two conspicuous phenotypes: very abnormal patterning (Fig. 1, A and B) and early chlorophyll accumulation (Fig. 1, D–H). Due to the latter phenotype, we named it green too early (gty). Siliques from heterozygous plants segregated approximately 25% (458 of 1,864; χ2 test P < 0.01) defective embryos, indicating that gty was a single recessive nuclear mutation. gty was mapped to the tip of chromosome I, close to marker nga59 and in the region of the known embryo-defective mutant dcl1 (At1g01040). gty failed to complement the null alleles dcl1-3 and dcl1-5 and the weak allele dcl1-9 (Schauer et al., 2002). Sequencing of the gty allele from genomic DNA uncovered a G-to-A transition at position 7,096 (counting from the ATG; chromosomal position 30,241). This mutation is predicted to change a conserved Gly to Glu in one of two active sites of the RNase IIIb domain (position 1,692), presumably affecting catalysis (Du et al., 2008; Supplemental Fig. S1). Therefore, we renamed gty as dcl1-15.
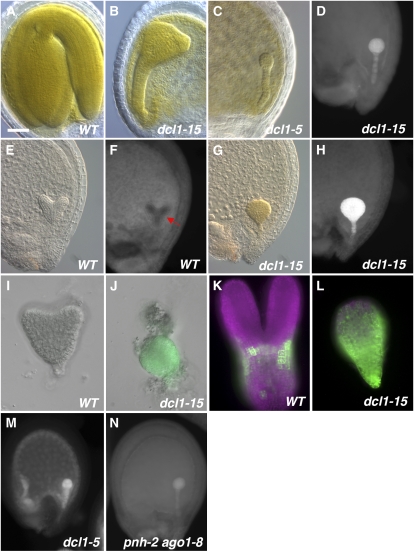
Figure 1.
Phenotypes of miRNA pathway mutants. A to C, Full-size embryos showing the late morphological phenotypes of the wild type (WT; A), dcl1-15 (B), and dcl1-5 (C). D, Chlorophyll autofluorescence in a midglobular stage dcl1-15 embryo. E to H, DIC optics and chlorophyll autofluorescence images of heart stage embryos. Wild-type embryos are white (E), and chlorophyll is starting to accumulate in the protodermis (arrow; F). dcl1-15 embryos at this stage are green (G) and show very strong fluorescence throughout (H). I to L, Expression of pAt2S3:GFP (green; magenta is chlorophyll autofluorescence). I, Wild-type heart stage embryos do not express the reporter. J, dcl1-15 early heart stage embryos show strong expression. K, In wild-type embryos, expression is first clearly observed around the early torpedo stage. L, In dcl1-15, early torpedo stage embryo expression is much higher. M, Chlorophyll autofluorescence in a dcl1-5 late globular embryo. N, pnh-2 ago1-8 embryos show chlorophyll autofluorescence at the late globular stage. Bar = 40 μm (A–H, M, and N) or 20 μm (I–L).
Because the morphology of the mutant embryos was altered, we staged them by referring to the wild-type embryos in the same silique. dcl1-15 embryos developed normally until the early globular stage. At this point, two distinct phenotypes were observed. The patterns of cell division became very abnormal, starting with misdivisions of the hypophysis, leading to mature embryos that had very disturbed tissue patterns (Fig. 1, A and B). The defects were not as severe as those of the null allele dcl1-5 (Fig. 1C; Schwartz et al., 1994), suggesting that dcl1-15 is not a null allele. The patterning defects will be discussed in detail in a separate paper. At the early globular stage, we also observed chlorophyll autofluorescence throughout the mutant embryo proper and suspensor (Fig. 1D). The fluorescence became more intense by the early heart stage, when dcl1-15 embryos turned green (compare Fig. 1, G and H, with E and F). This “early-fluorescence” phenotype was also observed in embryos mutant for the null allele dcl1-5 (Fig. 1M) but not in mutants for the weak allele dcl1-9. The early-fluorescence phenotype was not affected by crossing dcl1-15 into other genetic backgrounds (Landsberg erecta [Ler], Wassilewskija, or Columbia) for the purpose of generating double mutants or analyzing reporter genes (data not shown). dcl1-15 embryos aborted late in embryogenesis. Bent-cotyledon stage dcl1-15 embryos could not be rescued by culturing them on Murashige and Skoog (MS) agar plates (data not shown), suggesting that embryo abortion was not due to desiccation intolerance (Meinke et al., 1994).
The early accumulation of chlorophyll suggested early maturation of chloroplasts. We collected wild-type embryos at the heart stage (when they do not yet show chlorophyll autofluorescence) and dcl1-15 embryos from the same siliques (recognizable for their abnormal cell division patterns) and studied the morphology of the chloroplasts using transmission electron microscopy (TEM; 598 wild-type versus 310 dcl1-15 chloroplasts). We found that chloroplasts in dcl1-15 embryos were more developed than those in wild-type embryos (Fig. 2, A and B). They contained more stacks of grana per chloroplast (2.48 ± 0.08 versus 0.75 ± 0.04; ANOVA P < 0.001), and each stack had more thylakoids (2.67 ± 0.06 versus 1.53 ± 0.04; ANOVA P < 0.001). There were also more starch grains in chloroplasts per cell (0.62 ± 0.08 versus 0.04 ± 0.01; ANOVA P < 0.01). By the time they reached the bent-cotyledon stage, however, the chloroplasts in dcl1-15 and wild-type embryos were indistinguishable (Fig. 2, C and D), although the mutant embryos contained significantly lower amounts of total chlorophyll (0.21 ± 0.03 versus 0.83 ± 0.04 μg per 30 embryos; t test P < 0.001).
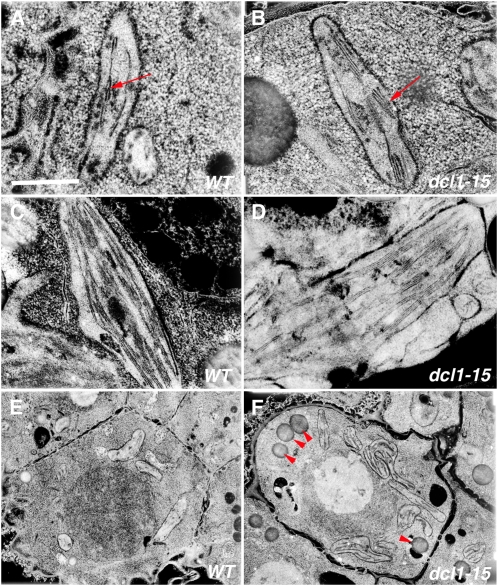
Figure 2.
TEM analysis of maturation phenotypes. A and B, Chloroplasts in wild-type (WT) heart stage embryos (A) are less developed than those of dcl1-15 embryos at the same stage (B). Arrows point to a thylakoid stack. C and D, Chloroplasts in wild-type (C) and dcl1-15 (D) bent-cotyledon stage embryos look very similar. E and F, Early accumulation of lipid bodies (arrowheads) is higher in dcl1-15 heart stage embryos (E) compared with wild-type embryos (F). Bar = 1 μm (A–D) or 5 μm (E and F).
