Abstract
We describe here the diversity of chloroplast proteins required for embryo development in Arabidopsis (Arabidopsis thaliana). Interfering with certain chloroplast functions has long been known to result in embryo lethality. What has not been reported before is a comprehensive screen for embryo-defective (emb) mutants altered in chloroplast proteins. From a collection of transposon and T-DNA insertion lines at the RIKEN chloroplast function database (http://rarge.psc.riken.jp/chloroplast/) that initially appeared to lack homozygotes and segregate for defective seeds, we identified 23 additional examples of EMB genes that likely encode chloroplast-localized proteins. Fourteen gene identities were confirmed with allelism tests involving duplicate mutant alleles. We then queried journal publications and the SeedGenes database (www.seedgenes.org) to establish a comprehensive dataset of 381 nuclear genes encoding chloroplast proteins of Arabidopsis associated with embryo-defective (119 genes), plant pigment (121 genes), gametophyte (three genes), and alternate (138 genes) phenotypes. Loci were ranked based on the level of certainty that the gene responsible for the phenotype had been identified and the protein product localized to chloroplasts. Embryo development is frequently arrested when amino acid, vitamin, or nucleotide biosynthesis is disrupted but proceeds when photosynthesis is compromised and when levels of chlorophyll, carotenoids, or terpenoids are reduced. Chloroplast translation is also required for embryo development, with genes encoding chloroplast ribosomal and pentatricopeptide repeat proteins well represented among EMB datasets. The chloroplast accD locus, which is necessary for fatty acid biosynthesis, is essential in Arabidopsis but not in Brassica napus or maize (Zea mays), where duplicated nuclear genes compensate for its absence or loss of function.
Chloroplasts play a central role in plant metabolism and in supporting the growth and differentiation of plant cells. For decades, plant biologists analyzed the functions of a limited number of chloroplast proteins by using a combination of genetic, physiological, and biochemical methods. Following completion of the Arabidopsis (Arabidopsis thaliana) genome sequence (Arabidopsis Genome Initiative, 2000), establishment of public resources for reverse genetics (Sessions et al., 2002; Alonso et al., 2003; Rosso et al., 2003; Kuromori et al., 2004), and development of improved methods for intracellular protein localization (Heazlewood et al., 2007; Sun et al., 2009), genetic dissection of chloroplast function expanded to include large-scale phenotyping of hundreds of insertion mutants disrupted in genes predicted to encode chloroplast-localized proteins. Results of two such projects in Arabidopsis have recently been published (Ajjawi et al., 2010; Myouga et al., 2010). Additional details can be accessed through public databases (www.plastid.msu.edu; http://rarge/psc.riken.jp/chloroplast).
We first became interested in the diversity of chloroplast functions required for plant viability through our efforts to isolate and characterize Arabidopsis mutants defective in seed development (Tzafrir et al., 2004; Meinke et al., 2008). Forward and reverse genetic screens in Arabidopsis have repeatedly shown that interfering with chloroplast functions can result in embryo lethality. Many such examples are included in the SeedGenes database of essential genes (Tzafrir et al., 2003; www.seedgenes.org). However, little attention has been given to establishing a comprehensive dataset of genes encoding chloroplast proteins required at different stages of the life cycle. This oversight is surprising given the important role that chloroplasts play in supporting plant growth and development.
In this report, we describe the results of a project designed to characterize the broad spectrum of nuclear genes that encode chloroplast proteins required for embryo development in Arabidopsis. We then compare this dataset with a complementary list of genes encoding chloroplast proteins with a mutant phenotype first detected at some other stage of the life cycle. We conclude that in Arabidopsis, eliminating biosynthetic functions within the chloroplast and interfering with expression of the chloroplast genome often result in embryo lethality. Disabling the photosynthetic machinery leads instead to reduced pigmentation and altered physiology. Interfering with other chloroplast functions can result in a wide range of mutant phenotypes detected under standard or specialized growth conditions. Gametophyte lethals are underrepresented among knockout collections of genes encoding chloroplast proteins. This indicates that mutant gametophytes can function in the absence of postmeiotic expression of most nuclear genes encoding chloroplast proteins.
Three major types of chloroplast-localized proteins appear to be most frequently associated with embryo lethality in Arabidopsis: (1) enzymes required for the biosynthesis of amino acids, vitamins, nucleotides, and fatty acids; (2) proteins required for the import, modification, and localization of essential proteins within the chloroplast; and (3) proteins required for chloroplast translation. Factors that determine whether disruption of a specific chloroplast protein results in embryo lethality or in defects observed at another stage of development are noted. In addition, we consider why expression of the chloroplast genome is required for embryo development in Arabidopsis and evaluate which proteins encoded by the chloroplast genome appear to be required for viability. Attention is focused on the chloroplast accD gene, which encodes one subunit of a multimeric acetyl-CoA carboxylase required for fatty acid biosynthesis. The presence of duplicated nuclear genes that compensate for the loss of accD function in maize (Zea mays) and Brassica napus may explain why interfering with chloroplast translation in some plant species does not result in embryo lethality.
RESULTS
Reverse Genetic Identification of Chloroplast-Localized EMBRYO-DEFECTIVE Proteins
We pursued a reverse genetic approach to the identification of additional EMBRYO-DEFECTIVE (EMB) genes encoding chloroplast-localized proteins in Arabidopsis based on published work from the laboratory of Kazuo Shinozaki at the RIKEN Plant Science Center in Japan. We focused on 106 insertion lines corresponding to 79 of 92 genes that appeared to lack insertion homozygotes and were designated AbH (for absence of homozygotes) in table 4 of Myouga et al. (2010). Lines that included an active resistance marker were analyzed first in order to facilitate selection strategies and the identification of desired heterozygotes (McElver et al., 2001). SALK lines (Alonso et al., 2003) with a silenced resistance marker were added later. Table I presents a broad overview of the results obtained. Additional details on the genes and insertion lines examined are shown in Supplemental Table S1.
