Abstract
Flavonoids are a class of metabolites found in many plant species. They have been reported to serve several physiological roles, such as in defense against herbivores and pathogens and in protection against harmful ultraviolet radiation. They also serve as precursors of pigment compounds found in flowers, leaves, and seeds. Highly methylated, nonglycosylated derivatives of the flavonoid myricetin flavonoid, have been previously reported from a variety of plants, but O-methyltransferases responsible for their synthesis have not yet been identified. Here, we show that secreting glandular trichomes (designated types 1 and 4) and storage glandular trichomes (type 6) on the leaf surface of wild tomato (Solanum habrochaites accession LA1777) plants contain 3,7,3′-trimethyl myricetin, 3,7,3′,5′-tetramethyl myricetin, and 3,7,3′,4′,5′-pentamethyl myricetin, with gland types 1 and 4 containing severalfold more of these compounds than type 6 glands and with the tetramethylated compound predominating in all three gland types. We have also identified transcripts of two genes expressed in the glandular trichomes and showed that they encode enzymes capable of methylating myricetin at the 3′ and 5′ and the 7 and 4′ positions, respectively. Both genes are preferentially expressed in secreting glandular trichome types 1 and 4 and to a lesser degree in storage trichome type 6, and the levels of the proteins they encode are correspondingly higher in types 1 and 4 glands compared with type 6 glands.
Flavonoids constitute a large and structurally diverse family of metabolites synthesized in plants. The core structure of flavonoids is a 2-phenylchromen-4-one (flavonoids), a 3-phenylchromen-4-one (isoflavonoids), or a 4-phenylcoumarin (neoflavonoids). The great structural diversity of flavonoids stems from the possible substitution on up to 10 carbons of the core structure. Some common functional group substitutions include hydroxylation, methylation, sulfonation, methylation, and (iso)prenylation (Ibrahim and Anzellotti, 2003; Ibrahim, 2005). In addition to these core substitutions, hydroxyl functional groups can be further modified by the addition of a wide range of different sugar moieties, which can be further modified themselves. Current estimates of the number of structurally distinct, plant-derived flavonoids probably exceed 9,000 (Williams and Grayer, 2004). This rich structural diversity extends well into the functional diversity of flavonoids. They play crucial roles in plants in pathogen and herbivore defense, protection from harmful UV radiation, and pigmentation of flowers, fruits, and seeds. They also act as plant-microbe signaling molecules, inhibitors in biochemical pathways, and developmental regulators (Taylor and Grotewold, 2005; Treutter, 2005; for review, see Buer et al., 2010).
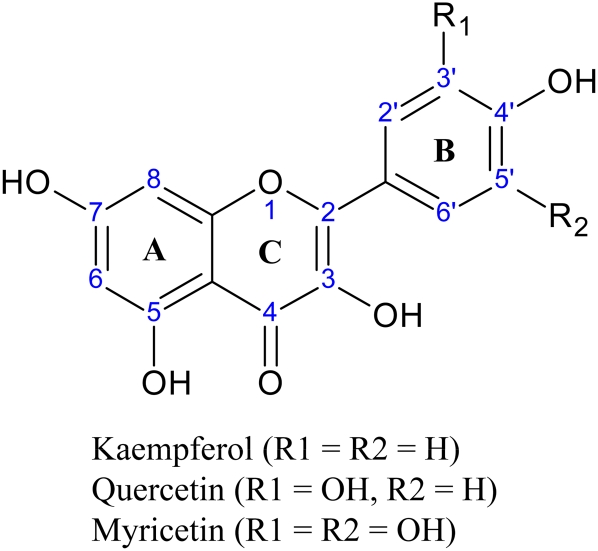
The flavonoid pathway in flowering plants can be traced back to the first plants to colonize land. The most primitive form of the pathway probably terminated at the production of flavonols (Rausher, 2008). Dihydroflavonols, the reduced forms of flavonols, represent an important step in the evolution of the structural and functional diversity of flavonoids seen in extant flowering plants. All anthocyanins, flavonols, and derivatives of these come from one of the three dihydroflavonols (dihydrokaempferol, dihydroquercetin, and dihydromyricetin), the latter being the most highly substituted, with hydroxyl groups on the 3, 5, 7, 3′, 4′, and 5′ carbons. The enzyme flavonol synthase converts the dihydroflavonoids to their corresponding flavonols, kaempferol, quercetin, and myricetin, by oxidation of the C2-C3 bond of the C ring (Fig. 1).
Figure 1.
Generic flavonol structure showing the lettering system for the three rings and the numbering system for the carbons. Addition of hydroxyl groups at the 3′ position or 3′ and 5′ positions designate quercetin and myricetin, respectively. [See online article for color version of this figure.]
In plants that synthesize methylated and glycosylated derivatives of myricetin, levels of unmodified myricetin are generally very low or not detectable (Stevens et al., 1995, 1996; Reynertson et al., 2008; Riihinen et al., 2008; Kumar et al., 2009; Michodjehoun-Mestres et al., 2009). Methylation has been reported at five of the six available hydroxyl groups: C3 (C ring), C7 (A ring), and C-3′, -4′, and -5′ of the B ring, but not on C5 (A ring), from a variety of different families of flowering plants (Jay et al., 1980; Kumari et al., 1984; Stevens et al., 1995; Jung et al., 2003). Glycosylation of myricetin occurs consistently at C3 of the C ring and appears to be reversible in vitro (Gerats et al., 1983; Kumari et al., 1984; Modolo et al., 2009; Singh et al., 2009). Glycosylation renders the flavonoids more water soluble and facilitates transport into the vacuole, where they are often stored (for review, see Vogt and Jones, 2000). Myricetin, myricetin methyl ethers, and 3-O-glycosylated myricetin derivatives have been reported in leaf tissues (Braca et al., 2001; Motta et al., 2005; Lee et al., 2006; Oliveira et al., 2007), fruits (Gorbatsova et al., 2007; Lako et al., 2007; Le et al., 2007; Riihinen et al., 2008), flowers (Tabart et al., 2006; Kumar et al., 2008; Liu et al., 2008; Wu et al., 2008), stems and bark (Min et al., 2003), and roots (Ojong et al., 2008).
In plants that synthesize highly methylated flavonols, the process occurs in a stepwise manner, with O-methylation at position 3 being the first step in the process (Macheix and Ibrahim, 1984; Thresh and Ibrahim, 1985; Ibrahim et al., 1987; Huang et al., 2004). In Chrysosplenium americanum, methylation of quercetin proceeds from 3-methylquercetin (3-MeQ) to 3,7-MeQ to 3,7,4′-MeQ. Several species of the genus Aeonium accumulate highly methylated quercetin and myricetin. In these species, the methylation pattern appears to follow the same stepwise addition of methyl groups beginning with position 3. The myricetin methyl ethers that accumulate in the leaves include 3,7,3′-trimethyl myricetin, 3,7,3′,4′-tetramethyl myricetin, and 3,7,3′,4′,5′-pentamethyl myricetin (Stevens et al., 1995). To date, an enzyme responsible for the synthesis of polymethylated myricetin in these species has not been identified. An enzyme isolated from Catharanthus roseus was shown to methylate free myricetin in vitro, and this reaction was hypothesized to occur in vivo prior to the further modifications of myricetin into the anthocyanins observed in the plant, but analysis of the kinetic parameters of the enzyme was not reported (Cacace et al., 2003).
Glandular trichomes are specialized storage and secreting organs that develop on the surface of aerial parts of a wide variety of different plant species (Wagner, 1991; Schilmiller et al., 2008). They synthesize, store, and secrete specialized metabolites important to plant defense and serve as the major source of essential oils (Iijima et al., 2004; Croteau et al., 2005; Ambrósio et al., 2008). They are a valuable resource for elucidating specialized biochemical pathways because they are biochemically highly active in select pathways, metabolite accumulation is species specific, and their metabolites and gene transcripts can be easily extracted and analyzed (Schilmiller et al., 2008, 2010). In the Solanum genus, glandular trichomes can be divided into two main groups, secreting glands and storage glands (Luckwill, 1943; Schilmiller et al., 2010). The secreting glands (of which there are two types, 1 and 4, with type 4 being shorter) are supported atop a relatively long multicellular stalk that varies in length, and the gland itself appears to be unicellular. Droplets rich in specialized metabolites are often observed on the surface of these glands or on the stalk near the gland. The storage glands, defined as type 6 glands, are multicellular and sit atop a relatively short multicellular stalk. The storage glands consist of four cells arranged such that each makes up one-quarter of the round structure.
