Abstract
Redox regulation based on disulfide-dithiol conversion catalyzed by thioredoxins is an important component of chloroplast function. The reducing power is provided by ferredoxin reduced by the photosynthetic electron transport chain. In addition, chloroplasts are equipped with a peculiar NADPH-dependent thioredoxin reductase, termed NTRC, with a joint thioredoxin domain at the carboxyl terminus. Because NADPH can be produced by the oxidative pentose phosphate pathway during the night, NTRC is important to maintain the chloroplast redox homeostasis under light limitation. NTRC is exclusive for photosynthetic organisms such as plants, algae, and some, but not all, cyanobacteria. Phylogenetic analysis suggests that chloroplast NTRC originated from an ancestral cyanobacterial enzyme. While the biochemical properties of plant NTRC are well documented, little is known about the cyanobacterial enzyme. With the aim of comparing cyanobacterial and plant NTRCs, we have expressed the full-length enzyme from the cyanobacterium Anabaena species PCC 7120 as well as site-directed mutant variants and truncated polypeptides containing the NTR or the thioredoxin domains of the protein. Immunological and kinetic analysis showed a high similarity between NTRCs from plants and cyanobacteria. Both enzymes efficiently reduced 2-Cys peroxiredoxins from plants and from Anabaena but not from the cyanobacterium Synechocystis. Arabidopsis (Arabidopsis thaliana) NTRC knockout plants were transformed with the Anabaena NTRC gene. Despite a lower content of NTRC than in wild-type plants, the transgenic plants showed significant recovery of growth and pigmentation. Therefore, the Anabaena enzyme fulfills functions of the plant enzyme in vivo, further emphasizing the similarity between cyanobacterial and plant NTRCs.
Hydrogen peroxide is a by-product of aerobic metabolism that, when accumulated at high levels, may cause oxidative damage to the cell. Despite this potential toxic effect, hydrogen peroxide is also an important second messenger, in particular in eukaryotic organisms (Veal et al., 2007; Toledano et al., 2010). Therefore, in order to balance the toxic and signaling effects, the intracellular hydrogen peroxide concentration needs to be tightly controlled. For that purpose, cells are equipped with different enzymatic systems for hydrogen peroxide reduction, including peroxiredoxins (Prxs), which are thiol-based peroxidases able to detoxify hydrogen peroxide, organic peroxides and peroxynitrite (Poole et al., 2004). Based on structural and catalytic properties, Prxs are classified into three types, 1-Cys Prxs, typical 2-Cys Prxs, and atypical 2-Cys Prxs (Wood et al., 2003b). In multicellular organisms, Prxs are encoded by small gene families. For example, mammals are equipped with six Prxs distributed in different cell compartments that include cytosol, endoplasmic reticulum, mitochondria, and peroxisomes (Rhee et al., 2005). In plants, the gene family encoding Prxs is even more complex, since it is formed by 10 genes in Arabidopsis (Arabidopsis thaliana) and at least eight genes in rice (Oryza sativa; Dietz, 2003). Prxs are also distributed in different cell compartments in plant cells: PrxII A through D are localized to the cytosol; PrxII F to the mitochondria; and 1-Cys Prx is also cytosolic (Dietz et al., 2006) but accumulates to high levels in the nucleus (Stacy et al., 1999; Pulido et al., 2009). It is remarkable that the chloroplast is the plant organelle with the highest content of Prxs. The Arabidopsis chloroplast is equipped with three types of Prxs: two almost identical typical 2-Cys Prxs, termed 2-Cys Prx A and B; PrxII E; and Prx Q (Dietz et al., 2006; Kirchsteiger et al., 2009).
Following each catalytic cycle of peroxide decomposition, a disulfide-reducing catalyst must regenerate the Prxs. Although typical and atypical 2-Cys Prxs can be recycled by cyclophilins and glutaredoxins, the most common reductant for Prxs are thioredoxins (Trxs; Dietz, 2003). The reducing power of Trxs is normally provided by NADPH in a reaction catalyzed by NADPH-dependent thioredoxin reductase (NTR; Florencio et al., 1988). Thus, the pathway of reducing power required to maintain the Prx-dependent detoxifying activity depends on NADPH and is formed by a two-component system, NTR and Trx. However, there are two remarkable exceptions. In bacteria, such as Salmonella typhimurium, the abundant typical 2-Cys Prx, termed AhpC, is recycled by a bimodular enzyme, AhpF, composed of a double Trx fold and an NTR domain (Poole et al., 2000). This enzyme uses NADH, not NADPH, as a source of reducing power (Poole et al., 2000; Reynolds and Poole, 2000). The other exception is a peculiar NTR with a joint Trx domain at the C terminus, termed NTRC, which is found exclusively in oxygenic photosynthetic organisms (Serrato et al., 2002, 2004). NTRC is able to conjugate NTR and Trx activities to reduce 2-Cys Prxs with a high catalytic efficiency (Moon et al., 2006; Pérez-Ruiz et al., 2006; Alkhalfioui et al., 2007; Pérez-Ruiz and Cejudo, 2009), thus serving as an antioxidant system of the chloroplast.
While all plant and algal genomes so far sequenced contain a single gene encoding NTRC, the distribution of NTRC in cyanobacteria is uneven. Besides cyanobacteria that contain a gene encoding NTRC, there are other cyanobacteria that lack this gene (Florencio et al., 2006; Pascual et al., 2010). Interestingly, in the cyanobacterium Thermosynechococcus elongatus, NTRC was identified as a component of a protein complex showing NAD(P)H oxidase activity. The other component of this complex, which is induced by oxidative stress, was identified as the 2-Cys Prx (Sueoka et al., 2009). The tight association of NTRC with 2-Cys Prx in this cyanobacterium suggests that, as occurs in plant chloroplasts, NTRC acts as an efficient reductant of 2-Cys Prxs, thus serving as an antioxidant system.
Eukaryotic 2-Cys Prxs become reversibly inactivated at elevated peroxide concentrations by overoxidation of the peroxidatic catalytic Cys residue to sulfinic acid. This is believed to facilitate the signaling of hydrogen peroxide in eukaryotes (Wood et al., 2003a). Recently, it was shown that cyanobacterial 2-Cys Prx also could undergo overoxidation (Pascual et al., 2010). The degree of overoxidation depends on the intracellular peroxide concentration but also on the efficiency of reduction of the 2-Cys Prx, since the disulfide-bonded form should be inert to peroxides. Interestingly, the 2-Cys Prx from Anabaena, a cyanobacterium harboring NTRC, is more sensitive to overoxidation than the enzyme from Synechocystis, which lacks NTRC. These cyanobacteria seem to have developed different strategies to cope with hydrogen peroxide. Anabaena, which is equipped with the NTRC-2-Cys Prx system but has low catalase activity, is more sensitive to hydrogen peroxide than Synechocystis, which lacks NTRC but has high catalase activity (Pascual et al., 2010).