Table IV. EMB and PDE genes encoding PPR proteins thought to be chloroplast localized.
| Phenotype Class | Locus | Gene Symbol | Alias | Scorea | Identity Statusb | Information Source |
| PDE | At1g05750 | PDE247 | CLB19 | 2 | C | SeedGenes; Chateigner-Boutin et al. (2008) |
| EMB | At1g10910 | EMB3103 | 1 | C | This report | |
| PDE | At1g15510 | AtECB2 | VAC1 | 4 | C | This report; Yu et al. (2009); Tseng et al. (2010) |
| EMB | At1g30610 | EMB2279 | EMB88 | 1 | C | SeedGenes; Meinke et al. (2009) |
| PDE | At1g74850 | PDE343 | PTAC2 | 4 | C | This report; Pfalz et al. (2006) |
| EMB | At2g01860 | EMB975 | 1 | C | SeedGenes | |
| EMB | At3g06430 | EMB2750 | AtPPR2 | 1 | C | SeedGenes; Williams and Barkan (2003) |
| EMB | At3g18110 | EMB1270 | 0c | NC | SeedGenes; Cushing et al. (2005) | |
| EMB | At3g49170 | EMB2261 | 1 | C | SeedGenes; Cushing et al. (2005) | |
| EMB | At3g49240 | EMB1796 | 1d | C | SeedGenes; Cushing et al. (2005) | |
| EMB | At4g20090 | EMB1025 | 1d | C | SeedGenes; Cushing et al. (2005) | |
| EMB | At4g20740 | EMB3131 | 1 | C | This report | |
| PDE | At4g34830 | PDE346 | MRL1 | 4 | NC | This report; Johnson et al. (2010)e |
| EMB | At4g39620 | EMB2453 | AtPPR5 | 0f | NC | SeedGenes; Beick et al. (2008) |
| EMB | At5g03800 | EMB1899 | EMB175 | 1 | C | SeedGenes; Cushing et al. (2005) |
| EMB | At5g04810 | AtPPR4 | 2 | C | Schmitz-Linneweber et al. (2006) | |
| EMB | At5g27270 | EMB976 | 1 | NC | SeedGenes | |
| EMB | At5g39980 | EMB3140 | 1 | C | This report | |
| EMB | At5g50280 | EMB1006 | 1 | NC | SeedGenes | |
| EMB | At5g50390 | EMB3141 | 1 | NC | This report | |
| EMB | At5g67570 | EMB1408 | EMB246; DG1 | 2 | C | SeedGenes; Chi et al. (2008) |
Likelihood of protein localization in the plastid: ranked from high (5) to low (0 or 1).
Gene identity confirmed (C) or not confirmed (NC) through allelism tests or molecular complementation.
Predicted by TargetP and Predotar to be chloroplast localized.
May be targeted to mitochondria; conflicting localization data.
Pale green seed phenotype identified here; absence of plant phenotype reported by Johnson et al. (2010).
Chloroplast localized (Beick et al., 2008).
Table I. Analysis of 92 Arabidopsis genes from the RIKEN collection reported to lack knockout homozygotes.
| Category Based on Mutant Phenotype | Genes | |
| Consistent seed phenotype observeda | 54 | |
| A | Present in SeedGenes database | 16 |
| B | EMB addition to SeedGenes | 29 |
| C | PDE addition to SeedGenes | 1 |
| D | Promising EMB candidate | 3 |
| E | Promising OVA candidate | 5 |
| Seed phenotype absent or problematic | 38 | |
| F | Not examined in this study | 9 |
| G | Unresolved; conflicting data | 15 |
| H | No seed phenotype observed | 14 |
The SeedGenes database includes genes with embryo-defective (emb) and pigment-defective embryo (pde) phenotypes. The ovule abortion (ova) phenotype can also be detected in heterozygous siliques. Locus and insertion line numbers for each category (A–H) are listed in Supplemental Table S1.
Eight genes were eliminated from further consideration because the locus was obsolete or defined a pseudogene at The Arabidopsis Information Resource (TAIR) or because the line examined at RIKEN appeared to have a chromosomal deletion or to produce unusually few aborted seeds. Another locus was excluded because it was a known gametophyte lethal (Pagnussat et al., 2005). Twelve genes were not examined because they represented confirmed essential genes (TIC110, PDE166, EMB2279, SCO1, ISE2, SUS2, EMB1956, AtGYRA, TOC75, EDD, HISN6A, and EMB2036) already in the SeedGenes database. Four others (EMB2394, EMB2458, EMB2750, and EMB1865) were previously represented in SeedGenes by a single mutant allele. In these cases, insertion lines from RIKEN were used to demonstrate allelism through genetic complementation tests, which confirmed the identity of the gene responsible for the seed phenotype.
For the remaining genes from the original list, we identified additional candidate alleles from insertion lines available through the Arabidopsis Biological Resource Center (ABRC) and the Nottingham Arabidopsis Stock Center (NASC). These lines were then grown along with counterparts from RIKEN. Consistent seed phenotypes were documented, percentages of mutant seeds in heterozygous siliques calculated, and segregating plants crossed to test for allelism. This enabled the identification of 29 additional EMB genes for inclusion in the SeedGenes database. With 18 of these loci, we confirmed gene identities through genetic complementation tests involving multiple alleles (Table II). Mutant phenotypes associated with several of these genes were also described in recent publications: EMB3146 (Kim et al., 2009), EMB3116 (Pignocchi et al., 2009), and EMB3138 (Bang et al., 2009; Chigri et al., 2009; Meinke et al., 2009; Garcia et al., 2010). Based on updated protein function and localization data, some of the genes included in Table II encode proteins that are not localized to chloroplasts. The original dataset from RIKEN, therefore, includes some Arabidopsis proteins that are instead found elsewhere in the cell.
Table II. EMB additions to the SeedGenes dataset based on mutants found in the RIKEN collection.