Our TEM analysis revealed that heart stage dcl1-15 embryos not only contained developmentally more advanced chloroplasts but also had a significant number of structures that looked like lipid bodies, which are not found in wild-type embryos until the torpedo stage (3.7 ± 0.37 versus 0.22 ± 0.04 per cell; ANOVA P < 0.001; Fig. 2, E and F). To see if the entire embryo maturation program was switched on earlier in dcl1-15, we studied the expression of the gene for the embryonic storage protein At2S3 using the reporter gene pAt2S3:GFP (Kroj et al., 2003). In wild-type embryos, pAt2S3:GFP was not expressed until the late heart stage and GFP did not accumulate significantly until the late torpedo stage (Fig. 1, I and K). dcl1-15 embryos, in contrast, showed GFP expression starting at the early heart stage (Fig. 1J), and by the late torpedo stage, they had much higher levels than wild-type embryos (Fig. 1L). These data indicate that the whole embryonic maturation program (chloroplast development and synthesis of storage products) was induced precociously in dcl1-15, making it a heterochronic mutant. These maturation events were induced in a normal sequence in dcl1-15, with chloroplast maturation occurring ahead of the accumulation of storage proteins and lipids.
Early Maturation in Other Mutants of the miRNA Pathway
The strong phenotype of dcl1-15 embryos and the location of the mutation in the active site of the enzyme suggested that the embryonic defects were due to a significant reduction in the levels of miRNAs, since miRNA synthesis appears to be the primary role of DCL1 (Laubinger et al., 2010). However, DCL1 also participates in the biosynthetic pathway for other small interfering RNAs (siRNAs), including 21- to 22-nucleotide transacting siRNAs (Peragine et al., 2004; due to the need for miRNAs to generate transacting siRNAs) and natural antisense siRNAs (Borsani et al., 2005). Unlike miRNA biogenesis, these other pathways also require RDR6. We distinguished between those options genetically by testing a variety of mutants in the miRNA pathway using the early-fluorescence phenotype as a proxy for early maturation. Mutations in most of the other members of the miRNA biosynthetic and utilization pathways either have no obvious embryonic defects (hyl1-2, hen1-1; Chen et al., 2002; Vazquez et al., 2004) or have phenotypes that are distinct from those of dcl1-15 (se-3, ago1-8, pnh-2; Lynn et al., 1999; Grigg et al., 2005). This is most likely due to redundancy, since there are several homologs of HYL1, HEN1, and AGO1 in Arabidopsis (Hiraguri et al., 2005; Vaucheret, 2008) and the levels of miRNAs are reduced but not eliminated in the single mutants (Vazquez et al., 2004; Grigg et al., 2005). Not surprisingly, hyl1-2, hen1-1, se-3, ago1-8, and pnh-2 single mutant embryos did not show early chlorophyll fluorescence. However, pnh-2 ago1-8 double mutant embryos did, although not as early as dcl1-15 or dcl1-5 embryos (late globular stage instead of early globular stage; Fig. 1N). Interestingly, the morphology of pnh-2 ago1-8 embryos is similar to that of mutants for strong dcl1 alleles (Lynn et al., 1999; Mallory et al., 2009). This result is consistent with other studies that indicate that AGO1 and PNH redundantly mediate the action of miRNAs (Lynn et al., 1999; Mallory et al., 2009). In addition, embryos of a functionally null RDR6 mutant (rdr6-11; Peragine et al., 2004) do not show early chlorophyll fluorescence. These data support the hypothesis that the dcl1-15 early maturation phenotype is most likely due to the misregulation of one or more miRNA targets.
Transcript Profiling of dcl1 Confirms the Heterochronic Phenotype
The mutation in dcl1-15 is predicted to significantly reduce the levels of all miRNAs present in embryos. There are more than 500 validated or predicted miRNA targets, and a significant proportion of these are transcription factors. Therefore, it seemed likely that many pathways would be altered in dcl1-15 embryos. In order to gain deeper insight into the observed phenotypes, we conducted a global transcriptional profiling experiment of wild-type and dcl1-15 torpedo stage embryos in triplicate using Affymetrix ATH1 microarrays. The torpedo stage was chosen because it was the earliest time point when wild-type and mutant embryos could be easily differentiated and isolated from the seed coat and endosperm in sufficient numbers.
Differential expression analysis by Limma found 2,960 increasing and 3,428 decreasing genes at a Benjamini-Hochberg multiple test correction (MTC) ≤ 0.05 (Supplemental Tables S2 and S3). As expected from the phenotypes described above, most of the genes involved in the synthesis of storage products were significantly up-regulated in dcl1-15 embryos (Table I), including all eight seed storage protein genes (most of them more than 12-fold), all 10 genes encoding the oil body proteome (CLO1 and HSD1 more than 140-fold), and the genes encoding enzymes involved in the synthesis of polyunsaturated fatty acids (FAD3 and FAD2), triacylglycerols (DAGAT and ECR), and very-long-chain fatty acids (FAE) storage lipids (Baud et al., 2008). Gene Ontogeny (GO) term and SP-PIR keyword enrichment analysis of the differentially expressed genes confirmed that genes involved in storage protein, storage lipid, and carbohydrate biosynthesis and localization are overrepresented in the genes up-regulated in dcl1-15 and that genes involved in cell cycle, basic cellular processes, and development are down-regulated (Supplemental Table S4).
Table I. Transcripts significantly misregulated in dcl1-15 torpedo stage embryos.
Within each category, genes are sorted by fold change (dcl1-15 versus the wild type).