As plant chloroplasts are equipped with NTRC and sensitive-2-Cys Prxs, but not with catalase, it was proposed that this antioxidant system evolved from the system present in cyanobacterial strains, such as Anabaena (Pascual et al., 2010). Indeed, the plant NTRC is involved in several functions that are associated with 2-Cys Prx reduction (Pérez-Ruiz et al., 2006; Stenbaek et al., 2008). However, the different phenotype of the NTRC knockout mutant of Arabidopsis, as compared with a 2-Cys Prx double mutant, suggested additional functions for NTRC unrelated to reduction of 2-Cys Prxs (Pulido et al., 2010). These functions include the redox regulation of starch synthesis (Michalska et al., 2009) and the metabolism of aromatic amino acids (Lepistö et al., 2009). These findings suggest that NTRC has evolved to adopt a wide variety of functions in plant chloroplasts, which might imply different properties of the enzymes from cyanobacteria and plants. However, our knowledge of NTRC from cyanobacteria is still very scarce. The objective of this work was to characterize the biochemical properties of a cyanobacterial NTRC and to perform a comparative analysis with the plant enzyme. For that purpose, we expressed in Escherichia coli and purified the NTRC from the cyanobacterium Anabaena and analyzed the interaction with 2-Cys Prxs from either cyanobacterial or plant origin. Furthermore, the ability of cyanobacterial NTRC to complement the phenotype of the NTRC-deficient mutant of Arabidopsis was analyzed.
RESULTS
NTRC from Anabaena Is a Bimodular Enzyme with NTR and Trx Activity
In plants, NTRC is encoded by a single gene, which produces a bimodular enzyme composed of an NTR domain at the N terminus, a Trx domain at the C terminus, and an N-terminal sequence serving as transit peptide to target the enzyme to the chloroplast (Serrato et al., 2004). With the aim of characterizing a cyanobacterial NTRC, we focused on the gene all0737 of Anabaena sp. PCC 7120, which encodes a protein showing 60.1% identity with the rice NTRC (OsNTRC). The deduced amino acid sequence of the Anabaena putative NTRC (Serrato et al., 2004), herein denoted AnabNTRC, showed that, as the plant enzyme, it is composed of NTR and Trx domains. Moreover, the characteristic motifs of the NTR domain, the FAD and NADPH binding sites, and the active sites of the NTR and Trx domains are highly conserved in the cyanobacterial enzyme (Serrato et al., 2004).
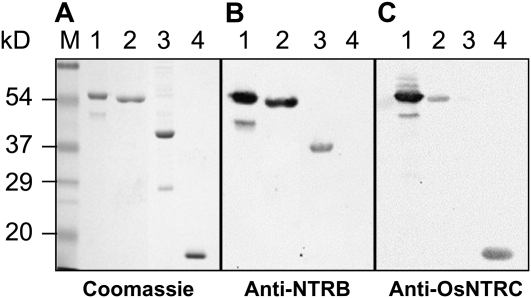
The coding sequence of the putative NTRC from Anabaena was cloned into the expression vector pQE30 so that the recombinant protein was produced in E. coli as an N-terminally His-tagged protein, following the strategy previously described for the rice enzyme, which was included in this study for comparative purposes (Fig. 1A, lanes 1 and 2). To test the functionality of the two domains, NTR and Trx, of the cyanobacterial enzyme, these were also produced as truncated His-tagged polypeptides (Fig. 1A, lanes 3 and 4). The full-length protein, AnabNTRC, and the rice enzyme were both efficiently detected by an anti-NTR antibody raised against wheat (Triticum aestivum) NTRB (Serrato et al., 2002). As expected, this antibody cross-reacted with the truncated polypeptide containing the NTR domain but not the Trx domain of the cyanobacterial enzyme (Fig. 1B). The anti-OsNTRC antibody, which was raised against the Trx domain of the rice enzyme, detected both the full-length enzyme and the Trx domain from Anabaena but not the NTR domain. However, this antibody detected the cyanobacterial NTRC less efficiently than the rice enzyme (Fig. 1C). Therefore, the immunological analysis confirmed that Anabaena NTRC, like the plant enzyme, is made up of two distinct domains, NTR and Trx.
Figure 1.
Immunological characterization of recombinant AnabNTRC and NTR and Trx domains. A, Purified His-tagged (1 μg of protein) OsNTRC (lane 1), AnabNTRC (lane 2), NTR domain (lane 3), or Trx domain (lane 4) of AnabNTRC were subjected to SDS-PAGE under reducing conditions. Proteins were stained with Coomassie Brilliant Blue R-250. Molecular markers were loaded, and their molecular mass, in kD, is indicated on the left. B and C, Replicates of this gel but loaded with 50 ng of the purified proteins were electrotransferred to nitrocellulose membranes and probed with anti-NTRB (B) and anti-OsNTRC (C) antibodies, as indicated.
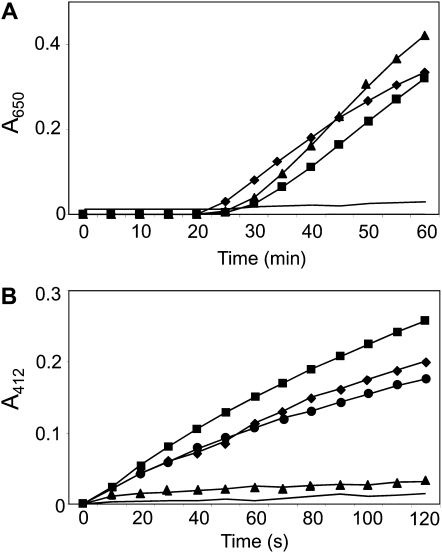
In order to characterize biochemically the cyanobacterial NTRC, the NTR and Trx activities of the full-length enzyme and the truncated versions were analyzed and compared with the previously reported rice enzyme (Serrato et al., 2004). The full-length AnabNTRC showed Trx activity, as determined by the dithiothreitol (DTT)-dependent insulin reduction assay, which was slightly lower than the activity of the rice enzyme (Fig. 2A). This activity was due to the Trx domain of the enzyme, which alone also showed DTT-dependent insulin reduction activity (Fig. 2A). Similarly, the full-length AnabNTRC showed NTR activity, assayed as NADPH-dependent reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), at a higher rate than the rice enzyme (Fig. 2B). As expected, this activity was due to the NTR domain of the enzyme, which showed NADPH-dependent reduction of DTNB, although at a lower rate than the full-length enzyme (Fig. 2B). Therefore, the biochemical analysis confirmed that the cyanobacterial NTRC, as the plant enzyme, is made up of two functional domains with the expected NTR and Trx activities, thus emphasizing the high similarity of the NTRCs from plant and cyanobacteria.
Figure 2.
Trx and NTR activity of recombinant AnabNTRC and the NTR and Trx domains. A, Insulin reduction catalyzed by the AnabNTRC polypeptide was performed in an incubation mixture containing 2 μm AnabNTRC (squares), 2 μm OsNTRC (diamonds), and 2 μm Trx domain truncated polypeptide (triangles) supplemented with 0.5 mm DTT. B, NADPH-dependent reduction of DTNB was assayed at room temperature in a buffer containing 0.1 μm AnabNTRC (squares), 0.1 μm OsNTRC (diamonds), 0.5 μm NTR domain truncated polypeptide (circles), or 1.0 μm Trx domain truncated polypeptide (triangles) in 100 mm potassium phosphate buffer, pH 7.0, 2 mm EDTA, 5 mm DTNB, and 150 μm NADPH. A negative control in the absence of enzymes was performed (solid line) for both panels. Assays were performed at least three times with similar results, and representative results are shown.