| Locus | Symbola | Statusb | Scorec | Predicted Protein Function | Allele 1 | Allele 2 | Allele 3 |
| At1g05600 | EMB3101 | C | X | Mitochondrial PPR protein | 53-3364-1 | 15-3350-1d | |
| At1g10910 | EMB3103 | C | 1 | Unknown | WiscDsLox241F03 | SALK-002449 | |
| At1g12410 | EMB3146a | C | 4 | CLP protease catalytic subunit | SALK-016774 | SALK-118775 | |
| At1g48350 | EMB3105 | NC | 4 | Chloroplast 50S ribosomal protein L18 | 52-0944-1e | ||
| At1g49880 | EMB3106 | C | X | Mitochondrial sulfhydryl oxidase | CSHL-ET6113 | SALK-110883d | SALK-001649d |
| At1g59990 | EMB3108 | NC | 2 | DEAD box RNA helicase | WiscDsLox457-460K23e | ||
| At2g01390 | EMB3111 | C | X | Mitochondrial PPR protein | 12-5583-1 | 13-2831-1 | 52-0110-1 |
| At2g30200 | EMB3147 | NC | 4 | ACP-S-malonyltransferase | GT_5_100190 | ||
| At2g33800 | EMB3113 | C | 4 | Chloroplast 30S ribosomal protein S5 | 11-2624-1 | SALK-095863d | |
| At2g36000 | EMB3114 | NC | 1 | mTERF domain protein | 11-0808-1e | ||
| At2g44190 | EMB3116a | C | X | Microtubule-associated protein | 11-5223-1 | 15-2706-1 | |
| At2g47940 | EMB3117 | NC | 4 | Chloroplast DegP2 protease | GT-5-53741f | ||
| At3g04790 | EMB3119 | C | 4 | Ribulose-5-phosphate isomerase | 11-0136-1 | SAIL-874-E07d | |
| At3g14900 | EMB3120 | C | 2 | Unknown | 16-2974-1 | SALK-123989d | |
| At3g17910 | EMB3121 | C | X | Cytochrome c oxidase assembly | 54-3828-1 | SALK_133094d | |
| At3g27750 | EMB3123 | C | 2 | VPS9 domain protein | 12-5139-1 | SALK-140334d | |
| At3g63490 | EMB3126 | C | 4 | Chloroplast 50S ribosomal protein L1 | 53-3245-1 | CSHL-GT6376 | GT-5-101962 |
| At4g00620 | EMB3127 | C | 4 | Folic acid biosynthesis | 12-1633-1 | GT-5-24037 | |
| At4g02790 | EMB3129 | C | 1 | Similar to ribosome biogenesis GTPase | WiscDsLox345-348J11 | SAIL-288-A03d | |
| At4g20740 | EMB3131 | C | 1 | PPR protein | 16-3060-1 | CSHL-GT6247 | |
| At5g11890 | EMB3135 | NC | 1 | Unknown | 13-6017-1f | ||
| At5g13510 | EMB3136 | NC | 4 | Chloroplast 50S ribosomal protein L10 | 51-2522-3e | ||
| At5g14320 | EMB3137 | C | 5 | Chloroplast 30S ribosomal protein S13 | 15-0663-1 | SALK-133412d | |
| At5g18570 | EMB3138a | C | 4 | Chloroplast Obg-like GTPase | emb269g | 52-0311-1 | GABI-387B03d |
| At5g39980 | EMB3140 | C | 1 | PPR protein | 11-2393-1 | SAIL-14-A10d | SALK-129401d |
| At5g50390 | EMB3141 | NC | 1 | PPR protein | WiscDsLox293-296invK21f | ||
| At5g51200 | EMB3142 | NC | X | Nuclear pore complex protein (Nup205) | 53-3083-1f | ||
| At5g58250 | EMB3143 | NC | 4 | Unknown | 53-4365-1e | ||
| At5g64580 | EMB3144 | C | 2 | AAA ATPase | 52-0428-1 | SAIL-74-G12d | SALK-113657d |
Alias symbols are as follows: EMB3146: CLPR2; EMB3116: EDE1; EMB3138: EMB269, AtOBGL, AtOBGC, CPSAR1.
Gene responsible for mutant phenotype confirmed (C) through allelism tests or not confirmed (NC) because a second mutant allele is not available.
Likelihood of protein localization in the chloroplast based on experimental data and prediction programs: from 5 (high) to 1 (low) or X (elsewhere).
Insertion line not in RIKEN database; requested instead from stock centers.
No additional insertion lines available for analysis.
No phenotype found in other insertion lines tested; presence of inserts in those lines not confirmed.
Allelic based on map location and genetic complementation tests (Meinke et al., 2009).
When only a single mutant allele was available or exhibited a seed phenotype, further evidence in support of gene tagging was provided by scoring plants that survived germination on selection medium for the presence of defective seeds. Results of these cosegregation studies are presented in Supplemental Table S2. One locus excluded from the list (At3g55250) exhibited a knockout phenotype affecting embryo pigmentation but not embryo development. Another locus (At1g08070) was excluded because the consistent seed phenotype identified for a single RIKEN line (13-2793-1) conflicted with the absence of a seed phenotype noted elsewhere for multiple insertion lines (Okuda et al., 2010). Phenotypic and genetic segregation data for insertion lines included in Table II are presented in Supplemental Table S3. Terminal phenotypes of mutant seeds typically extended from the globular to cotyledon stages of embryo development. Most of the mutant seeds and embryos appeared white or pale yellow before desiccation. Lines 11-5049-1 (At1g79790) and 51-2522-3 (At5g13510) segregated for two different mutations affecting seed development. In both cases, a single mutant phenotype was associated with the insertion.
Mutant alleles disrupted in the same gene often exhibited similar terminal embryo phenotypes. One exception was EMB3113 (At2g33800), which encodes a chloroplast ribosomal protein. A SALK line with an insertion in the 5′ untranslated region had a seed pigment phenotype, whereas a RIKEN line with an insertion in the second (last) exon resulted in early embryo lethality. Seeds with one copy of each mutant allele, formed through genetic complementation tests, exhibited the later phenotype, indicating that the SALK allele had sufficient residual activity to rescue embryo development. In another case (EMB3123; At3g27750), the pigment defect observed in a SALK line resulted from an insertion near the 3′ end of the coding region, whereas the embryo defect found in a second (RIKEN) allele was associated with an insertion near the 5′ end. Terminal phenotypes of mutant embryos can be influenced by multiple factors, including level of gene redundancy and location of the mutation site. The range of phenotypes observed here is consistent with other reports in the literature of seed defects found in Arabidopsis mutants altered in chloroplast proteins.
We identified a knockout seed phenotype and obtained supporting genetic data for 45 of the 92 loci that initially appeared to lack insertion homozygotes (Myouga et al., 2010). Sixteen of these genes (about 35%) were already included in the SeedGenes database. This degree of overlap with existing mutant collections is comparable to the estimated current level of saturation (30%) for EMB genes in Arabidopsis (Meinke et al., 2009). Another eight genes listed in Table I (classes D and E) are potentially associated with an emb or ovule abortion (ova) phenotype but will require further analysis to confirm. One such gene (At3g58140) encodes an aminoacyl-tRNA synthetase that is targeted to both mitochondria and chloroplasts and exhibits a knockout phenotype resembling that of other aminoacyl-tRNA synthetase proteins with similar patterns of localization (Berg et al., 2005). Another gene (At4g04780; MED21) was reported elsewhere to encode a protein not localized to chloroplasts and to exhibit an embryo-lethal knockout phenotype (Dhawan et al., 2009). We found instead an ova phenotype. This suggests that structures labeled as aborted seeds in that publication are aborted ovules. We also detected reduced transmission of the mutant allele through male gametes, providing further evidence that MED21 is required for normal gametophyte function.
