Abstract
Large-scale phenotypic screening presents challenges and opportunities not encountered in typical forward or reverse genetics projects. We describe a modular database and laboratory information management system that was implemented in support of the Chloroplast 2010 Project, an Arabidopsis (Arabidopsis thaliana) reverse genetics phenotypic screen of more than 5,000 mutants (http://bioinfo.bch.msu.edu/2010_LIMS; www.plastid.msu.edu). The software and laboratory work environment were designed to minimize operator error and detect systematic process errors. The database uses Ruby on Rails and Flash technologies to present complex quantitative and qualitative data and pedigree information in a flexible user interface. Examples are presented where the database was used to find opportunities for process changes that improved data quality. We also describe the use of the data-analysis tools to discover mutants defective in enzymes of leucine catabolism (heteromeric mitochondrial 3-methylcrotonyl-coenzyme A carboxylase [At1g03090 and At4g34030] and putative hydroxymethylglutaryl-coenzyme A lyase [At2g26800]) based upon a syndrome of pleiotropic seed amino acid phenotypes that resembles previously described isovaleryl coenzyme A dehydrogenase (At3g45300) mutants. In vitro assay results support the computational annotation of At2g26800 as hydroxymethylglutaryl-coenzyme A lyase.
Large-scale genetic screening projects are now possible in Arabidopsis (Arabidopsis thaliana; Boyes et al., 2001; Van Eenennaam et al., 2003; Jander et al., 2004; Myouga et al., 2010) and other model plant species (Schauer et al., 2006; Yu et al., 2008) due to the availability of large collections of immortal genetic resources including homozygous gene knockouts (Alonso et al., 2003; O’Malley and Ecker, 2010; Williams-Carrier et al., 2010), randomly generated mutant families (Menda et al., 2004), nearly isogenic germplasm (Eshed and Zamir, 1995; http://zamir.sgn.cornell.edu/Qtl/il_story.htm), and populations of natural variants or breeding lines (Yu et al., 2008). It is possible to screen thousands of plants using modern high-throughput techniques ranging from quantitative phenotypes such as small molecule or gene expression analysis to qualitative traits including assessment of morphology under varied environmental conditions. The potential for discovery using this approach is great, but the informatics challenges of conducting this type of research are formidable due to the enormous numbers of samples analyzed and the amount of data generated (Baxter et al., 2007).
There are several major challenges in large-scale phenotypic screens that do not arise in more traditional genetics. The most important is developing a laboratory information management system (LIMS) and laboratory workflow ensuring that data are accurately recorded and correctly associated with the plant sample analyzed. The second is to implement relevant statistical analysis of the mass of data collected over months or years. Perhaps the biggest challenge is presentation of diverse types of phenotypic data to researchers in a way that enables them to make scientific discoveries. We present here a description of a software suite designed to assist users in collecting and analyzing such data sets.
This software was developed to support a study of several thousand nuclear genes, most of which are predicted to encode proteins targeted to the chloroplast (Chloroplast 2010 Project; http://www.plastid.msu.edu/; Lu et al., 2008; Ajjawi et al., 2010). The goal of the Chloroplast 2010 Project was to obtain phenotypic information that would inform the creation of hypotheses on the function of as many of these genes as possible using parallel phenotypic analysis of homozygous sequence-indexed T-DNA insertion lines (O’Malley and Ecker, 2010). Both descriptive morphological and quantitative metabolite screens were used to determine the effects of each gene disruption. To maximize the number of lines screened, each assay was limited to two biological replicates per line, making the collection of high-quality data even more important. The collected data include controlled text descriptions, photographs, and quantitative data of multiple mutant plants from each mutant line, and each combined plant and seed sample was ultimately associated with 85 discrete observations.
Scaling the number of samples processed in a laboratory from hundreds to tens of thousands requires more than hiring personnel and purchasing additional equipment. The data collection methods that work well in the typical academic laboratory do not scale to handle the larger sample numbers. The repetitive nature of the work typically results in an unacceptably high error rate due to user fatigue. The labeling of tubes, transfer of materials between containers, as well as addition of reagents to containers is error prone under any circumstance and even more so under these conditions. Recording large volumes of instrument data into laboratory notebooks or spreadsheets creates many errors. The lack of controlled vocabularies results in difficulties comparing data across multiple investigators and over prolonged periods of time. The distribution of data across many notebooks or computer files impedes the analysis of the collected data.
Our design goal for this project was to implement workstations integrated with a Web-based LIMS to minimize these problems. By integrating the workflow for each specific task into a custom-designed LIMS and designing the physical environment to assist the workers in managing the flow of materials through each task, we believe that we have significantly improved our ability to collect error-free data.
The second informatics challenge for this project was to present these very large data sets in a form that allows researchers who are neither skilled in computer programming nor familiar with the intricacies of the project to identify interesting phenotypes. This was addressed primarily by allowing the user to sort and filter the data using a custom query tool and by presenting the large number of assays for each mutant plant using an efficient and flexible graphical interface. This interface uses features of the Ruby on Rails framework (RoR; Thomas et al., 2006) and Flash objects to coordinate the presentation of controlled vocabulary terms, images, graphs, and chromatographs. The major design goal for this interface was to minimize the number of interactions with the system required to obtain a subset of the data needed to develop a hypothesis. However, the interface needs to present sufficient data for the user to evaluate the results while at the same time see larger scale interactions or trends that may be present in the data. We used animated graphs and coordinated changes in the set of displayed data to achieve these conflicting design goals. An example is presented where the data-analysis tools led to the discovery that mutants in several steps of Leu catabolism accumulate multiple amino acids in the seed.
