Abstract
l-Proline (Pro) catabolism is activated in plants recovering from abiotic stresses associated with water deprivation. In this catabolic pathway, Pro is converted to glutamate by two reactions catalyzed by proline dehydrogenase (ProDH) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH), with Δ1-pyrroline-5-carboxylate (P5C) as the intermediate. Alternatively, under certain conditions, the P5C derived from Pro is converted back to Pro by P5C reductase, thus stimulating the Pro-P5C cycle, which may generate reactive oxygen species (ROS) as a consequence of the ProDH activity. We previously observed that Pro biosynthesis is altered in Arabidopsis (Arabidopsis thaliana) tissues that induce the hypersensitive response (HR) in response to Pseudomonas syringae. In this work, we characterized the Pro catabolic pathway and ProDH activity in this model. Induction of ProDH expression was found to be dependent on salicylic acid, and an increase in ProDH activity was detected in cells destined to die. To evaluate the role of ProDH in the HR, ProDH-silenced plants were generated. These plants displayed reduced ROS and cell death levels as well as enhanced susceptibility in response to avirulent pathogens. Interestingly, the early activation of ProDH was accompanied by an increase in P5C reductase but not in P5CDH transcripts, with few changes occurring in the Pro and P5C levels. Therefore, our results suggest that in wild-type plants, ProDH is a defense component contributing to HR and disease resistance, which apparently potentiates the accumulation of ROS. The participation of the Pro-P5C cycle in the latter response is discussed.
Proline dehydrogenase (ProDH; EC 1.5.99.8) is a flavoprotein that catalyzes the first and rate-limiting step of two reactions converting Pro into Glu at the mitochondria. This enzyme is located at the matrix side of the inner mitochondrial membrane, where it oxidizes Pro into Δ1-pyrroline-5-carboxylate (P5C) and transfers electrons to the primary mitochondrial electron transport chain (mETC; Boggess and Koeppe, 1978; Huang and Cavalieri, 1979; Elthon and Stewart, 1981). Subsequently, in the same compartment, P5C is transformed into Glu by Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH; Hare and Cress, 1997).
ProDH activity and ProDH gene expression are modified during plant development, with this activity being usually enhanced in tissues supporting cell growth, such as meristems, pollen, pistils, flowers, and siliques (Dashek and Harwood, 1974; Skubatz et al., 1989; Hare and Cress, 1997; Nakashima et al., 1998; Verbruggen and Hermans, 2008). Then, the enzyme may supply reducing power to the mitochondria, thereby contributing to the generation of ATP, and may also provide nitrogen from Pro. Moreover, by working together with P5CDH and Glu dehydrogenase, ProDH may contribute carbon to the tricarboxylic acid cycle (Hare and Cress, 1997; Szabados and Savouré, 2010).
ProDH is also sensitive to environmental stresses. Dehydration, extreme temperatures, and other abiotic injuries repress ProDH and stimulate Pro synthesis, thus leading to net Pro accumulation. Once the stress is relieved, ProDH becomes activated to consume the stored Pro (Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996; Verbruggen and Hermans, 2008; Szabados and Savouré, 2010). Here again, the enzyme is suspected of supplying energy and nutrients for stress recovery.
Interestingly, under certain conditions, the catabolism of Pro may produce accumulation of reactive oxygen species (ROS). This effect is specifically associated with the activation of the Pro-P5C cycle, which involves the action of ProDH and Δ1-pyrroline-5-carboxylate reductase (P5CR), with the latter enzyme catalyzing the second step in the biosynthesis of Pro from Glu taking place in the cytosol (Hare and Cress, 1997). This cycle operates when the increase in ProDH activity does not correspond to a rise in P5CDH activity, resulting in an incomplete oxidation of Pro and accumulation of P5C at the mitochondria. In this case, P5C is exported to the cytosol to be converted into Pro by P5CR, with Pro then being incorporated into mitochondria to restart the cycle (Phang et al., 2008). The generation of ROS by the Pro-P5C cycle was initially demonstrated in animal cells (Hagedorn and Phang, 1983), where the overexpression of ProDH (Pro oxidase [POX]) generates both mitochondrial ROS accumulation (Donald et al., 2001; Liu et al., 2005) and apoptosis (Maxwell and Davis, 2000; Maxwell and Rivera, 2003; Phang et al., 2008) in a Pro-dependent manner. In plants, the export of P5C from mitochondria has also been suggested, and activation of the Pro-P5C cycle was described in ProDH-overexpressing or p5cdh Arabidopsis (Arabidopsis thaliana) mutant plants, with stimulation of the cycle leading to an increase of mitochondrial ROS, apparently by raising the electron flow to mETC (Miller et al., 2009).