Cyanobacterial and Plant NTRCs Are Efficient Reductants of 2-Cys Prxs from Plant and Anabaena But Not from Synechocystis
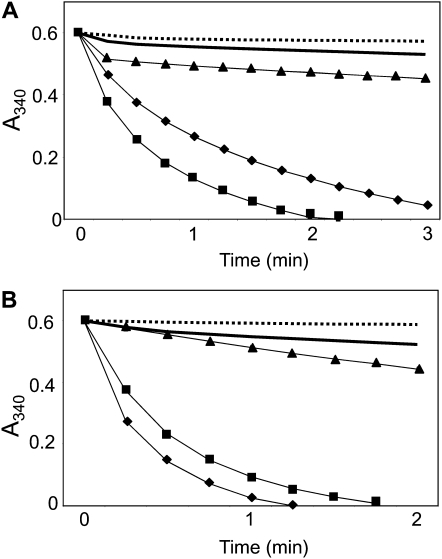
Comparative analyses of the reactivity of plant and cyanobacterial NTRCs were carried out with 2-Cys Prxs from either plant or cyanobacteria. To that end, 2-Cys Prxs from the cyanobacterial strains Anabaena and Synechocystis as well as from rice were expressed in E. coli as N-terminally His-tagged polypeptides (Pérez-Ruiz et al., 2006; Pascual et al., 2010). Anabaena NTRC was an efficient reductant of the 2-Cys Prx from Anabaena and, to a somewhat lower extent, of the 2-Cys Prx from rice. However, it failed to reduce the 2-Cys Prx from Synechocystis (Fig. 3A). Similarly, the rice NTRC efficiently reduced 2-Cys Prx from rice and Anabaena but not from Synechocystis (Fig. 3B). The kinetic analysis of AnabNTRC with 2-Cys Prxs from plants (rice and Arabidopsis) or Anabaena showed slightly lower Km and higher Kcat for 2-Cys Prx from Anabaena as compared with the values obtained with the plant 2-Cys Prxs (Table I). As a consequence, AnabNTRC showed better catalytic efficiency in terms of Kcat/Km with the 2-Cys Prx from Anabaena (Table I). Therefore, these results underscore the similarity of the rice and Anabaena NTRC. Furthermore, the results display a clear difference between the 2-Cys Prx from Anabaena, which behaves as a plant enzyme, and the Synechocystis 2-Cys Prx, which shows no reactivity with NTRC from either source.
Figure 3.
Reactivity of cyanobacterial and plant NTRC with 2-Cys Prxs from different sources. Assays were performed in reaction mixtures containing 100 mm potassium phosphate buffer, pH 7.0, 2 mm EDTA, 0.25 mm NADPH, 0.5 mm hydrogen peroxide, and purified enzymes at the following concentrations. For A, 4 μm AnabNTRC plus 8 μm 2-Cys Prx from Anabaena (squares), rice (diamonds), and Synechocystis (triangles). Controls were performed with 8 μm Anabaena 2-Cys Prx without AnabNTRC (dotted line) or with 4 μm AnabNTRC without 2-Cys Prx (solid line). For B, 2 μm OsNTRC plus 4 μm 2-Cys Prx from Anabaena (squares), rice (diamonds), and Synechocystis (triangles). Controls were performed with 4 μm rice 2-Cys Prx without OsNTRC (dotted line) or 2 μm OsNTRC without 2-Cys Prx (solid line). Assays were performed at least three times with similar results, and representative results are shown.
Table I. Kinetic parameters of the interaction of Anabaena NTRC with 2-Cys Prx from different sources.
Reactions were performed at a fixed concentration of AnabNTRC (2 μm) and variable concentrations of the 2-Cys Prxs in the presence of 0.25 mm NADPH. Data are means ± sd of three determinations.
| Source | Vmax | Km | Kcat | Kcat/Km |
| μmol min−1 | μm | s−1 | μm−1 s−1l | |
| Anabaena 2-Cys Prx | 169 ± 6.4 | 2.84 ± 0.3 | 2.81 ± 0.1 | 0.99 |
| Rice 2-Cys Prx | 170.1 ± 6.98 | 7.9 ± 0.4 | 1.42 ± 0.06 | 0.18 |
| Arabidopsis 2-Cys Prx A | 138 ± 2.9 | 5.9 ± 0.4 | 1.15 ± 0.1 | 0.19 |
| Arabidopsis 2-Cys Prx B | 114.3 ± 0.7 | 6.3 ± 0.2 | 0.95 ± 0.007 | 0.15 |
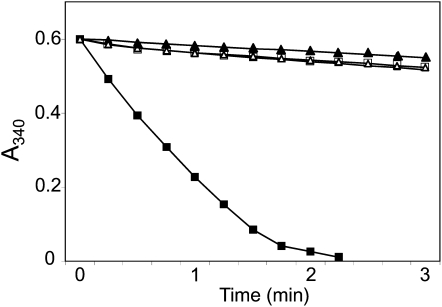
To further characterize the biochemical properties of the Anabaena NTRC, mutant variants of the enzyme were produced by a Cys-to-Ser mutation at the active site of both domains: C170S mutant in the NTR domain, and C411S mutant in the Trx domain. Neither of these mutants showed significant activity when assayed in the presence of the Anabaena 2-Cys Prx (Fig. 4). In addition, the wild-type AnabNTRC showed almost negligible activity when assayed with NADH as electron donor (Fig. 4).
Figure 4.
Effect of mutation of the Anabaena NTRC active sites. The activity of the NTRC-2-Cys Prx system was assayed as oxidation of NAD(P)H in a reaction mixture containing 100 mm potassium phosphate buffer, pH 7.0, 2 mm EDTA, 0.5 mm hydrogen peroxide, 8 μm Anabaena 2-Cys Prx, and 2 μm wild-type AnabNTRC supplemented with 0.25 mm NADPH (black squares) or 0.25 mm NADH (white squares). The effect of mutations at the active sites of the NTR and Trx domains of the Anabaena NTRC was assayed replacing the wild-type enzyme by mutants AnabNTRC (C170S) (black triangles) and AnabNTRC (C411S) (white triangles), respectively. Assays were performed at least three times with similar results, and representative results are shown.