RESULTS AND DISCUSSION
Data Entry
The high-throughput functional genomics project served by this database (www.plastid.msu.edu) required the planting, growth, and leaf and seed harvesting of more than 5,000 mutant lines, resulting in more than 1.5 million discrete observations. Given the magnitude of the project, it was critical to simplify the workflow while reducing the error rate of sample tracking and phenotypic data entry to the lowest possible level (for summary schematic representations of the laboratory workflows and quality assurance/quality control approaches, see Supplemental Fig. S1).
This challenge was approached in two related ways: by careful design of the work areas and processes as well as through software design. The laboratory workstation was designed to work with the data entry software to minimize common process errors such as switching samples and to create a consistent and logical workflow. Barcodes, checkboxes, and drop-downs were employed wherever possible to avoid the diverse types of errors and inconsistencies that can arise from hand-typed data entry (Figs. 1 and 2). The data entry interfaces were designed to resemble the laboratory environment; for example, showing a representation of the flat of 32 pots (Fig. 2). Barcoded pots, flats, and sample containers were used throughout the workflow, eliminating the need to choose specific prelabeled containers. Instead, the database prompts the user to scan a barcode on a seed tube, pot containing a plant, or tube containing a tissue sample and then scan the barcode on a randomly chosen empty recipient container. Because the sample associated with the first barcode has an identity in the database, the LIMS system creates an association with the sample now in the previously empty target container. User log-ins and data entry time stamps were used to identify operator errors or causes of other systematic errors such as time of day or order in which manipulations were made (see below). This information is vital to improving the design of the laboratory process and software and was collected for all data entry sessions during the project.
Figure 1.
Examples of pull-down menus used to capture qualitative phenotypic data in the LIMS.
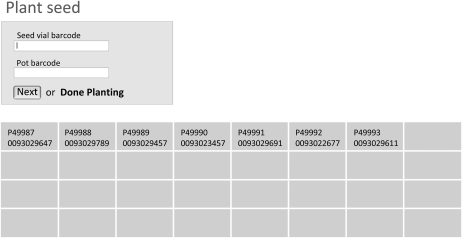
Figure 2.
LIMS interface for planting seeds. To ensure correct labeling of the plant pot, (1) the Seed vial barcode is scanned to identify the seeds being planted, and (2) the Pot barcode is next scanned to create an association between seed stock and the plant that will grow. Once the operation is completed, the pot and seed stock identities are recorded in a cell of the 8 × 4 matrix, which shows the technician where the completed pot should be placed within the flat. Once 32 pots are planted to a flat, a new flat barcode must be scanned before more pots can be planted.
As an example of the type of workflow designed for the project, Figure 2 and Supplemental Figure S2 show how the interface is used when planting seeds into a pot. The session is initiated by establishing a database identity for the 32-place flat by scanning a barcode and recording the type of assays to be performed on the plants as well as the growth chamber in which the plants will be grown. The barcode on the first seed vial is scanned to identify the seed stock (“Seed vial barcode” in Fig. 2), and the barcode on an unplanted pot (“Pot barcode” in Fig. 2) is scanned next; this creates an association in the database between the two barcodes. Once the scanned data are submitted, the planting flat template is filled with the pot and seed identifiers to indicate where to place the pot that was just planted and to confirm the association for the user.
An error-checking process is initiated by the database each time a barcode is scanned. During planting, the software checks the database to verify that the seed stock was not already used during the current session and that the pot barcode was not previously scanned. This ensures that each sample association is unique; if this rule is violated, the LIMS generates an error message on the user interface and prevents the input of data into the database. In addition, the format of each barcode is checked to be sure that the correct types of containers are entered and associated; for example, a pot barcode cannot be entered in the flat barcode data entry window. Similar error checks were designed for every type of data entry, ensuring unique data associations for all seeds, plants, or samples.
Separation of the sequence of user actions is a critical element in workflow design, which is complementary to the database error prevention features (Supplemental Fig. S2). Three key areas must be present and physically separated in the work area: (1) containers or samples not yet entered in the database; (2) those objects currently being manipulated; and (3) items that are entered and finished. This configuration minimizes mistakes where database error checks cannot: skipped samples, for example, or accidental use of a container other than the one scanned. We found that this arrangement works best when data are entered by two users working in tandem, since it allows collaborative error avoidance and checking.
Capture and Analysis of Qualitative Data
Whole plant, seed, and chloroplast morphological characteristics were captured by complementary approaches. The software allows the user to record observations about morphology using checkboxes and pull-down menus at the same time that photographs are taken (Fig. 1). This enforces the use of a controlled vocabulary, which permits data to be compared and queried consistently (see “Querying the Data” below). Image files are created offline, creating an increased possibility of sample mistracking. To reduce this hazard, file upload to the database is facilitated by establishing image file names based upon the sample container barcode identifier. For example, an image file of a plant is named by scanning the pot barcode for that plant’s pot, and the LIMS system converts the pot name into a photograph identifier.
Once acquired, the images can be redisplayed to create a visual representation of the cohort of samples (flat of plants, box of seeds, set of samples processed for chloroplast morphology analysis), and our project used this capability to allow quality assessment. For example, an individual familiar with the assay can review the controlled vocabulary assessments of a relatively inexperienced student or technician and make changes in annotation (Supplemental Fig. S3). The ability to review all data helps to ensure that the quality of the annotation is uniform over time and across different annotators. Both time and user information are stored for the most recent edit.