| Gene Name | Locus Identifier | Fold Change | P | Reference |
| Seed storage proteins | ||||
| CRUCIFERIN2 (CRU2) | At1g03890 | 261 | 6 × 10−6 | Baud et al., 2008 |
| CRUCIFERINB (CRB) | At1g03880 | 148 | 0.0012 | Baud et al., 2008 |
| CRUCIFERINC (CRC) | At4g28520 | 98 | 0.016 | Baud et al., 2008 |
| At2S5 | At5g54740 | 57 | 0.0353 | Baud et al., 2008 |
| Vicillin-like | At2g18540 | 42 | 7 × 10−6 | Baud et al., 2008 |
| At2S2 | At4g27150 | 23 | 0.0126 | Baud et al., 2008 |
| At2S4 | At4g27170 | 15 | 0.0259 | Baud et al., 2008 |
| CRUCIFERINA1 (CRA1) | At5g44120 | 13 | 0.0001 | Baud et al., 2008 |
| At2S1 | At4g27140 | 12 | 0.0023 | Baud et al., 2008 |
| At2S3 | At4g27160 | 8.07 | 0.0090 | Baud et al., 2008 |
| Globulin-like | At1g07750 | 4.79 | 0.0036 | Baud et al., 2008 |
| Globulin-like | At4g36700 | 3.52 | 0.0003 | Baud et al., 2008 |
| Oil body proteome | ||||
| HYDROXYSTEROID DEHYDROGENASE (HSD1) | At5g50600/700 | 196 | 0.0006 | Baud et al., 2008 |
| CALEOSIN1 (CLO1) | At4g26740 | 149 | 8 × 10−6 | Baud et al., 2008 |
| GPI-anchored | At1g54860 | 12 | 0.0005 | Baud et al., 2008 |
| S1/OLEOSIN5 | At3g01570 | 6.01 | 0.0002 | Baud et al., 2008 |
| BETA-TONOPLAST INTRINSIC PROTEIN (TIP3.2) | At1g17810 | 3.66 | 0.0017 | Baud et al., 2008 |
| S4/OLEOSIN2 | At5g40420 | 2.84 | 0.0004 | Baud et al., 2008 |
| S3/OLEOSIN1 | At4g25140 | 2.42 | 0.0013 | Baud et al., 2008 |
| S2/OLEOSIN4 | At3g27660 | 1.69 | 0.0092 | Baud et al., 2008 |
| S5/OLEOSIN3 | At5g51210 | 1.58 | 0.0193 | Baud et al., 2008 |
| Enzymes involved in the synthesis of storage lipids | ||||
| FATTY ACID ELONGATION1 (FAE1) | At4g34520 | 13 | 9 × 10−5 | Baud et al., 2008 |
| FATTY ACID DESATURASE3 (FAD3) | At2g29980 | 3.04 | 0.0003 | Baud et al., 2008 |
| DIACYLGLYCEROL ACYL-TRANSFERASE (DAGAT) | At3g51520 | 1.55 | 0.0198 | Baud et al., 2008 |
| AtGLB1 | At4g01900 | 1.52 | 0.040 | Baud et al., 2010 |
| FATTY ACID DESATURASE2 (FAD2) | At3g12120 | 1.51 | 0.055 | Baud et al., 2008 |
| ENOYL-COA REDUCTASE (ECR) | At3g55360 | 1.44 | 0.0329 | Baud et al., 2008 |
| Positive regulators of maturation (known or proposed) | ||||
| PEI1 | At5g07500 | 15 | 1 × 10−5 | Li and Thomas, 1998 |
| AtbZIP67 | At3g44460 | 14 | 0.0014 | Yamamoto et al., 2009 |
| MYB118 | At3g27785 | 7.85 | 7 × 10−5 | Wang et al., 2009 |
| MYB33 | At5g06100 | 4.68 | 0.0035 | Reyes and Chua, 2007 |
| AGAMOUS-LIKE18 (AGL18) | At3g57390 | 3.42 | 0.0042 | Zheng et al., 2009 |
| MYB115 | At5g40360 | 2.98 | 0.0011 | Wang et al., 2009 |
| MYB101 | At2g32460 | 2.67 | 0.0014 | Reyes and Chua, 2007 |
| FUSCA3 (FUS3) | At3g26790 | 2.61 | 0.0224 | Meinke et al., 1994 |
| MYB44 | At5g67300 | 2.18 | 0.0035 | Kirik et al., 1998 |
| AtbZIP12/ENHANCED EM LEVEL (EEL) | At2g41070 | 1.84 | 0.0048 | Bensmihen et al., 2002 |
| LEAFY COTYLEDON1-LIKE (L1L) | At5g47670 | 1.84 | 0.0054 | Yamamoto et al., 2009 |
| AtbZIP10 | At4g02640 | 1.75 | 0.0492 | Alonso et al., 2009 |
| LEAFY COTYLEDON2 (LEC2) | At1g28300 | 1.71 | 0.0798 | Meinke et al., 1994 |
| NF-YC6 | At5g50480 | 1.67 | 0.0127 | Yamamoto et al., 2009 |
| WRINKLED1 (WRI1) | At3g54320 | 1.57 | 0.0223 | Cernac and Benning, 2004 |
| NF-YC2 | At1g56170 | 1.42 | 0.0453 | Yamamoto et al., 2009 |
| LEAFY COTYLEDON1 (LEC1) | At1g21970 | −2.02 | 0.0059 | Meinke et al., 1994 |
| GOLDEN-LIKE2 (GLK2) | At5g44190 | −2.82 | 0.0044 | Waters et al., 2008 |
| ABA-INSENSITIVE4 (ABI4) | At2g40220 | −3.15 | 0.0016 | Brocard-Gifford et al., 2003 |
| Negative regulators of maturation (known or proposed) | ||||
| ENHANCER(ZESTE)A1/SWINGER (EZA1/SWN) | At4g02020 | 2.32 | 0.0197 | Makarevich et al., 2006 |
| HISTONE DEACETYLASE-TUIN1 (HDT1) | At3g44750 | −1.38 | 0.0430 | Zhou et al., 2004 |
| HISTONE DEACETYLASE10 (HDA10) | At3g44660 | −1.53 | 0.0312 | Hollender and Liu, 2008 |
| CURLY LEAF (CLF) | At2g23380 | −1.69 | 0.0357 | Makarevich et al., 2006 |
| HISTONE DEACETYLASE6 (HDA6/SIL1) | At5g63110 | −1.83 | 0.0042 | Tanaka et al., 2008 |
| HISTONE DEACETYLASE9 (HDA9) | At3g44680 | −1.94 | 0.0299 | Hollender and Liu, 2008 |
| ARABIDOPSIS 6B-INTERACTING PROTEIN 1-LIKE2 (ASIL2) | At3g14180 | −1.97 | 0.0096 | Gao et al., 2009 |
| ARABIDOPSIS 6B-INTERACTING PROTEIN 1-LIKE1 (ASIL1) | At1g54060 | −2.30 | 0.0123 | Gao et al., 2009 |
| HISTONE DEACETYLASE5 (HDA5) | At5g61060 | −2.73 | 0.0005 | Hollender and Liu, 2008 |
| APETALA2 (AP2) | At4g36920 | −6.28 | 0.0001 | Ohto et al., 2005 |
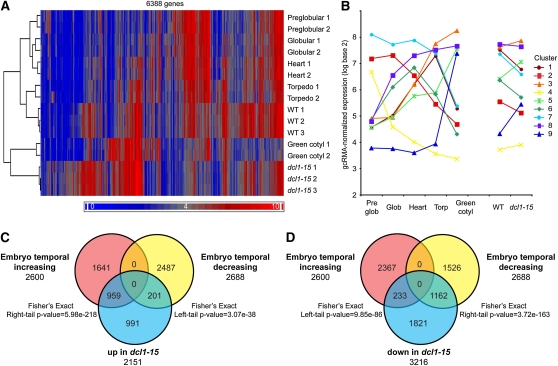
The phenotypes of dcl1-15 mutants suggest that they are maturing faster than normal embryos. We tested this idea further using a recently generated microarray data set of laser capture-microdissected preglobular, globular, heart, torpedo, and green cotyledon stage embryos from the Goldberg and Harada laboratories (http://www.seedgenenetwork.net/arabidopsis). Hierarchical clustering of these arrays and our arrays using the 6,388 genes differentially expressed in dcl1-15 found that, for the best clustering pattern (as determined using average silhouette widths; Supplemental Fig. S3), wild-type samples clustered with the torpedo arrays from the public data set, consistent with the timing of their development. The dcl1-15 samples, however, clustered with the more mature green cotyledon arrays, further suggesting that dcl1-15 is a heterochronic mutation (Fig. 3A). We then identified temporally expressed genes in embryos, genes whose expression levels vary at different developmental stages, using the Goldberg-Harada data set and compared these genes with those differentially expressed in dcl1-15 torpedo embryos. A total of 6,139 probe sets that fell into nine clusters of expression patterns were temporally expressed in the Goldberg-Harada data set (Fig. 3B; Supplemental Table S5). We found that 32.4% of the genes up-regulated in dcl1-15 were genes more highly expressed in developmentally older embryos (temporally increasing genes) and that 33.9% of the genes down-regulated in dcl1-15 were genes more highly expressed in developmentally younger embryos (temporally decreasing genes; Fig. 3, C and D; Supplemental Tables S6 and S7). This supports the conclusion that the expression profile of dcl1-15 torpedo stage embryos resembles more the expression profile of developmentally older wild-type embryos.