Absence of a Consistent Seed Phenotype in Lines Thought to Lack Insertion Homozygotes
Another 29 genes examined here were not associated with a knockout seed phenotype or gave conflicting results when screened for defects in seed development (Table I; Supplemental Table S1). SALK lines were often involved, which we and others have found can lack the predicted insert (Ajjawi et al., 2010; Myouga et al., 2010). With At1g55380 and At4g24860, seed phenotypes were found in two insertion lines associated with the same locus, but the mutants were not allelic in genetic complementation tests. Several genes without a seed defect found here are known from the literature to exhibit other mutant phenotypes. Examples include HCF173, At1g16720 (Schult et al., 2007); CSLD4, At4g38190 (Bernal et al., 2008); RPH1, At2g48070 (Belhaj et al., 2009); MTO1, At3g01120 (Inaba et al., 1994); CSR1, At3g48560 (Haughn and Somerville, 1990); BYPASS1, At1g01550 (Van Norman et al., 2004); and ELF7, At1g79730 (He et al., 2004). Two of these loci (MTO1 and CSR1) are associated with gain-of-function mutations; the others are defined by loss-of-function phenotypes. Why insertion homozygotes for these genes were not found in the initial screen at RIKEN remains unresolved. We thought at first that mutants lacking both insertion homozygotes and a seed phenotype might have defects in male gametophyte development, which would escape detection in developing siliques. Further analysis of selected lines did not support this explanation. Because our primary objective was to maximize the identification of EMB genes, we decided against a more detailed characterization of lines that failed to exhibit a consistent seed phenotype. Nevertheless, we found a substantial number of problematic cases where the apparent absence of insertion homozygotes reported by Myouga et al. (2010) could not be confirmed or explained. This highlights once again the challenges faced when undertaking large-scale reverse genetic screens in Arabidopsis (O’Malley and Ecker, 2010).
Screens of Reported Seedling Mutants for Defects in Seed Pigmentation
Because many defects in seedling pigmentation can first be detected by screening immature siliques of heterozygotes for the presence of pale seeds, we decided to determine whether insertion lines with an albino or pale seedling phenotype (Myouga et al., 2010) also exhibited reduced seed pigmentation. We had identified a number of these pigment-defective embryo (pde) mutants in the past using a combination of forward and reverse genetics. These mutants are of particular interest to us because, although they are not embryo lethal, they do exhibit a phenotype that can be detected during embryo development. We identified a seed phenotype in 28 pigment mutants associated with 25 different genes in the RIKEN collection (Supplemental Table S4). This dataset includes seven genes with confirmed identities, 15 with identities not confirmed, and three that exhibited a consistent seed pigment phenotype but lacked supporting cosegregation data. Another eight genes assigned to the seedling pigment classes by Myouga et al. (2010) remained unresolved or failed to exhibit a seed phenotype.
Updating Protein Localization Information for a Comprehensive EMB Gene Dataset
In addition to defining confidence levels for the identity of each gene responsible for an observed mutant phenotype, we decided to evaluate, based on several different criteria, the likelihood that the corresponding gene product was indeed localized to chloroplasts. We then combined genes identified here with those already in the SeedGenes database to establish an updated dataset of EMB genes encoding chloroplast-localized proteins in Arabidopsis. The original collection of 4,273 insertion lines analyzed by Myouga et al. (2010) was based on a published dataset of 2,090 predicted chloroplast proteins derived from examination of N-terminal sequences in the Arabidopsis proteome (Richly and Leister, 2004). Following publication of that dataset, additional methods and databases were established to track proteins localized to chloroplasts. Because there is no definitive dataset of every chloroplast protein in Arabidopsis and because prediction and localization strategies can give conflicting results, we ranked each EMB protein based on whether it was included in different proteome datasets. A single point was awarded for inclusion in the predicted chloroplast proteome of Richly and Leister (2004) and for mass spectrometry or GFP evidence supporting chloroplast localization in the SUBA database (Heazlewood et al., 2007). Two points were given for inclusion in the curated Plant Proteome Database (Sun et al., 2009). Protein functions obtained from TAIR and the literature were manually curated, evaluated to determine whether they conflicted with chloroplast localization, and then assigned to general classes.
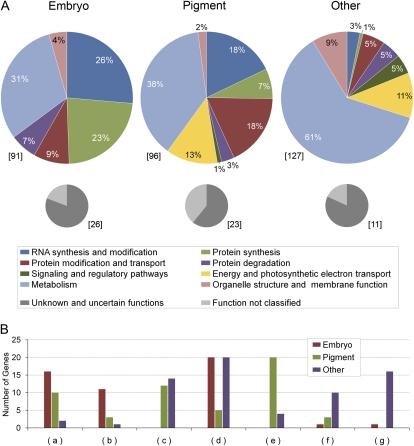
Using this approach, we identified a total of 119 EMB genes encoding proteins thought to be localized to chloroplasts. This represents 30% of the EMB genes identified to date. The dataset includes gene identities classified as either confirmed (92 entries) or not confirmed (27 entries) and with chloroplast localization ratings from least (1) to most (5) confident. Thirteen functional categories and 18 subcategories were developed to capture relevant information. Protein function assignments for the dataset are summarized in Figure 1 and Supplemental Table S5. Details for individual genes are presented in Supplemental Table S6. Genes with known functions that conflicted with chloroplast localization were removed, whereas genes encoding unknown proteins with questionable chloroplast localization were retained. All of these EMB genes are included in a recent update of the SeedGenes database.
Figure 1.
