DNA-containing cellular compartments in plant cells are the nucleus, plastids, and mitochondria. The nuclear genome, encoding approximately 29,000 to 32,000 genes, is the most common target for biotechnological applications. The number of genes encoded by the plastid and mitochondrial genomes is much smaller, approximately 120 and approximately 60, respectively. The number of processes that may be modified by engineering the organellar genomes is much higher than the number of genes would suggest. That is because approximately 10% of the nuclear gene products are targeted to plastids and approximately 10% to mitochondria (Leister, 2003), making these nucleus-encoded processes amenable to manipulation by plastome engineering. Traits that may be engineered by plastid genome manipulation are not restricted by the plant’s gene content, because incorporating new genes or gene clusters from heterologous sources may expand the plastid’s biosynthetic repertoire beyond what is provided by nature.
Engineering of the plastid genomes (plastomes; plastid DNA or ptDNA) was first accomplished in Chlamydomonas reinhardtii, a unicellular green alga (Boynton et al., 1988), followed by plastid transformation in tobacco (Nicotiana tabacum), a flowering plant species (Svab et al., 1990). Since 1988, plastid transformation has been expanded to a diverse group of species (see below). However, commercial applications are lagging behind, and currently no crops are grown commercially utilizing this technology. This review focuses on the principles of plastome engineering and highlights recent developments. Most examples will be described in tobacco, which is the model species of plastid engineering. Additional information on the biotechnological applications of plastid transformation can be found in recent reviews (Bock, 2007; Daniell et al., 2009; Cardi et al., 2010).
GENOME SORTING TO OBTAIN GENETICALLY STABLE TRANSPLASTOMIC PLANTS
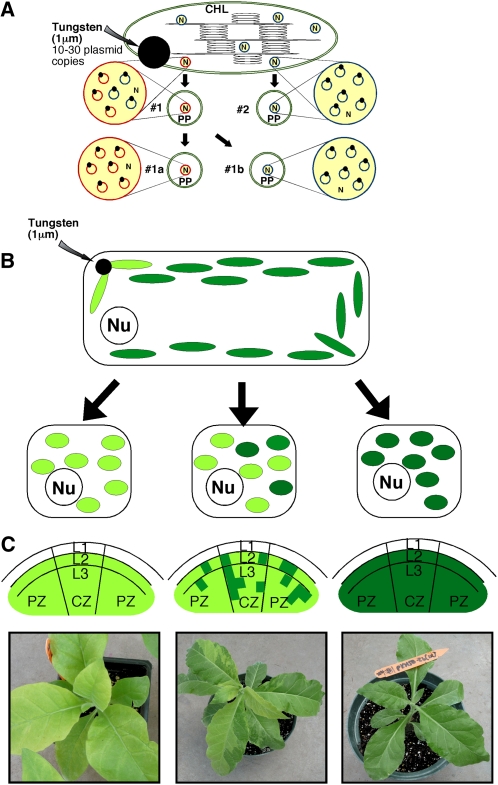
The target of the transforming DNA, the plastid genome, is highly polyploid. The number of plastids per cell and the number of ptDNA copies per plastid is dependent on the species and the cell type. For example, an Arabidopsis (Arabidopsis thaliana) leaf mesophyll cell contains about 120 chloroplasts, and these harbor approximately 1,000 to 1,700 copies of the approximately 154-kb plastid genome (Zoschke et al., 2007). Tobacco leaf cells contain a comparable number (approximately 100) of chloroplasts harboring approximately 10,000 copies of the approximately 156-kb ptDNA (Shaver et al., 2006). The ptDNA in chloroplasts is localized to membranes in clusters of approximately 10 ptDNA copies, referred to as nucleoids (Fig. 1A; Maliga, 2004). Obtaining plants with a genetically uniform population of transformed plastid genomes (transplastomes or T-ptDNA) is still the bottleneck that hinders rapid extension of the technology to new crops. Figure 1 depicts the stages of plastid sorting that ultimately yield genetically stable transplastomic plants and are discussed below.
Figure 1.
Sorting of T-ptDNA at the organelle and cellular levels yields homoplastomic plants. A, Replication and sorting of T-ptDNA at the organelle level yields homoplastomic organelles. Sorting is facilitated by the conversion of chloroplasts (CHL) to proplastids (PP), which contain only one to two nucleoids instead of 10. Wild-type ptDNA and T-ptDNA (blue circles and red circles, respectively) are anchored to membranes by proteins (black dots) in nucleoids (N). Sorting of ptDNA and T-ptDNA in heteroplastomic nucleoids (#1) yields nucleoids with only T-ptDNA (#1a) and wild-type ptDNA (#1b). For details, see Maliga (2004). B, Division and sorting of plastids yields genetically stable transplastomic plants. Sorting is accelerated by reduction from approximately 100 chloroplasts in leaf cells to approximately 10 to 14 proplastids in meristematic cells. In the cells, the nucleus (Nu) is also marked. C, The plastid genotype of long-term stem cells in the three layers (L1, L2, L3) of the shoot apex determines the plastid genotype in leaves. PZ and CZ are the peripheral and central zones, respectively. On the left is a shoot apex with T-ptDNA in all three layers and a homoplastomic plant carrying only T-ptDNA encoding the aurea spectinomycin resistance (aadA) gene. The variegated plant in the middle has cells with wild-type ptDNA and T-ptDNA in its shoot apex. The regenerated plant on the right has only wild-type ptDNA. C is modified from Lutz and Maliga (2008); the plants were described by Tungsuchat-Huang et al. (2011).
When the transforming DNA is introduced on the surface of microscopic particles (Fig. 1, A and B), only one or a few chloroplasts in a leaf cell may be damaged by the impact, and only a few of the approximately 100 ptDNA copies incorporate the transforming DNA. However, the transforming DNA carries antibiotic-detoxifying genes conferring a selective advantage to plastids that carry the T-ptDNA. Because the selective agent can most conveniently be administered in the tissue culture environment, selective enrichment for the T-ptDNA is carried out in tissue culture cells. The tissue culture medium triggers cell division, yielding meristematic cells with 10 to 14 proplastids, each of which carries only one or two nucleoids. Reduction of plastid number from 100 to 10 to 14 greatly accelerates plastid sorting during cell division, during which plastids carrying the T-ptDNA are dividing at a faster rate (Fig. 1B). Plastids carrying only the wild-type ptDNA are ultimately lost by dilution during cell division. Depicted in Figure 1 is a selection for an aurea construct that confers a pale pigment phenotype to plants (Lutz and Maliga, 2008; Tungsuchat-Huang et al., 2011).