Capture and Analysis of Quantitative Data
Three assays in the project produce quantitative data collected from different analytical instruments. To minimize transcription errors, samples are processed for metabolite analysis using barcoded containers and procedures similar to those described above. For example, plant identity is captured by scanning a pot barcode, a leaf is removed and placed directly on an analytical balance, and the sample mass is recorded directly to the database. The leaf is then moved to a barcoded tube whose identity is established by the LIMS by scanning. This one-to-one chain of identity is established for the sample based on the barcode each time a laboratory manipulation requires that it is moved to a new container all the way until quantitative metabolite data are obtained. This ensures that users are never required to transfer a sample to a prespecified “correct” container.
As is customary with most instruments used for analysis of high-throughput metabolite analysis, the gas chromatography-flame ionization and liquid chromatography-tandem mass spectrometry instruments employ autosamplers. A barcode scanner is used to construct the sample list for the autosampler, and the software used by the analytical instruments associates the metabolite data with the sample identifiers. Loading software scripts are employed to process the data generated from the instruments and directly load the results into the database.
To permit comparisons of results from plants grown in the microenvironments of different flats, we converted quantitative data to z-scores based on median absolute deviation of data from the flat in which a plant was grown (Rousseeuw and Croux, 1993; Lu et al., 2008). This statistic was chosen for two main reasons. First, using z-scores, which normalize the median absolute deviation, allows comparisons across plants grown in different flats and at different times, simplifying database queries. Second, a median-based statistic reduces the impact of individual samples from a flat that have one or more values dramatically different from others in its cohort, either due to mutation or environmental effects.
Querying the Data
One of the biggest challenges of a project that generates a large amount of diverse phenotypic data is to give consumers of the information an efficient mechanism to make discoveries about potentially interesting plants. Among the hurdles confronting the researcher are the following: (1) evaluating noisy data due to biological and process variation that inevitably occurs over large numbers of samples; (2) the variable number of samples analyzed for different mutants (two for most lines and larger numbers for process controls and lines of interest to project personnel and collaborators); (3) the different numbers of homozygous alleles available for different target genes (Ajjawi et al., 2010); and (4) variations in T-DNA insertion locations for different alleles.
Two general approaches are provided to explore the phenotypic data, and the search page for both is found by open-access login through http://bioinfo.bch.msu.edu/2010_LIMS. The first query method is “gene centric,” allowing the researcher to examine information for mutants defective in one or more specified genes or harboring particular mutations (“Search by Query Terms”). The second is to conduct an in silico “mutant screen” by searching for plants that are aberrant for one trait or a combination of phenotypes (“Search by Assay Results”). This query interface provides the researcher with the opportunity to choose the level of stringency of the query (Fig. 3). The main choices are (1) whether multiple biological replicates need to fit the criteria; (2) the selection of specific controlled vocabulary terms for morphology and chlorophyll fluorescence measurements; and (3) setting the statistical threshold for quantitative assays. In addition, simple Boolean logic is available for creating more complex queries with multiple search terms (e.g. looking for lines with pleiotropic phenotypes).
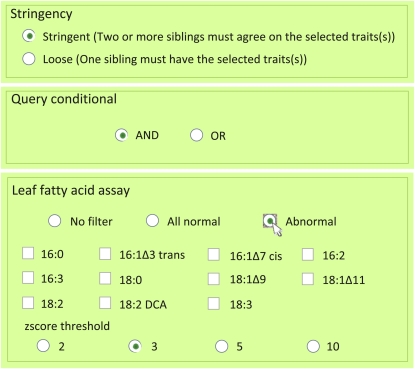
Figure 3.
Menus used to conduct a search for potential mutants in a quantitative phenotype. Illustration of features available for searching quantitative phenotypic data. Users of the search tools can select the search stringency (“Stringency”) and Boolean logic (“AND/OR”). Choosing “Abnormal” allows the user to specify one or more analytes and/or the z-score cutoff value.
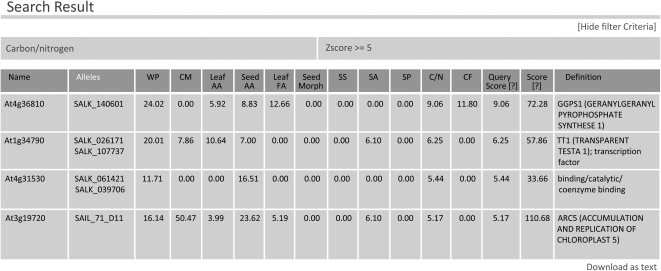
As shown in Figure 4, a table is returned. This page summarizes the results of a query, with information about the number of alleles for which one or more plants had a phenotype that matched that query. The query terms may be shown (“Show filter criteria”) or hidden. The table contains phenotypic severity scores for each assay: “Query Score” includes a summation of the severity of phenotypes selected in the query, and “Score” takes into account the results of all assays. Each score reflects both the consistency with which the siblings score in the assay as well as the rarity of the change in the full data set. The “Definition” column contains the “top line” annotation from The Arabidopsis Information Resource (www.arabidopsis.org). The entire table may be downloaded in Excel-compatible format (“Download as text”). Clicking on the Arabidopsis Genome Initiative locus designation (“Name”) opens a new page that displays data when there is at least one confirmed homozygous mutation in that locus (Fig. 5).
Figure 4.