Here, we report the identification of polymethylated myricetin from isolated types 1, 4, and 6 glandular trichomes from the wild tomato Solanum habrochaites. We also report the identification and the biochemical characterization of two myricetin O-methyltransferases (OMTs) encoded by transcripts found in the S. habrochaites glandular trichomes and show that one of them, S. habrochaites myricetin O-methyltransferase 1 (ShMOMT1), is likely responsible for O-methylation of the 3′ and 5′ hydroxyl groups and the second, ShMOMT2, is likely responsible for O-methylation of the 7 and 4′ hydroxyl groups.
RESULTS
Glandular Trichomes of S. habrochaites Contain Methylated, Nonglycosylated Myricetin
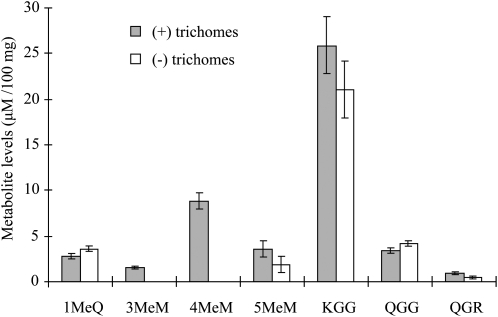
In an initial screen for flavonoids present in leaves of S. habrochaites (accession LA1777), whole leaves were ground and extracted with methyl tert-butyl ether and the extract was analyzed by liquid chromatography-mass spectrometry (LC-MS). This analysis revealed that the leaves contain several glycosylated flavonoids, mostly kaempferol diglucoside but also rutin and quercerin diglucoside (Fig. 2). In addition, several nonglycosylated flavonoids were detected, including 3,7,3′-trimethyl myricetin (3,7,3′-MeM), 3,7,3′,5′-MeM, 3,7,3′,4′,5′-MeM, and 3-MeQ (Fig. 2). When the trichomes were physically removed before the leaves were extracted, the levels of kaempferol diglucoside, rutin, quercetin diglucoside, and 3-MeQ detected remained similar, but no 3,7,3′-MeM, 3,7,3′,5′-MeM, and approximately half the levels of 3,7,3′,4′,5′-MeM were detected, suggesting that these compounds were completely or mostly located in the trichomes (Fig. 2).
Figure 2.
Levels of flavonoid compounds extracted from S. habrochaites leaf with trichomes and with trichomes removed. Flavonoid compounds detected were as follows: 3-methyl quercetin (1MeQ), 3,7,3′-trimethyl myricetin (3MeM), 3,7,3′,5′-tetramethyl myricetin (4MeM), 3,7,3′,4′,5′-pentamethyl myricetin (5MeM), kaempferol diglucoside (KGG), quercetin diglucoside (QGG), and rutin (QGR). Results are averages of three biological replicates ± se.
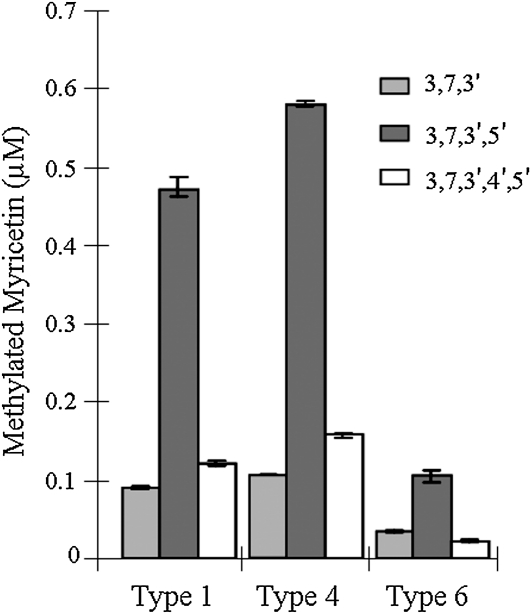
To examine the relative distribution of the nonglycosylated myricetins in specific types of trichomes, secreting glands and storage glands were collected individually from leaves of S. habrochaites for metabolic profiling (Fig. 3; Supplemental Fig. S1). Secreting glands (types 1 and 4) contained higher levels per gland of all three methylated myricetins compared with storage glands (type 6). Levels of 3,7,3′,5′-MeM were greatest in both secreting and storage gland types compared with the 3,7,3′-MeM and 3,7,3′,4′,5′-MeM. However, the levels of all myricetin methyl ethers were 5- to 6-fold greater in secreting type 1 and 4 glands compared with the corresponding levels in storage glands. Also, in secreting glands, the levels of myricetin pentamethyl ether were slightly higher than levels of myricetin trimethyl ether, whereas in storage glands, levels of the trimethylated and pentamethylated myricetins were not significantly different.
Figure 3.
Levels of O-methylated myricetin compounds measured in extracts from 50 glandular trichomes from S. habrochaites leaf. O-Methylated myricetin compounds detected were as follows: 3,7,3′-trimethyl myricetin (3,7,3′), 3,7,3′,5′-tetramethyl myricetin (3,7,3′,5′), and 3,7,3′,4′,5′-pentamethyl myricetin (3,7,3′,4′,5′). Results are averages of three biological replicates ± se.
Characterization of the Substrate Specificity of ShMOMT1 and ShMOMT2
We have recently constructed EST libraries from the secreting and storage glands (types 1 and 4 and type 6, respectively) of S. habrochaites leaves (http://www.trichome.msu.edu/; McDowell et al., 2011). A bioinformatics search of these libraries using BLAST sequence comparisons with known OMT sequences identified three OMT sequences in S. habrochaites trichomes. All three cDNAs were expressed in Escherichia coli, and the crude extracts were tested for OMT activity with a battery of substrates (Table I) and [methyl-14C]S-adenosyl-l-methionine ([14C]SAM) as the methyl donor. One cDNA encoded a protein with similarity to plant N-methyltransferases and had no methylating activity with myricetin, quercetin, kaempferol, or any other flavonol tested in this investigation (for a list of tested compounds, see Table I). Consequently, it was not investigated further. A second cDNA encoded a protein, subsequently named ShMOMT1, with methylating activity toward myricetin and quercetin but not kaempferol, suggesting that this protein has 3′/5′-OMT activity. A third cDNA encoded a protein, subsequently named ShMOMT2, with methylating activity against all three of these flavonols.
Table I. Relative activity of ShMOMT1 with flavonol and O-methyl flavonol substrates.
Additional substrates tested include caffeic acid, eugenol, isoeugenol, chavicol, orcinol, and several different flavones, isoflavones, and anthocyanidins. Substrate concentration was 200 μm, and incubation time was 30 min.
| Substrate | Relative Activitya | ShMOMT1Product | Relative Activitya | ShMOMT2Product |
| % | % | |||
| Flavonols | ||||
| Kaempferolb | ≤1 | – | 117 | 4′-Methyl kaempferol |
| Quercetinc | 51 | 3′-Methyl quercetin | 119 | 7-Methyl quercetin |
| Myricetind | 100 | 3′-Methyl myricetin | 100 | 7-Methyl myricetine |
| O-Methyl flavonols | ||||
| 4′-Methyl kaempferolf | ≤1 | – | 105 | 7,4′-Dimethyl kaempferol |
| 3,7,4′-Trimethyl kaempferol | ≤1 | – | 3 | – |
| 3′-Methyl querceting | ≤1 | – | 23 | 3′,4′-Dimethyl quercetin |
| 3-Methyl quercetin | 61 | 3,3′-Dimethyl quercetin | 19 | 3,7-Dimethyl quercetin |
| 7-Methyl quercetinh | 70 | 7,3′-Dimethyl quercetin | 120 | 7,4′-Dimethyl quercetin |
| 3,7,3′,4′-Tetramethyl quercetin | ≤1 | – | 3 | – |
| 3′-Methyl myricetini | 64 | 3′,5′-Dimethyl myricetin | 5 | 3′,4′-Dimethyl myricetin |
| 3′,5′-Dimethyl myricetinj | ≤1 | – | 11 | 7,3′,5′-Trimethyl myricetin |
| 3′,4′,5′-Trimethyl myricetin | ≤1 | – | 36 | 7,3′,4′,5′-Tetramethyl myricetin |
One hundred percent relative activity represents total methylating activity (ShMOMT1, 3.4 nmol min−1 mg−1; ShMOMT2, 0.02 nmol min−1 mg−1) of the recombinant proteins assayed with myricetin as substrate.