In tobacco, selection for T-ptDNA is carried out on a shoot induction medium. Cells carrying T-ptDNA are relieved from inhibition by the toxic selective agent and will regenerate shoots on the selective medium (Fig. 2). Cells in the shoot derive from three developmental layers, each of which is the progeny of two to three slowly dividing long-term stem cells (Fig. 1C). Genetic uniformity of a regenerated plant is ensured when each of the long-term stem cells in the three developmental layers carries the same T-ptDNA. Shoots emerging from a bombarded leaf are chimeric, consisting of transplastomic and wild-type sectors. The transplastomic sectors, often very small, can be visualized by the expression of aurea transgenes that confer a golden-yellow phenotype to leaves (Fig. 1C, bottom middle). The pigment phenotype is due to posttranscriptional interference with the plastid clpP1 (ATP-dependent protease proteolytic subunit) gene expression by the aurea aadAau (aminoglycoside 3′′-adenylyltransferase, aurea) transgene. Wild-type sectors in the shoot are present because transplastomic sectors protect wild-type cells against antibiotics in culture. Cross-protection enables wild-type shoots to form on a selective medium even after two cycles of shoot regeneration. Genetically stable transplastomic plants can be obtained by collecting seeds from plants with a uniform phenotype after two cycles of plant regeneration or from branches that carry transplastomic sectors in the second leaf layer, the source of germline cells (Lutz and Maliga, 2008; Tungsuchat-Huang et al., 2011). Because heteroplastomic cells are rare even in variegated plants and are localized at the edge of sectors, the aurea and green colors typically identify homoplastomic transgenic and wild-type sectors, respectively. Plants in Figure 1C were transformed with an aurea aadA gene that is selectable in culture and gives a visual phenotype in leaves (Tungsuchat-Huang et al., 2011). In the absence of a visual marker, DNA gel-blot analysis is employed to identify plants with a uniform population of T-ptDNA molecules (Maliga, 2004).
Figure 2.
Transplastomic clones are identified as green shoots in bombarded tobacco leaf culture on spectinomycin medium. The aurea aadAau gene (Tungsuchat-Huang et al., 2011) enables greening and shoot regeneration in the culture shown here and causes intense golden-yellow leaf pigmentation in plants (Fig. 1C).
THE TECHNOLOGY OF PLASTID TRANSFORMATION
Incorporation of foreign DNA is based on homologous recombination between the targeting region of the vector and the ptDNA (Fig. 3). The transformation vectors are Escherichia coli plasmids that do not replicate in plastids; thus, the marker gene encoded in the vector will be stably expressed only if incorporated in the plastid genome by homologous recombination. The transforming DNA is introduced on the surface of microscopic (0.4–1.0 μm) gold or tungsten particles or by polyethylene glycol treatment. Biolistic DNA delivery is used when the targets are plastids in intact tissue; polyethylene glycol treatment is used for DNA introduction into protoplasts (Dix and Kavanagh, 1995). The most commonly used selective marker gene is aadA, encoding spectinomycin resistance (Svab and Maliga, 1993). Kanamycin (Carrer et al., 1993), chloramphenicol (Li et al., 2011), and the amino acid analogs 4-methylindole and 7-methyl-dl-Trp (Barone et al., 2009) have also been successfully employed as selective agents.
Figure 3.
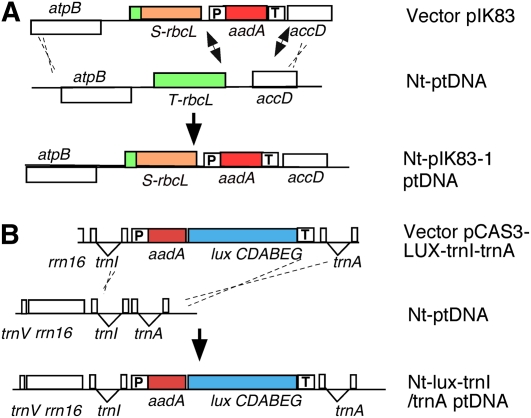
Plastid genome manipulation is based on homologous recombination between ptDNA and the targeting regions in the vector. A, Replacement of the tobacco rbcL gene (T-rbcL) with the sunflower homolog (S-rbcL). The sunflower S-rbcL is incorporated in the tobacco ptDNA only if recombination is via the atpB and accD genes (dotted lines). Recombination adjacent to aadA (arrows) confers spectinomycin resistance, but the tobacco T-rbcL is retained. Out of six transplastomic lines, three carried aadA only, two incorporated S-rbcL, and one had recombination within rbcL (Kanevski et al., 1999). B, Insertion of the lux operon in the trnI/trnA intergenic region. Note that the lux operon is transcribed from the aadA promoter and the gene cluster has only a single 3′ UTR (Krichevsky et al., 2010).
The types of plastid genome manipulations include knockout of plastid genes to probe function, replacement of plastid genes with mutant forms, and insertion of transgenes to confer novel functions. Replacement of the tobacco rbcL plastid gene (T-rbcL), encoding the Rubisco large subunit, with the sunflower (Helianthus annuus) large subunit (S-rbcL) is shown in Figure 3A (Sharwood et al., 2008). The example shown in Figure 3B is insertion of aadA and six genes (approximately 6.5 kb) of the luciferase (lux) operon in the plastid genome in the trnI-trnA intergenic region (Krichevsky et al., 2010). Thus far, this is the highest number of genes inserted in the ptDNA. In both cases, the genes of interest (S-rbcL and lux operon) were incorporated in the plastid genome by homologous recombination via the homologous flanking (“targeting”) sequences (dashed lines in Fig. 3). The recovery of T-ptDNA was subsequently facilitated by selection for the linked spectinomycin resistance (aadA) marker gene.
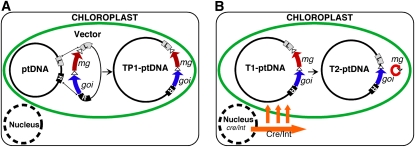
Once a uniform population of engineered T-ptDNA has been obtained, the marker genes are no longer necessary to maintain the T-ptDNA. Excision of the marker gene enables multistep transformation with the same marker gene. The metabolic burden from high-level expression of the marker gene and opposition to antibiotic resistance markers in field crops are additional reasons that make the removal of marker genes desirable. Protocols for marker gene removal employ the Cre and Int phage site-specific recombinases that excise the marker genes via flanking recombinase target sites (Fig. 4; Lutz and Maliga, 2007). Alternatively, the marker gene may be removed by recombination via flanking direct repeats (Fig. 5; Kode et al., 2006).