The Synechocystis 2-Cys Prx Quaternary Structure Is Different from That of Anabaena and Plant Enzymes
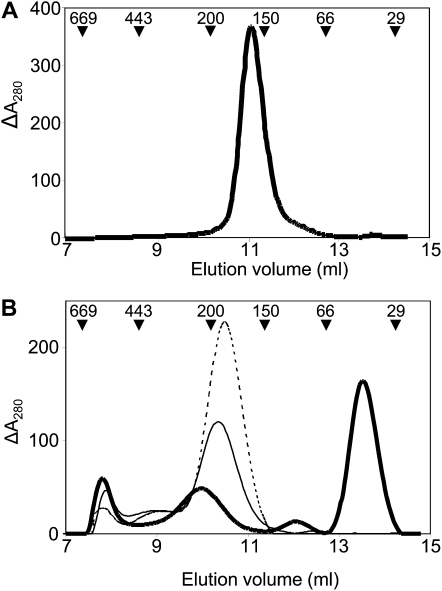
Finally, the quaternary structure of the components of the NTRC-2-Cys Prx system was analyzed by exclusion gel chromatography. In contrast to NTRC from rice, which oligomerized in the absence of NADPH (Pérez-Ruiz et al., 2009), the Anabaena enzyme eluted as a dimer regardless of the presence of NADPH (Fig. 5A). Concerning 2-Cys Prxs, the enzyme from Anabaena showed an elution profile almost identical to the rice 2-Cys Prx, the most abundant form of the protein eluting at a volume consistent with an octamer or a decamer (Fig. 5B). In contrast, the elution profile of the Synechocystis 2-Cys Prx was remarkably different. The enzyme eluted predominantly as a dimer and showed different intermediary forms, including the decamer, all of which were present in lower amounts. Thus, despite the high sequence similarity between Anabaena and Synechocystis 2-Cys Prxs (Pascual et al., 2010), both the quaternary structure and the reactivity with NTRC indicate clearly different properties of these enzymes.
Figure 5.
Analysis of the oligomeric state of AnabNTRC and 2-Cys Prx from rice and cyanobacteria. Purified His-tagged AnabNTRC (0.5 mg; A) and purified His-tagged 2-Cys Prx (0.5 mg; B) from rice (thin line), Anabaena (dotted line), and Synechocystis (thick line) were subjected to Superdex 200 gel filtration chromatography in 20 mm potassium phosphate buffer, pH 7.4, 150 mm NaCl. Molecular mass markers, in kD, are indicated.
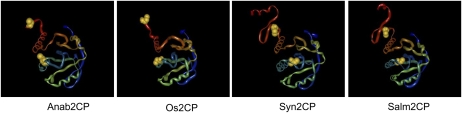
Structure modeling of the 2-Cys Prxs from Synechocystis and Anabaena predicted a very similar structure for both enzymes (Fig. 6). However, there is one significant difference affecting the location of the peroxidatic Cys residue in the reduced monomeric form, which is predicted to be more exposed in the Anabaena than in the Synechocystis 2-Cys Prx (Fig. 6). Interestingly, the model of the rice 2-Cys Prx predicts a structure highly similar to the Anabaena enzyme, with the peroxidatic Cys residue more exposed. In contrast, the structure deduced for the 2-Cys Prx from S. typhimurium, a prokaryotic enzyme insensitive to overoxidation (Wood et al., 2003a), predicted a more buried position of the peroxidatic Cys residue, resembling the structure of the Synechocystis enzyme (Fig. 6). Therefore, the modeling of the 2-Cys Prxs from these different sources suggests that the structural determinants around the peroxidatic Cys may be critical to the properties of these enzymes, apart from previously recognized structural features located at the C terminus (Wood et al., 2003a).
Figure 6.
Comparison of the structures of 2-Cys Prxs of cyanobacteria, plant, and enteric bacteria. The structural models of 2-Cys Prxs from cyanobacteria (Anabaena and Synechocystis), enteric bacteria (S. typhimurium), and plant (rice) were produced with the PHYRE (http://www.sbg.bio.ic.ac.uk/phyre/html/index.html) modeling server using the following templates: bovine mitochondrial 2-Cys Prx III (c1zyeB) for Anab2CP, human Trx peroxidase B from red blood cells (d1qmva) for Syn2CP, and alkyl hydroperoxide reductase (AhpC) from Helicobacter pylori (c1zofA) for Os2CP. Models were also produced with Swiss-model, obtaining essentially the same results. The positions of Cys residues at the active site are indicated in yellow. The program did not allow the modeling of the C-terminal region of the Anabaena and rice enzymes.
Expression of NTRC from Anabaena in the Arabidopsis NTRC Knockout Mutant
As described above, NTRC from Anabaena is remarkably similar to the plant enzyme, a notion further supported by the high efficiency of the cyanobacterial enzyme to reduce 2-Cys Prxs of plant origin in vitro. These results prompted us to analyze whether the cyanobacterial NTRC is able to carry out the functions of the plant enzyme in vivo. To test this possibility, the coding sequence of the Anabaena NTRC was fused to the putative chloroplast transit peptide of the enzyme from Arabidopsis to target the expressed protein to the chloroplast. The resulting gene was expressed in the Arabidopsis NTRC knockout mutant (ntrc) and wild-type plants under the control of the cauliflower mosaic virus 35S promoter. Northern-blot analysis revealed a high expression of the transgene in the wild-type background (Supplemental Fig. S1, WT_AnabNTRC lines) but a much lower expression in the ntrc mutant plants (Supplemental Fig. S1, ntrc_AnabNTRC lines). Of the transgenic lines obtained, WT_AnabNTRC lines 3.7.1.3 and 3.7.3.2 and ntrc_AnabNTRC lines 3.3.6.1 and 3.3.6.2 were chosen for further analysis. For comparison, transgenic plants previously described (Pérez-Ruiz et al., 2006) expressing the wild-type enzyme from Arabidopsis under the control of the 35S promoter in the wild type (WT_AtNTRC) and the ntrc mutant background (ntrc_AtNTRC) were included in these studies.
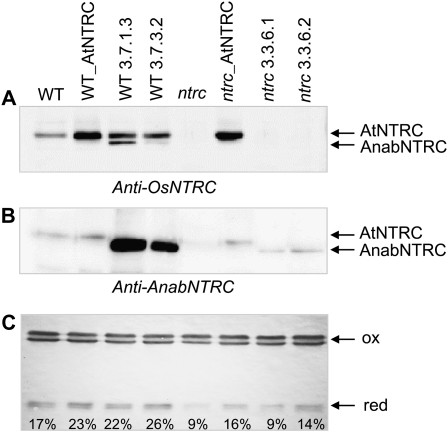
The content of Anabaena NTRC in the transgenic plants was examined by western-blot analysis of stromal extracts from isolated chloroplasts probed with anti-NTRC antibodies raised against the rice and the Anabaena enzymes (Fig. 7, A and B). As expected, the anti-OsNTRC antibody showed the presence of the endogenous enzyme in stromal fractions from the wild type, transgenic plants in the wild-type background, and the transgenic line (ntrc_AtNTRC) expressing the plant enzyme in the mutant background (Fig. 7A). However, this antibody failed to detect the cyanobacterial enzyme in any of the transgenic lines in the mutant background and only detected the Anabaena enzyme, with an electrophoretic mobility reflecting a lower Mr, in the WT_AnabNTRC line 3.7.1.3 (Fig. 7A), in agreement with the high content of transcripts shown by the northern-blot analysis (Supplemental Fig. S1). The failure of the anti-OsNTRC antibody to detect AnabNTRC in the transgenic lines was not surprising, since this antibody cross-reacted poorly with recombinant purified AnabNTRC (Fig. 1C). To overcome this problem, western blots were probed with an antibody that detected the Anabaena enzyme (Fig. 7B). This antibody showed the presence of AnabNTRC in the transgenic lines generated either in the wild type or the ntrc mutant background, although the level of AnabNTRC in the ntrc background plants was much lower than in wild-type background plants (Fig. 7B), in agreement with the mRNA content revealed by the northern-blot analysis (Supplemental Fig. S1). Therefore, these results show that the cyanobacterial NTRC is expressed and correctly targeted to chloroplasts in the Arabidopsis ntrc background, although the content of the enzyme is lower than in wild-type plants. In agreement with the lower content of cyanobacterial NTRC, the redox status of the chloroplast 2-Cys Prxs was not fully recovered in the transgenic lines expressing the cyanobacterial NTRC in the ntrc background, as compared with wild-type plants and the transgenic lines expressing the plant enzyme or the cyanobacterial enzyme in the wild-type background (Fig. 7C).