Structure in Figure 1.
Structure in Figure 1.
Structure in Figure 1.
After overnight incubation (more than 10 h), the main product is 7,4′-dimethyl myricetin.
Kaempferide.
Isorhamnetin.
Rhamnetin.
Laricitrin.
Syringetin.
ShMOMT1 and ShMOMT2 were further tested with a range of substrates related to myricetin that could be obtained in sufficient concentrations for these assays. ShMOMT1 catalyzed the transfer of a methyl group to the 3′ hydroxyl of myricetin, as indicated by comigration with an authentic standard of 3′-methyl myricetin in radioactive thin-layer chromatography (TLC; Supplemental Fig. S2) and by LC-MS (Supplemental Fig. S3; the 3′ and 5′ hydroxyl positions in this compound are equivalent, and by convention, the product is designated as 3′-methyl myricetin, or laricitrin) and the equivalent position of several other related compounds, including quercetin, 3-methyl quercetin, and 7-methyl quercetin (i.e. rhamnetin; Table I). When the 3′ hydroxyl of the substrate was already methylated, as in laricitrin, ShMOMT1 transferred a methyl group to the 5′ hydroxyl, as determined by comigration with an authentic standard of 3′,5′-methyl myricetin in radioactive TLC and by LC-MS (Table I; Supplemental Figs. S2 and S3). When both 3′ and 5′ hydroxyls were already methylated, for example in the substrate 3′,5′-dimethyl myricetin (i.e. syringetin), ShMOMT1 could not transfer a methyl group to any other hydroxyl (Table I).
ShMOMT2 transferred a methyl group to the 4′ hydroxyl of kaempferol but to the 7 position of quercetin and myricetin (Table I). When the hydroxyl at the 7 position was already methylated, it transferred a methyl to the 4′ hydroxyl (e.g. with substrate 7-methyl quercetin), and when the 4′ hydroxyl was already methylated, ShMOMT2 transferred the methyl group to the hydroxyl at the 7 position (e.g. with the substrate 3′,4′,5′-trimethyl myricetin). It did not transfer a methyl group to any hydroxyl other than at the 7 or 4′ position (Table I). Radioactive TLC of the reaction with myricetin revealed a single product that migrated between myricetin and 3′-methyl myricetin (Supplemental Fig. S2). This product was identified by LC-MS as 7-methyl myricetin. When myricetin was incubated with ShMOMT2 overnight, the product obtained was 7,4′-methyl myricetin (Supplemental Fig. S4).
The Structural Relatedness of ShMOMT1 and ShMOMT2 to Other OMTs
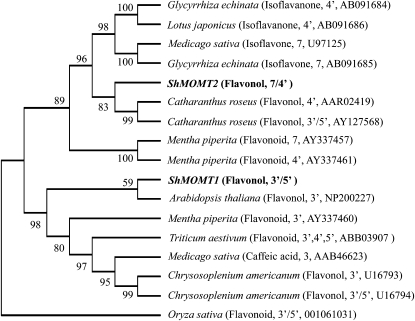
The protein encoded by the ShMOMT1 cDNA is 362 amino acids long, with a calculated molecular mass of 40.7 kD, and it contains all of the recognized plant OMT domains known or hypothesized to be involved in binding to SAM and metal cofactors (Ibrahim, 1997; Joshi and Chiang, 1998; data not shown). ShMOMT1 is most similar (40%–49% identity) to a number of mostly 3′ and 3′/5′-OMTs (Fig. 4), consistent with its regiospecificity for the 3′ and 5′ positions.
Figure 4.
Analysis of the level of relatedness of the ShMOMT1-encoded protein and the ShMOMT2-encoded protein to other plant OMTs. For each protein, descriptions in parentheses indicate the chemical class of the preferred substrate, the position(s) that are O-methylated by the enzyme, and the accession number of the sequence. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to each branch (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method and are in units of the number of amino acid substitutions per site (Zuckerkandl and Pauling, 1965). Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
The protein encoded by the ShMOMT2 cDNA is 355 amino acids long, with a calculated molecular mass of 39.4 kD, and it also contains all of the recognized plant OMT domains known or hypothesized to be involved in binding to SAM and metal cofactors (data not shown). ShMOMT2 is most similar (29%–47% identity) to several OMTs identified (with one exception) as specific for the 7 and/or 4′ position (Fig. 4), consistent with its regiospecificity for these positions. ShMOMT2 is only 27% identical to ShMOMT1.
Distribution of ShMOMT1 and ShMOMT2 Transcripts and Protein in Trichome Glands
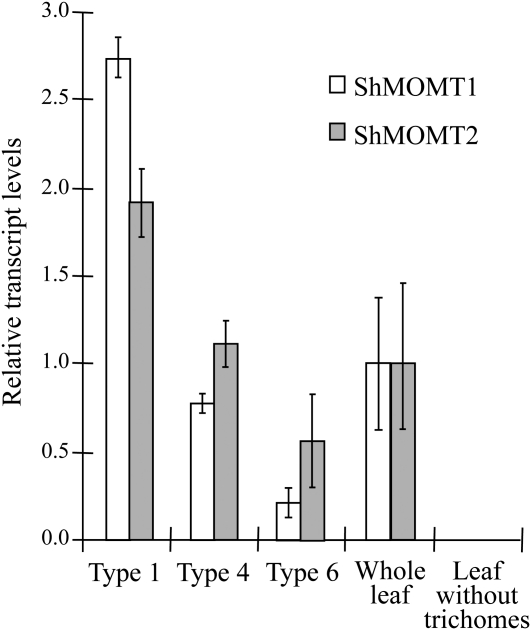
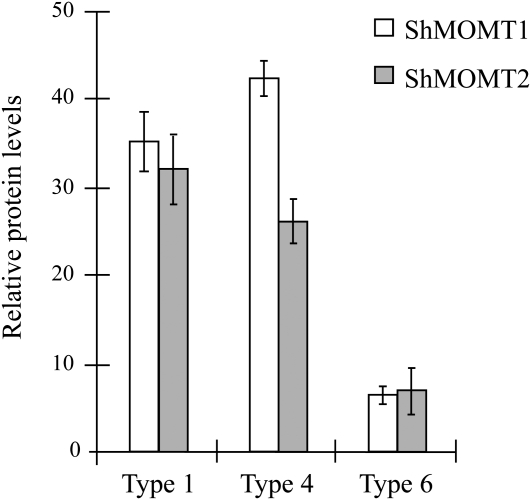
We used quantitative reverse transcription (qRT)-PCR and western-blot analyses to localize ShMOMT1 and ShMOMT2 transcripts, and ShMOMT1 and ShMOMT2 proteins, respectively, in the different types of trichome glands. Extracts of collections of individual types of glands were compared with whole leaf extracts in both types of experiments. ShMOMT1 transcript levels were 3.5- to 12.5-fold higher in secreting glands from types 4 and 1 trichomes, respectively, compared with storage glands of type 6 trichomes (Fig. 5). ShMOMT2 transcript levels were 2- to 4-fold higher in secreting glands from types 4 and 1 trichomes, respectively, compared with storage glands of type 6 trichomes (Fig. 5). Comparison of transcript levels from leaf tissue with trichomes versus leaf tissue from which the trichomes had been mechanically removed indicated that transcripts of both ShMOMT1 and ShMOMT2 are present exclusively in trichomes (Fig. 5). Protein-blot analysis indicated that levels of ShMOMT1 protein were 7- to 8.6-fold higher in secreting glands compared with storage glands and 1.9- to 2.4-fold higher compared with whole leaf extracts (Fig. 6). Levels of ShMOMT2 protein were 5- to 6.6-fold higher in secreting glands compared with storage glands, while ShMOMT2 was not detectable in whole leaf extracts (Fig. 6).
Figure 5.
Relative levels of ShMOMT1 and ShMOMT2 transcript measured in trichomes, leaves, and leaves with trichomes removed by qRT-PCR. Results are averages of three biological replicates ± se.
Figure 6.
Relative levels of ShMOMT1 and ShMOMT2 protein measured in trichomes. Levels were determined by quantitative western-blot analysis with anti-ShMOMT1 or anti-ShMOMT2 and anti-α-tubulin. Results are averages of three biological replicates ± se.