Distribution of protein functions in three datasets of Arabidopsis mutants defective in chloroplast-localized proteins. Genes were classified as having an embryo-defective, pigment-defective, or other loss-of-function mutant phenotype. A, Percentages of genes with assigned (colored) and unassigned (gray) protein functions. Gene totals are indicated in brackets. B, Examples of function subclasses that differ widely between the three datasets: (a) PPR and RNA-binding proteins; (b) ribosomal proteins; (c) photosynthesis; (d) biosynthesis of amino acids, vitamins, nucleotides, and fatty acids; (e) biosynthesis of chlorophyll, carotenoids, and terpenoids; (f) biosynthesis of lipids; modification of fatty acids and lipids; (g) biosynthesis and modification of complex carbohydrates. Gene totals for each dataset are indicated. See Supplemental Table S5 for additional details.
Datasets of Genes Encoding Chloroplast-Localized Proteins with Other Mutant Phenotypes
While assembling the dataset of EMB proteins, we realized it would be helpful to have a comparable dataset of chloroplast proteins with mutant phenotypes detected at other stages of the life cycle. In order to complete this task, we made use of an ongoing project in our laboratory to update the list of Arabidopsis genes with a loss-of-function phenotype of any kind that we published several years ago (Meinke et al., 2003). We started with information from TAIR about genes thought to be associated with phenotype information, queried the recent literature in PubMed (http://www.ncbi.nlm.nih.gov/pubmed), and identified genes with mutant phenotypes resulting from the loss of chloroplast functions using the approach outlined above. This resulted in three additional datasets: one for gametophyte lethals, one for mutants defective in pigmentation (Supplemental Table S7), and another for mutants with other phenotypes (Supplemental Table S8).
Several interesting differences can be found between the embryo and pigment datasets. Thirty-three percent of genes with assigned functions in the pigment dataset encode proteins involved in photosynthesis or the production of chlorophyll, carotenoids, or terpenoids. None of the EMB proteins is included in these functional classes. Embryo development, therefore, can proceed to completion regardless of what individual components of the photosynthetic system are disrupted. Twenty-three percent of chloroplast-localized EMB proteins function in translation within the chloroplast, far more than the 7% of proteins found in the pigment dataset. A complete disruption of chloroplast gene function, therefore, results in embryo lethality. Ten examples of chloroplast-localized ribosomal proteins known to be associated with an embryo-defective phenotype are shown in Table III. Five of these mutants were first characterized here. Knocking out chloroplast-localized aminoacyl-tRNA synthetases also results in embryo lethality (Berg et al., 2005). In addition, at least 17 genes encoding chloroplast-localized pentatricopeptide repeat (PPR) proteins, which often target specific RNAs for modification (Schmitz-Linneweber and Small, 2008), are required for embryo development in Arabidopsis and several more for normal seed pigmentation (Table IV). Five of the corresponding mutants were first characterized here.
Table III. Chloroplast-localized ribosomal proteins with a knockout phenotype in Arabidopsis.
| Phenotype Class | Locus | Gene Symbol | Ribosomal Protein | Identity Statusa | Information Source |
| EMB | At2g33800 | EMB3113 | S5 | C | This report |
| EMB | At1g74970 | TWN3 | S9 | NC | SeedGenes |
| EMB | At5g14320 | EMB3137 | S13 | C | This report |
| Pigment | At1g79850 | PDE347; ORE4 | S17 | C | Woo et al. (2002) |
| Pigment | At3g27160 | GHS1 | S21 | C | Morita-Yamamuro et al. (2004) |
| EMB | At3g63490 | EMB3126 | L1 | C | This report |
| EMB | At1g07320 | EMB2784 | L4 | NC | SeedGenes |
| EMB | At1g05190 | EMB2394 | L6 | NC | SeedGenes |
| EMB | At5g13510 | EMB3136 | L10 | NC | This report |
| Pigment | At1g32990 | PAM14 | L11 | C | Pesaresi et al. (2001) |
| EMB | At1g78630 | EMB1473 | L13 | NC | SeedGenes |
| EMB | At1g48350 | EMB3105 | L18 | NC | This report |
| EMB | At1g75350 | EMB2184 | L31 | NC | SeedGenes |
Identity confirmed (C) or not confirmed (NC) through allelism tests or molecular complementation.
Major disruptions of chloroplast function, such as blocking protein import from the cytosol, can also result in embryo lethality, although less severe perturbations often result in reduced embryo pigmentation. Other chloroplast-localized proteins are required not just for chloroplast function but also for general cell growth. For example, 22% of chloroplast EMB proteins function in the biosynthesis of amino acids, vitamins, nucleotides, or fatty acids, consistent with the chloroplast localization of these pathways. Embryo lethality in these mutants is caused by the absence of an essential compound that the chloroplast provides to the plant cell. In some cases, the underlying cause of embryo lethality remains unclear. One example involves the large collection of chloroplast-localized EMB proteins of unknown function. These genes represent promising targets for additional biochemical and physiological studies, particularly if weak alleles that support continued development can be identified or RNA interference strategies are employed to circumvent lethality.
Remarkably, of the 115 gametophyte lethals that can be associated with reasonable confidence to a single gene disruption in our phenotype datasets, only three genes (2.6%) appear to encode chloroplast-localized proteins: HISN8 (His biosynthesis), GPT1 (plastid Glc importer), and PUR4 (purine biosynthesis), which is targeted to both chloroplasts and mitochondria. Gametophyte lethals, therefore, are underrepresented among knockouts of chloroplast proteins. Several chloroplast-localized proteins are required for both embryo development and male gametophyte functions. Two of these (HISN3 and HISN4) are involved in His biosynthesis and ATP recycling (Muralla et al., 2007).
One hundred thirty-eight chloroplast-localized proteins with other knockout phenotypes are included in Figure 1. Sixty-one percent of the proteins with assigned functions are known to be involved in plant metabolism, consistent with the impressive diversity of biosynthetic pathways found in chloroplasts. Most enzymes that modify fatty acids and lipids or function in the biosynthesis or modification of complex carbohydrates are included in this “other” phenotype class. Disruption of some biosynthetic pathways for essential amino acids and vitamins can also result in seedling lethality rather than embryo lethality. Often, successful completion of embryo development appears to result from a partial loss of gene function or from redundant genes or biosynthetic pathways. Compared with the embryo and pigment datasets, few of the genes with other mutant phenotypes function in chloroplast translation or RNA modification. Each of the phenotype datasets (embryo, pigment, other), therefore, has a distinctive but somewhat overlapping profile of protein functions, consistent with the underlying biological processes involved.