Selected results returned from a search for genes with mutants assayed as having seed carbon-nitrogen z-scores ≥ 5 based on the flat cohort median. Summary scores indicate consistency of phenotypes across samples as follows: WP, whole plant; CM, chloroplast morphology; Leaf and Seed AA, leaf and seed amino acids; Leaf FA, leaf fatty acid methylesters; Seed Morph, seed morphology; SS, seed excess starch; SA, high early photoperiod leaf starch; SP, low later photoperiod starch; C/N, seed carbon-nitrogen ratio; CF, abnormal chlorophyll fluorescence measurement(s); Query Score, score for phenotypic query (in this case C/N ≥ 5); Score, general assessment of overall phenotypic abnormality of one or more mutant alleles (higher scores typically suggest the possibility of pleiotropy).
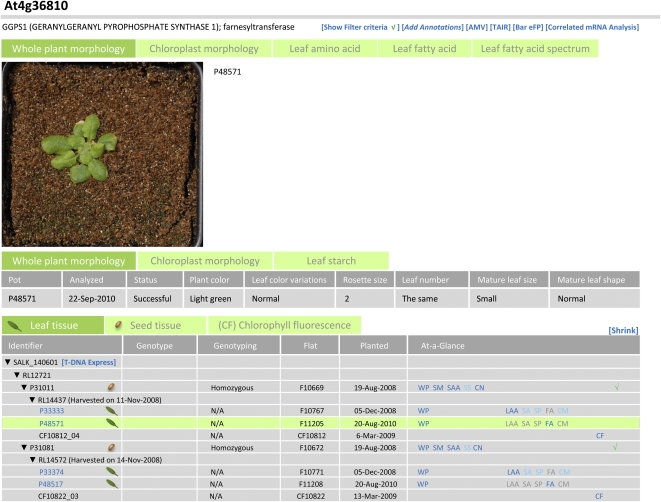
Figure 5.
Example of a Gene Page display. Top, photograph of selected plant P48571; middle, controlled vocabulary descriptions of whole plant morphology captured for this plant; bottom, table showing pedigree relationships and summary information about genotyping results and other metadata for the plant, seed, and chlorophyll fluorescence samples. Clicking on the blue links (e.g. P33333) in the “Identifier” column of the bottom table causes images and other data for that plant to be displayed in the middle and top sections of the page. For more information about the behavior of Gene Pages, see Supplemental Figure S4 and main text.
Interacting with Project Data Using Flexible Displays
The interface that displays mutant data was designed to provide flexibility to the researcher in a relatively simple and interactive layout (Fig. 5). The data are presented in three functionally linked display elements, and the researcher can make selections in each of these. Images and graphs of quantitative data are displayed on top. Controlled vocabulary and seed carbon-nitrogen ratio data are displayed in the center panel. The bottom window contains an overall summary of all individuals grown and assays performed for alleles of the given gene. This table also provides information about the pedigrees of assayed lines, planting date, cohort, and genotyping results. For genes where many alleles or individuals were grown, the table window may be expanded (“Expand”) to show all data or shrunk (“Shrink”) to keep more visual elements on one screen. Alternatively, branches of the pedigree tree can be collapsed or expanded to hide or view samples of interest by clicking on the tree arrows. “At-a-Glance” provides a visual summary of the phenotypic assays represented by abbreviations defined by “mousing over” the acronym. Data types (leaf or seed) are grouped together to permit trends in the phenotypic data to be easily identified. Gray text indicates the absence of assay data, and light blue text designates assay data not deviating from normal. Dark blue text indicates assay data not normal for controlled vocabulary-type data or exceeding the default z-score cutoffs defined in the Search by Assay query (Fig. 3). A green check mark indicates individuals meeting the original search filter criteria.
Navigation within the Gene Page is performed in two ways: (1) by selecting data types via tabs located at the top of each display panel; and (2) by choosing specific plants to view within the bottom summary panel. Selecting “Leaf Tissue,” “Seed Tissue,” or “Chlorophyll Fluorescence” tabs on the bottom allele summary window controls which data types are available for display in the top and middle windows (summarized in Supplemental Fig. S4). Next, selection of one of the tabs on the top window is coordinated with the associated data type displayed in the middle panel. For example, selection of “Leaf Tissue” in the bottom window and “Chloroplast Morphology” in the top window displays micrographs and associated controlled vocabulary descriptions. Data types not associated with an image or graph, such as leaf starch or seed carbon-nitrogen ratio, can be viewed by choosing the appropriate center panel tab.
The types of data can also be selected independently to look for pleiotropy. For example, selection of controlled vocabulary for leaf starch and chloroplast morphology images reveals the influence of starch-excess mutations on the chloroplast. All data for each individual plant may be viewed by selecting the relevant sample names (“P” and “CF” numbers), which become active in the bottom summary table depending on which tab is chosen. Researchers can assess the consistency of the data for different biological replicate siblings or across independent alleles of the same gene by viewing each individual. Links to external resources are also provided to allow access to a wide variety of data related to the gene of interest.
When large numbers of phenotypes are evaluated in a functional genomics project, it is common to see outlier samples caused by biological and process variation or severity of alleles. The data displays were designed to provide several ways for researchers to compare data sets quickly and evaluate the consistency of results. Summary scores in the search result tables allow researchers to compare the strength of the phenotypic syndromes of different genes (Fig. 4), and At-a-Glance (Fig. 5) summaries are useful for seeing trends in all the assays for a single gene. On the Gene Page, graphs are animated to allow the researcher to evaluate the consistency of metabolite data. Flash player animation allows viewing of consecutive images displaying a set of quantitative leaf or seed amino acids or leaf fatty acid methylesters for all samples assayed. It is possible to adjust the speed at which the player moves from displaying one sample to another and the z-score scale and threshold. Individual z-scores can be seen by mousing over metabolite bars. After reviewing the full set of data, any interesting patterns can be examined in detail by clicking on individual samples in the bottom window summary table.