Figure 7.
Analysis of the content of Anabaena NTRC and the redox state of the 2-Cys Prxs in chloroplast stroma from Arabidopsis transgenic lines. Chloroplasts were isolated from leaves of wild-type (WT), ntrc knockout mutant, and independent transgenic lines of Arabidopsis, as indicated. Chloroplasts were lysed by hypoosmotic shock, and the level of endogenous and Anabaena NTRC was analyzed in stromal fractions. A and B, Protein samples (30 μg of protein) were subjected to SDS-PAGE under reducing conditions, blotted onto nitrocellulose membranes, and probed with polyclonal antibodies raised against the rice or Anabaena NTRC, as indicated. C, The redox state of the 2-Cys Prxs was analyzed on protein samples (15 μg of protein), which were fractionated by SDS-PAGE under nonreducing conditions, blotted onto nitrocellulose membranes, and probed with an anti-2-Cys Prx antibody. Band intensities were quantified with the ImageJ software, and the percentage of reduced 2-Cys Prx per line is indicated at the bottom. ox, Oxidized enzyme; red, reduced enzyme.
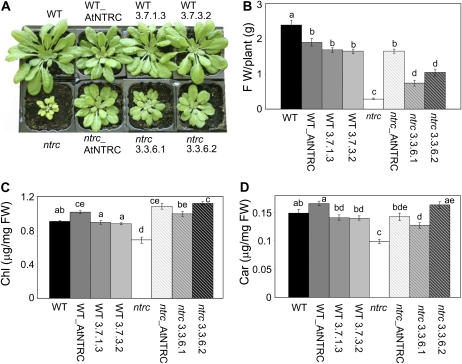
To test whether the cyanobacterial enzyme complements the phenotypic effects caused by the deficiency of NTRC, transgenic plants were grown under a short-day photoperiod, which causes more severe phenotypic effects in the ntrc mutant (Pérez-Ruiz et al., 2006; Lepistö et al., 2009). Despite the low content of cyanobacterial NTRC in the ntrc background transgenic plants (Fig. 8A, lines 3.3.6.1 and 3.3.6.2), these plants showed a partial but significant recovery of the wild-type phenotype, which was confirmed by the fresh weight of the rosette leaves (Fig. 8B). Moreover, the content of photosynthetic pigments, chlorophyll and carotenoids, which are reduced in the ntrc mutant, were fully recovered (Fig. 8, C and D). These results show that the cyanobacterial NTRC is able to carry out, at least partially, the functions of the plant enzyme in vivo.
Figure 8.
Analysis of the complementation of the Arabidopsis ntrc mutant phenotype by expression of the Anabaena NTRC. A, Arabidopsis plants, wild type (WT), ntrc mutant, and transgenic lines expressing either the Arabidopsis or the Anabaena NTRC in the wild-type or mutant background, as indicated, were grown under short-day conditions for 52 d. B to D, Fresh weight (FW) of the rosette leaves (B), content of total leaf chlorophyll (C), and leaf carotenoids (D) were determined. The experiment was repeated three times, and se values are indicated. Values with different letters are significantly different (P ≤ 0.05) as determined by Tukey’s multiple range test.
DISCUSSION
NTRC is an enzyme exclusive for photosynthetic organisms encoded by a single gene in all plant and algal genomes so far sequenced. However, it is found in some, but not all, cyanobacteria (Serrato et al., 2004; Pascual et al., 2010). Previously reported analysis showed the close phylogenetic relationship between plant and cyanobacterial NTRCs, thus suggesting a cyanobacterial origin for the plant enzyme (Serrato et al., 2004; Alkhalfioui et al., 2007). If this was the case, a high similarity between NTRC from plants and cyanobacteria should be expected. However, a comparison between these enzymes has hitherto not been possible due to the limited knowledge of cyanobacterial NTRC. The objective of this work was to perform a biochemical characterization of a cyanobacterial NTRC, from Anabaena sp. PCC 7120, and to carry out a comparative analysis with a plant enzyme.
The truncated polypeptide containing the NTR domain of the cyanobacterial enzyme was efficiently detected with an anti-NTR antibody raised against NTRB from wheat (Serrato et al., 2002), which showed a similar cross-reactivity with the NTR domain of the plant enzyme (Fig. 1B; Serrato et al., 2004). Similarly, the anti-OsNTRC antibody, which was raised against the Trx domain of the rice enzyme, detected the truncated polypeptide containing the Trx domain of the Anabaena enzyme. Therefore, the Anabaena NTRC, as the rice enzyme, may be considered as a functional NTR with a joint Trx domain at the C terminus. However, this antibody showed a poor detection of the full-length NTRC from Anabaena as compared with the rice enzyme (Fig. 1C). This result suggests a somehow different conformation of AnabNTRC limiting the cross-reactivity with the antibody. In this regard, it should be noted that NTRC from Anabaena is a homodimer regardless of the reducing conditions (Fig. 5A). This feature constitutes a remarkable difference with respect to the enzyme from plants, which is a homodimer in its catalytically active form (Pérez-Ruiz and Cejudo, 2009) but shows a high tendency to aggregate under oxidizing conditions (Pérez-Ruiz et al., 2009). The examination of the amino acid sequence of the plant enzymes (Serrato et al., 2004) reveals the presence of three Cys residues (positions 187, 499, and 525 in the Arabidopsis enzyme) that are absent from the Anabaena NTRC. To determine whether these Cys residues play any role in the redox-sensitive tendency of the plant enzyme to aggregate requires further study.
Our results from analyses of the recombinant full-length enzyme, as well as truncated polypeptides containing either the NTR or the Trx domains of the Anabaena enzyme, clearly show that the cyanobacterial NTRC may be considered as a bimodular protein formed by two domains, both of which show the expected NTR and Trx activities (Fig. 2). Therefore, the cyanobacterial enzyme is highly similar to the eukaryotic counterparts, such as the enzymes from rice (Serrato et al., 2004) or the green alga Chlorella (Machida et al., 2007). It should be noted that the full-length AnabNTRC showed a higher rate of NADPH-dependent reduction of DTNB than the truncated NTR domain of the enzyme (Fig. 2B); therefore, the Trx domain, which has no NTR activity, does contribute to the NTR activity of the full-length enzyme, most probably because this domain is important for the dimeric conformation of NTRC. Indeed, when truncated NTR and Trx domains of rice NTRC were incubated together, the activity was much lower than that of the full-length enzyme (Pérez-Ruiz et al., 2006). Thus, both domains must be part of a single polypeptide chain to show full activity. This bimodular enzyme, encoded by a single gene, may be evolutionarily advantageous because of its higher catalytic efficiency as compared with the two-component system formed by separate NTR and Trx enzymes, and thus encoded by two genes, which will require coordinated expression.