DISCUSSION
We present in this report the identities of 119 nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Many different protein functions are represented in this collection, from biosynthetic enzymes and components of protein modification and import systems to factors required for proper expression of the chloroplast genome. In addition, we demonstrate that a wide range of mutant phenotypes can result from the disruption of chloroplast-localized proteins. One of the initial goals of research with embryo-lethal mutants of Arabidopsis was to determine what types of genes underlie this common phenotype (Meinke and Sussex, 1979). After several decades of mutant analysis, we have finally established the robust dataset needed to document that interfering with chloroplast functions is frequently involved. Among the 400 EMB genes of Arabidopsis identified to date, roughly 30% are thought to encode chloroplast-localized proteins. Embryo phenotypes are consistent with the known role that chloroplasts play in biosynthetic pathways required to support embryo development beyond the globular stage. Two common features of embryo lethality in Arabidopsis, reduced seed pigmentation and globular embryo arrest, therefore, can often be explained as a consequence of the disruption of essential chloroplast functions. The cellular roles of a number of chloroplast-localized EMB proteins with unknown functions nevertheless remain to be determined. Because most proteins involved in basic metabolism should resemble known proteins characterized in other model organisms, we believe that many EMB proteins of unknown function act in specialized, plant-specific complexes or processes that either support chloroplast translation or interact with other proteins that perform essential functions within the chloroplast.
Knowledge of which chloroplast-localized proteins are essential for plant growth and development has the added benefit of facilitating the analysis of mutants disrupted in related chloroplast functions. For example, a seedling mutant that represents a null allele and is defective in a chloroplast ribosomal protein encoded by a nonredundant gene provides indirect evidence that the protein in question is not required for basal ribosome function, because the complete elimination of chloroplast translation results in embryo lethality. Similarly, disruption of a chloroplast PPR protein should result in embryo lethality only if the function of that protein extends beyond photosynthesis. Several factors contribute to the observed differences in mutant phenotypes when similar chloroplast functions are disrupted: redundancy of genes, metabolic pathways, and cellular processes; differences in gene expression patterns and strengths of mutant alleles; and variations in the contributions of different proteins to the same fundamental process. As described in this report, the establishment of an initial dataset of mutants disrupted in chloroplast-localized proteins should help to illuminate the significance of different chloroplast functions throughout plant growth and development.
Developmental Significance of the Chloroplast accD Gene Product
One important issue that remains to be addressed concerns the identity of chloroplast genes required for embryogenesis in Arabidopsis. If interfering with chloroplast translation results in embryo lethality, then one or more chloroplast gene products must play an essential role. In other words, the lethality observed in knockouts of chloroplast ribosomal proteins must result from a failure to produce certain chloroplast proteins. The identity of these essential proteins has remained elusive, despite continued advances in our understanding of chloroplast gene expression. Surprisingly, interfering with chloroplast translation in barley (Hordeum vulgare), maize, and B. napus has a different impact on development: embryogenesis proceeds and albino seedlings are formed instead (Hess et al., 1994; Zubko and Day, 1998; Asakura and Barkan, 2006). What underlies these striking differences in the developmental significance of chloroplast translation in different plant species?
When evaluating the different phenotypes observed with maize and Arabidopsis mutants defective in plastid RNA processing, Asakura and Barkan (2006) focused attention on three genes found in the chloroplast genome of Arabidopsis but missing from that of maize: accD, which encodes a component of the heteromeric, plastid acetyl-CoA carboxylase that functions in fatty acid biosynthesis; and two large genes of unknown function, ycf1 and ycf2. These same three genes were also highlighted in a recent review of PPR proteins (Schmitz-Linneweber and Small, 2008). The essential nature of all three genes is known from tobacco (Nicotiana tabacum), where targeted gene disruptions were used to demonstrate that homoplastomic leaves lacking a functional version of any one of these genes were not obtained, presumably because they were inviable (Drescher et al., 2000; Kode et al., 2005). A similar approach was used to demonstrate that another protein (clpP1) encoded by the chloroplast genome of tobacco is also essential (Kuroda and Maliga, 2003). Therefore, it seems likely that in maize, the functions of these essential chloroplast proteins are bypassed or replaced with other proteins encoded in the nucleus. Further evidence can be found in the chloroplast genome of the parasitic plant Epifagus virginiana, which has lost most of its coding capacity but still retains accD, ycf1, and ycf2 (Wolfe et al., 1992). Something about these genes has ensured their continued presence, although isolated examples of gene loss have been documented in certain plant lineages (Jansen et al., 2007).
Several lines of evidence suggest that accD is the single, most important chloroplast gene required for embryo development in Arabidopsis. The key point is that plant species that can tolerate the loss of chloroplast gene expression appear to have evolved a compensation mechanism that allows an alternate form of the enzyme produced in the cytosol to carry out fatty acid biosynthesis in the chloroplast. Plant cells contain two different forms of acetyl-CoA carboxylase: a heteromeric plastid enzyme that functions in the initial stages of fatty acid biosynthesis and is composed of four different polypeptides, three of which are typically encoded by the nuclear genome; and a large, homomeric cytosolic enzyme that functions more in secondary plant metabolism (Sasaki and Nagano, 2004). The inability of malonyl-CoA, the product of acetyl-CoA carboxylase activity, to be readily transported across the chloroplast membrane appears to explain the requirement for two distinct but related enzymes in plant metabolism. A nuclear gene duplication event associated with the evolution of grasses enabled the appearance of a modified homomeric enzyme targeted to plastids that ensures the production of malonyl-CoA in the absence of a functional plastid genome (Chalupska et al., 2008). This duplication also made possible the development of a novel class of herbicides that inhibits the plastid-targeted homomeric enzyme in grasses while not interfering with the heteromeric enzyme found in most other plants (Liu et al., 2007).
A similar phenomenon has been described in B. napus, where treatment of germinating seedlings with spectinomycin, which interferes with chloroplast translation, leads to plants with albino leaves devoid of chloroplast ribosomes (Zubko and Day, 1998). In contrast to maize, the essential accD gene is retained in the chloroplast genome of Brassica. However, gene duplication has once again allowed the cytosolic production of a modified homomeric enzyme that is targeted to the chloroplast (Schulte et al., 1997). A similar duplication is found in the nuclear genome of Arabidopsis. Disruption of the gene (At1g36160) encoding a cytosolic enzyme (ACC1) results in defects in embryo development (emb22, gurke, pas3) that we first described as resembling a “green blimp” (Meinke, 1985). This odd phenotype was later shown to result from alterations in the homomeric acetyl-CoA carboxylase (Baud et al., 2004). The second member of this tandem duplication (At1g36180) is predicted to encode a plastid-localized protein (ACC2). However, this gene appears to be poorly expressed, particularly in developing siliques (Yanai et al., 1995; Baud et al., 2003). The inability of this gene to rescue disabled plastids unable to produce their own acetyl-CoA carboxylase likely explains why interfering with chloroplast translation in Arabidopsis results in embryo lethality.