Figure 4.
Excision of marker genes by site-specific recombinase enzymes. A, Marker genes in the plastid transformation vectors are flanked by loxP or attP/attB sequences (triangles) that are the targets for site-specific recombinases. B, The marker genes are efficiently removed when a gene encoding a plastid-targeted Cre or Int recombinase is introduced into the nucleus by transformation or pollination (Lutz and Maliga, 2007). T1-ptDNA and T2-ptDNA refer to the marker-containing and marker-free transplastomes.
Figure 5.
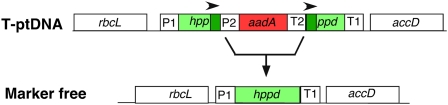
Marker-free plastids by repeat-mediated deletion of the marker gene. In the transplastome (T-ptDNA), the aadA marker gene, expressed in the P2/T2 cassette, disrupts the hppd herbicide tolerance gene encoding 4-hydroxyphenylpyruvate dioxygenase (HPPD), an enzyme in the tocopherol biosynthetic pathway. The hppd coding region is flanked by the P1/T1 cassette but is not expressed due to disruption by aadA. Note the 403-nucleotide duplicated segment (darker color) flanking the aadA. Deletion of aadA via the 403-nucleotide repeats (arrowheads) reconstitutes a functional hppd gene enabling the expression of herbicide resistance in seedlings (Dufourmantel et al., 2007; for review, see Kode et al., 2006).
Extension of the technology of plastid transformation to new crops has been more challenging than nuclear gene transformation. Although there are reports of partial success in many species, reproducible protocols for plastid transformation have been described only in tobacco (Svab and Maliga, 1993), tomato (Solanum lycopersicum; Ruf et al., 2001), petunia (Petunia hybrida; Zubko et al., 2004), potato (Solanum tuberosum; Valkov et al., 2011), soybean (Gycine max; Dufourmantel et al., 2004), lettuce (Lactuca sativa; Kanamoto et al., 2006), and cabbage (Brassica oleracea; Liu et al., 2007). Monocots as a group appear to be the most recalcitrant species.
ENGINEERING OF PLASTID TRANSGENES FOR HIGH-LEVEL PROTEIN EXPRESSION
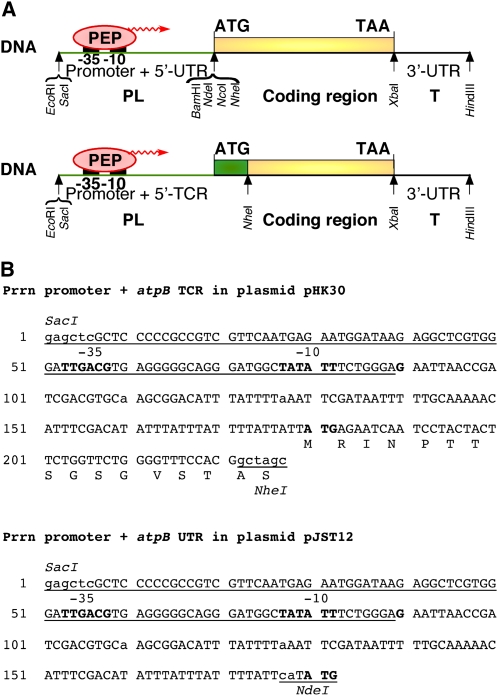
Expression of transgenes is facilitated by the availability of expression cassettes, into which coding regions can be inserted (Fig. 6). These cassettes are available in plastid transformation vectors that also function as E. coli cloning vectors, so that transformation-ready vectors can be obtained in one cloning step. The 5′ regulatory regions are provided by a PL cassette, which includes a promoter (P) and translation control sequences (leader [L]). The translation control sequences may be the mRNA 5′ untranslated region (5′ UTR) or the 5′ translation control region (5′ TCR) that includes the 5′ UTR and the coding region’s N terminus. The role of the 5′ UTR is to stabilize the mRNA and to facilitate loading of mRNAs onto the prokaryote-type 70S ribosomes. Loading of the mRNAs is facilitated by mRNA-16S rRNA interactions by a variant (GGAGG, GGAG) of the prokaryotic Shine-Dalgarno sequence found upstream of the AUG (or, much less frequently, GUG) translation initiation codon. A significant number of mRNAs, such as atpB, have no recognizable Shine-Dalgarno sequence, and 5′ UTR-binding proteins are thought to facilitate translation initiation in these reading frames. Efficient translation of some of the mRNAs is dependent on processing (Yukawa et al., 2007). The 3′ regulatory region or T cassette encodes the mRNA 3′ UTR, which typically harbors a stem-loop-type RNA secondary structure. The 3′ regulatory region is important for mRNA stability (Monde et al., 2000).
Figure 6.
Transgene assembly in cassettes for protein expression. A, Shown are schematic maps of transgenes transcribed from a PL-UTR (top) and a PL-TCR (bottom) cassette. BamHI (GGATCC), NcoI (CCATGG), and NheI (GCTAGC) restriction sites may be present in the same cassette (GGATTCCATGGCTAGC), while the NcoI and NdeI (CATATG) sites, each of which includes the translation initiation codon (ATG; in boldface), are incompatible. B, DNA sequence of the Prrn promoter with the atpB UTR (plasmid pJST12; Tregoning et al., 2003) and TCR (plasmid pHK30; Kuroda and Maliga, 2001b). Note that expression in the PL-TCR cassette yields a fusion protein with 14 amino acids derived from the plastid atpB gene and two amino acids encoded in the NheI restriction site, whereas the PL-UTR transgene encodes an unmodified protein. In boldface are shown the Prrn promoter conserved −35 (TTGACG) and −10 (TATATT) promoter elements.