Characterization of the Kinetic Parameters of ShMOMT1 and ShMOMT2
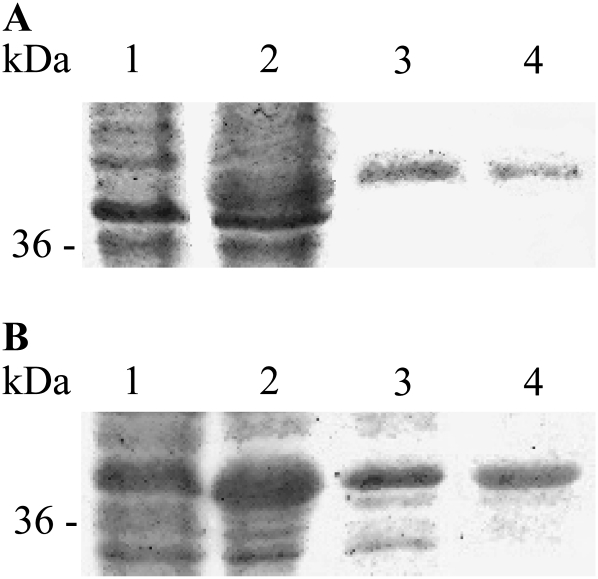
ShMOMT1 and ShMOMT2 were expressed in E. coli BL21 (DE3) cells, and the recombinant proteins were purified to near homogeneity by two successive anion-exchange chromatography steps (Fig. 7). The purified ShMOMT1 protein catalyzed the formation of laricitrin (3′-methyl myricetin) from myricetin, with an apparent Km value of 0.46 μm and an apparent Kcat value of 1.59 s−1. An apparent Km value of 0.21 μm was measured for ShMOMT1 with laricitrin (giving the product syringetin [3′,5′-dimethyl myricetin]) as the substrate, with an apparent Kcat value of 0.45 s−1. The apparent Km value for SAM with myricetin as cosubstrate was 16.64 μm, with an apparent Kcat value of 0.47 s−1 (Table II; Supplemental Fig. S5).
Figure 7.
Purification of E. coli-produced ShMOMT1 (A) and ShMOMT2 (B). Lane 1, noninduced crude extract E. coli cells carrying the ShMOMT1 or ShMOMT2 expression vector but not induced with isopropylthio-β-galactoside; lane 2, induced crude extract; lane 3, the fractions eluting from the DE53 column (A, 2 μg; B, 10 μg) with the highest MOMT activity; lane 4, the fractions eluting from the HiTrap Q column (A, 0.25 μg; B, 5 μg) with the highest MOMT activity. SDS-PAGE results were visualized with Coomassie Brilliant Blue.
Table II. Kinetic parameters of ShMOMT1 with myricetin, laricitrin, and SAM as substrates.
Km and Kcat ± se values are averages of three assays.
Purified ShMOMT2 catalyzed methylation of the 7 hydroxyl group of myricetin, the 7 hydroxyl group of kaempferide (4′-methyl kaempferol), and the 4′ hydroxyl group of rhamnetin (7-methyl quercetin). An apparent Km of 1.68 μm was determined for myricetin, with an apparent Kcat value of 7.4 × 10−3 s−1. An apparent Km of 2.27 μm was determined for kaempferide, with an apparent Kcat value of 5.76 × 10−3 s−1. And an apparent Km of 2.30 μm was determined for rhamnetin, with an apparent Kcat value of 6.40 × 10−3 s−1. The apparent Km value for SAM with kaempferide as cosubstrate was 18.71 μm, with an apparent Kcat value of 1.64 × 10−2 s−1 (Table III; Supplemental Fig. S6).
Table III. Kinetic parameters of ShMOMT2 with myricetin, kaempferide, rhamnetin, and SAM as substrates.
Km and Kcat ± se values are averages of three assays.
| Substrate | Km | Kcat | Kcat/Km |
| μm | s−1 | μm−1 s−1 | |
| Myricetina | 1.68 ± 0.23 | 7.40 × 10−3 ± 9.03 × 10−4 | 4.41 × 10−3 ± 5.74 × 10−4 |
| 4’-Methylkaempferolb | 2.27 ± 0.37 | 5.76 × 10−3 ± 8.89 × 10−4 | 2.53 × 10−3 ± 3.27 × 10−4 |
| 7-Methylquercetinc | 2.30 ± 0.20 | 6.40 × 10−3 ± 5.22 × 10−4 | 2.78 × 10−3 ± 3.06 × 10−4 |
| SAMd | 18.71 ± 2.51 | 1.64 × 10−2 ± 2.33 × 10−3 | 8.75 × 10−4 ± 1.73 × 10−4 |
Characterization of optimal conditions for catalysis revealed that both ShMOMT1 and ShMOMT2 do not require the addition of Mg2+ or Mn2+ for activity. At levels below 2.5 mm, Mg2+ had little negative effect on activity (10% or less); however, concentrations above 2.5 mm had increasing inhibitory effects on activity with myricetin. Similarly, addition of Mn2+ to enzyme assays, using myricetin as substrate, had little negative effect (10% or less) on activity until levels exceeded 2.5 mm. ShMOMT1 activity with myricetin was observed in the pH range of 6.0 to 8.5, with optimal activity observed at pH 7.5. And ShMOMT2 activity with myricetin was observed in the pH range of 6.0 to 9.0, with optimal activity observed at pH 8.0.
DISCUSSION
S. habrochaites Glandular Trichomes Contain Methylated, Nonglycosylated Myricetin
Our metabolic profiling of trichome glands from S. habrochaites leaf identified three forms of O-methylated myricetin species: 3,7,3′-MeM, 3,7,3′,5′-MeM, and 3,7,3′,4′,5′-MeM. These three compounds have previously been shown to accumulate in tissues of several other plants (Stevens et al., 1995; Dachriyanus et al., 2004; Ariyanathan et al., 2010), but they have not yet been reported to be present in trichomes. By isolating individual types of glands, we were able to show that these compounds are found in three types of glandular trichomes, types 1, 4, and 6 (Fig. 3), although they are most abundant in the secreting glands (types 1 and 4).
All of the myricetin methyl ethers that we detected in the glands of glandular trichomes were methylated at the 3 position (in the C ring). This position is often glycosylated, and the glycosylated form is then transported to the vacuole (Vogt and Jones, 2000). We did not find any glycosylated myricetin in the trichomes or myricetin species that are not modified at the 3 position, suggesting that the 3-OMT responsible for this methylation reaction is quite efficient. However, our attempts to detect OMT activity in crude extracts of glands or whole leaves capable of adding a methyl group to the 3-hydroxyl position of myricetin was unsuccessful, nor could we identify a cDNA encoding such an enzyme in our EST databases. To our knowledge, no 3-OMTs capable of methylating myricetin or any other flavonols have been identified from any plant, although a cellular activity capable of methylating quercetin at the 3 position has been reported (De Luca and Ibrahim, 1985; Huang et al., 2004).
Our analyses of S. habrochaites leaves with trichomes and leaves with trichomes removed revealed that 3,7,3′-MeM and 3,7,3′,5′-tetramethyl myricetin were found in trichome gland cells only, and 3,7,3′,4′,5′-pentamethyl myricetin was found in both trichomes and the rest of the leaf organ (Fig. 2). All other flavonol compounds were apparently confined mostly to nontrichome leaf cells, since their levels did not decrease significantly when trichomes were removed (Fig. 2). The presence of 3,7,3′,4′,5′-pentamethyl myricetin, whose 3,7,3′,5′-tetramethyl myricetin precursor is found only in the trichomes, outside the trichomes is most likely due to secretion, since our analysis indicates that ShMOMT1 and ShMOMT2 are not expressed in nontrichome leaf cells (Fig. 5).
ShMOMT1 Is a 3′/5′-Myricetin Methyltransferase and ShMOMT2 Is a 7/4′-Myricetin Methyltransferase
The characterization of the enzymatic properties of ShMOMT1 in vitro showed that it has high affinity for both myricetin and 3′-methyl myricetin, and its products are 3′-methyl myricetin (laricitrin) and 3′,5′-dimethyl myricetin (syringetin; Supplemental Fig. S3). In previous studies, OMTs have been identified that can methylate myricetin at these positions, but in all such cases, myricetin was not the best substrate for the enzyme and the tissue source of the enzyme did not actually contain methylated myricetin but only related compounds, such as quercetin, kaempferol, tricin, tricetin, and luteolin (Muzac et al., 2000; Zhou et al., 2006; Lee et al., 2008). The catalytic efficiency of ShMOMT1 with both myricetin and laricitrin is significantly higher than for such 3′/5′-OMTs (Table IV), and these enzymes had higher affinity to the substrates whose methylation led to the compounds actually observed in the plant.