Treatment with exogenous Pro activates ProDH expression (Kiyosue et al., 1996; Verbruggen et al., 1996) and causes toxicity in plant and animal cells (Hellmann et al., 2000; Maxwell and Davis, 2000: Deuschle et al., 2001; Donald et al., 2001; Ayliffe et al., 2002; Mani et al., 2002). This adverse effect could be due to different causes. One possibility is that at high concentrations, Pro is toxic by itself, producing an amino acid imbalance, an effect observed for most amino acids except Gln at millimolar concentrations (Bonner et al., 1996). Consistent with this possibility, Arabidopsis ProDH-deficient plants are hypersensitive to Pro, and ProDH overexpressors have enhanced tolerance to this treatment (Mani et al., 2002; Nanjo et al., 2003; Funck et al., 2010). On the other hand, Pro supply stimulates the catabolic pathway; thus, it may produce P5C accumulation (Hellmann et al., 2000; Deuschle et al., 2001, 2004). This derivative induces apoptosis in animal tumor cell lines (Maxwell and Davis, 2000) and may have similar effects on plants. In agreement with this, Arabidopsis p5cdh knockout plants are hypersensitive to exogenous Pro and carry enhanced P5C levels, while P5CDH-overexpressing plants are tolerant to this treatment (Deuschle et al., 2004). Finally, Pro toxicity could also result from the accumulation of ROS at the mitochondria as a consequence of ProDH activation, an effect that may be potentiated by stimulation of the Pro-P5C cycle (Hare and Cress, 1997). Consistent with this latter possibility, exogenous Pro leads to ROS accumulation in p5cdh plants (Deuschle et al., 2004), with these ROS being apparently located at the mitochondria (Miller et al., 2009).
While the Pro metabolism has been widely studied in response to abiotic stresses (Hare and Cress, 1997; Verbruggen and Hermans, 2008; Szabados and Savouré, 2010), few investigations have characterized it under conditions of pathogen attack. At the molecular level, the flax fis1 gene homolog to P5CDH is locally induced by compatible, but not by incompatible, interactions with Melampsora lini (Ayliffe et al., 2002). However, as transgenic flax plants reducing P5CDH expression show normal responses to virulent and avirulent rust infections, the function of fis1 and Pro catabolism in rust disease remains unclear (Mitchell et al., 2006). At the biochemical level, several works have reported the accumulation of Pro in tissues infected with viral (Mohanty and Sridhar, 1982; Radwan et al., 2007) and bacterial (Meon et al., 1978; Fabro et al., 2004) pathogens. In tobacco (Nicotiana tabacum), Pro accumulation is related to susceptibility to Agrobacterium tumefaciens, as Pro antagonizes plant defenses by interfering with the γ-aminobutyrate-mediated degradation of bacterial quorum-sensing signals that normally increase pathogen spread (Haudecoeur et al., 2009). Whether, in this case, the accumulation of Pro occurring on tumors was stimulated by the pathogen is still not determined. In Arabidopsis, avirulent, but not virulent, races of Pseudomonas syringae pv tomato (Pst) trigger Pro accumulation in close association with development of the hypersensitive response (HR; Fabro et al., 2004). The HR is a local defense, triggered by recognition of pathogen effectors by plant disease resistance proteins (R), which ends with the generation of a dry and sharp-edged lesion at the infection site, resulting from programmed cell death (Stakman, 1915), and confers race-specific resistance (Chisholm et al., 2006; Bent and Mackey, 2007).