Most biotechnological applications utilize the promoter (P) upstream of the plastid rRNA (rrn) operon. Prrn is the strongest plastid promoter, but its native transcripts are not translated. To enable translation, Prrn is fused with the 5′ UTR or 5′ TCR of plastid or other prokaryote-type genes. Expression of a protein using a PL-UTR cassette enables expression of the protein with its native (unmodified) N and C termini, because the expression signals are linked up with the coding region via restriction sites comprising the translation initiation and termination codons (Fig. 6B). Sometimes, using a PL-TCR fusing the protein of interest with an N-terminal peptide is the only approach that yields high-level protein expression. Examples are expression of the EPSPS enzyme from the CP4 coding region (Ye et al., 2001) or the human papillomavirus L1 capsid protein (Lenzi et al., 2008; Fig. 6B). The engineered plastid 5′ UTR (5′ TCR) is typically a truncated and mutant form of the corresponding native sequence: the length of the 5′ UTR is reduced to minimize the probability of unwanted homologous recombination (with the native UTR copy), and point mutations are introduced to remove undesirable restriction sites. For example, the highly expressed Prrn:LrbcL PL-TCR cassette (typically giving approximately 10% of total soluble protein) contains only 57 of the 189-nucleotide rbcL leader sequence and carries a point mutation to eliminate an EcoRI restriction site (Kuroda and Maliga, 2001b). The importance of the 5′ UTR/5′ TCR was shown by protein accumulation varying as much as 10,000-fold depending on the choice of translation control signals (Maliga, 2002). The most efficient translation control sequences derive from the E. coli phage T7 gene10 (T7g10; Kuroda and Maliga, 2001a), the Bacillus thuringiensis (Bt) cry9Aa2 (Chakrabarti et al., 2006), and the plastid rbcL gene in its engineered form (Kuroda and Maliga, 2001b). The highest level of protein expression in chloroplasts on record is greater than 70% of total soluble protein of a highly stable protein antibiotic in a PrrnT7g10-UTR/TrbcL cassette (Oey et al., 2009a; see below). Translation of mRNAs, at least in some cases, is dependent on intercistronic transcript processing (i.e. cleavage of polycistronic into monocistronic mRNAs). Where processing of polycistronic transcripts is required, a short sequence encoding an intercistronic expression element may be inserted to trigger intercistronic cleavage (Zhou et al., 2007).
The choice of the insertion site in the plastome may have a profound effect on the level of protein accumulation. Inserting a transgene in the repeated region of the ptDNA doubles the number of transgene copies per genome, as compared with insertions in unique regions. Insertion of transgenes between genes of a heavily transcribed operon will further increase the level of translatable mRNA, typically yielding higher protein levels (De Cosa et al., 2001).
APPLICATIONS IN AGRICULTURE
Currently, no transplastomic crops are grown commercially. However, proof of concept exists in tobacco for many potential applications. Commercial exploitation is hindered by the lack of the technology in major field crops such as maize (Zea mays), wheat (Triticum aestivum), and rice (Oryza sativa) or only recent implementation of the technology (e.g. in soybean).
The first example demonstrating the great potential of the transplastomic technology came from the expression of the Bt cry1A(c) insecticidal protein in chloroplasts (McBride et al., 1995). The coding region of the bacterial Bt genes, when expressed in the plant nucleus, yielded extremely low protein levels. It was noted that the Bt genes are AT rich as compared with plant nuclear genes, which led to the speculation that low expression was due to premature transcription termination, aberrant mRNA splicing, mRNA instability, and/or inefficient codon usage. Expression of Bt proteins from a synthetic coding region with an increased GC content dramatically increased protein yield, from undetectable to 0.2% to 0.3% of total soluble cellular protein. However, when the unmodified bacterial coding region was expressed in chloroplasts, the mRNA was stable and the cry1A(c) protein accumulated to 3% to 5% of the total soluble cellular protein, which was considered extraordinary at the time. In the meantime, several Bt proteins have been expressed in chloroplasts with a similar outcome: high-level accumulation of the Bt protein from the bacterial coding sequence. The protein levels obtained were greater than 10% in tobacco (Chakrabarti et al., 2006) and cabbage (Liu et al., 2008), and when including two open reading frames upstream of the cry2Aa2 operon, one of which encodes a putative chaperonin, the protein accumulated to up 45% of the total soluble protein (De Cosa et al., 2001).
Herbicide resistance is one of the most common commercial transgenic traits. Herbicide resistances in commercial crops are encoded in the nucleus, unavoidably allowing for occasional dissemination of the herbicide resistance by pollen. By now, resistances to the same set of herbicides have been obtained by expression of transgenes in plastids. Examples include resistance to glyphosate (Ye et al., 2001), Bialaphos/Liberty (Iamtham and Day, 2000; Lutz et al., 2001), isoxaflutole (Dufourmantel et al., 2007), and sulfonylurea herbicides (Shimizu et al., 2008). If these plastid transgenes are incorporated in commercial crops, plastid localization provides an efficient containment tool (Ruf et al., 2007; Svab and Maliga, 2007). Expression of transgenes in the plastid genome to confer herbicide resistance is an example for modification of a nucleus-encoded trait (i.e., a nucleus-encoded metabolic pathway that is sensitive to an herbicide) by plastome engineering.
Improving the efficiency of photosynthesis by engineering of the photosynthetic machinery is the holy grail of plant breeding. While the technology is available, thus far very little progress has been made. The only exception is Rubisco, nature’s main CO2-fixing enzyme, the large (catalytic) subunit of which is encoded in the plastome. Exploration of strategies has begun to examine the feasibility of supercharging photosynthesis by Rubisco engineering to drive a new green revolution (Whitney et al., 2011).