Figure 3.
Microarray analysis of torpedo stage dcl1-15 embryos reveals heterochronic changes in gene expression patterns. A, Hierarchical clustering of wild-type (WT) and dcl1-15 microarrays with arrays of preglobular, globular, heart, torpedo, and green cotyledon stage embryos from the Goldberg and Harada laboratories using the genes differentially expressed between wild-type and dcl1-15 embryos. Wild-type samples cluster with the same stage embryos (torpedo), but dcl1-15 samples cluster with developmentally more mature embryos (green cotyledon). B, The 7,146 temporally expressed genes (genes whose expression varies with embryonic stage) identified from the Goldberg-Harada data set fall into nine clusters of unique expression patterns based on Pearson’s dissimilarity. Each line shows the average expression pattern for all genes in that cluster. Values from replicate arrays were averaged. For all clusters with significant differences between torpedo and green cotyledon samples, the dcl1-15 samples looked more like the green cotyledon samples when all temporally expressed genes were considered. C, There is a significant positive enrichment for temporally increasing genes in dcl1-15 up-regulated genes (significant Fisher’s exact right-tail P value) and a significant absence of temporally decreasing genes (significant Fisher’s exact left-tail P value). Only genes expressed in both data sets were considered in C and D. D, There is a significant positive enrichment for temporally decreasing genes in dcl1-15 down-regulated genes and a significant absence of temporally increasing genes. Temporally increasing genes increase in expression with stage from preglobular through green cotyledon, while temporally decreasing genes decrease in expression with stage from preglobular through green cotyledon.
Differential Regulation of miRNA Targets in the Seed
To look at the effect of dcl1-15 on miRNA target expression, we compiled a list of validated or predicted miRNA targets using public lists from the Arabidopsis Small RNA Project at Oregon State University (http://asrp.cgrb.oregonstate.edu/) and the Arabidopsis Next-Gen Sequence Databases at the University of Delaware (http://mpss.udel.edu/), the latter of which includes the results of the parallel analysis of RNA ends sequencing data from German et al. (2008). The ATH1 array has 499 probe sets that recognize miRNA targets on this list, with 148 of these being temporally expressed in embryos (Supplemental Tables S8 and S9). Of the 499 probe sets on the array, 409 passed the MAS5.0 filter in our arrays, with 67 up-regulated (targeted by 35 miRNAs, including 18 temporally expressed genes), 80 down-regulated (targeted by 34 miRNAs, including 48 temporally expressed genes), and 262 not changing in dcl1-15 (Supplemental Tables S10 and S11). Interestingly, in many cases, different targets of the same miRNA are up-regulated while others are down-regulated, further suggesting that the regulation of the steady-state levels of miRNA targets is complex.
Known Regulators of the Seed Maturation Program Are Regulated by DCL1
The observed heterochronic phenotype and gene expression patterns of dcl1-15 suggest that one or more DCL1-generated miRNAs and their target genes regulate the timing of maturation, possibly by altering the expression of the master regulators of maturation. Consistent with this hypothesis, L1L and FUS3 transcripts were significantly higher in the microarray (1.8- and 2.6-fold, respectively) and LEC2 transcripts were higher, although just below the statistical cutoff (1.7-fold; P = 0.079). WRI1, encoding a transcription factor that induces the expression of the genes involved in the synthesis of storage lipids, was also significantly up-regulated in the mutant (1.57-fold; Table I).
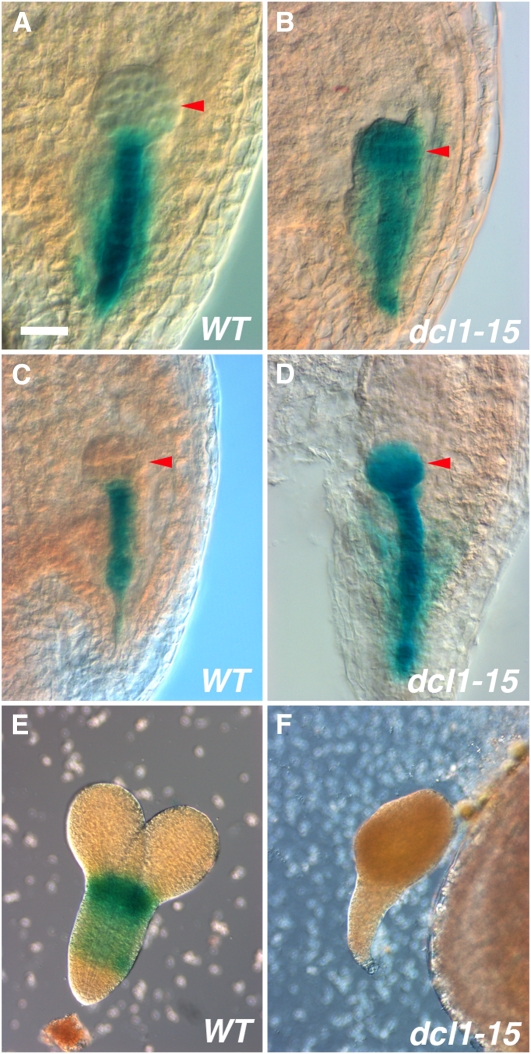
To strengthen the inferences suggested by the microarray analysis, we decided to study the expression of FUS3 and LEC2 using reporter genes. This method allowed us to check the spatial expression of the genes at different stages of embryogenesis. In wild-type embryos, both pFUS3:GUS and pLEC2:GUS are expressed from early embryogenesis in the suspensor and the hypophysis (Fig. 4, A and C). At the late globular/early heart stage, expression spreads to encompass the whole embryo (although pLEC2:GUS is weaker in the upper half of the embryo; Kroj et al., 2003). In dcl1-15 early to midglobular stage embryos, both reporter genes were expressed throughout the embryo proper (also weaker in the upper tier for pLEC2:GUS), in a fashion similar to later stage wild-type embryos (Fig. 4, B and D). After the early heart stage, both wild-type and dcl1-15 embryos stained similarly. We conclude that DCL1 represses FUS3 and LEC2 in the embryo proper during early embryogenesis and that the loss of miRNAs leads to their ectopic expression.
Figure 4.
Expression of master regulators of maturation in dcl1-15. A and B, pFUS3:GUS at the midglobular stage is expressed only in the suspensor in wild-type (WT) embryos (A) but throughout the embryo proper in dcl1-15 (B). C and D, pLEC2:GUS shows similar expression to pFUS3:GUS at the midglobular stage in wild-type (C) and dcl1-15 (D) embryos. E and F, At the midtorpedo stage, pLEC1:GUS is expressed in wild-type (E) but not in dcl1-15 (F) embryos. Arrowheads point to the embryo proper. Bar = 20 μm (A–d) or 40 μm (E and F).
The microarray also suggested that LEC1 expression is reduced in dcl1-15. pLEC1:GUS expression in wild-type embryos is first detected at the early heart stage, increasing until the late torpedo stage and then decreasing (Lotan et al., 1998; Fig. 4E). Consistent with the microarray results, we were not able to detect pLEC1:GUS expression in dcl1-15 embryos at any stage (Fig. 4F).