ROS and salicylic acid (SA) are two positive regulators of the HR that concentrate in the center of the lesion and signal numerous defense responses (Alvarez, 2000; Chisholm et al., 2006; Bent and Mackey, 2007; Vlot et al., 2008). Interestingly, at an advanced stage of HR (24 h post infiltration [hpi]), the infected tissues activate the expression of the Pro biosynthetic gene AtP5CS2 as well as the accumulation of Pro, with both being responses stimulated by ROS and SA (Fabro et al., 2004). By analyzing the Pro metabolism in the early stages of HR, we detected activation of the catabolic pathway at 6 hpi. In this study, we describe the spatiotemporal expression pattern of Arabidopsis ProDH genes during HR development and assess the requirement of SA for induction of these genes. We also demonstrate activation of the ProDH enzyme in HR-developing cells and position this response with respect to early HR components. Furthermore, using ProDH-silenced plants, we provide evidence for a positive role of ProDH in the activation of both the HR and resistance to avirulent pathogens.
RESULTS
Topology of ProDH mRNA Accumulation in the HR Lesion
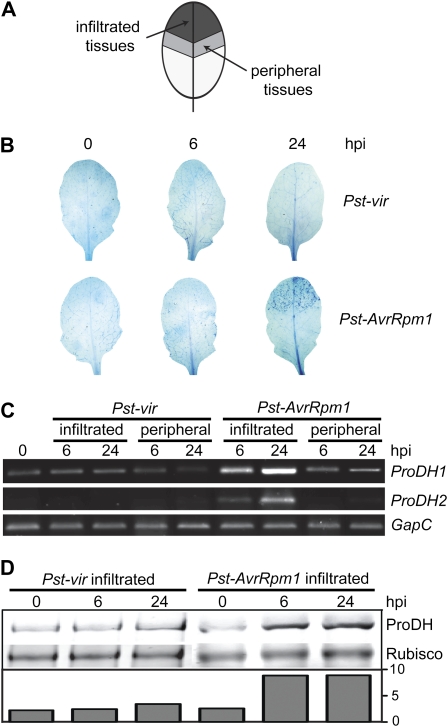
The observation that ProDH transcripts accumulate in leaves treated with avirulent bacteria (Fabro et al., 2004) prompted us to investigate whether this response was specific to cells destined to die. To address this question, we infiltrated Pst-AvrRpm1 in the top of the leaves and collected the inoculated and peripheral tissues (Fig. 1A) to quantify the abundance of ProDH1 (At3g30775) and ProDH2 (At5g38710) mRNAs. Samples were isolated at two stages, one preceding and the other succeeding cell death (6 and 24 hpi, respectively). These time points were selected by staining treated leaves with the vital dye trypan blue (Fig. 1B). In these experiments, the Pst virulent strain (Pst-vir) pathogen was used as a control, because although it activates defense responses, it is unable to trigger hypersensitive cell death (Fig. 1B).
Figure 1.
Early accumulation of ProDH at the center of the HR lesion. A, Sampling scheme. Suspensions of Pst-vir or Pst-AvrRpm1 isolates were locally inoculated (5 × 106 cfu mL−1) at the ends of Arabidopsis leaves to then identify infiltrated and noninfiltrated peripheral tissues. B, Trypan blue staining of leaves excised at 6 or 24 hpi with pathogens. C, Quantification of ProDH1 and ProDH2 transcripts in pairs of samples (infiltrated, peripheral) isolated from pathogen-treated leaves as described in A. The assays involve semiquantitative, two-step, reverse transcription-PCR. The constitutively expressed GapC gene (At3g04120) was used as a control for cDNA content in the reactions. D, Western-blot analysis comparing the ProDH content in naive tissues and Pst-vir- or Pst-AvrRpm1-infiltrated tissues. The ProDH content is indicated at the bottom after normalization to Rubisco signal quantified by Ponceau S staining. The antibodies were generated against a peptide conserved between both ProDH isoforms, so that they may detect both proteins with similar sensitivity in these assays. One representative of three independent infection experiments is shown for each assay. Each experiment included six leaves (B) or four leaves (C and D) isolated from three different plants per time point. [See online article for color version of this figure.]