METABOLIC PATHWAY ENGINEERING
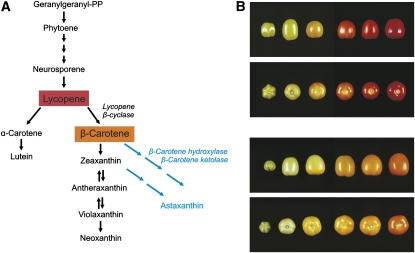
Metabolic pathway engineering offers great potential for improving the yield and nutritional quality of foodstuffs and animal feed. Moreover, it can provide an inexpensive and renewable source of raw materials for industrial processes (“green chemicals”). Attempts to engineer the carotenoid biochemical pathway by using plastid transformation have been particularly successful (Wurbs et al., 2007; Hasunuma et al., 2008; Apel and Bock, 2009; Fig. 7). Carotenoids are important antioxidants and represent an essential component of the human diet. The carotenoid β-carotene (also referred to as provitamin A) is of special importance because it provides the precursor for vitamin A, a lipid-soluble vitamin essential to all vertebrates. Vitamin A deficiency is a global health problem; therefore, increasing the provitamin A content of crops represents an important goal of breeding and genetic engineering efforts. Taking advantage of the availability of a plastid transformation protocol for tomato, a plant that accumulates massive amounts of carotenoids in fruits during the ripening process, provitamin A production in transplastomic tomatoes was attempted. Lycopene, the major storage carotenoid of ripe red tomato fruits, can be converted into β-carotene in a single-step enzymatic reaction catalyzed by lycopene β-cyclases (Fig. 7A). Lycopene β-cyclase genes from various carotenoid-synthesizing source organisms (bacteria, fungi, and plants) were fused to plastid expression signals and tested as transgenes in transplastomic tomato plants (Wurbs et al., 2007; Apel and Bock, 2009). Synthesis of β-carotene was most efficient with a plant cyclase gene borrowed from daffodil (Narcissus pseudonarcissus), with provitamin A levels reaching as much as 1 mg g−1 dry weight of the fruit (while wild-type fruits have less than 200 ng provitamin A g−1; Apel and Bock, 2009; Fig. 7B). Interestingly, expression of the daffodil lycopene β-cyclase from the tomato plastid genome not only resulted in efficient conversion of lycopene to β-carotene, it also led to a 50% increase in total carotenoid content, thus providing a welcome additional improvement in the nutritional quality of the fruits. This finding suggests that the lycopene β-cyclase reaction represents an important control point in tomato carotenoid biosynthesis, which, at least in part, determines the total flux through the pathway in the fruit.
Figure 7.
Metabolic engineering of the carotenoid pathway in transplastomic plants. A, Schematic overview of the carotenoid biosynthetic pathway. Lycopene, the major storage carotenoid in tomato, is indicated in red, and β-carotene (provitamin A) is indicated in orange. Enzymes that have been used for plastid genome engineering are given in italics. Parts of the pathway not occurring in higher plants are shown in blue. Multiple arrows denote conversions involving multiple enzymatic steps. The reversible reactions of the xanthophyll cycle are indicated by double arrows. B, Phenotypes of fruits from transplastomic tomato plants expressing a lycopene β-cyclase transgene from daffodil. Fruits from a wild-type plant (top two panels) and a transplastomic line (bottom two panels) were harvested at different ripening stages and photographed from the side and from the bottom. The orange color of the ripe transplastomic tomatoes comes from the efficient conversion of the red storage carotenoid lycopene into the orange provitamin A (β-carotene). The provitamin A levels reached 1 mg g−1 dry weight, while wild-type fruits had less than 200 ng provitamin A g−1. This figure is modified from Apel and Bock (2009).
The possibility of producing novel carotenoids was assessed in a proof-of-concept study in tobacco (Hasunuma et al., 2008). The ketocarotenoid astaxanthin is a high-value carotenoid that is extensively used as a food and feed additive (e.g. in salmon farming, where it is responsible for the orange color of the meat and accounts for up to 25% of the total costs). It also has many applications in the pharmaceutical and cosmetic industry, mainly due to its much higher antioxidant activity compared with most other carotenoids. Astaxanthin is not synthesized by higher plants but accumulates in some marine bacteria and algae. Its biosynthesis utilizes β-carotene and/or zeaxanthin as precursors and involves two enzymes: β-carotene ketolase and β-carotene hydroxylase (Fig. 7A). Coexpression of genes for the two enzymes (taken from a marine bacterium of the genus Brevundimonas) in tobacco chloroplasts resulted in astaxanthin accumulation to more than 0.5% of the dry weight of the leaves, indicating that chloroplasts can accommodate significant amounts of novel carotenoid species. Similar to lycopene β-cyclase expression in tomato, this also resulted in a substantially increased total carotenoid content in the transplastomic tobacco plants (Hasunuma et al., 2008).
Because of their ability to express operons, chloroplasts were a natural choice for the production of polyhydroxybutyric acid (PHB), requiring the expression of three bacterial genes. In the most successful case, the Ralstonia eutropha phbC-phbB-phbA genes were expressed from the T7g10 promoter PT7g10 (Lössl et al., 2005). The polycistronic mRNA was transcribed by a nucleus-encoded, plastid-targeted T7 RNA polymerase. The first gene, phbC, was translated from the T7g10 leader; phbB and phbA were translated from the bacterial intergenic sequences. Production of PHB in chloroplasts indicates that there is sufficient conservation between the plastid and prokaryotic translation machineries to translate the bacterial proteins from the native mRNAs. Toxicity associated with PHB production was overcome by expressing the plastid-targeted T7 RNA polymerase from an ethanol-inducible promoter.
Construction of truly autonomously luminescent plants required the expression of a fully functional bacterial luciferase pathway consisting of six genes (luxCDABEG; Krichevsky et al., 2010). Again, luxC, the first gene of the lux operon, was expressed from the plastid PrrnLrbcL promoter/leader cassette, and the rest of the lux operon genes were expressed from their native translation control sequences. Emission of visible light detectable by the naked eye attested to the plastid’s ability to properly interpret the prokaryotic expression signals.
Part of lipid biosynthesis takes place inside plastids, making it amenable to engineering by plastid transformation. Overexpression of the plastid accD gene, encoding the β-carboxyl transferase subunit of acetyl-CoA carboxylase, increased leaf lipid content (Madoka et al., 2002). Expression of a Δ9-desaturase gene in tobacco chloroplasts from either the wild potato species Solanum commersonii or the cyanobacterium Anacystis nidulans resulted in altered fatty acid profiles and an increase in their unsaturation level both in leaves and seeds (Craig et al., 2008). Given the interest in biofuel production, increasing lipid content by plastid engineering will remain a subject of exploration.
CHLOROPLASTS AS BIOREACTORS FOR MOLECULAR FARMING
Arguably, the most alluring feature of transplastomic plants is their enormous capacity to accumulate foreign proteins (Maliga, 2004; Bock, 2007; Daniell et al., 2009). This is, at least in part, due to the high number of chloroplasts per cell, the large volume of the cell occupied by the chloroplast compartment, and the high copy number of the plastid genome, with hundreds or even thousands of copies being present in a single cell. The carbon dioxide-fixing enzyme Rubisco provides a case in point for the high protein accumulation capacity of the chloroplast. Its large subunit is encoded in the plastid genome, and in C3 plants, the Rubisco enzyme can accumulate to more than 20% to 30% of the total soluble protein in leaves (Whitney et al., 2011).