Process Improvements Enabled by the Database
In addition to allowing data to be interrogated by scientists with access to the internet, having the full data set with time-stamp metadata provided the opportunity to check data consistency throughout the project and make process improvements during early stages (Baxter, 2010). For example, because transitory starch accumulation begins immediately upon the shift from dark to light, we were concerned that the time required to harvest leaves in the morning would influence the results of the starch assay. As shown in Supplemental Figure S5, there is no correlation between the number of hits (i.e. leaves scored as having higher than normal staining for morning leaf starch) and the order in which samples were harvested.
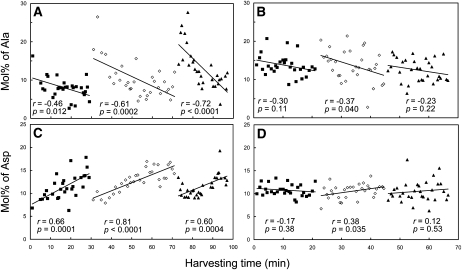
In contrast, a striking example of the need to review results to improve data quality is shown in Figure 6. It was noted that dramatic changes in values for leaf Ala (Fig. 6A) and Asp (Fig. 6C) occurred during harvesting. After approximately 30 samples (during 20–30 min of harvest time), the values reverted to the high (Ala) or low (Asp) end of the range and then rapidly drifted again. The periodicity corresponded to the time it took to harvest a flat, suggesting that removing the flat from the growth chamber to the laboratory bench was causing rapid shifts in leaf amino acid levels. Maintaining the plants under a bank of fluorescent lamps similar to growth chamber light levels during leaf harvest largely reversed this drifting behavior and improved data quality for these two amino acids (Fig. 6, B and D).
Figure 6.
Changes in leaf free Ala and Asp under laboratory low light are reduced under supplementary light. A and B, Mol % of leaf Ala of samples harvested under laboratory room light (approximately 10 μmol photons m−2 s−1; A) or room light supplemented with cool-white fluorescent lights (approximately 100 μmol photons m−2 s−1; B). C and D, Mol % of leaf Asp of samples harvested under approximately 10 μmol photons m−2 s−1 (C) or approximately 100 μmol photons m−2 s−1 (D). Samples in A and C were harvested on July 20, 2006, and samples in B and D were harvested on July 21, 2006. Harvesting was done flat by flat on the bench next to the growth chamber. Alike symbols (black squares, white diamonds, and black triangles) represent samples from the same flat harvested consecutively. Black lines represent linear regression for samples harvested from a single flat over time. Pearson correlation coefficient r and P values are shown below the data for each flat. Maintaining plants at approximately 100 μmol photons m−2 s−1 on the bench minimized the decrease of Ala and the increase of Asp during the harvest.
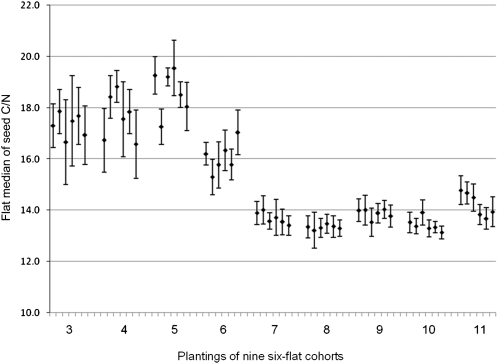
Uniform plant growth conditions are difficult to achieve even in the relatively well-controlled environment of a modern growth chamber. Even knowing when improvement in culture conditions will lead to higher quality data can require elaborate experimental designs (Massonnet et al., 2010). Seed carbon and nitrogen levels are highly dependent on available light levels during seed development (Li et al., 2006); therefore, metadata about growth chamber light levels were collected in the database. As shown in Figure 7, unsatisfactorily large variance in the ratio of seed carbon and nitrogen from one experimental set to another, despite uniform light levels, indicated that more improvements in growth conditions were needed. First, a more uniform growth medium was selected. Second, a strict water/fertilizer regime was adopted. Flats were filled with water or fertilizer so that all pots could uniformly absorb solution for 1 to 2 h, and then excess solution was poured off. This eliminated disparities that may be caused by growth chamber shelves not being perfectly level, resulting in unequal moisture levels in different parts of a single flat or between different flats over time. Finally, at each watering, the orientation and position of flats in the growth chamber were changed to provide more uniform light, temperature, and air flow. Analysis of the effects of these growth condition improvements on other metabolites showed that these process improvements were also effective in reducing variance in seed amino acid levels (Supplemental Fig. S6).
Figure 7.
Improvement in seed carbon-nitrogen data quality informed by analysis of early data sets. Values are displayed as flat median ± 1 median absolute deviation. High variance within plantings (six-flat cohorts) and flat median values (black diamonds) were found during the first six plantings (data for third through sixth plantings shown). Changes in plant culture conditions resulted in more consistency in the data (data for seventh through 11th plantings shown).
Using the Database to Discover Phenotypes: Pleiotropy of High Seed Branched-Chain Amino Acid Mutants
To illustrate how the data analysis tools can be used to find mutants with complex phenotypes, we searched for plants with altered levels of free seed branched-chain amino acids. When seed amino acid results were queried for alternations in Ile, Leu, and Val greater than a z-score of 5, the four genes with the highest query and overall scores were At2g26800, At1g03090, and At4g34030 along with the previously described high seed amino acid mutant coenzyme A dehydrogenase (ivd1-2), defective in gene At3g45300 (Gu et al., 2010). These mutants were among several hundred lines included in an effort to increase the coverage of amino acid metabolic processes in the chloroplast and other subcellular compartments.