FUS3, LEC2, and AGL23 Are Required for the Early Maturation Phenotype
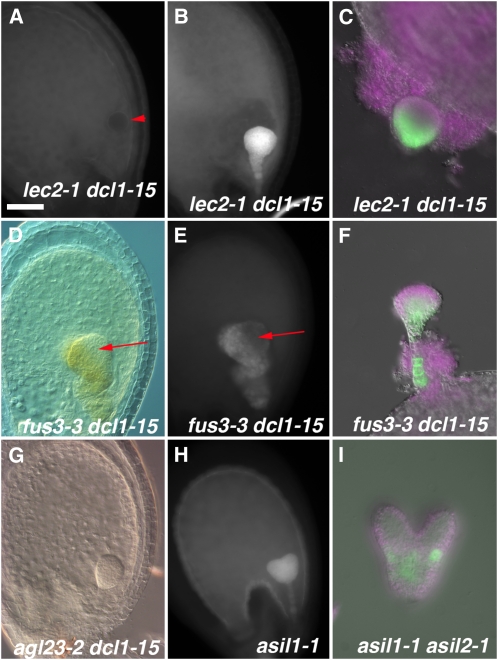
The analysis of reporter genes indicated that both FUS3 and LEC2 are negatively regulated by miRNA pathways during early embryogenesis. We wondered whether the early maturation seen in dcl1-15 embryos required the presence of FUS3 and LEC2 proteins or whether miRNA target genes triggered the program independently of these regulators. Previous studies have shown that the timing of embryo maturation in fus3-3 and lec2-1 embryos is normal, based on the stages of appearance of chlorophyll, storage proteins, and the expression of pAt2S3:GFP. In both mutants, the levels of chlorophyll and pAt2S3:GFP could be significantly reduced, although with pAt2S3 expression more severely affected in fus3-3 (Meinke et al., 1994; To et al., 2006). To distinguish between these hypotheses, we generated plants heterozygous for dcl1-15 and homozygous for pAt2S3:GFP and fus3-3 or lec2-1. We evaluated the presence of chlorophyll autofluorescence and the expression of pAt2S3:GFP in fus3-3 dcl1-15 and lec2-1 dcl1-15 double mutant embryos. Preliminary observations suggest that lec2-1 does not rescue the morphological defects of dcl1-15 embryos while fus3-3 enhances them (data not shown).
lec2-1 dcl1-15 embryos did not show chlorophyll autofluorescence or pAt2S3:GFP expression until the heart stage (n = 29 and 13, respectively), a bit earlier than the wild type but later than dcl1-15 (Fig. 5A). After the heart stage, the double mutant embryos showed intense chlorophyll autofluorescence and pAt2S3:GFP expression, although the reporter was concentrated on the basal part of the embryo (n = 52 and 14, respectively; Fig. 5, B and C). A functional LEC2 protein, therefore, is required for dcl1-15 embryos to mature before the heart stage but becomes dispensable afterward.
Figure 5.
Phenotypes of mutants that regulate maturation downstream of dcl1-15. A to C, lec2-1 dcl1-15 double mutant embryos. No chlorophyll autofluorescence is observed at the globular stage (arrowhead points at the embryo proper; A) but is intense at the late heart stage (B). pAt2S3:GFP expression is seen at the late heart stage (C). D to F, fus3-3 dcl1-15 double mutant embryos. DIC optics (D) and chlorophyll autofluorescence (E) images of a heart stage embryo show a patch without chlorophyll (arrows). pAt2S3:GFP expression is seen at the late heart stage (F). G, An agl23-2 dcl1-15 heart stage embryo showing dcl1-15-like morphology but no chlorophyll. H, Early chlorophyll autofluorescence throughout an early heart stage asil1-1 embryo. I, Early pAt2S3:GFP expression in a heart stage asil1-1 asil2-1 embryo. In C, F and I the magenta color indicates chlorophyll autofluorescence. Bar = 40 μm (A–H) or 20 μm (I).
In contrast, fus3-3 dcl1-15 embryos were quite variable in their early maturation phenotypes. The proportion of fus3-3 dcl1-15 embryos that had chlorophyll autofluorescence before the heart stage varied from silique to silique, even in the same plant, from 0% to 100%. On average, 68% of the double mutant embryos in a silique fluoresced (n = 122 embryos, eight siliques) or expressed pAt2S3:GFP (n = 29) earlier than normal. The proportion of fluorescing (n = 74) or pAt2S3:GFP-expressing (n = 9) fus3-3 dcl1-15 embryos increased to 88% by the heart stage. In some of these embryos, there were patches (toward the apical end) that had no chlorophyll fluorescence and were not green (Fig. 5, D and E). pAt2S3:GFP was also expressed in a patchy way, mostly in the basal part of the embryo (Fig. 5F). These data indicate that FUS3, like LEC2, is necessary for the early maturation phenotypes of dcl1-15 but not required after the heart stage. The variable and patchy nature of the phenotype suggests redundancy with other factors, as has been proposed previously (To et al., 2006).
We also analyzed whether AGL23 was required for the dcl1-15 phenotypes. AGL23 is one of the few known transcription factors that specifically regulate chloroplast maturation during embryogenesis. agl23-2 embryos are white or very pale green with underdeveloped chloroplasts (Colombo et al., 2008) but accumulate storage proteins normally (Supplemental Fig. S2). Homozygotes are seedling lethal (Colombo et al., 2008). In our microarrays, AGL23 was not significantly expressed, consistent with the observation that its expression decreases after the early torpedo stage (Colombo et al., 2008). We wanted to test whether AGL23 was required for establishing the early chloroplast maturation we saw in dcl1-15. We looked at the progeny of DCL1/dcl1-15 AGL23/agl23-2 self-pollinated plants. We found that 4% of dcl1-15 heart stage embryos (n = 162) were not green nor had chlorophyll fluorescence (Fig. 5G). A similar percentage of pale dcl1-15 embryos was observed at the bent-cotyledon stage (n = 511). These percentages are not significantly different from those expected for the dcl1-15/dcl1-15 agl23-2/agl23-2 class (χ2 test, P = 0.2 and 0.46, respectively, for heart and bent-cotyledon stage, assuming 60% female transmission of agl23-2 as described by Colombo et al. [2008]). Our conclusion is that agl23-2 is epistatic to dcl1-15; therefore, a functional AGL23 protein is required for the heterochronic maturation of chloroplasts.
ASIL1, ASIL2, and HDA6/SIL1 Repress the Maturation Program during Early Embryogenesis
Our transcript profiling data also revealed that a handful of the genes that have been shown to repress the expression of the embryonic program after germination were down-regulated in torpedo stage dcl1-15 mutants (Table I). These were CLF, HDT1, HDA6/SIL1, and ASIL1. We also saw down-regulation of several other histone deacetylases (HDA5, -9, and -10, none of which is known to affect embryogenesis; Hollender and Liu, 2008) and of the closest homolog of ASIL1, At3g14180 (ASIL2 hereafter).
We hypothesized that since these genes are involved in repressing the maturation program postgermination, they might also be involved in repressing it during early embryogenesis downstream of one or more miRNA targets. To test this idea, we looked at asil1-1 (Gao et al., 2009), asil2-1 (line SAIL_258_F06 from the Arabidopsis Biological Resource Center [ABRC]), and sil1-1 (Furner et al., 1998) embryos in both single and double mutant combinations. The results are summarized in Table II. We observed no obvious morphological defects in the single or double mutant embryos. All the homozygous single mutant plants were viable and fertile (data not shown). Of the three single mutants, only a fraction (9.2%) of asil2-1 embryos showed early chlorophyll fluorescence during the globular stage, in a similar pattern to that seen in dcl1-15 embryos. By the early heart stage, that fraction had increased to 27.6%. At this stage, 43.5% of asil1-1 embryos also start to show chlorophyll fluorescence. In both these cases, the fluorescence is localized throughout the embryo (Fig. 5H), which is distinct from the protodermal localization seen in wild-type heart stage embryos (Fig. 1F). sil1-1 embryos do not mature early, at least by this measure.