High yields of recombinant protein are particularly crucial to the production of pharmaceutical proteins in plants, an area of biotechnology commonly referred to as molecular farming. Proteinaceous biopharmaceuticals include, for example, antibodies, vaccines, and various antimicrobials. Pharmaceutical proteins can either be purified from the plant or, in the case of vaccines, expressed in edible plants and administered orally. In both cases, high expression levels are of pivotal importance. The costs of downstream processing usually make up by far the largest fraction of the costs for plant-made pharmaceuticals, and protein purification costs are, in general, negatively correlated with expression levels. High protein accumulation levels are absolutely essential for the production of edible vaccines, because stimulation of the mucosal immune system generally requires much higher doses of the antigen than conventional vaccination by injection into the bloodstream.
With the concept of oral vaccination in mind, expression of a large number of antigens has been attempted in chloroplasts (for recent reviews, see Bock and Warzecha, 2010; Cardi et al., 2010). Expression levels have been quite variable, ranging from nearly undetectable to as much as 40% of the total soluble protein (Tregoning et al., 2003; Zhou et al., 2008). However, so far, very few of the chloroplast-produced antigens have successfully been tested in oral vaccination experiments (Tregoning et al., 2003; Davoodi-Semiromi et al., 2010). Moreover, all studies conducted so far have been confined to small experimental animals (usually mice), and the concept of oral vaccination with transplastomic plants still awaits its validation in large mammals and ultimately in humans.
Extraordinarily high expression levels have recently been obtained with protein antibiotics derived from phage lytic proteins (so-called endolysins). These proteins efficiently digest the cell wall of pathogenic bacteria and, for this reason, have considerable potential as next-generation antibiotics suitable to control pathogens that have acquired resistance to most conventional antibiotics that are currently in clinical use. The chloroplast may be an ideal site for the production of these protein antibiotics because (1) chloroplasts do not have a cell wall and, therefore, should be able to accommodate large amounts of these proteins and (2) chloroplasts possess a very similar set of proteases as bacteria. The latter is important, considering that phage endolysin proteins have evolved high levels of resistance to degradation by prokaryotic proteases. When expressed from the tobacco plastid genome, endolysins (targeted against pathogenic streptococci, including Streptococcus pneumoniae, the causal agent of pneumonia) indeed turned out to be extremely stable (Oey et al., 2009a, 2009b). Maximum protein accumulation reached unprecedented levels of up to more than 70% of the total soluble protein of the plant (Oey et al., 2009a, 2009b). Importantly, the plastid-produced endolysins were highly active and efficiently killed the target pathogens.
The attainable high expression levels also make transplastomic plants an attractive production platform for other high-value proteins, such as industrial enzymes. With the growing interest in biofuels, enzymes and enzyme cocktails that are potentially suitable to digest lignocellulosic biomass into fermentable sugars have received particular attention. Several recent studies have demonstrated that the chloroplast compartment can accumulate very high levels of these enzymes (including endocellulases, exocellulases, β-glucosidases, xyloglucanases, pectate lyases, and cutinases; Yu et al., 2007; Verma et al., 2010; Kim et al., 2011), although extreme overaccumulation can cause mutant phenotypes (Petersen and Bock, 2011). For industrial enzymes expressed to high levels in plants, protein purification to homogeneity is often unnecessary and crude protein extracts can be used. Therefore, transplastomic plants expressing cell wall-degrading enzymes can potentially provide a cheap source of enzymes for the production of cellulosic ethanol. However, key issues in the conversion of lignocellulosic biomass into fuels remain to be addressed in order to make biofuels an economically viable solution. First and foremost, the requirement for expensive (e.g. thermoacidic) pretreatments of the biomass needs to be bypassed. This will require efficient methods for lignin decomposition and subsequent removal or degradation of the released phenolic compounds.
EXPRESSION OF RECOMBINANT PROTEINS IN CHLAMYDOMONAS CHLOROPLASTS
A number of recent studies have explored the potential in molecular farming of the unicellular green alga Chlamydomonas (Coragliotti et al., 2011). Although microbial cultivation in bioreactors is considerably more expensive than growth of higher plants in soil, these extra costs are less of an issue in molecular farming compared with the food sector, especially if one considers that pharmaceutical proteins are usually high-value products and are needed in limited quantities. Moreover, production in fermenters offers significant advantages, especially by (1) providing a fully contained production facility, (2) relying on established procedures in microbial biotechnology, and (3) potentially posing lower regulatory hurdles. Considerable progress has recently been made with expressing pharmaceutical proteins from the chloroplast genome of Chlamydomonas. In general, the capacity of the Chlamydomonas chloroplast to synthesize and/or accumulate foreign proteins appears to be much lower than that of higher plants. Unlike in flowering plants, codon optimization of transgene-coding regions is essential (Franklin et al., 2002). Levels of protein expression are 2% to 3% of total soluble cellular protein (Rasala et al., 2010), although 10% and 20% protein expression levels were also obtained from the psbA expression signals in nonphotosynthetic psbA knockout algae (Manuell et al., 2007; Surzycki et al., 2009). High-level recombinant protein expression in the psbA knockout plastids may be due to compensatory overactivation of transgene expression by psbA-specific trans-acting factor(s) and/or the lack of competition for these factors by the native mRNA. Incorporation of psbA at an ectopic location and expression from heterologous signals enabled photosynthetic competence and accumulation of the recombinant protein at a somewhat reduced level (Manuell et al., 2007). Current efforts focus on the construction of Chlamydomonas strains in which limitations of protein expression have been removed by nuclear mutations (Neupert et al., 2009; Michelet et al., 2011).
FUTURE DIRECTIONS
Plastid engineering is currently pursued in three distinct systems: microalgae, bryophytes, and flowering plants. Plastid transformation was first implemented in a microalga, Chlamydomonas. However, biotechnology-oriented research with Chlamydomonas was initiated relatively late, when recognition of the importance of codon modification enabled the expression of recombinant proteins at commercially viable levels (Franklin et al., 2002). Chlamydomonas is likely to remain a useful expression vehicle for pharmaceutical proteins. At the same time, the hunt is on for new species that may be more suitable for biofuel production.
Plastid transformation in the bryophytes Physcomitrella patens (Sugiura and Sugita, 2004) and Marchantia polymorpha (Chiyoda et al., 2007) is relatively recent and has not yet been explored for the expression of recombinant proteins. However, by targeted nuclear gene replacement, Physcomitrella strains were created with nonimmunogenic humanized glycan patterns and the moss is grown in a contained tissue culture system for recombinant protein production. These photobioreactors were proven to be superior to currently used mammalian cell lines in producing antibodies with enhanced effectiveness (Decker and Reski, 2007). Thus, a contained production system is already available. Plastid-based protein expression will further enhance the utility of the system.