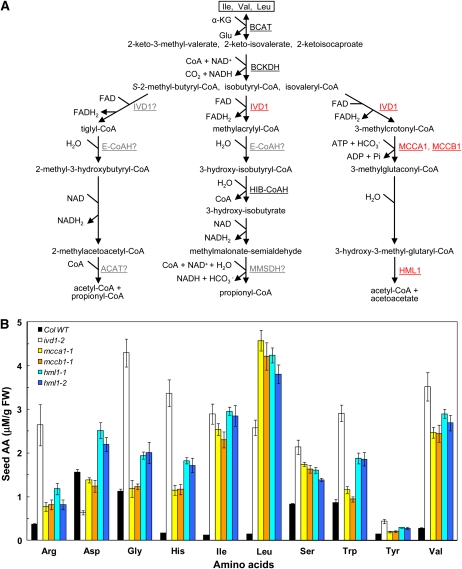
All four genes had alleles with consistently high soluble Ile, Leu, and Val, and results on T-DNA Express (http://signal.salk.edu/cgi-bin/tdnaexpress) suggested that these alleles are likely to cause reduced gene expression (for schematic diagrams of the insertion sites, see Supplemental Fig. S7A). Mutants mcca1-1 and mccb1-1 carry T-DNA insertions in the α- and β-subunits of the mitochondrial 3-methylcrotonyl-CoA carboxylase (At1g03090 and At4g34030), respectively. Consistent with the large increase in Leu accumulation in this mutant, the heteromeric mitochondrial 3-methylcrotonyl-CoA carboxylase catalyzes the fourth step of Leu catabolism in mammals and higher plants (Fig. 8A; Binder et al., 2007; Binder, 2010). Mutants hml1-1 and hml1-2 carry T-DNA insertions in the first and third introns of At2g26800, which is annotated as a putative mitochondrial hydroxymethylglutaryl (HMG)-CoA lyase (HML1; Fig. 8A). Consistent with the hypothesis that the four mutants are loss-of-function alleles, quantitative reverse transcription-PCR analysis confirmed that these lines have dramatically reduced steady-state levels of mRNA (Supplemental Fig. S7B). Follow-up studies were conducted to extend the initial observations from the pipeline data.
Figure 8.
Mutations in IVD1, MCCA1, MCCB1, and HML1 cause changes in seed free amino acids. A, Pathways of degradation of Ile, Leu, and Val in plant mitochondria. IVD1, MCCA1, MCCB1, and HML1 are highlighted in red. Hypothesized enzymes are shown in gray with question marks. ACAT, Acetyl-CoA acetyltransferase; α-KG, α-ketoglutarate; BCAT, branched-chain aminotransferase; E-CoAH, enoyl-CoA hydratase; HIB-CoAH, hydroxy-isobutyryl-CoA hydrolase; MCCA, methylcrotonyl-CoA carboxylase α-subunit; MCCB, methylcrotonyl-CoA carboxylase β-subunit; MMSDH, methylmalonate semialdehyde dehydrogenase. B, Seed free amino acids (AA) in ivd1-2, mcca1-1, mccb1-1, hml1-1, and hml1-2 mutants. Mature seeds were harvested from plants grown under a 16/8-h photoperiod and were analyzed for free amino acids using liquid chromatography-tandem mass spectrometry. Values are shown as means ± se (n = 5). FW, Fresh weight; WT, wild type.
Because annotation of the HMG-CoA lyase gene in The Arabidopsis Information Resource 9 was based on sequence similarity to the mammalian enzyme without published experimental evidence, we tested whether the protein product has the expected enzyme activity. Open reading frames for the two largest annotated splice variants (At2g26800.1 and At2g26800.2, predicted to differ in the length of the N-terminal open reading frame) were expressed in Escherichia coli and the products were assayed for the ability to hydrolyze HMG-CoA. The rapid in vitro production of acetyl-CoA by HML1 proteins is consistent with the hypothesis that At2g26800 encodes HMG-CoA lyase (Supplemental Fig. S7C).
To confirm the high seed amino acid phenotype, more replicates (n = 5) of the four mutants were grown outside of the Chloroplast 2010 pipeline, along with the ecotype Columbia wild type and positive control line ivd1-2, a previously characterized high seed branched-chain amino acid mutant defective in the catabolic enzyme isovaleryl-CoA dehydrogenase (At3g45300; Gu et al., 2010). Liquid chromatography-tandem mass spectrometry analysis of seed amino acids (Gu et al., 2007) from these mutants confirmed the initial phenotype (Fig. 8B; Supplemental Table S1). The mcca1-1, mccb1-1, hml1-1, and hml1-2 mutants have coordinate increases in seed Ile (18–23 times), Leu (28–32 times), and Val (9–10 times), despite their presumptive roles in the degradation of only Leu (Fig. 8). This is reminiscent of the high-Ile, -Leu, and -Val phenotype of ivd1 mutants (Fig. 8B; Gu et al., 2010), defective in an enzyme that is thought to function in Leu and Val catabolism (Däschner et al., 2001).