ProDH1 and ProDH2 mRNAs were only found to accumulate in the Pst-AvrRpm1-infiltrated tissues. This was visible at 6 hpi and became more pronounced at 24 hpi (Fig. 1C). No significant increase was detected in the ProDH1/2 transcripts in the tissues surrounding the Pst-AvrRpm1 infiltration (peripheral) site or in the infiltrated or peripheral tissues of Pst-vir-treated leaves (Fig. 1C).
We next examined if there was any accumulation of the protein in the Pst-AvrRpm1-challenged tissues. Using western-blot assays, the abundance of ProDH in nontreated and Pst-AvrRpm1-treated tissues was compared, including Pst-vir-treated samples again as a control. At 6 and 24 hpi, the avirulent bacteria induced a 3.5 times increase in the ProDH basal content, whereas the virulent bacteria did not alter this value (Fig. 1D). Neither were the protein levels modified by the virulent pathogen after 24 hpi (data not shown).
Therefore, the accumulation of ProDH induced by Pst was associated with the activation of cell death, with the increase in protein content seeming to be caused by transcriptional activation of both ProDH1 and ProDH2 genes. Although these results suggested that HR-developing tissues increased the ProDH activity, this could not be determined in extracts from leaf tissues. As described below, ProDH activity was later quantified using another system.
Other Early Alterations in Pro Metabolism That Take Place in the HR
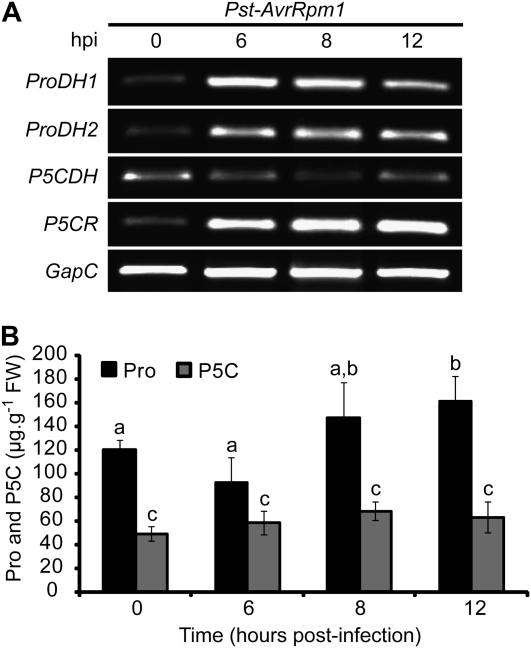
To analyze other features of Pro metabolism in HR-developing tissues, we quantified the abundance of P5CR and P5CDH transcripts during the initial 12 hpi with Pst-AvrRpm1. In parallel, the expression of ProDH genes was evaluated. We noticed that at 6, 8, and 12 hpi, the infected tissues accumulated not only ProDH but also P5CR transcripts (Fig. 2A). In contrast, no increase or even slight reduction of P5CDH mRNAs was found in these tissues.
Figure 2.
Pro metabolic changes associated with ProDH induction in the HR. A, Quantification of ProDH1, ProDH2, P5CDH, and P5CR transcripts levels in leaves isolated at 0, 6, 8, and 12 hpi with the avirulent bacterium Pst-AvrRpm1. Pathogen infiltration and transcript quantification were performed as described in Figure 1. One representative of three independent infection experiments (each one including at least three plants and four leaves per time point) is shown. B, Free Pro and P5C content (μg g−1fresh weight [FW]) was determined in aliquots of the leaf samples described in A. Data represent means ± sd of Pro values in five independent experiments (three leaves from three plants each). Statistically significant differences (P < 0.01, ANOVA) are shown using different lowercase letters.