A major task in flowering plants will be the implementation of the technology in major crops, a requirement for agronomic applications. Implementation in new crops will depend on the availability of a genetic line that is suitable for repeated cycles of plant regeneration (to attain homoplasmy) and a suitable selectable marker gene, as discussed above. As far as industrial and pharmaceutical applications are concerned, in view of cost considerations and scalability, flowering plans will be the likely source of bulk industrial enzymes. The tobacco plastid expression system is already suitable to perform this role. However, it is not optimal for the production of edible vaccines due to its alkaloid content. From the available choices, the efficient lettuce system could serve as a vehicle for the expression of oral vaccines (Ruhlman et al., 2007). There is a need for robust, high-level regulated gene expression so that vegetative growth and the production phases can be separated and the production of proteins induced prior to harvest. Prototypes of such regulatory elements are metabolite-activated riboswitches (Verhounig et al., 2010) and other inducible expression systems (Tungsuchat et al., 2006), which, however currently still lack the robustness that is required for practical applications.
References
- Apel W, Bock R. (2009) Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol 151: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone P, Zhang XH, Widholm JM. (2009) Tobacco plastid transformation using the feedback-insensitive anthranilate synthase [alpha]-subunit of tobacco (ASA2) as a new selectable marker. J Exp Bot 60: 3195–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. (2007) Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol 18: 100–106 [DOI] [PubMed] [Google Scholar]
- Bock R, Warzecha H. (2010) Solar-powered factories for new vaccines and antibiotics. Trends Biotechnol 28: 246–252 [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al. (1988) Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538 [DOI] [PubMed] [Google Scholar]
- Cardi T, Lenzi P, Maliga P. (2010) Chloroplasts as expression platforms for plant-produced vaccines. Expert Rev Vaccines 9: 893–911 [DOI] [PubMed] [Google Scholar]
- Carrer H, Hockenberry TN, Svab Z, Maliga P. (1993) Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol Gen Genet 241: 49–56 [DOI] [PubMed] [Google Scholar]
- Chakrabarti SK, Lutz KA, Lertwiriyawong B, Svab Z, Maliga P. (2006) Expression of the cry9Aa2 B.t. gene in tobacco chloroplasts confers resistance to potato tuber moth. Transgenic Res 15: 481–488 [DOI] [PubMed] [Google Scholar]
- Chiyoda S, Linley PJ, Yamato KT, Fukuzawa H, Yokota A, Kohchi T. (2007) Simple and efficient plastid transformation system for the liverwort Marchantia polymorpha L. suspension-culture cells. Transgenic Res 16: 41–49 [DOI] [PubMed] [Google Scholar]
- Coragliotti AT, Beligni MV, Franklin SE, Mayfield SP. (2011) Molecular factors affecting the accumulation of recombinant proteins in the Chlamydomonas reinhardtii chloroplast. Mol Biotechnol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig W, Lenzi P, Scotti N, De Palma M, Saggese P, Carbone V, McGrath Curran N, Magee AM, Medgyesy P, Kavanagh TA, et al. (2008) Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res 17: 769–782 [DOI] [PubMed] [Google Scholar]
- Daniell H, Singh ND, Mason H, Streatfield SJ. (2009) Plant-made vaccine antigens and biopharmaceuticals. Trends Plant Sci 14: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi-Semiromi A, Schreiber M, Nalapalli S, Verma D, Singh ND, Banks RK, Chakrabarti D, Daniell H. (2010) Chloroplast-derived vaccine antigens confer dual immunity against cholera and malaria by oral or injectable delivery. Plant Biotechnol J 8: 223–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Reski R. (2007) Moss bioreactors producing improved biopharmaceuticals. Curr Opin Biotechnol 18: 393–398 [DOI] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix PJ, Kavanagh TA. (1995) Transforming the plastome: genetic markers and DNA delivery systems. Euphytica 85: 29–34 [Google Scholar]
- Dufourmantel N, Dubald M, Matringe M, Canard H, Garcon F, Job C, Kay E, Wisniewski JP, Ferullo JM, Pelissier B.et al (2007) Generation and characterization of soybean and marker-free tobacco plastid transformants over-expressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol J 5: 118–133 [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garçon F, Peltier G, Ferullo JM, Tissot G. (2004) Generation of fertile transplastomic soybean. Plant Mol Biol 55: 479–489 [DOI] [PubMed] [Google Scholar]
- Franklin S, Ngo B, Efuet E, Mayfield SP. (2002) Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J 30: 733–744 [DOI] [PubMed] [Google Scholar]
- Hasunuma T, Miyazawa S, Yoshimura S, Shinzaki Y, Tomizawa K, Shindo K, Choi SK, Misawa N, Miyake C. (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55: 857–868 [DOI] [PubMed] [Google Scholar]
- Iamtham S, Day A. (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18: 1172–1176 [DOI] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa K. (2006) Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res 15: 205–217 [DOI] [PubMed] [Google Scholar]
- Kanevski I, Maliga P, Rhoades DF, Gutteridge S. (1999) Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol 119: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kavas M, Fouad WM, Nong G, Preston JF, Altpeter F. (2011) Production of hyperthermostable GH10 xylanase Xyl10B from Thermotoga maritima in transplastomic plants enables complete hydrolysis of methylglucuronoxylan to fermentable sugars for biofuel production. Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. (2006) Isolation of precise plastid deletion mutants by homology-based excision: a resource for site-directed mutagenesis, multi-gene changes and high-throughput plastid transformation. Plant J 46: 901–909 [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Meyers B, Vainstein A, Maliga P, Citovsky V. (2010) Autoluminescent plants. PLoS ONE 5: e15461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2001a) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2001b) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol 125: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Lenzi P, Scotti N, Alagna F, Tornesello ML, Pompa A, Vitale A, De Stradis A, Monti L, Grillo S, Buonaguro FM.et al (2008) Translational fusion of chloroplast-expressed human papillomavirus type 16 L1 capsid protein enhances antigen accumulation in transplastomic tobacco. Transgenic Res 17: 1091–1102 [DOI] [PubMed] [Google Scholar]
- Li W, Ruf S, Bock R. (2011) Chloramphenicol acetyltransferase as selectable marker for plastid transformation. Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Liu CW, Lin CC, Chen JJ, Tseng MJ. (2007) Stable chloroplast transformation in cabbage (Brassica oleracea L. var. capitata L.) by particle bombardment. Plant Cell Rep 26: 1733–1744 [DOI] [PubMed] [Google Scholar]