To our knowledge, the only NTRC so far described from cyanobacteria is the Thermosynechococcus elongatus enzyme (Sueoka et al., 2009). In this thermophilic cyanobacterium, NTRC was identified as a component of an NAD(P)H oxidase complex induced by oxidative stress, which also contained 2-Cys Prx. This result indicates that the cyanobacterial enzyme might act as a reductant of 2-Cys Prx and, thus, may function as an antioxidant system, as initially proposed for the enzyme in plant chloroplasts. However, the NTRC from T. elongatus seems to have a reaction mechanism different from that of the plant enzyme, as suggested by the fact that a mutant variant at the active site of the Trx domain of this enzyme was active (Sueoka et al., 2009). In contrast with this result, the analysis of mutant variants at the active site of either the NTR or Trx domain of the Anabaena enzyme completely lost activity (Fig. 4), thus showing the same behavior as the plant enzyme. Moreover, whereas the T. elongatus NTRC was reported to oxidize NADH (Sueoka et al., 2009), the Anabaena NTRC shared with the plant enzyme its specificity for NADPH as a source of reducing power (Fig. 4), once again emphasizing its close relationship with the plant enzyme. Based on these results, we propose that the reaction mechanism of the Anabaena NTRC is very similar to that previously reported for the plant enzyme. The different properties of the T. elongatus enzyme might be indicative of the existence of other forms of NTRC in cyanobacteria, but the clarification of this question awaits the characterization of NTRC from additional cyanobacterial sources.
The kinetic analysis of the interaction of NTRC with 2-Cys Prxs revealed a high catalytic efficiency of the Anabaena NTRC as a reductant of 2-Cys Prxs from either cyanobacterial or plant origin (Table I). Surprisingly, neither the Anabaena nor the plant NTRC was able to reduce the 2-Cys Prx from Synechocystis (Fig. 3). Because Synechocystis 2-Cys Prx shows a dimeric conformation and a low tendency to form decamers, a possibility to be taken into account is that NTRC has affinity for decamers rather than for dimers. However, the rice NTRC was shown to be active with the dimeric form of the 2-Cys Prx (Pérez-Ruiz and Cejudo, 2009). Previous analyses have revealed important differences between the 2-Cys Prx from Anabaena and Synechocystis (i.e. the Anabaena enzyme is more sensitive to overoxidation and requires a lower concentration of DTT for its reduction in vitro; Pascual et al., 2010). Thus, the inability of the NTRCs from either cyanobacteria or plants to reduce the Synechocystis 2-Cys Prx underscores the different properties of this enzyme as compared with the plant or Anabaena counterpart. These results lend further support to the proposal that an ancestral cyanobacterium resembling the modern Anabaena harbored the original components of the chloroplast hydrogen peroxide detoxification system formed by NTRC, 2-Cys Prx and sulfiredoxin (Deusch et al., 2008; Pascual et al., 2010), and are in agreement with additional phylogenetic analyses of 2-Cys Prxs from cyanobacteria, algae, and plants (Baier and Dietz, 1997; Pitsch et al., 2010). The inability of NTRC to reduce the Synechocystis 2-Cys Prx does not have physiological implications, since this cyanobacterium lacks NTRC. Moreover, it was established that 2-Cys Prx reduction in this cyanobacterium may be catalyzed by the simple-module Trxs m, x, and y (Pérez-Pérez et al., 2009).
Despite the high sequence similarity (75% identity) of the 2-Cys Prxs from Anabaena and Synechocystis (Pascual et al., 2010), modeling of their tridimensional structure revealed a remarkable difference between these enzymes affecting the peroxidatic Cys residue, which is predicted to be more exposed to the protein surface in the reduced monomeric form of the Anabaena enzyme (Fig. 6). Interestingly, the model of the Anabaena 2-Cys Prx predicted a high similarity to the rice homolog, whereas the Synechocystis enzyme was predicted to be similar to the prokaryotic enzyme from Salmonella. In agreement with this prediction, the Anabaena and rice 2-Cys Prxs show an almost identical quaternary structure, the enzyme being detected predominantly in oligomeric form (Fig. 5B). The oligomer-dimer transition allows the switch between the two activities of this enzyme: the low-molecular-weight form functions as a peroxidase, the high-molecular-weight form as a chaperone (Jang et al., 2004). Interestingly, the Synechocystis enzyme has a rather poor capacity to oligomerize (Fig. 5B), suggesting that this enzyme is unable to switch between peroxidase and chaperone activity. The crystallization and structure analysis of the 2-Cys Prxs from Anabaena and Synechocystis might help clarify the molecular evolution of these enzymes, which have been proposed to play an important function in signaling in eukaryotic organisms (Wood et al., 2003a; Dietz et al., 2006; Woo et al., 2010).
The similar biochemical properties of the Anabaena and plant NTRCs suggested the possibility that the Anabaena NTRC might carry out the functions of the plant enzyme in vivo. This was addressed by the expression of the Anabaena NTRC in the Arabidopsis ntrc knockout mutant. Although the cyanobacterial enzyme was correctly targeted to the chloroplast in the transgenic plants, for unknown reasons, the transgene was poorly expressed in the ntrc mutant background, in contrast to its high expression in the wild-type background (Supplemental Fig. S1). In agreement with the low expression of the transgene, the content of the Anabaena NTRC in the transgenic plants, based on western-blot analysis with either anti-OsNTRC or anti-AnabNTRC antibody, was lower than that of the endogenous enzyme in wild-type plants (Fig. 7, A and B). Nevertheless, the transgenic plants showed a partial recovery of the redox status of the 2-Cys Prxs as compared with the NTRC knockout plants (Fig. 7C), which is indicative of the activity of the cyanobacterial enzyme in the context of the plant chloroplast. It was previously proposed that NTRC exerts functions associated with its ability to reduce 2-Cys Prx (Pulido et al., 2010), including the protection against oxidative stress of the enzyme magnesium-protoporphyrin monomethylester cyclase, which is involved in the synthesis of chlorophylls (Stenbaek et al., 2008). The recovery of the content of photosynthetic pigments in the transgenic plants (Fig. 8, C and D) suggests that the capacity of the cyanobacterial NTRC to reduce the Arabidopsis 2-Cys Prxs, although incomplete, is sufficient to complement this function. However, the transgenic plants did not display a fully recovered phenotype, as shown by the reduced growth and lower fresh weight of leaves as compared with the wild-type plants (Fig. 8, A and B). This result suggests that additional functions of NTRC in plants, unrelated to 2-Cys Prx reduction, might not be efficiently performed by the cyanobacterial enzyme. However, this might also be due, at least in part, to the lower content of this enzyme in the transgenic plants. The additional functions of plastid NTRC include redox regulation of starch synthesis (Michalska et al., 2009) or synthesis of aromatic amino acids and auxin (Lepistö et al., 2009), although other functions not yet known are also likely to be controlled by NTRC. Based on these results, we propose that the plant NTRC originated from the cyanobacterial endosymbiont, probably as part of an antioxidant system through its ability to reduce 2-Cys Prxs. However, in plants, NTRC may have evolved to assume new targets and thus to participate in the redox regulation of other processes.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana ecotype Columbia) wild type and the previously reported NTRC knockout mutant, T-DNA insertion line SALK_012208 (Serrato et al., 2004), were grown in soil supplemented with Hoagland medium in culture chambers. Plants were grown under a short-day photoperiod of 14 h of darkness at 20°C/10 h of light at 22°C. The light intensity was set at 140 μmol m−2 s−1. For production of Arabidopsis transgenic lines, the gene encoding NTRC of Anabaena sp. PCC 7120 (open reading frame all0737) was amplified from genomic DNA by PCR using the gene-specific oligonucleotides AnaNTRC1 (5′-GATCTAGAACTGTAGAAAACTTAGTC-3′) and AnaNTRC2 (5′-CCGTCGACCTAAAGATTACC-3′), which added XbaI and SalI restriction sites (underlined) at the 5′ and 3′ ends, respectively. The putative signal peptide of the NTRC cDNA from Arabidopsis (78 N-terminal amino acids) was directly produced by PCR using as template the full-length NTRC cDNA clone from Arabidopsis (DNA stock no. U-14278), obtained from the Arabidopsis Biological Resource Center, with oligonucleotides 5′-CAGGTACCATGGCTGCGTCTC-3′ and 5′-GAGATCTAGACTCGCCTCCTGAAGAAG-3′, which added KpnI and XbaI restriction sites (underlined) at the 5′ and 3′ ends, respectively.