These results suggest that the accumulation of Leu catabolic intermediates directly or indirectly regulates the activity of one or both of the two enzymes common to catabolism of all three branched-chain amino acids: branched-chain aminotransferase and branched-chain keto acid dehydrogenase (BCKDH) complex (Fig. 8A). In mammalian species, the activity of the large multiprotein BCKDH complex is inhibited by phosphorylation and activated by dephosphorylation (Brosnan and Brosnan, 2006). The phosphatase that activates mammalian BCKDH is, in turn, inhibited by the Leu and Val catabolic intermediates isovaleryl-CoA and isobutyryl-CoA (Damuni and Reed, 1987). Our working hypothesis is that, as in mammals, accumulation of one or more CoA ester intermediates in the branched-chain amino acid catabolic mutants inhibits BCKDH in developing Arabidopsis seeds, causing the buildup of unusually high levels of branched-chain amino acids. Consistent with this hypothesis, it was recently reported that the ivd1-2 mutant accumulates the Leu catabolic intermediate isovaleryl-CoA during artificially induced senescence (Araújo et al., 2010; note that ivd1-2 is called ivdh-1 in this publication).
CONCLUSION
The transition from low- to moderate-scale experiments common in biological research laboratories to higher throughput projects requires researchers to confront unfamiliar problems. The data collection methods typically used in a biological laboratory cannot be adapted to these large-scale experiments, nor can the data be analyzed without computational assistance. Recording data in a laboratory notebook and hand labeling sample containers will result in high error rates and difficulty retrieving data. If the data are then entered into a computer-readable form, more data entry errors will occur. User fatigue becomes a much larger source of error in these large-scale projects, and they require more robust error reduction protocols.
For these reasons, a high-throughput project needs software specifically designed for the task (Baxter et al., 2007; Baxter, 2010). Developing the workstations for processing the biological material in conjunction with software used to gather the data has the potential for reducing the error rate and providing data in a form that is more amendable to computational analysis. We have presented one solution using the RoR Web application framework. The availability of such software allowed rapid development of data collection and analysis systems and, due to the modular nature of the frameworks, promotes the distribution and modification of these systems. A Web interface to these large data sets enables rapid distribution of the data and connects the producers and consumers of such data sets.
Large data sets such as that from the Chloroplast 2010 Project are useful for developing hypotheses but require in-depth follow-up. This is because of challenges associated with any large data set and specific issues related to screening mutant germplasm. For example, inconsistent phenotypes can result from diverse types of alleles: loss-of-function mutations range from insertions that abolish gene expression (amorphic alleles) to weak loss of function due to T-DNA in promoters, introns, or at the end of genes (hypomorphic alleles). Although less common, increased expression due to the 35S enhancer in the T-DNA alleles has also been found (Ajjawi et al., 2010). Another cause of inconsistent phenotypes in seemingly allelic mutants is the widespread occurrence of secondary mutations in T-DNA lines (discussed in Ajjawi et al., 2010). Large data sets are also subject to the multiple test problem due to random variation in data. Finally, run-to-run process variation over months and years is unavoidable. The database and analysis tools developed for this project were employed to reduce the impact of these influences. In addition, the data analysis tools developed for this project are designed to allow the researcher to identify these potential problems when exploring phenotypic data.
MATERIALS AND METHODS
Database Design
A major design goal was to make the phenotypic data available to the scientific community. For that reason, the software was implemented as a Web application utilizing RoR and Adobe Flash. A Web application allows widespread use of the software without the inherent problems of distributing an application. The use of asynchronous updates of the Web page results in fast response times of the software, which obviates the speed disadvantage of typical Web-based tools. The database management software is Oracle 10g. The main Web application is implemented using the RoR framework (Thomas et al., 2006). Ruby is employed because it is a dynamically typed object-oriented interpreted language that allows for rapid development. The RoR framework has a model (view) controller architecture that promotes good separation of the application functions and allows for easier maintenance of the application’s code base. RoR also implements the necessary code to enable Ajax (asynchronous JavaScript and XML) functionality. Additionally, RoR has an object-to-relational database mapping layer that reduces the amount of SQL code that must be written. RoR also reduces the amount of JavaScript code that needs to be written. The result of this built-in functionality results in more of the application being written in a single programming language (Ruby) than is typical for most Web applications. The standardization imposed by the RoR framework allows efficient collaboration by multiple programmers working on the project and allows users with RoR experience to quickly understand our application logic.
The presentation of graphs on Web applications can be problematic. Adobe Flash Player is used to display charts using the charting functions of Adobe Flash Builder 3.0. The data for the graph are rendered into an XML stream, and the particular data to be displayed in the graph are selected using E4X (ECMAScript for XML), which is supported by Adobe ActionScript 3.0 used to program the Flash Player. The Flash Player is freely available at http://www.adobe.com/.
Plant Materials and Metabolite Analyses
T-DNA lines of Arabidopsis (Arabidopsis thaliana) used in the branched-chain amino acid case study were in the Columbia background. The mcca1-1, mccb1-1, hml1-1, and hml1-2 mutants were obtained from the Arabidopsis Biological Resource Center (stock numbers SALK_137966C [mcca1-1], SALK_117349C [mccb1-1], SALK_014207C [hml1-1], and SALK_145226C [hml1-2]). The stocks that were generated from these mutants in the course of this study were redeposited to the Arabidopsis Biological Resource Center as accession numbers CS66518 (mcca1-1), CS66519 (mccb1-1), CS66520 (hml1-1), and CS66521 (hml1-2). Seeds for amino acid assay and carbon and nitrogen analysis were harvested from plants grown under the 16/8-h photoperiod. Seed amino acid and carbon and nitrogen analyses were performed as described previously (Lu et al., 2008).