- Liu CW, Lin CC, Yiu JC, Chen JJ, Tseng MJ. (2008) Expression of a Bacillus thuringiensis toxin (cry1Ab) gene in cabbage (Brassica oleracea L. var. capitata L.) chloroplasts confers high insecticidal efficacy against Plutella xylostella. Theor Appl Genet 117: 75–88 [DOI] [PubMed] [Google Scholar]
- Lössl A, Bohmert K, Harloff H, Eibl C, Mühlbauer S, Koop HU. (2005) Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol 46: 1462–1471 [DOI] [PubMed] [Google Scholar]
- Lutz KA, Knapp JE, Maliga P. (2001) Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol 125: 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Maliga P. (2007) Construction of marker-free transplastomic plants. Curr Opin Biotechnol 18: 107–114 [DOI] [PubMed] [Google Scholar]
- Lutz KA, Maliga P. (2008) Plastid genomes in a regenerating tobacco shoot derive from a small number of copies selected through a stochastic process. Plant J 56: 975–983 [DOI] [PubMed] [Google Scholar]
- Madoka Y, Tomizawa KI, Mizoi J, Nishida I, Nagano Y, Sasaki Y. (2002) Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol 43: 1518–1525 [DOI] [PubMed] [Google Scholar]
- Maliga P. (2002) Engineering the plastid genome of higher plants. Curr Opin Plant Biol 5: 164–172 [DOI] [PubMed] [Google Scholar]
- Maliga P. (2004) Plastid transformation in higher plants. Annu Rev Plant Biol 55: 289–313 [DOI] [PubMed] [Google Scholar]
- Manuell AL, Beligni MV, Elder JH, Siefker DT, Tran M, Weber A, McDonald TL, Mayfield SP. (2007) Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol J 5: 402–412 [DOI] [PubMed] [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P. (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology (NY) 13: 362–365 [DOI] [PubMed] [Google Scholar]
- Michelet L, Lefebvre-Legendre L, Burr SE, Rochaix JD, Goldschmidt-Clermont M. (2011) Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol J (in press) [DOI] [PubMed] [Google Scholar]
- Monde RA, Greene JC, Stern DB. (2000) The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation, and translation in tobacco chloroplasts. Plant Mol Biol 44: 529–542 [DOI] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. (2009) Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J 57: 1140–1150 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. (2009a) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Scharff LB, Kreikemeyer B, Bock R. (2009b) Plastid production of protein antibiotics against pneumonia via a new strategy for high-level expression of antimicrobial proteins. Proc Natl Acad Sci USA 106: 6579–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Bock R. (2011) High-level expression of a suite of thermostable cell wall-degrading enzymes from the chloroplast genome. Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Lee PA, Jager M, Cardoso RM, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M.et al (2010) Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J 8: 719–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. (2001) Stable genetic transformation of tomato plastids: foreign protein expression in fruit. Nat Biotechnol 19: 870–875 [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. (2007) Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA 104: 6998–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. (2007) Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts: oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J 5: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM. (2008) The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ. (2006) Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224: 72–82 [DOI] [PubMed] [Google Scholar]
- Shimizu M, Goto M, Hanai M, Shimizu T, Izawa N, Kanamoto H, Tomizawa K, Yokota A, Kobayashi H. (2008) Selectable tolerance to herbicides by mutated acetolactate synthase genes integrated into the chloroplast genome of tobacco. Plant Physiol 147: 1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura C, Sugita M. (2004) Plastid transformation reveals that moss tRNA(Arg)-CCG is not essential for plastid function. Plant J 40: 314–321 [DOI] [PubMed] [Google Scholar]
- Surzycki R, Greenham K, Kitayama K, Dibal F, Wagner R, Rochaix JD, Ajam T, Surzycki S. (2009) Factors effecting expression of vaccines in microalgae. Biologicals 37: 133–138 [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. (1990) Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA 87: 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. (2007) Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA 104: 7003–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning JS, Nixon P, Kuroda H, Svab Z, Clare S, Bowe F, Fairweather N, Ytterberg J, van Wijk KJ, Dougan G.et al (2003) Expression of tetanus toxin fragment C in tobacco chloroplasts. Nucleic Acids Res 31: 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungsuchat T, Kuroda H, Narangajavana J, Maliga P. (2006) Gene activation in plastids by the CRE site-specific recombinase. Plant Mol Biol 61: 711–718 [DOI] [PubMed] [Google Scholar]
- Tungsuchat-Huang T, Slivinski KM, Sinagawa-Garcia SR. (2011) Visual spectinomycin resistance gene for facile identification of transplastomic sectors in tobacco leaves. Plant Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Valkov VT, Gargano D, Manna C, Formisano G, Dix PJ, Gray JC, Scotti N, Cardi T. (2011) High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5′ and 3′ regulatory sequences. Transgenic Res 20: 137–151 [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R. (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Kanagaraj A, Jin S, Singh ND, Kolattukudy PE, Daniell H. (2010) Chloroplast-derived enzyme cocktails hydrolyse lignocellulosic biomass and release fermentable sugars. Plant Biotechnol J 8: 332–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. (2011) Advancing our understanding and capacity to engineer nature’s CO2 sequestering enzyme, Rubisco. Plant Physiol 155: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R. (2007) Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J 49: 276–288 [DOI] [PubMed] [Google Scholar]
- Ye GN, Hajdukiewicz PTJ, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25: 261–270 [DOI] [PubMed] [Google Scholar]
- Yu LX, Gray BN, Rutzke CJ, Walker LP, Wilson DB, Hanson MR. (2007) Expression of thermostable microbial cellulases in the chloroplasts of nicotine-free tobacco. J Biotechnol 131: 362–369 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Kuroda H, Sugiura M. (2007) A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. Plant J 49: 367–376 [DOI] [PubMed] [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AM, Maloney AP, Kavanagh TA, Gray JC, Bock R. (2008) High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6: 897–913 [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R. (2007) Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J 52: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]
- Zubko MK, Zubko EI, van Zuilen K, Meyer P, Day A. (2004) Stable transformation of petunia plastids. Transgenic Res 13: 523–530 [DOI] [PubMed] [Google Scholar]