This model is further supported by evidence presented here that a mutant (emb3147) unable to complete the next step in fatty acid biosynthesis, catalyzed by the product of At2g30200, also exhibits embryo lethality. Disruption of a second component of the heteromeric plastid enzyme (At5g16390) also results in embryo lethality (Li et al., 2011). The conversion of acetyl-CoA into fatty acids within the chloroplast, therefore, is required for plant embryo development. The precise functions of ycf1 and ycf2 remain to be determined, although they likely enhance either the production or activity of the heteromeric acetyl-CoA carboxylase, in part because they do not appear to be duplicated in the nuclear genome of Brassica, whose seedlings can survive without their expression, and also because their presence is often linked to that of the heteromeric enzyme in different plant lineages (Jansen et al., 2007; Guisinger et al., 2010).
Abundance of EMB Genes Encoding PPR Proteins
A second question that needs to be addressed concerns the molecular functions of essential, chloroplast-localized PPR proteins in Arabidopsis. This large family of proteins has been the topic of widespread interest in recent years (Lurin et al., 2004; Cushing et al., 2005; Schmitz-Linneweber and Small, 2008). Multiple examples of PPR proteins with mutant phenotypes are presented here and elsewhere. Why do some PPR knockouts result in embryo lethality while others permit embryo development to proceed and result instead in defects in plant pigmentation?
One explanation that can be rejected is that all chloroplast-localized PPR proteins required for embryo development edit transcripts that encode essential proteins (accD, ycf1, ycf2), whereas those required for seedling pigmentation edit transcripts involved with photosynthesis. Remarkably, none of the known editing mutants exhibits embryo lethality. At least 34 edited sites have been found in the transcripts of chloroplast genes in Arabidopsis (Chateigner-Boutin and Small, 2007). None of these is located in ycf1 or ycf2. Three sites are in genes encoding ribosomal proteins (rps12 intron, rps14 and rpl23 coding region). PPR proteins required for editing these transcripts (OTP81, OTP86, OTP80) have been identified, but the corresponding insertion mutants exhibit a normal phenotype (Hammani et al., 2009). The rps12 protein is known to be essential because a failure to process its transcript causes embryo lethality in Arabidopsis (Asakura and Barkan, 2006). Disruption of the PPR protein (CLB19; PDE247) required for editing of the conserved clpP1 site results in pigmentation defects, not embryo lethality (Chateigner-Boutin et al., 2008). This observation remains to be reconciled with the apparent requirement of clpP1 for shoot development in tobacco (Kuroda and Maliga, 2003). Two edited sites in the accD transcript have been examined in detail. One of these is located in the coding region and requires a PPR protein (RARE1) for modification. Disruption of this locus (At5g13270) results in a normal phenotype, which is unexpected because the site affected was thought to be required for carboxylase activity (Robbins et al., 2009). Another PPR protein (VAC1; AtECB1) is required for a second editing site located in the 3′ untranslated region (Yu et al., 2009; Tseng et al., 2010). Disruption of this locus (At1g15510) results in albino seedlings, presumably from reduced levels of functional protein.
Most chloroplast-localized PPR proteins encoded by EMB genes are likely to function instead in ensuring the production of essential gene products, including accD, ycf1, ycf2, and components of the translational machinery. PPR proteins are known to have a variety of important functions associated with RNA binding (Schmitz-Linneweber and Small, 2008). Some of these functions are clearly required for effective translation of the chloroplast genome. This has been demonstrated by Alice Barkan and colleagues, who found that one EMB gene product (AtPPR4), orthologous to a well-characterized protein in maize, is involved in trans-splicing of chloroplast rps12 transcripts, another (AtPPR5; EMB2453) is required for tRNA stabilization in chloroplasts, and a third (AtPPR2; EMB2750) is needed to establish the chloroplast translation machinery (Williams and Barkan, 2003; Schmitz-Linneweber et al., 2006; Beick et al., 2008). Based on this evidence, it seems likely that a number of EMB genes encoding PPR proteins have similar roles in chloroplast gene function. The phenotype datasets presented here should continue to provide a valuable point of reference for future studies on the cellular functions of essential PPR proteins in Arabidopsis and the developmental significance of different chloroplast proteins in flowering plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Many of the insertion lines described in this report were isolated in the laboratory of Kazuo Shinozaki at the RIKEN Plant Science Center. Preliminary phenotype information is presented in table 4 of Myouga et al. (2010). We requested a number of SALK (Alonso et al., 2003), SAIL (Sessions et al., 2002), GABI (Rosso et al., 2003), CSHL (Martienssen, 1998), Wisconsin (Sussman et al., 2000), and JIC (Sundaresan et al., 1995) insertion lines from the Arabidopsis (Arabidopsis thaliana) seed stock centers (ABRC and NASC). Internal seed stocks were used for a separate collection of Syngenta mutants defective in embryo development (McElver et al., 2001). Duplicate stocks of these mutants can be obtained through the ABRC. Mature seeds were first germinated in culture (Meinke et al., 2009) to track germination rates, observe seedling phenotypes, and produce uniform plant populations. Seedlings were later transplanted to pots containing a mixture of soil, sand, and vermiculite, placed under fluorescent lights (16-h-light/8-h-dark cycles) in a growth room maintained at room temperature (23°C), and watered daily with a nutrient solution as described by Berg et al. (2005).
Genetic and Phenotypic Characterization of Insertion Mutants
Methods used to phenotype mutant seeds and embryos are described at the tutorial section of the SeedGenes Web site. Genetic complementation tests were performed as detailed by Meinke et al. (2009). Selection agents used for genetic cosegregation studies were added to germination medium containing Murashige and Skoog salts, 3% (w/v) Glc, and 0.8% (w/v) agar. The final concentrations of selection agents were as follows: 50 mg L−1 kanamycin for JIC and CSHL lines, 50 mg L−1 Basta for SAIL and Wisconsin lines, 30 mg L−1 hygromycin for RIKEN lines, and 5.2 mg L−1 sulfadiazine for GABI lines.