Table II. Fractions of embryos showing early chlorophyll fluorescence in single and double mutants of putative repressors of the maturation program.
n.a., Not applicable.
| Genotype of Selfed Parental Plant | Fraction of Fluorescent Embryos: Globular Stage | Fraction of Fluorescent Embryos: Early Heart/Heart Stage | Expected Fraction of Double Mutant Embryos |
| Wild type | 0% (n > 100) | 0% (n > 100) | n.a. |
| sil1-1/sil1-1 | 0% (n > 100) | 0% (n > 100) | n.a |
| asil1-1/asil1-1 | 0% (n > 100) | 43.5% (n = 230) | n.a |
| asil2-1/asil2-1 | 9.2% (n = 261) | 27.6% (n = 87) | n.a |
| ASIL1/asil1-1 SIL1/sil1-1 | 27% (n = 209) | 38% (n = 279) | 6.25% |
| asil1-1/asil1-1 SIL1/sil1-1 | 41.7% (n = 48) | 70% (n = 80) | 25% |
| ASIL2/asil2-1 SIL1/sil1-1 | 5.2% (n = 328) | 27% (n = 271) | 6.25% |
| ASIL1/asil1-1 ASIL2/asil2-1 | 25% (n = 354) | 60% (n = 319) | 6.25% |
The phenotypes of double mutant combinations strongly suggest that the timing of maturation is controlled at least in part by the concerted action of ASIL1, ASIL2, and HDA6/SIL1. Double mutant embryos of all combinations phenocopy the early maturation phenotype of dcl1-15. The fraction of embryos that show chlorophyll fluorescence at the globular stage indicates that these three factors regulate the timing of maturation in a dose-dependent manner, dominantly enhancing each other, because many embryos that are homozygous for one mutation and heterozygous for the other show the phenotype (Table II).
We were also able to analyze the expression of pAt2S3:GFP in asil1-1 and asil1-1 asil2-1 embryos. In both cases, we observed expression as early as the early heart stage, phenocopying dcl1-15 embryos, although the levels and extent of expression were lower than in dcl1-15 embryos (Fig. 5I). These data support the conclusion that these mutants are also heterochronic.
DISCUSSION
miRNAs Regulate Embryonic Maturation
The maturation program was a key innovation of seed plants (Vicente-Carbajosa and Carbonero, 2005). Like any other important developmental process, this program is tightly regulated. Several of the mechanisms involved in inducing maturation during embryogenesis and repressing it after germination have been worked out (Baud et al., 2008; Santos-Mendoza et al., 2008; Zhang and Ogas, 2009). However, other pieces of the puzzle are missing, in particular the genes that repress maturation during early embryogenesis. The data presented here strongly suggest that miRNAs are these key negative regulators of the maturation program. In dcl1-15 embryos, we observed early chloroplast maturation and early accumulation of starch grains, lipid bodies, and storage proteins. Our conclusion is reinforced by the transcriptional profile of dcl1-15 torpedo stage embryos, which resembles that of older embryos. Similarly, Nodine and Bartel (2010) described the early accumulation of OLEOSIN transcripts and a heterochronic change in the transcriptional profile in dcl1-5 embryos. Interestingly, even though the maturation program starts too early, the timing of the different components of the program seems to be unaffected. In particular, we still see accumulation of chlorophyll preceding the expression of the seed storage protein At2S3, as in wild-type embryos. It is tempting to speculate that miRNAs regulate the initiation, but not the progression, of maturation.
Positive Regulators of Maturation Are Downstream of miRNAs
The key positive regulators of the maturation program are the LEC genes, probably assisted by several bZIP and MYB transcription factors (Baud et al., 2008). All of these bind to the promoters of seed storage protein genes and genes involved in the synthesis of storage lipids (Lara et al., 2003; Kagaya et al., 2005; Braybrook et al., 2006; Santos-Mendoza et al., 2008). We found that several of these genes were up-regulated in the dcl1-15 mutant: FUS3, LEC2, L1L, and several MYBs and bZIPs (Table I). In contrast, ABI3 was unaffected and LEC1 was down-regulated.
We observed that in the wild type, the FUS3 and LEC2 transcriptional reporter genes are excluded from the embryo proper until the early heart stage, just before the beginning of maturation. In dcl1-15, they are expressed in the embryo proper at least by the early globular stage, coinciding with the onset of chlorophyll accumulation in the mutant. Our analysis of lec2-1 dcl1-15 and fus3-3 dcl1-15 double mutants confirms that both proteins are required for the early maturation phenotype, further supporting their position downstream of miRNAs. The requirement for FUS3 was less pronounced, and it has been suggested previously that it acts redundantly with LEC2 (To et al., 2006). This redundancy, and the fact that FUS3 positively regulates itself, likely explain the patchy appearance (in terms of chlorophyll and pAt2S3:GFP expression) of fus3-3 dcl1-15 embryos. We propose, therefore, that miRNA targets are responsible, directly or indirectly, for repressing FUS3, LEC2, and possibly other inducers of maturation in the embryo proper until they are required. The ectopic expression of these factors is sufficient to induce maturation even in the absence of LEC1. Interestingly, after the heart stage, fus3-3 dcl1-15 and lec2-1 dcl1-15 embryos have high levels of chlorophyll and pAt2S3:GFP, something not seen in either fus3-3 or lec2-1 embryos (Meinke et al., 1994; Kroj et al., 2003; To et al., 2006), suggesting that after that stage, other miRNA-regulated factors can compensate for the absence of LEC2 or FUS3.
Identification of Repressors of the Maturation Program during Embryogenesis
One lingering mystery has been the identity of the genes directly repressing the maturation program early in development. Our microarray analysis indicated that ASIL1, HDA6/SIL1, and CLF were down-regulated in dcl1-15. These genes have previously been shown to repress the transcription of the LEC genes and other maturation-related genes in seedlings. Interestingly, several other histone deacetylases were down-regulated (HDA5, -9, -10, and HDT1) as well as the closest homolog to ASIL1 (ASIL2). It has already been shown that ASIL1 binds to a GT-box, which is present in both the promoters of the LEC genes and of the genes encoding storage products, regulating the program at least at two levels (Gao et al., 2009). We analyzed single and double mutant combinations of asil1-1, asil2-1, and sil1-1 and found that these genes redundantly repress the maturation program during early embryogenesis, downstream of miRNA targets. Thus, our studies demonstrate, to our knowledge for the first time, that the same genetic mechanism negatively regulates seed storage genes during early embryogenesis and seedling development.
miRNA Targets as Key Regulators of Maturation
Based on our findings, we suggest that during early embryogenesis, specific miRNAs down-regulate one or more miRNA targets to both promote the repressors and repress the inducers of maturation. Later in embryonic development, a reduction in those miRNAs leads to the induction of maturation. We cannot deduce from our data whether this is a linear pathway (i.e. miRNA targets repress the repressors, which in turn silence the activators) or whether miRNA targets act at more than one level.