Both fragments were digested, ligated, and cloned into the pGEMt vector (Promega), producing a 1.76-kb fragment, which was sequenced in both strands. The construct was then inserted into the binary vector pBIB-A7 (Becker, 1990) and integrated into the Arabidopsis wild type and T-DNA insertion mutant SALK_012208 (ntrc) by Agrobacterium tumefaciens (C58pMP90)-mediated transformation using the floral dip method (Clough and Bent, 1998). Transformants were selected on plates with Murashige and Skoog medium supplemented with 25 mg L−1 hygromycin. Transgenic lines with a single insertion, and homozygous for the transgene, were selected for further characterization.
Expression in Escherichia coli and Purification of AnabNTRC and Mutant Variants
NTRC from Anabaena, AnabNTRC, was expressed in E. coli as an N-terminally His-tagged polypeptide. The coding sequence was amplified from genomic DNA by PCR using the oligonucleotides AnaNTRC1 and AnaNTRC2, above described, which included XbaI and SalI restriction sites at the 5′ and 3′ ends, respectively. The PCR fragment was digested with XbaI and SalI, subcloned into the pQE-30 expression vector (Qiagen), and introduced into E. coli XL1-Blue. For the expression of the recombinant protein, cells were grown at 37°C with shaking in Luria-Bertani broth medium supplemented with 100 μg mL−1 ampicillin until a cell density (optical density at 600 nm) of 0.6 was reached. Isopropyl-l-d-thiogalactose was then added at a final concentration of 1 mm, and incubation was continued for an additional 1 h. Cells were then collected by centrifugation at 5,000g for 10 min at 4°C and resuspended in buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, 10% [v/v] glycerol, and 1 mm phenylmethylsulfonyl fluoride). Cells were broken by sonication on ice using four 30-s bursts at 35% intensity with a 30-s cooling period between each burst. Cell debris was removed by centrifugation at 10,000g for 20 min, and the recombinant protein was purified from the supernatant by Hi-Trap affinity chromatography (GE Healthcare) according to the manufacturer’s instructions. The NTR domain of AnabNTRC (residues 1–370) was amplified with oligonucleotides 5′-GTGGCTCAAGCTTGTAGATG-3′ and 5′-CATCAAATCAAGCTTCTTATTCCGC-3′. The Trx domain (residues 372–483) was amplified with oligonucleotides 5′-AACCGAAGCTTAAATGGAAGCG-3′ and 5′-GTAGAGATAAGCTTACCCAATCCC-3′. Both of these truncated polypeptides were produced in E. coli as described for the full-length protein.
Mutant variants of AnabNTRC were produced replacing Cys residues of the NTR and Trx domains of the enzyme by Ser. Mutations were introduced by PCR using pQE30-AnabNTRC as the template DNA. The following sequences were used for the mutations: C170S mutant (forward, 5′-CGGCTTGTGCAATCTCTGATGGTGCAACCC-3′; reverse, 5′-GGGTTGCACCATCAGAGATTGCACAAGCCG-3′) and C411S mutant (forward, 5′-GGTTGTGGGCCTTCCCATACCCTCAAGCC-3′; reverse, 5′-GGCTTGAGGGTATGGGAAGGCCCACAACC-3′). The mutant variants (C170S and C411S) were amplified with oligonucleotides 5′-CGACCATGGCCACTGTAGAAAACTTAGTC-3′ and 5′-CCCTCGAGAAGATTACCTTCAATC-3′, which added an NcoI restriction site at the 5′ end and an XhoI restriction site (underlined) and removed the stop codon at the 3′ end. The PCR fragment was digested with NcoI and XhoI and subcloned into the pET28a expression vector (Qiagen). The correct introduction of the desired mutations and the absence of additional mutations were checked by sequencing the final constructs. These plasmids were then introduced into E. coli BL21 (DE3), and the expression and purification of the recombinant proteins were performed as described for the wild-type enzyme.
NTR, Trx, and Prx Activity Assays
Prx activity was determined as oxidation of NADPH following the A340 in a reaction mixture containing 100 mm potassium phosphate buffer (pH 7.0), 2 mm EDTA, 0.25 mm NADPH, 0.5 mm hydrogen peroxide, and purified enzymes at the concentrations indicated in the figure legends. Trx activity was determined by the DTT-dependent reduction of insulin as described by Serrato et al. (2001). The reaction mixture contained 100 mm phosphate buffer, pH 7.0, 2 mm EDTA, 0.5 mg mL−1 bovine insulin, and 2 μm purified enzymes. The reaction was initiated by the addition of 0.5 mm DTT, and the increase in A650 was monitored. NTR activity was determined by the reduction of DTNB according to the method described by Holmgren and Björnstedt (1995). The reaction was performed in 100 mm potassium phosphate buffer, pH 7.0, 2 mm EDTA, 5 mm DTNB, 150 μm NADPH, and purified enzymes at the concentrations indicated in the figure legends. The reduction of DTNB was monitored by the increase in A412.
Determination of Photosynthetic Pigments
Photosynthetic pigments (total chlorophyll and carotenoids) were extracted from leaf discs from plants that were grown for 52 d under short-day conditions with 100% methanol, and the content was determined according to Lichtenthaler and Wellburn (1983).