We also determined the Pro and P5C levels in the same set of samples and observed that Pro and P5C remained almost constant until 8 hpi (Fig. 2B). At 12 hpi, P5C still maintained its basal levels, whereas Pro displayed a slight increase.
These results are consistent with the possibility that at the early stages of HR, the tissues activate not only ProDH but also P5CR, without a corresponding rise in P5CDH activity.
Sensitivity of ProDH Genes to SA
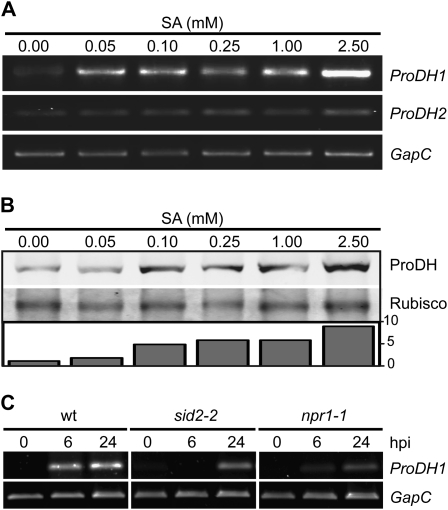
We examined if the expression of ProDH genes was modulated by SA, a main regulator of defenses against biotrophic pathogens including the HR (Alvarez, 2000; Vlot et al., 2008). First, we investigated how exogenous SA affects transcript and protein levels in wild-type plants. To carry this out, leaves were infiltrated with SA solutions (0.05, 0.10, 0.25, 1.00, and 2.50 mm) and were collected at 3 h post treatment (hpt) to evaluate the abundance of both ProDH1/2 transcripts and protein. We found that SA stimulated ProDH1 mRNA accumulation at all tested doses (Fig. 3A). In sharp contrast, SA had no effect on the ProDH2 mRNA level, even at 2.50 mm. As SA increased by four to five or eight times the basal protein content at 0.10 to 1.00 or 2.50 mm concentration, respectively (Fig. 3B), the accumulation of ProDH generated by SA treatment seemed to result from transcriptional activation of ProDH1.
Figure 3.
SA stimulates the expression of ProDH1 in HR. A and B, Effects of exogenous SA on ProDH mRNA and protein levels in wild-type plants. Leaves infiltrated with the indicated SA concentrations were excised at 3 h post treatment to monitor the abundance of ProDH1 and ProDH2 transcripts (A) as well as ProDH protein (B). C, Requirement of SID2 and NPR1 for ProDH1 activation by Pst-AvrRpm1. Wild-type (wt), sid2-2, and npr1-1 leaves were infiltrated with Pst-AvrRpm1 and excised at 6 and 24 hpi to determine the abundance of ProDH1 transcript in all three plants. Pathogen infiltration and transcript and protein quantification were performed as described in Figure 1. One representative from three (C) or four (A and B) independent experiments is shown for each assay. Each experiment included at least three plants and at least three leaves isolated from different plants per time point.
We then evaluated if endogenous SA was required for ProDH1 transcript accumulation in response to Pst-AvrRpm1 by quantifying the abundance of the transcript in pathogen-treated plants deficient in the SA pathway. We used the SA induction-deficient2 (sid2-2) mutant, unable to generate SA by isochorismate synthase in response to pathogens (Wildermuth et al., 2001), and the nonexpressor of PR genes1 (npr1-1) mutant, deficient in the activation of SA-sensitive defense genes (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997). As shown in Figure 3C, sid2-2 plants failed to accumulate ProDH1 transcripts at 6 hpi and could only do so weakly at 24 hpi. In turn, npr1-1 contained lower ProDH1 mRNA levels than wild-type plants at 6 and 24 hpi. Therefore, the early accumulation of ProDH1 transcripts detected in HR-developing tissues (6 hpi) in wild-type plants seemed to be signaled, at least partially, through the SA pathway, with SID2 and NPR1 being positive regulators of this response. In contrast, at later time points (24 hpi), ProDH1 activation appeared to be signaled by SID2- and/or NPR1-independent pathways.