Quantitative Reverse Transcription-PCR Analysis
Developing siliques with embryos at torpedo and walking-stick stages were chosen for mRNA analysis. Total RNA was extracted as described previously (Takaha et al., 1993), digested with RNase-free DNase I (Roche), and reverse transcribed with oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Promega). Gene-specific primers were designed to span two or three exons as listed in Supplemental Table S2. Primers HML_L and HML_R were used in hml1-1 mutants, and primers HML_L2 and HML_R2 were used in hml1-2 mutants. Quantitative PCR was performed on a 7500 Real-Time PCR system with Power SYBR Green PCR master mix (Applied Biosystems).
Expression and Purification of Recombinant HML1 Proteins in Escherichia coli
Total RNA was extracted, digested with DNase, and reverse transcribed with oligo(dT)15 primers as described above. Full-length HML1 coding regions according to gene models At2g26800.1 and At2g26800.2 were amplified using the mRNA:cDNA hybrid, Pfu DNA polymerase (Promega) with forward primers HML1_BamH1_ATG and HML1_BamH1_ATG2, and reverse primer HML1_Xho1_TAA (Supplemental Table S2). The resulting PCR products were AT cloned into pGEM-T Easy vector and sequenced to confirm the absence of PCR errors. XhoI/BamHI-digested HML1 fragments were subcloned into pET28a expression vector (Novagen) and expressed in E. coli strain BL21 (DE3) (Stratagene). Expression of recombinant proteins was induced with 1 mm isopropyl β-d-thiogalactoside, and cells were grown at 30°C overnight. Recombinant proteins were affinity purified with nickel-nitrilotriacetic acid agarose resins under native conditions according to the QIAexpressionist protocol (Qiagen). Protein concentration was determined using bicinchoninic acid assay.
Activity Assay of Recombinant HMG-CoA Lyase
Recombinant HML1 (0.7 μm) was incubated with 0.5 mm HMG-CoA at 30°C for 0, 5, and 10 min in 0.2 m Tris-HCl buffer (pH 8.2) containing 0.2 mm EDTA and 20 mm MgCl2 (van der Heijden et al., 1994). Samples were then treated with 0.2 volume of 2 n perchloric acid and neutralized with 3 m KHCO3. The production of acetyl-CoA by HML1 was determined by PicoProbe acetyl-CoA assay kit (BioVision) according to the manufacturer’s protocols. Negative controls included recombinant enzyme preparations inactivated by 100°C treatment for 5 min and the full reaction without added recombinant enzyme.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Summary of workflows for the Chloroplast 2010 Project.
Supplemental Figure S2. Schematic representation of how the laboratory information management system and laboratory work space are used to reduce error.
Supplemental Figure S3. Time stamp-derived reconstructed image of plants grown together in a flat and photographed separately.
Supplemental Figure S4. Schematic diagram describing how the selection of green tabs controls the behavior of the Gene Page displays.
Supplemental Figure S5. The distribution of putative hits from a morning leaf starch assay.
Supplemental Figure S6. Improvement in seed free amino acid data consistency informed by analysis of early data sets.
Supplemental Figure S7. Decreased Leu catabolic enzyme mRNA accumulation in mcca1-1, mccb1-1, hml1-1, and hml1-2 mutants.
Supplemental Figure S8. Database schema.
Supplemental Table S1. Seed amino acid contents.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank all the principal investigators and other participants of the Chloroplast 2010 Project. Special thanks to Kathleen Imre and Imad Ajjawi for work designing the various assays, workflows, and database tools used in the project and database. We also thank Cheng Peng for help with acetyl-CoA assays and David Hall for nonpipeline amino acid analysis.
References
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL. (2010) Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA.et al (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I. (2010) Ionomics: the functional genomics of elements. Brief Funct Genomics 9: 149–156 [DOI] [PubMed] [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. (2007) Purdue Ionomics Information Management System: an integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi: 10.1043/tab.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J. (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan JT, Brosnan ME. (2006) Branched-chain amino acids: enzyme and substrate regulation. J Nutr (Suppl) 136: 207S–211S [DOI] [PubMed] [Google Scholar]
- Damuni Z, Reed LJ. (1987) Purification and properties of the catalytic subunit of the branched-chain alpha-keto acid dehydrogenase phosphatase from bovine kidney mitochondria. J Biol Chem 262: 5129–5132 [PubMed] [Google Scholar]
- Däschner K, Couée I, Binder S. (2001) The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2007) LC-MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Anal Chem 79: 8067–8075 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL. (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening: evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber AP.et al (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Myouga F, Akiyama K, Motohashi R, Kuromori T, Ito T, Iizumi H, Ryusui R, Sakurai T, Shinozaki K. (2010) The Chloroplast Function Database: a large-scale collection of Arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J 61: 529–542 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR. (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Rousseeuw P, Croux C. (1993) Alternatives to the median absolute deviation. J Am Stat Assoc 88: 1273–1283 [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Takaha T, Yanase M, Okada S, Smith SM. (1993) Disproportionating enzyme (4-alpha-glucanotransferase; EC 2.4.1.25) of potato: purification, molecular cloning, and potential role in starch metabolism. J Biol Chem 268: 1391–1396 [PubMed] [Google Scholar]
- Thomas DH, Hansson D, Breedt L, Clark M, Davidson JD, Gehtland J, Schwartz A. (2006) Agile Web Development with Rails, Ed 3. Pragmatic Bookshelf, Raleigh, NC [Google Scholar]
- van der Heijden R, de Boer-Hlupá V, Verpoorte R, Duine JA. (1994) Enzymes involved in the metabolism of 3-hydroxy-3-methylglutaryl-coenzyme A in Catharanthus roseus. Plant Cell Tissue Organ Cult 38: 345–349 [Google Scholar]
- Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, et al. (2003) Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell 15: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A. (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]