Table IV. Kinetic parameters of ShMOMT1 with myricetin as a substrate compared with other 3′,5′-OMT enzymes that exhibit activity with myricetin.
ShMOMT2 is most similar to some enzymes characterized as 4′-methyltransferases and some characterized as 7-methyltransferases, with one exception (Fig. 4). This exception is C. roseus flavonol 3′/5′-OMT (Cacace et al., 2003), which is very similar to C. roseus flavonol 4′-OMT (Schröder et al., 2004; Fig. 4) and may represent a recent case of gene duplication and divergence. When ShMOMT2 was incubated with kaempferol, a substrate missing both a 3′- and a 5′-hydroxyl, it added a methyl group to the 4′-hydroxyl (Table I). However, with either quercetin or myricetin, ShMOMT2 initially added a methyl group to the 7-hydroxyl, suggesting that 3′- and/or 5′-hydroxyls might inhibit its activity with the 4′-hydroxyl. This is consistent with the observation that 3,7,3′-MeM is found in the glands, but no 3,3′,4′-MeM was observed (Fig. 3). Thus, it appears that after the 3-hydroxyl is methylated, the next hydroxyls to be methylated are at the 7 and 3′ positions, although which of these two is methylated first cannot yet be resolved. This is also consistent with what has been shown in Chrysosplenium americanum, where methylation of quercetin proceeds first to 3-MeQ and then to 3,7-dimethyl quercetin (De Luca and Ibrahim, 1985). It can be deduced that the next hydroxyl to be methylated is at the 5′ position, since we see accumulation of 3,7,3′,5′-tetramethyl myricetin but no 3,7,3′,4′-tetramethyl myricetin and also because it appears that ShMOMT1 is not active with a substrate that has a methyl group at both the 3′ and 4′ positions (Table I). ShMOMT2 clearly is capable of methylating the 4′-hydroxyl after it methylated the 7-hydroxyl (tested with 7-methyl quercetin for lack of 7-methyl myricetin [Table I] and also by incubating myricetin with ShMOMT2 for an extended period [more than 10 h], after which the major product is 7,4′-myricetin [Supplemental Fig. S4]). However, it seems to be less efficient at methylating the 4′-hydroxyl once the 3′- and/or the 5′-hydroxyls have been methylated (Table I), consistent with the lower levels of 3,7,3′,4′,5′-pentamethyl myricetin observed in the trichomes.
A caveat for the kinetic analysis of ShMOMT1 and ShMOMT2 presented here is that, for lack of availability, we were not able to test them with 3-methyl myricetin or other combinations of polymethylated myricetin with one methyl group at the 3 position (for example, 3,7-dimethyl myricetin). However, we did obtain and test both enzymes with 3-methyl quercetin. The results with ShMOMT1 indicated that it had higher activity with the 3-methyl quercetin than with quercetin, although ShMOMT2 had lower activity (Table I). It has been shown for many OMTs that they are regiospecific but not substrate specific, meaning that their specificity is determined by the part of the molecule that is modified by their catalytic activity (Vogt, 2004). However, we note that the turnover rates of ShMOMT2 with the in vitro substrates tested were substantially lower than the turnover rates observed for ShMOMT1 (Tables II and III). This might indicate that ShMOMT2 activity could possibly be rate limiting in the synthesis of polymethylated myricetins in the trichomes. Alternatively, this enzyme may be more sensitive to the lack of correct functional groups in the in vitro-tested substrates. It is also possible that it may have additional, non-myricetin-related substrates in the cell with which it is more efficient.
ShMOMT1 and ShMOMT2 Are Expressed in Three Different Glandular Trichomes
Our data indicate that ShMOMT1 and ShMOMT2 transcripts and proteins are found in all three types of glandular trichomes in S. habrochaites that are metabolically active. The levels of the transcripts and proteins in these gland types (1, 4, and 6) correlate well with the amount of methylated myricetin found in them, with type 6 glands containing 1 order of magnitude less of each compared with type 1 and 4 glands, with the exception that type 4 glands have somewhat reduced amounts of both transcripts compared with type 1 glands. However, the levels of ShMOMT1 and ShMOMT2 transcripts in type 4 glands are still 4- and 2-fold higher, respectively, than that found in type 6 glands. In addition to localizing ShMOMT1 and ShMOMT2 in glandular trichomes, we have detected transcripts of putative genes involved in flavonoid and flavonol biosynthesis in our EST databases created from isolated trichome glands (types 1, 4, and 6; Supplemental Table S1). Transcripts of both flavonol 3′-hydroxylase and flavonol 3′,5′-hydroxylase, required for the synthesis of myricetin, were detected in these databases, with highest representation in type 1 glands.
Possible Roles of Methylated Myricetins in Tomato Glandular Trichomes
Flavonoids in general have been hypothesized to act as UV protectants, chemical defense compounds, and in plant-insect, plant-microbe, plant-pathogen, and plant-plant interactions (for review, see Treutter, 2005). While evidence for some of these roles (e.g. in plant-microbe interactions) is strong, other roles are still tentative (Treutter, 2005). Furthermore, since flavonoids often occur as a mixture, assigning roles to specific compounds is difficult. Currently, no physiological function has been postulated specifically for laricitrin and syringetin in plants or for the more highly methylated myricetins found in the tomato trichomes. Laricitrin and syringetin, but not the more highly methylated myricetin ethers, are found in red grape (Vitis vinifera) and are probably responsible, along with several other flavonols and methylated derivatives, for the antioxidant potency of red grapes and wine (Mattivi et al., 2006). However, there is no evidence to support specific roles for these compounds in grape. Myricetin has also been linked to radical-scavenging activity, xanthine oxidase inhibitory activity, and antioxidant activity in extracts of Ginkgo biloba leaves and Bridelia ferruginea stem bark (Kobus et al., 2009; Cimanga et al., 2001), but in both investigations the tested mixture contained several other flavonols and methylated derivatives.
Thus, we can only hypothesize that in tomato trichomes, the methylated myricetins contribute to some of the general roles postulated for flavonoids. Their synthesis and accumulation in glandular trichomes along with their relatively lipophilic nature suggest that they are likely targeted to the cuticular space surrounding the secretory cells. In this location, they are well placed to serve roles in chemical defense against herbivores, as UV protectants, or as radical scavengers to aid in preventing the peroxidation of lipids.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Solanum habrochaites (accession LA1777) seeds were obtained from the C.M. Rick Tomato Genetics Resource Center (University of California at Davis). The seeds were germinated on sterile filter paper in germination boxes and kept for approximately 5 to 7 d before transfer of seedlings to soil. Plants were grown in a mixture of regular soil:fine sand (3:1, v/v) in a growth chamber under a 14-h-light/10-h-dark photoperiod. Temperature was maintained at 22°C throughout the light period and 18°C during the dark period.
Gland cells were collected from glandular trichomes by hand with micropipettes using a dissecting microscope (Leica MZ6). Micropipettes were hand pulled and shaped from either 9-inch disposable Pasteur pipettes or 1.8-mm × 100-mm capillary tubes. The micropipettes were approximately 6 cm in length and shaped to taper from one end, approximately 1.5 to 2.0 mm, down to approximately 0.25 mm diameter at the opposite end. Both ends of the pipette were flame sealed to prevent capillary action. Gland cells were picked from the top of glandular trichome structures using the thin tip of the micropipette. The cells adhered to the tip until being put into an appropriate buffer for downstream analyses. Trichomes were removed from leaf material using the same type of micropipettes, except that they were lightly scraped across the leaf surface in order to remove the bulk of trichomes without disturbing the leaf surface.
Chemicals
All chemicals were from Sigma-Aldrich unless otherwise noted. Flavonols and methyl flavonols were purchased from Extrasynthese, with the exception of the flavonol kaempferol, which was purchased from Indofine Chemical. Deuterium-labeled SAM was purchased from C/D/N Isotopes. Methanol, 88% formic acid, and acetonitrile were purchased from VWR Scientific.