In a report published while this article was in preparation, Nodine and Bartel (2010) demonstrated that globular stage dcl1-5 embryos show heterochronic gene expression. Their genetic data also suggest that the increased expression of the miR156 targets SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE10 (SPL10) and SPL11 is partly responsible for the early onset of the maturation program in the mutant, despite the fact that the RNA levels of these genes do not change significantly during embryogenesis, according to the Goldberg-Harada data set. Another SPL gene, SPL5, does significantly increase in expression during embryogenesis in this data set, however. Thus, it might be possible that multiple miR156-targetted SPL genes redundantly regulate the same targets, with the developmental increase in SPL5 expression allowing for the threshold for maturation to be crossed. In such a model, removing the functions of both SPL10 and SPL11 may have lowered the overall SPL activity below the threshold for maturation.
While Nodine and Bartel (2010) demonstrated, as we did, that DCL1 regulates the onset of maturation and identified some of the miRNA targets involved in the process, they did not explore their connection with the components of the maturation pathway. We, on the other hand, placed several known and novel regulators of maturation downstream of unidentified miRNA targets. If their inferences about the SPL genes are correct, it is possible that these proteins positively regulate maturation by repressing ASIL1 and other negative regulators, since SPL10 and SPL11 (and nine other miRNA targets) are predicted to contain transcriptional repression domains (Mitsuda and Ohme-Takagi, 2009). Further studies are now required to integrate both sets of observations and to fully uncover the pathway by which miRNAs control the seed maturation program during embryogenesis.
MATERIALS AND METHODS
Plant Material
Our genetic screen was performed in a mixed Wassilewskija/Ler background of Arabidopsis (Arabidopsis thaliana). Heterozygous mutant plants were back-crossed four times to Ler before further analysis. Most mutants and reporter lines have been described before: pAT2S3:GFP, pLEC2:GUS, and pFUS3:GUS (Kroj et al., 2003), pLEC1:GUS (Siefers et al., 2009), dcl1-3, dcl1-5, and dcl1-9 (Schauer et al., 2002), pnh-2 and ago1-8 (Lynn et al., 1999), se-3 (Grigg et al., 2005), hen1-1 (Chen et al., 2002), hyl1-2 (Vazquez et al., 2004), rdr6-11 (Peragine et al., 2004), fus3-3 and lec2-1 (Meinke et al., 1994), agl23-2 (Colombo et al., 2008), asil1-1 (Gao et al., 2009), and sil1-1 (Furner et al., 1998). asil2-1 is line SAIL_258_F06 (ABRC stock no. CS812086). Seeds were obtained from the ABRC or other researchers. Plants were grown in a greenhouse or growth chamber in MetroMix 360 (SunGro Horticulture) at 20°C to 24°C with 16 h of light at 150 μE m−2 s−1. For transgenic selection, seeds were surface sterilized and grown on sterile medium containing 4.4 g L−1 MS salts, 1× Gamborg’s vitamins, 0.5 g L−1 MES, 10 g L−1 Suc, and 7.5 g L−1 tissue culture agar (Carolina Biological), pH 5.7, plus the appropriate selective agent. To generate fus3-3/fus3-3 and lec2-1/lec2-1 plants, homozygous bent-cotyledon stage embryos were germinated on MS plates and then transplanted to soil. All reagents were from Sigma-Aldrich unless indicated. Primers used for PCR genotyping of mutants are detailed in Supplemental Table S1.
Optical Microscopy and Histochemistry
For morphological characterization, whole developing seeds were cleared in Hoyer’s solution (70% chloral hydrate, 4% glycerol, and 5% gum arabic) and examined with differential interference contrast (DIC) optics. To prevent extraction of chlorophyll, cleared embryos were analyzed within a few hours. To stain whole seeds for GUS, the developing seeds were incubated in 90% acetone for 15 min at room temperature, washed with water, and then transferred to GUS staining solution (100 mm phosphate buffer, pH 7, 1 mm EDTA, 1% Tween 20, 2.5 mm potassium ferrocyanide/ferricyanide, and 1 mg mL−1 X-GlcA [Sigma-Aldrich]). After 1 to 2 h at 37°C, the seeds were washed with water and cleared in Hoyer’s solution. For GUS staining or epifluorescence of isolated embryos, they were squeezed out of the seed in either GUS staining solution and incubated at 37°C or in 10% glycerol and observed immediately. All optical microscopy was done on a Leica DMRB microscope (Leica Microsystems). For observation of GFP and chlorophyll fluorescence, the excitation/emission wavelengths were 480/535 nm and 560/645 nm, respectively. Images were acquired with either a ProgRes MFcool or a ProgRes C5 camera (Jenoptik). Images taken on different channels were merged with the ProgRes software.
Total chlorophyll content in mature embryos was measured as described (Albrecht et al., 2008).
TEM
Developing seeds were fixed overnight at 4°C in 10 mm sodium phosphate, pH 6.8, 100 mm sodium chloride, 4% paraformaldehyde, 2% glutaraldehyde, and 0.1% Triton X-100. After washing with 10 mm sodium phosphate and 100 mm sodium chloride, the seeds were postfixed with 1% osmium tetroxide for 1 h, washed with water, and stained with 1% aqueous uranyl acetate. The stained seeds were gradually dehydrated with a dilution series of ethanol and acetone and embedded in Spurr’s resin according to the manufacturer’s instructions. After thin sectioning (80 nm), the sections were stained with uranyl acetate in 50% ethanol, washed with water, and treated with Reynold’s lead citrate. Thin sections were visualized on a JEOL JEM-100SX electron microscope (JEOL USA). Photographs were taken at 10,000× or 50,000× on Kodak 4489 film. All the reagents for TEM were from Electron Microscopy Sciences.
Microarray Experiments
Sample Collection, RNA Isolation, and Labeled Target Preparation
Three hundred torpedo stage mutant embryos from siliques of DCL1/dcl1-15 plants and 300 wild-type torpedo stage embryos from DCL1/DCL1 siblings were collected and ground in liquid nitrogen. RNA was extracted from the tissue using the RNeasy Micro kit (Qiagen) according to the directions for extracting RNA from tissues, with modifications as follows. The samples were ground in 350 μL of RLT buffer alone and then with 25 μL of Plant RNA Isolation Aid (Ambion) added. The samples were homogenized by passing the material over a Qiashredder column (Qiagen). The rest of the RNeasy Micro protocol was performed according to the instructions, including the on-column DNA digestion. The eluted RNA was precipitated overnight at −20°C in 0.1 μg μL−1 glycogen, 0.6 m ammonium acetate, and 70% ethanol, spun at 4°C for 50 min at 18,000g, and resuspended in 10 μL of water. The RNA concentration and purity were verified using a Nanodrop spectrophotometer (Thermo Fisher Scientific).
Biotin-labeled cDNA targets for hybridization to Affymetrix Arabidopsis ATH1 microarrays were made from 50 ng of starting RNA that was reverse transcribed and amplified using the Ovation RNA Amplification System V2 and fragmented and labeled using the FL-Ovation cDNA Biotin Module V2 (NuGEN Technologies) per the manufacturer’s instructions. Three biological replicates each of wild-type and dcl1-15 embryos were performed. The University of Pennsylvania Microarray Core Facility hybridized the arrays. Raw data have been deposited at the Gene Expression Omnibus under accession number GSE24887.