Establishment and Analysis of Gene Datasets
We started with the known collection of genes with a mutant seed phenotype included in the SeedGenes database (seventh release, December 2007) and then supplemented this dataset with additional genes identified in our laboratory and through recent literature searches. The final dataset of EMB and PDE genes described here is consistent with that found in the updated SeedGenes release (December 2010). Genes encoding chloroplast-localized proteins with knockout phenotypes detected at other stages of the life cycle were identified from a comprehensive dataset of genes with any loss-of-function phenotype being developed in our laboratory. Because a substantial number of gametophyte mutants identified through large-scale screens for insertion lines with altered transmission of selectable markers appear to contain chromosomal aberrations, we focused on those gametophyte mutants that had either been complemented with a wild-type transgene or had flanking sequence information for both sides of the insert, demonstrating that a single gene disruption was likely the cause of the mutant phenotype. This enabled a more direct comparison between large datasets of mutants with defects in embryo and gametophyte development. When assembling the dataset of mutants with other phenotypes, we excluded mutants altered only in the modification of transcripts, proteins, or protein complexes because we felt that such a subtle modification did not constitute a mutant phenotype. However, we included mutants with no morphological defects but with altered physiology or biochemical profiles that required special instrumentation to identify and mutants with phenotypes detected only under certain environmental conditions. We realize that our datasets are incomplete and that some genes with published mutant phenotypes escaped our attention. However, the information presented should be representative of the full dataset that may eventually be assembled once saturation for gene knockouts in Arabidopsis has been reached.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Genes and insertion lines examined for defects in seed development.
Supplemental Table S2. Cosegregation of resistance marker and seed phenotype in lines with a single mutant allele.
Supplemental Table S3. Genetic and phenotypic characterization of EMB genes identified.
Supplemental Table S4. PDE genes with seed pigment phenotypes represented in the RIKEN collection.
Supplemental Table S5. Chloroplast protein functions represented in Arabidopsis loss-of-function mutant collections.
Supplemental Table S6. EMB genes encoding chloroplast-localized proteins in Arabidopsis.
Supplemental Table S7. Genes encoding chloroplast proteins with a pigment-defective phenotype in Arabidopsis.
Supplemental Table S8. Genes encoding chloroplast proteins with other mutant phenotypes in Arabidopsis.
Acknowledgments
We thank Ian Small for valuable input on the identity of essential chloroplast genes and Rosanna Muralla for contributing to the dataset of genes with mutant phenotypes. Author contributions: D.M. designed the research and wrote the paper. N.B., J.L., C.S., and D.M. performed the research. D.M. and J.L. analyzed the data. F.M. contributed seeds and information on insertion lines in advance of publication.
References
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL. (2010) Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang WY, Hata A, Jeong IS, Umeda T, Masuda T, Chen J, Yoko I, Suwastika IN, Kim DW, Im CH, et al. (2009) AtObgC, a plant ortholog of bacterial Obg, is a chloroplast-targeting GTPase essential for early embryogenesis. Plant Mol Biol 71: 379–390 [DOI] [PubMed] [Google Scholar]
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C. (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep 5: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C. (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj K, Lin B, Mauch F. (2009) The chloroplast protein RPH1 plays a role in the immune response of Arabidopsis to Phytophthora brassicae. Plant J 58: 287–298 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D. (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WG. (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P. (2008) Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA 105: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. (2007) A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res 35: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. (2008) The pentatricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F, Sippel C, Kolb M, Vothknecht UC. (2009) Arabidopsis OBG-like GTPase (AtOBGL) is localized in chloroplasts and has an essential function in embryo development. Mol Plant 2: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM. (2005) Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221: 424–436 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T, Jr, Carrer H, Bock R. (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Garcia C, Khan NZ, Nannmark U, Aronsson H. (2010) The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J 63: 73–85 [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Somerville CR. (1990) A mutation causing imidazolinone resistance maps to the Csr1 locus of Arabidopsis thaliana. Plant Physiol 92: 1081–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Hoch B, Zeltz P, Hübschmann T, Kössel H, Börner T. (1994) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S. (1994) Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine (temporal and spatial patterns of soluble methionine accumulation). Plant Physiol 104: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. (2010) MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, van Wijk KJ. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ. (2011) Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol 155: 293–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Harrison DK, Chalupska D, Gornicki P, O’Donnell CC, Adkins SW, Haselkorn R, Williams RR. (2007) Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA 104: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA. (1998) Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA 95: 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Meinke D, Sweeney C, Muralla R. (2009) Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE 4: e7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. (1985) Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor Appl Genet 69: 543–552 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Meinke LK, Showalter TC, Schissel AM, Mueller LA, Tzafrir I. (2003) A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol 131: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Sussex IM. (1979) Embryo-lethal mutants of Arabidopsis thaliana: a model system for genetic analysis of plant embryo development. Dev Biol 72: 50–61 [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Tanaka A, Yamaguchi J. (2004) Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol 45: 781–788 [DOI] [PubMed] [Google Scholar]
- Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. (2007) Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol 144: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Akiyama K, Motohashi R, Kuromori T, Ito T, Iizumi H, Ryusui R, Sakurai T, Shinozaki K. (2010) The Chloroplast Function Database: a large-scale collection of Arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J 61: 529–542 [DOI] [PubMed] [Google Scholar]
- Okuda K, Hammani K, Tanz SK, Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. (2010) The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J 61: 339–349 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR. (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. (2001) Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J 27: 179–189 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, Lloyd CW, Doonan JH, Hills MJ. (2009) ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21: 90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR. (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y. (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P. (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte W, Töpfer R, Stracke R, Schell J, Martini N. (1997) Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA 94: 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CC, Sung TY, Li YC, Hsu SJ, Lin CL, Hsieh MH. (2010) Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant. Plant Mol Biol 73: 309–323 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Frederick RL, Sieburth LE. (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol 14: 1739–1746 [DOI] [PubMed] [Google Scholar]
- Williams PM, Barkan A. (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36: 675–686 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Goh CH, Park JH, Teyssendier de la Serve B, Kim JH, Park YI, Nam HG. (2002) Extended leaf longevity in the ore4-1 mutant of Arabidopsis with a reduced expression of a plastid ribosomal protein gene. Plant J 31: 331–340 [DOI] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Zubko MK, Day A. (1998) Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J 15: 265–271 [DOI] [PubMed] [Google Scholar]