Metabolic Profiling of Leaf and Trichome Gland Cells and Metabolite Identification
Approximately 50 mg fresh weight of leaf material was extracted in 100 μL of ice-cold acetonitrile:isopropanol:water (3:3:2, v/v/v) at room temperature overnight. Samples were evaporated to near dryness and resuspended in 50% methanol (v/v) for LC-MS analysis. For leaf material with trichomes removed, a glass probe (described previously) was used to gently scrape trichomes from the surface of the leaf prior to extraction with the 3:3:2 solvent mixture.
A total of 50 gland cells from each type of glandular trichome (types 1, 4, and 6) were collected with micropipettes and extracted in 50 μL of ice-cold acetonitrile:isopropanol:water (3:3:2, v/v/v). Samples were stored overnight at −20°C, evaporated to near dryness, and resuspended in 50% methanol (v/v) for LC-MS analysis.
Samples were analyzed on a QTRAP 3200 mass spectrometer from Applied Biosystems/MDS Sciex coupled to a Shimadzu UFLC LC-20AD system and SIL-HTc autosampler. Separation was achieved with a Thermo β-basic C18 column (150 mm × 1.0 mm, 5 μm) at 30°C. The mobile phases were 0.5% formic acid (A) and 0.5% formic acid in 60% methanol + 40% acetonitrile (B). A 15-min reverse phase gradient at a flow rate of 0.100 mL min−1 was used for separation. The linear gradient elution program was as follows: 10% B for 0.3 min, 40% B and linear increase to 100% from 0.31 to 8.5 min, followed by an isocratic hold at 100% B for 2.5 min. At 11 min, B was returned to 10% and the column was equilibrated for 4 min before the next injection.
The mass spectrometer was operated in the positive ion mode with a TurboIonSpray source. Enhanced product ion (MS/MS) scanning was accomplished with dynamic fill time and was used for ion detection at 40-V collision energy. The other ionization parameters were as follows: curtain gas, 10; ion source gas 1, 12; ion source gas 2, 30; source temperature, 400°C; entrance potential, 10 V; collision-activated dissociation high; ion spray voltage, 5,500 V. The mass spectrometer and the HPLC system were controlled by Analyst 1.4.2 software from Applied Biosystems/MDS Sciex.
All flavonol glycosides observed in leaf dip extracts were glycosylated in the 3 position judged by the high abundance of radical anion aglycone fragment ions in negative ion MS/MS spectra (Cuyckens and Claeys, 2005). Authentic standards of most aglyconic polymethylated myricetin metabolites were not available from commercial sources. Owing to the substantial number of methylated isomers, their low levels in plant tissues, and their coelution with other metabolites, comparisons with UV spectra of standards was not feasible, nor were sufficient amounts of purified metabolites available for detailed NMR structure determination. In view of these limitations, positions of methyl groups in methylated myricetins were assigned based on coelution of plant metabolites with authentic standards when available, produced semisynthetically from standards of myricetin or methylated myricetins when possible, predictions of relative LC retention times based on the ease of formation of intramolecular hydrogen bonds in some isomers, and on MS/MS product ion spectra that showed positional isomer-selective differences in fragmentation behavior. In the latter case, ion structure assignments were aided through enzymatic synthesis of individual O-d3-methylated myricetin derivatives from d3-SAM. Product ion MS/MS spectra of [M+H]+ ions derived from methylated myricetins yielded evidence for position-selective fragmentation chemistry. Abundances of fragments arising from loss of a methyl radical (−15 D) relative to [M+H]+ ions varied among methylated myricetins, and relative yields of these fragments decreased based on methyl position as 3 > 4′ > 3′ = 5′ > > 7. Assignments of myricetins methylated on the A ring (either the 5 or 7 position) were facilitated by observations of characteristic fragment ion masses observed in the MS/MS product ion spectra. In the absence of methylation in these positions, the fragment ion derived from the A ring group (designated as 1,3A+) appears at mass-to-charge ratio 153, but when either the 5 or 7 position is methylated, this fragment mass shifts upward in mass by 14 D to appear at mass-to-charge ratio 167. We considered methylation at the 5 position unlikely because this is a rare metabolite, and none of the metabolites gave MS/MS fragments suggestive of two methyl groups on the A ring. One additional feature, the loss of 16 D from the [M+H]+ precursor, was shown using deuterium labeling to specifically occur when at least two methyl ether groups were present on the B ring (3′, 4′, or 5′ position). The combinations of these features in the MS/MS spectra allowed us to use a process of elimination to generate unambiguous evidence for the assignments of methyl group positions in methylated myricetin metabolites.
RNA Isolation
Total RNA was extracted from 100 mg fresh weight of young leaf material or young leaf material from which trichomes had been removed. Tri Reagent (Molecular Research Center) was used in accordance with the manufacturer’s instructions to extract total RNA from leaves and from leaves with trichomes removed. First-strand cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) using an anchored poly-T primer supplied by the manufacturer.
qRT-PCR
Total RNA from young leaf material and young leaf material with trichome removed was extracted as described above and then treated with DNase using the DNA-free kit (Ambion). SuperScript II reverse transcriptase (Invitrogen) and an anchored poly-T primer were used for first-strand cDNA synthesis. A negative control sample was run in parallel without reverse transcriptase added to the reaction mixture. All samples were normalized to the amplification of a Solanum lycopersicum actin gene (accession no. BT013707). Quantitative expression analysis was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems). The Fast SYBR Green Master Mix (Applied Biosystems) reagent was used according to the manufacturer’s instructions in preparation of the qPCR. The cycling conditions were as follows: 40 times for 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Cycling was followed by a melting stage that ramped up from 55°C to 95°C with an increasing gradient of 0.5°C and a 10-s pause at each temperature. The entire experiment was performed in triplicate starting with total RNA isolation from gland cells, leaves, or leaves with trichomes removed. The threshold cycle values from each experiment were averaged, and the relative expression level of ShMOMT1 in each tissue was calculated using the comparative threshold cycle method (Schmittgen and Livak, 2008). The results were expressed relative to expression levels of ShMOMT1 or ShMOMT2 in leaf material with trichomes.
Isolation, Expression, and Purification of Recombinant ShMOMT1 and ShMOMT2
The full-length ShMOMT1 and ShMOMT2 open reading frames were cloned from cDNA made from S. habrochaites leaf RNA. Tri Reagent (Molecular Research Center) was used to extract total RNA from approximately 100 mg of material, and SuperScript II reverse transcriptase (Invitrogen) was used to synthesize first-strand cDNA. ShMOMT1 sequence was amplified using KOD Hot Start DNA polymerase (Novagen) from first-strand cDNA and ligated into the pGEM-T Easy vector (Promega), grown in Escherichia coli Top 10 cells, and full-length cDNAs were verified by DNA sequencing. The full-length open reading frame was amplified from the pGEM-T Easy vector using KOD Hot Start DNA Polymerase (Novagen), gel-purified using MinElute (Qiagen), and inserted into the pEXP5-CT/TOPO expression vector (Invitrogen) with the native stop codon intact. The correct pEXP5-CT/TOPO construct was verified by DNA sequencing, isolated using the QIAprep Spin Miniprep kit (Qiagen), and transformed into E. coli BL21(DE3)pLysS cells (Invitrogen). A colony carrying the correct construct was isolated and grown in Luria-Bertani medium containing 100 μg mL−1 ampicillin and 50 μg mL−1 chloramphenicol at 37°C to an optical density at 600 nm of 0.5 to 0.8. Cultures were induced with 1 mm isopropylthio-β-galactoside and grown at 18°C for an additional 4 h.
Induced cultures were pelleted by centrifugation, resuspended in one-tenth volume lysis buffer (50 mm Tris, 10 mm NaCl, 1 mm EDTA, 10% glycerol, and 14 mm β-mercaptoethanol, pH 8.0), and lysed at 4°C by sonication. The cell lysate was cleared by centrifugation, and the supernatant was partially purified with DE53 anion exchanger (Whatman International). ShMOMT1 and ShMOMT2 were each purified by anion-exchange chromatography on a HiTrap Q HP column (GE Healthcare). A linear gradient (10–1,000 mm) of NaCl in lysis buffer was used for the initial purification on the DE53 anion exchanger, and a linear gradient (250–500 mm) of NaCl in lysis buffer was used for the second round of purification on the HiTrap Q HP anion exchanger. ShMOMT1 eluted in the 400 to 500 mm fraction and ShMOMT2 eluted in the 300 to 400 mm fraction from the DE53 anion exchanger; ShMOMT1 eluted in the 350 to 400 mm fraction and ShMOMT2 eluted in the 300 to 350 mm fraction from the HiTrap Q HP anion exchanger. The active fractions were identified by radiochemical enzyme assays as described above using myricetin as substrate. SDS-PAGE was used to visualize the degree of homogeneity of the active fractions.