Normalization and Identification of Differentially Expressed Genes
The microarrays were gcRMA normalized in R and filtered using MAS5.0 presence/absence calls to remove any probe sets not expressed in at least one sample. The remaining 17,159 probe sets were tested for differential expression in R using Limma with a Benjamini-Hochberg MTC ≤ 0.05. Quality control verification was done using the simpleaffy package in R (http://www.bioconductor.org/). Principal components analysis performed in Partek Genomics Suite 6.5 showed that the replicates clustered together well. GO term and SP-PIR keyword enrichment was performed using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/home.jsp; Dennis et al., 2003; Huang et al., 2009).
Meta-Analysis
We compared our data set with a set of 10 ATH1 microarrays of laser-capture microdissection samples of preglobular, globular, heart, linear cotyledon (torpedo), and mature green cotyledon stage Arabidopsis embryos (two replicates of each) from the laboratories of Robert Goldberg (University of California Los Angeles) and John Harada (University of California Davis; http://www.seedgenenetwork.net/arabidopsis). To identify temporally expressed genes in embryos, genes whose expression varies with embryonic stage, the 10 arrays were normalized, as described above, but all probe sets not expressed based on MAS5.0 presence/absence calls in at least two arrays were filtered out. Differential expression analysis was done using Limma on the remaining 15,010 probe sets, as above, with all pairwise contrasts between different stages. The union of the significantly different genes in these different contrasts (Benjamini-Hochberg MTC ≤ 0.01) was used as a unified list of temporally expressed genes in embryos. These genes were then clustered in Partek into nine partitions using Pearson’s dissimilarity, four temporally increasing (2,654 total genes increasing in subsequent developmental stages), three temporally decreasing (2,825), one peaking in heart stage embryos (335), and one peaking in torpedo stage embryos (325). Overlaps between differentially expressed genes in dcl1-15 embryos and these data sets were tested for statistical significance by a Fisher’s exact test using only the sets of genes expressed in both experiments by MAS5.0 presence/absence calls. For the purposes of hierarchical clustering (in Partek) with our own arrays, these were renormalized with our six arrays, with all probe sets not expressed based on MAS5.0 presence/absence calls in at least two arrays filtered out. To verify that we identified the most appropriate hierarchical clustering pattern, trees were drawn using Euclidean or Pearson’s dissimilarity measures of distance with single, complete, or average linkage measures. All six trees were evaluated in R by calculating the average silhouette widths at each node, which is a classic method for evaluating dendrograms (Rousseeuw, 1987). The tree with the highest average silhouette widths was that produced using Pearson’s dissimilarity with average linkage (Supplemental Fig. S3). Silhouette widths range from −1 (definitely wrong cluster assignment) to +1 (absolutely correct cluster assignment), with 0 being neutral. Values above 0 suggest proper assignment.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mutations in DCL1.
Supplemental Figure S2. Accumulation of seed storage proteins in wild-type and agl23-2 mature seeds.
Supplemental Figure S3. Hierarchical clustering validation of the cladogram in Figure 3 using average silhouette widths.
Supplemental Table S1. PCR primers used for genotyping.
Supplemental Table S2. Genes up-regulated in dcl1-15 torpedo stage embryos, based on Limma, with a Benjamini-Hochberg adjusted P < 0.05.
Supplemental Table S3. Genes down-regulated in dcl1-15 torpedo stage embryos, based on Limma, with a Benjamini-Hochberg adjusted P < 0.05.
Supplemental Table S4. GO term and SP-PIR keyword enrichment of genes up-regulated and down-regulated in dcl1-15 torpedo stage embryos.
Supplemental Table S5. Genes temporally expressed in embryos based on the Goldberg-Harada data set.
Supplemental Table S6. Genes up-regulated in dcl1-15 torpedo stage embryos and temporally increasing in embryos.
Supplemental Table S7. Genes down-regulated in dcl1-15 torpedo stage embryos and temporally decreasing in embryos.
Supplemental Table S8. Probe sets on the ATH1 array recognizing miRNA targets.
Supplemental Table S9. Temporally expressed miRNA targets with probe sets on the ATH1 array.
Supplemental Table S10. miRNA targets up-regulated in dcl1-15 torpedo stage embryos with temporal genes designated.
Supplemental Table S11. miRNA targets down-regulated in dcl1-15 torpedo stage embryos with temporal genes designated.
Acknowledgments
We are grateful to the following researchers who provided seeds: François Parcy (pAT2S3:GFP, pLEC2:GUS, pFUS3:GUS), Stewart Gillmor (pnh-2, ago1-8), and Ben Holt (pLEC1:GUS). We thank Rob Jinks for TEM training and advice, Qi Zheng for advice on the silhouette approach, Jim Engelman for expert plant care, and Stewart Gillmor for comments on the manuscript.
References
- Albrecht V, Ingenfeld A, Apel K. (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Leipiniec L. (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113, doi/10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Feria Bourrellier AB, Azzopardi M, Berger A, Dechorgnat J, Daniel-Vedele F, Lepiniec L, Miquel M, Rochat C, Hodges M, et al. (2010) PII is induced by WRINKLED1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J 64: 291–303 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F. (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR. (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L. (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Dennis GJ, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Du Z, Lee JK, Tjhen R, Stroud RM, James TL. (2008) Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci USA 105: 2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner IJ, Sheikh MA, Collett CE. (1998) Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics 149: 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M-J, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, Rozwadowski K. (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21: 54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. (2005) SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. (2005) Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57: 173–188 [DOI] [PubMed] [Google Scholar]
- Hollender C, Liu Z. (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50: 875–885 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. (1994) Arabidopsis. Bard J, , Embryos: Color Atlas of Development. Wolfe Publishing, London, pp 7–21 [Google Scholar]
- Jurkuta RJ, Kaplinsky NJ, Spindel JE, Barton MK. (2009) Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 21: 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kirik V, Kölle K, Miséra S, Bäumlein H. (1998) Two novel MYB homologues with changed expression in late embryogenesis-defective Arabidopsis mutants. Plant Mol Biol 37: 819–827 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F. (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kaminuma E, Matsui A, Kawashima M, Tanaka M, Morosawa T, Ishida J, Mochizuki Y, Shinozaki K, Toyoda T, et al. (2009) Transcriptome analyses revealed diverse expression changes in ago1 and hyl1 Arabidopsis mutants. Plant Cell Physiol 50: 1715–1720 [DOI] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J. (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Buechel S, Sachsenberg T, Wang J-W, Rätsch G, Weigel D. (2010) Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc Natl Acad Sci USA 107: 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL. (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C. (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Hinze A, Tucker MR, Bouché N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. (2009) Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet 5: e1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. (1994) leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 50: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24: 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua N-H. (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49: 592–606 [DOI] [PubMed] [Google Scholar]
- Ronemus MJ, Vaughn MW, Martienssen RA. (2006) MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ. (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20: 53–65 [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis Development 120: 3235–3245 [DOI] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, IV, Tayrose G, Holt BF., III (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang X, Niu Q-W, Teng C, Li C, Mu J, Chua N-H, Zuo J. (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19: 224–235 [DOI] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ogas J. (2009) An epigenetic perspective on developmental regulation of seed genes. Mol Plant 2: 610–627 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cao G, Qu L-J, Gu H. (2009) Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep 28: 337–346 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K. (2004) Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J 38: 715–724 [DOI] [PubMed] [Google Scholar]