Activation of ProDH in the HR Context
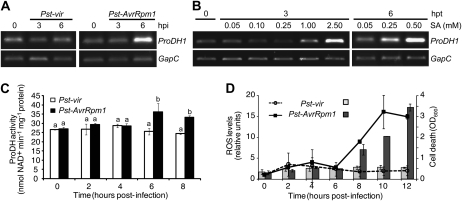
We investigated how activation of the ProDH1 gene was temporarily placed in the HR network and how the ProDH activity was modified in cells triggering HR, by using a cell culture system allowing characterization of HR components. This system included photosynthetic Arabidopsis cells, which elicit basal and race-specific defenses and trigger responses remarkably similar to that of green plant tissues in response to Pst-AvrRpm1 (Cecchini et al., 2009). The expression of ProDH1 was evaluated in nontreated cultured cells and cells treated with Pst-AvrRpm1, Pst-vir, or exogenous SA, collected at 3 and 6 hpt. As in the case of plant tissues, the cultured cells activated the expression of ProDH1 in response to Pst-AvrRpm1 at 6 hpt but did not respond to Pst-vir at this (Fig. 4A) or later time points (data not shown). Cells also induced ProDH1 in response to SA (Fig. 4B), requiring longer times than leaf tissues (Fig. 4A) to activate transcript accumulation by low hormone doses.
Figure 4.
Activation of ProDH in the HR context. Arabidopsis cultured cells were treated with virulent or avirulent races of Pst (Pst-vir and Pst-AvrRpm1, respectively) as described by Cecchini et al. (2009). In parallel, cultures were treated with the indicated final concentrations of SA. Samples collected from these cultures at the different times post treatment (hpt) were used for the following studies. A and B, ProDH1 expression evaluated as in Figures 1C and 2A. C, ProDH activity in protein extracts from naive and pathogen-treated cultured cells quantified according to Lutts et al. (1999) and Kant et al. (2006). D, ROS and cell death levels, determined by H2DCFDA (lines) and Evans blue staining (bars), respectively, with respect to untreated cells as described by Cecchini et al. (2009). One representative from three independent experiments is shown for each assay. Each experiment included three technical replicates (1-mL aliquot) isolated from treated cultures at each indicated time point. OD, Optical density. In C and D, data represent means ± sd of three replicates. In C, statistically significant differences (P < 0.01, ANOVA) are shown using different lowercase letters.
It was possible to quantify the ProDH activity in extracts obtained from cultured cells. This was monitored in samples from naive and pathogen-treated cells, determining in parallel ROS and cell death levels in these samples. An increase in ProDH activity was found in Pst-AvrRpm1-treated cells at 6 and 8 hpt (Fig. 4C), corresponding to 25% to 30% of the basal activity. This response was consistently detected in many different experiments and was always specifically induced by the avirulent bacteria. Interestingly, at 6 hpi with Pst-AvrRpm1, the ProDH activity was enhanced, whereas ROS and cell death levels remained unaltered (Fig. 4D). At 8 hpt with Pst-AvrRpm1, cultures began to accumulate ROS and generate cell death in response to the avirulent pathogen. In turn, cultures treated with Pst-vir neither modified the ROS or cell death levels nor the ProDH activity. Thus, the in vitro system, which reproduced the ProDH1 gene expression activation detected in planta, revealed that ProDH activity was specifically enhanced by avirulent pathogen before the onset of the oxidative burst and cell death.
Generation of Arabidopsis ProDH-Silenced Plants
In order to analyze whether ProDH has functional implications in HR development, we generated Arabidopsis ecotype Columbia (Col-0) transgenic plants by silencing the expression of the ProDH1 and ProDH2 genes (see “Materials and Methods”). Twenty-nine primary transformed (T0) plants were obtained and propagated. The abundance of ProDH transcripts was evaluated in the transgenic plants by using leaf tissues isolated at 6 and 24 hpi with Pst-AvrRpm1, including an identical set of samples obtained from wild-type plants as controls. Plants silencing ProDH expression were selected from the T1 and T2 populations. Unfortunately, some of the plants having the strongest silencing levels had a reduced or null seed production and could not be maintained (data not shown). Two T3 plant lines derived from the transformation experiments were characterized in detail. These were siPD B8, carrying a single insertion of the transgene in homozygosis, and siPD U9, containing multiple insertions of the construct. Besides the sterility found in the strongly silenced plants, we did not observe any other phenotypes in untreated ProDH-silenced plants.