Enzyme Assays and Product Identification
Radiochemical enzyme assays consisted of 50 mm Tris-HCl (pH 7.5 for ShMOMT1 or pH 8.0 for ShMOMT2), 5 μg of recombinant ShMOMT1 or ShMOMT2, 250 μm substrate dissolved in a 1:1 mixture of dimethyl sulfoxide:distilled, deionized water, and 200 μm SAM (Perkin-Elmer Instruments) in a final volume of 50 μL. Assays were incubated at room temperature for 30 min and stopped by the addition of 2 n HCl. Reaction products were extracted with 200 μL of ethyl acetate and counted in a scintillation counter (model LS6500; Beckman Coulter). Kinetic analyses were carried out within the linear range of reaction velocity by adjusting the concentration of recombinant protein in the assay. Raw data (cpm) were converted to pkat as described previously by D’Auria et al. (2002).
To produce ample product for LC-MS analyses, enzyme assays were performed using nonradiolabeled SAM and 10-fold reaction volumes. A continuous extraction assay method was designed to optimize product accumulation in these assays. Reactions were performed in 1.5-mL glass vials (Supelco Analytical; 27080-U) in a final volume of 500 μL (aqueous). A layer of 100% ethyl acetate (500 μL) was carefully applied over the aqueous assay volume to serve as a nonpolar extraction phase. The reactions were set up on ice and mixed briefly before addition of the ethyl acetate layer. Reactions were sealed with a screw-top septum and incubated overnight at room temperature. The ethyl acetate was removed and evaporated, and the residue was resuspended in 50 μL of ethanol:distilled, deionized water (1:1).
Metabolite identities were determined using TLC following the method of Owens and McIntosh (2009) with the following modifications: Polygram Sil G/UV254 plastic sheets (Macherey-Nagel) and running buffer of toluene:ethyl formate:formic acid (5:4:1, v/v/v). Metabolite identities were also determined by LC-MS using three different criteria: accurate mass, measured with time-of-flight mass spectrometry; retention time comparison with authentic standards; and comparison of mass spectra fragmentation patterns (see above; Supplemental Figs. S1, S3, and S4).
Protein-Blot Analysis
Total protein was extracted from collections of 500 gland cells from each of the different types of glandular trichomes (types 1, 4, and 6) in 50 μL of SDS-PAGE sample buffer (100 mm Tris, 2% SDS, 5% β-mercaptoethanol, 15% glycerol, and 0.1% bromphenol blue). Total protein extraction from leaves followed the protocol given by Dudareva et al. (1996). Polyclonal antibodies to ShMOMT1 or ShMOMT2 were generated at Cocalico Biologicals in rabbit from recombinant ShMOMT1 or ShMOMT2 protein (Supplemental Fig. S7). Anti-α-tubulin was from Sigma-Aldrich and served as an internal control to standardize samples from gland cells and leaves. All antibodies (anti-ShMOMT1, anti-ShMOMT2, and anti-α-tubulin) were used at a 1:3,000 dilution and incubated with gel blots for 1 h. All other conditions of the protein gel blotting were as described previously (Dudareva et al., 1996).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JF499656 and JF499657 for ShMOMT1 and ShMOMT2, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mass fragmentation patterns of methyl myricetin compounds in S. habrochaites trichomes.
Supplemental Figure S2. Radioactive thin-layer chromatography of products from enzyme assays.
Supplemental Figure S3. Mass fragmentation patterns of authentic standards and products of ShMOMT1.
Supplemental Figure S4. Mass fragmentation patterns of products from enzyme assays of ShMOMT2.
Supplemental Figure S5. Kinetic measurements of ShMOMT1.
Supplemental Figure S6. Kinetic measurements of ShMOMT2.
Supplemental Figure S7. Immunoblot detection of ShMOMT1 and ShMOMT2 in trichome and leaf extracts.
Supplemental Table S1. Representative cDNA sequences of flavonoid and flavonol biosynthetic enzymes in S. habrochaites trichome EST databases.
Acknowledgments
We thank Drs. Robert Last and Anthony Schillmiller and Ms. Jeongwoon Kim (Michigan State University) for sharing results with us prior to publication and for helpful advice.
References
- Ambrósio SR, Oki Y, Heleno VCG, Chaves JS, Nascimento PG, Lichston JE, Constantino MG, Varanda EM, Da Costa FB. (2008) Constituents of glandular trichomes of Tithonia diversifolia: relationships to herbivory and antifeedant activity. Phytochemistry 69: 2052–2060 [DOI] [PubMed] [Google Scholar]
- Ariyanathan S, Saraswathy A, Rajamanickam GV, Connolly JD. (2010) Polyphenols from the roots of Plumbago rosea. Indian J Chem Sect B Org Chem Incl Med Chem 49: 386–389 [Google Scholar]
- Braca A, Bilia AR, Mendez J, Morelli I. (2001) Myricetin glycosides from Licania densiflora. Fitoterapia 72: 182–185 [DOI] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Cacace S, Schröder G, Wehinger E, Strack D, Schmidt J, Schröder J. (2003) A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry 62: 127–137 [DOI] [PubMed] [Google Scholar]
- Cimanga K, Ying L, De Bruyne T, Apers S, Cos P, Hermans N, Bakana P, Tona L, Kambu K, Kalenda DT, et al. (2001) Radical scavenging and xanthine oxidase inhibitory activity of phenolic compounds from Bridelia ferruginea stem bark. J Pharm Pharmacol 53: 757–761 [DOI] [PubMed] [Google Scholar]
- Croteau RB, Davis EM, Ringer KL, Wildung MR. (2005) (−)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 92: 562–577 [DOI] [PubMed] [Google Scholar]
- Cuyckens F, Claeys M. (2005) Determination of the glycosylation site in flavonoid mono-O-glycosides by collision-induced dissociation of electrospray-generated deprotonated and sodiated molecules. J Mass Spectrom 40: 364–372 [DOI] [PubMed] [Google Scholar]
- Dachriyanus, Fahmi R, Sargent MV, Skelton BW, White AH. (2004) 5-Hydroxy-3,3',4',5',7-pentamethoxyflavone (combretol). Acta Crystallogr Sect E Struct Rep Online 60: O86–O88 [Google Scholar]
- D’Auria JC, Chen F, Pichersky E. (2002) Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiol 130: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, Ibrahim RK. (1985) Enzymatic synthesis of polymethylated flavonols in Chrysosplenium americanum. I. Partial purification and some properties of S-adenosyl-L-methionine:flavonol 3-, 6-, 7-, and 4′-O-methyltransferases. Arch Biochem Biophys 238: 596–605 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gerats AGM, Wallroth M, Donkerkoopman W, Groot SPC, Schram AW. (1983) The genetic control of the enzyme UDP-glucose-3-O-flavonoid-glucosyltransferase in flowers of Petunia hybrida. Theor Appl Genet 65: 349–352 [DOI] [PubMed] [Google Scholar]
- Gorbatsova J, Lõugas T, Vokk R, Kaljurand M. (2007) Comparison of the contents of various antioxidants of sea buckthorn berries using CE. Electrophoresis 28: 4136–4142 [DOI] [PubMed] [Google Scholar]
- Huang TS, Anzellotti D, Dedaldechamp F, Ibrahim RK. (2004) Partial purification, kinetic analysis, and amino acid sequence information of a flavonol 3-O-methyltransferase from Serratula tinctoria. Plant Physiol 134: 1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim RK. (1997) Plant O-methyltransferase signatures. Trends Plant Sci 2: 249–250 [Google Scholar]
- Ibrahim RK. (2005) A forty-year journey in plant research: original contributions to flavonoid biochemistry. Can J Bot 83: 433–450 [Google Scholar]
- Ibrahim RK, Anzellotti D. (2003) The enzymatic basis of flavonoid biodiversity. In Romeo JT, ed, Integrative Phytochemistry: From Ethnobotany to Molecular Ecology. Pergamon, Amsterdam, pp 1–36 [Google Scholar]
- Ibrahim RK, De Luca V, Khouri H, Latchinian L, Brisson L, Charest PM. (1987) Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry 26: 1237–1245 [Google Scholar]
- Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. (2004) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 134: 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay M, Voirin B, Hasan A, Gonnet JF, Viricel MR. (1980) Chemotaxonomic study of vascular plants. 43. Chemosystematic experiment on tribe Loteae. Biochem Syst Ecol 8: 127–132 [Google Scholar]
- Joshi CP, Chiang VL. (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol Biol 37: 663–674 [DOI] [PubMed] [Google Scholar]
- Jung HA, Kim JE, Chung HY, Choi JS. (2003) Antioxidant principles of Nelumbo nucifera stamens. Arch Pharm Res 26: 279–285 [DOI] [PubMed] [Google Scholar]
- Kobus J, Flaczyk E, Siger A, Nogala-Kalucka M, Korczak J, Pegg RB. (2009) Phenolic compounds and antioxidant activity of extracts of Ginkgo leaves. Eur J Lipid Sci Technol 111: 1150–1160 [Google Scholar]
- Kumar A, Malik AK, Tewary DK. (2009) A new method for determination of myricetin and quercetin using solid phase microextraction-high performance liquid chromatography-ultraviolet/visible system in grapes, vegetables and red wine samples. Anal Chim Acta 631: 177–181 [DOI] [PubMed] [Google Scholar]
- Kumar N, Bhandari P, Singh B, Gupta AP, Kaul VK. (2008) Reversed phase-HPLC for rapid determination of polyphenols in flowers of rose species. J Sep Sci 31: 262–267 [DOI] [PubMed] [Google Scholar]
- Kumari GNK, Rao LJM, Rao NSP. (1984) Myricetin methyl ethers from Solanum pubescens. Phytochemistry 23: 2701–2702 [Google Scholar]
- Lako J, Trenerry VC, Wahlqvist M, Wattanapenpaiboon N, Sotheeswaran S, Premier R. (2007) Phytochemical flavonols, carotenoids and the antioxidant properties of a wide selection of Fijian fruit, vegetables and other readily available foods. Food Chem 101: 1727–1741 [Google Scholar]
- Le K, Chiu F, Ng K. (2007) Identification and quantification of antioxidants in Fructus lycii. Food Chem 105: 353–363 [Google Scholar]
- Lee TH, Liu DZ, Hsu FL, Wu WC, Hou WC. (2006) Structure-activity relationships of five myricetin galloylglycosides from leaves of Acacia confusa. Botanical Studies 47: 37–43 [Google Scholar]
- Lee YJ, Kim BG, Chong Y, Lim Y, Ahn JH. (2008) Cation dependent O-methyltransferases from rice. Planta 227: 641–647 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li WJ, Ling XM, Lai XY, Li YZ, Zhang QY, Zhao YY. (2008) Simultaneous determination of the active ingredients in Abelmoschus manihot (L.) Medicus by CZE. Chromatographia 67: 819–823 [Google Scholar]
- Luckwill L. (1943) The genus Lycopersicon: an historical, biological, and taxonomic survey of the wild and cultivated tomatoes. PhD thesis. Aberdeen University, Aberdeen, UK [Google Scholar]
- Macheix JJ, Ibrahim RK. (1984) The O-methylation system of apple fruit cell-suspension. Biochem Physiol Pflanz 179: 659–664 [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. (2006) Metabolite profiling of grape: flavonols and anthocyanins. J Agric Food Chem 54: 7692–7702 [DOI] [PubMed] [Google Scholar]
- McDowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Descour A, Shi F, Larson M, Schilmiller A, An L, et al. (2011) Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol 155: 524–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michodjehoun-Mestres L, Souquet JM, Fulcrand H, Bouchut C, Reynes M, Brillouet JM. (2009) Monomeric phenols of cashew apple (Anacardium occidentale L.). Food Chem 112: 851–857 [Google Scholar]
- Min BS, Lee SY, Kim JH, Lee JK, Kim TJ, Kim DH, Kim YH, Joung H, Lee HK, Nakamura N, et al. (2003) Anti-complement activity of constituents from the stem-bark of Juglans mandshurica. Biol Pharm Bull 26: 1042–1044 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Li LN, Pan HY, Blount JW, Dixon RA, Wang XQ. (2009) Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol 392: 1292–1302 [DOI] [PubMed] [Google Scholar]
- Motta LB, Kraus JE, Salatino A, Salatino MLF. (2005) Distribution of metabolites in galled and non-galled foliar tissues of Tibouchina pulchra. Biochem Syst Ecol 33: 971–981 [Google Scholar]
- Muzac I, Wang J, Anzellotti D, Zhang H, Ibrahim RK. (2000) Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch Biochem Biophys 375: 385–388 [DOI] [PubMed] [Google Scholar]
- Ojot PB, Njiti V, Guo ZB, Gao M, Besong S, Barnes SL. (2008) Variation of flavonoid content among sweetpotato accessions. J Am Soc Hortic Sci 133: 819–824 [Google Scholar]
- Oliveira I, Sousa A, Valentao P, Andrade PB, Ferreira I, Ferreres F, Bento A, Seabra R, Estevinho L, Pereira JA. (2007) Hazel (Corylus avellana L.) leaves as source of antimicrobial and antioxidative compounds. Food Chem 105: 1018–1025 [Google Scholar]
- Owens DK, McIntosh CA. (2009) Identification, recombinant expression, and biochemical characterization of a flavonol 3-O-glucosyltransferase clone from Citrus paradisi. Phytochemistry 70: 1382–1391 [DOI] [PubMed] [Google Scholar]
- Rausher MD. (2008) The evolution of flavonoids and their genes. Grotewold E, , Science of Flavonoids. Springer, New York, pp 175–211 [Google Scholar]
- Reynertson KA, Yang H, Jiang B, Basile MJ, Kennelly EJ. (2008) Quantitative analysis of antiradical phenolic constituents from fourteen edible Myrtaceae fruits. Food Chem 109: 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riihinen K, Jaakola L, Karenlampi S, Hohtola A. (2008) Organ-specific distribution of phenolic compounds in bilberry (Vaccinium myrtillus) and ‘northblue’ blueberry (Vaccinium corymbosum × V. angustifolium). Food Chem 110: 156–160 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Daniel Jones A, Last RL. (2010) Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E. (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schröder G, Wehinger E, Lukacin R, Wellmann F, Seefelder W, Schwab W, Schröder J. (2004) Flavonoid methylation: a novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 65: 1085–1094 [DOI] [PubMed] [Google Scholar]
- Singh AP, Luthria D, Wilson T, Vorsa N, Singh V, Banuelos GS, Pasakdee S. (2009) Polyphenols content and antioxidant capacity of eggplant pulp. Food Chem 114: 955–961 [Google Scholar]
- Stevens JF, Hart H, Elema ET, Bolck A. (1996) Flavonoid variation in Eurasian Sedum and Sempervivum. Phytochemistry 41: 503–512 [Google Scholar]
- Stevens JF, Hart HT, Wollenweber E. (1995) The systematic and evolutionary significance of exudate flavonoids in Aeonium. Phytochemistry 39: 805–813 [Google Scholar]
- Tabart J, Kevers C, Pincemail J, Defraigne JO, Dommes J. (2006) Antioxidant capacity of black currant varies with organ, season, and cultivar. J Agric Food Chem 54: 6271–6276 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Thresh K, Ibrahim RK. (1985) Are spinach chloroplasts involved in flavonoid O-methylation?. Z Naturforsch C 40: 331–335 [Google Scholar]
- Treutter D. (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7: 581–591 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2004) Regiospecificity and kinetic properties of a plant natural product O-methyltransferase are determined by its N-terminal domain. FEBS Lett 561: 159–162 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P. (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Wagner GJ. (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol 96: 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CA, Grayer RJ. (2004) Anthocyanins and other flavonoids. Nat Prod Rep 21: 539–573 [DOI] [PubMed] [Google Scholar]
- Wu JH, Huang CY, Tung YT, Chang ST. (2008) Online RP-HPLC-DPPH screening method for detection of radical-scavenging phytochemicals from flowers of Acacia confusa. J Agric Food Chem 56: 328–332 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Gold ND, Martin VJJ, Wollenweber E, Ibrahim RK. (2006) Sequential O-methylation of tricetin by a single gene product in wheat. Biochim Biophys Acta 1760: 1115–1124 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. (1965) Evolutionary divergence and convergence in proteins. Bryson V, Vogel HJ, , Evolving Genes and Proteins. Academic Press, New York, pp 97–166 [Google Scholar]