Gel Filtration Chromatography
The oligomeric state of the recombinant proteins was analyzed by gel filtration chromatography in Superdex_200 prep grade columns (Amersham Biosciences). The chromatography was performed with 20 mm potassium phosphate buffer, pH 7.4, supplemented with 0.15 m NaCl at a constant flow rate of 0.5 to 1 mL min−1. The elution profile was monitored at 280 nm, and proteins used as standards (Sigma Chemical) were thyroglobulin (669 kD), apoferritin (443 kD), β-amylase (200 kD), alcohol dehydrogenase (150 kD), bovine serum albumin (67 kD), and carbonic anhydrase (29 kD).
Isolation of Intact Chloroplasts
Arabidopsis chloroplasts were isolated from approximately 10 g of leaves using the chloroplast isolation kit (Sigma Chemical). Leaves were homogenized with a mixer in 80 mL of ice-cold chloroplast isolation buffer, provided by the manufacturer, supplemented with 50 mm ascorbic acid. Homogenates were then filtered through two layers of nylon mesh (20 μm) and centrifuged for 7 min at 1,000g. Chloroplasts were purified by centrifugation on a 40% to 80% Percoll gradient, washed with 3 volumes of chloroplast isolation buffer, and lysed by hypoosmotic shock by resuspension in 62.5 mm Tris-HCl, pH 7.5, 2 mm MgCl2. After centrifugation at 14,000g, the supernatant was analyzed as the stromal soluble fraction.
SDS-PAGE and Western-Blot Analysis
Proteins were fractionated by SDS-PAGE (10%–12% polyacrylamide gels) and electrotransferred onto nitrocellulose sheets, which were then probed with the following antibodies: anti-OsNTRC (Serrato et al., 2004), anti-NTRB (Serrato et al., 2002), or anti 2-Cys Prx from rice (Oryza sativa; Pérez-Ruiz et al., 2006). NTRC from Anabaena was detected with an antibody raised against purified N-terminal His-tagged NTR encoded by the gene alr2204 from Anabaena, which shows a sequence identity with the NTR domain of NTRC of 64% (M.E. Pérez-Pérez and F.J. Florencio, unpublished data). Purified protein was used to immunize rabbits at the Servicio de Proteccion Animal, Seville University. Protein content was estimated by the Bradford method (Bradford, 1976) using bovine serum albumin as a standard.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Northern-blot analysis of NTRC transcript content in Arabidopsis transgenic lines.
References
- Alkhalfioui F, Renard M, Vensel WH, Wong JH, Tanaka CK, Hurkman WJ, Buchanan BB, Montrichard F. (2007) Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol 144: 1559–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Dietz K-J. (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Becker D. (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deusch O, Landan G, Roettger M, Gruenheit N, Kowallik KV, Allen JF, Martin W, Dagan T. (2008) Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol 25: 748–761 [DOI] [PubMed] [Google Scholar]
- Dietz K-J. (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54: 93–107 [DOI] [PubMed] [Google Scholar]
- Dietz K-J, Jacob S, Oelze M-L, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Florencio FJ, Pérez-Pérez ME, López-Maury L, Mata-Cabana A, Lindahl M. (2006) The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth Res 89: 157–171 [DOI] [PubMed] [Google Scholar]
- Florencio FJ, Yee BC, Johnson TC, Buchanan BB. (1988) An NADP/thioredoxin system in leaves: purification and characterization of NADP-thioredoxin reductase and thioredoxin h from spinach. Arch Biochem Biophys 266: 496–507 [DOI] [PubMed] [Google Scholar]
- Holmgren A Björnstedt M. (1995) Thioredoxin and thioredoxin reductase. Methods Enzymol 252: 199–208 [DOI] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al. (2004) Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635 [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González MC, Cejudo FJ. (2009) NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E. (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. (1983) Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 603: 591–592 [Google Scholar]
- Machida T, Kato E, Ishibashi A, Ohashi N, Honjoh K, Miyamoto T. (2007) Molecular characterization of low-temperature-inducible NTR-C in Chlorella vulgaris. Nucl Acid Symp Series 51: 463–464 [DOI] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, Lee JR, Lee SY, Jung YJ, Shin MR, Lim HS, Chung WS, Yun DJ, et al. (2006) The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem Biophys Res Commun 348: 478–484 [DOI] [PubMed] [Google Scholar]
- Pascual MB, Mata-Cabana A, Florencio FJ, Lindahl M, Cejudo FJ. (2010) Overoxidation of 2-Cys peroxiredoxin in prokaryotes: cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J Biol Chem 285: 34485–34492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Mata-Cabana A, Sánchez-Riego AM, Lindahl M, Florencio FJ. (2009) A comprehensive analysis of the peroxiredoxin reduction system in the cyanobacterium Synechocystis sp. strain PCC 6803 reveals that all five peroxiredoxins are thioredoxin dependent. J Bacteriol 191: 7477–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Cejudo FJ. (2009) A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Lett 583: 1399–1402 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, González MC, Spínola MC, Sandalio LM, Cejudo FJ. (2009) The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Mol Plant 2: 457–467 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsch NT, Witsch B, Baier M. (2010) Comparison of the chloroplast peroxidase system in the chlorophyte Chlamydomonas reinhardtii, the bryophyte Physcomitrella patens, the lycophyte Selaginella moellendorffii and the seed plant Arabidopsis thaliana. BMC Plant Biol 10: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Godzik A, Nayeem A, Schmitt JD. (2000) AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39: 6602–6615 [DOI] [PubMed] [Google Scholar]
- Poole LB, Karplus PA, Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol 44: 325–347 [DOI] [PubMed] [Google Scholar]
- Pulido P, Cazalis R, Cejudo FJ. (2009) An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. Plant J 57: 132–145 [DOI] [PubMed] [Google Scholar]
- Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ. (2010) Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot 61: 4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CM, Poole LB. (2000) Attachment of the N-terminal domain of Salmonella typhimurium AhpF to Escherichia coli thioredoxin reductase confers AhpC reductase activity but does not affect thioredoxin reductase activity. Biochemistry 39: 8859–8869 [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chae HZ, Kim K. (2005) Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signalling. Free Rad Biol Med 38: 1543–1552 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ. (2001) Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol 46: 361–371 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Cejudo FJ. (2002) Cloning of thioredoxin h reductase and characterization of the thioredoxin reductase-thioredoxin h system from wheat. Biochem J 367: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Nordeng TW, Culiáñez-Macià FA, Aalen RB. (1999) The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J 19: 1–8 [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz K-J, Jensen PE. (2008) NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett 582: 2773–2778 [DOI] [PubMed] [Google Scholar]
- Sueoka K, Yamazaki T, Hiyama T, Nakamoto H. (2009) The NADPH thioredoxin reductase C functions as an electron donor to 2-Cys peroxiredoxin in a thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. Biochem Biophys Res Commun 380: 520–524 [DOI] [PubMed] [Google Scholar]
- Toledano MB, Planson AG, Delaunay-Moisan A. (2010) Reining in H2O2 for safe signaling. Cell 140: 454–456 [DOI] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. (2007) Hydrogen peroxide sensing and signaling. Mol Cell 26: 1–14 [DOI] [PubMed] [Google Scholar]
- Woo HA, Yim SH, Shin DH, Kang D, Yu D-Y, Rhee SG. (2010) Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140: 517–528 [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. (2003a) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653 [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schröder E, Robin Harris J, Poole LB. (2003b) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40 [DOI] [PubMed] [Google Scholar]