We explored if siPD B8 and siPD U9 plants had lower ProDH activity than wild-type plants. As this activity could not be determined on leaf extracts (see above), a functional assay was applied.
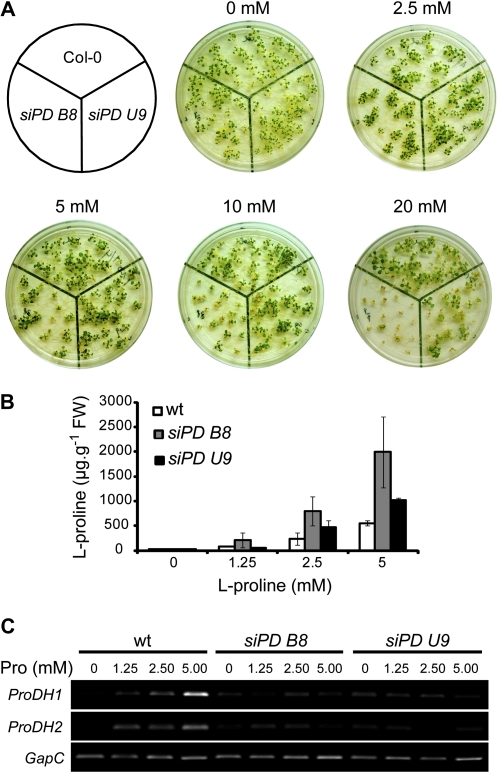
During seedling development, treatment with exogenous Pro is toxic for wild-type plants and is even more harmful for ProDH-deficient plants, with this effect being apparently due to exacerbated free Pro accumulation (Mani et al., 2002; Nanjo et al., 2003; Funck et al., 2008, 2010). Then, we compared the sensitivity of transgenic and wild-type plants to Pro supply. Seeds were sown on synthetic medium either lacking exogenous Pro or supplemented with different concentrations of the amino acid (2.5, 5.0, 10.0, and 20.0 mm). At the age of 3 weeks, plantlet growth and free Pro content in aerial tissues were assessed. In the absence of Pro, all three genotypes grew to a similar extent. In contrast, in Pro-supplemented medium (≥5 mm), the transgenic plants showed a reduced growth and chlorosis compared with wild-type plants (Fig. 5A). Differences in growth were detected after cotyledon emergence, whereas chlorosis was seen after true leaf appearance (data not shown). At high Pro concentrations (≥10 mm), siPD B8 showed a greater sensitivity to Pro than siPD U9 plants. In addition, both transgenic plants accumulated more Pro than wild-type plants, with siPD B8 again harboring the highest amino acid levels (Fig. 5B). These results suggest that both transgenic plants contained lower ProDH activity than wild-type plants. In agreement, the accumulation of ProDH transcripts in Pro-supplemented medium (5 mm) was reduced in these transgenic plants (Fig. 5C). These results also point to a stronger ProDH deficiency in siPD B8 than in siPD U9 plants.
Figure 5.
Phenotypes of siPD B8 and siPD U9 transgenic plants. A, Sensitivity of siPD B8 and siPD U9 plants to exogenous Pro. Seeds were sown on synthetic medium supplemented with Pro (2.5, 5.0, 10.0, and 20.0 mm). Images were taken at 3 weeks after sowing. The experiment was repeated three times with similar results. B, Free Pro content (μg g−1 fresh weight [FW]) in the aerial tissues of the plantlets shown in A. Samples corresponding to 20 mm Pro treatment were not analyzed for Pro content due to reduced plant viability. Values represent means ± sd of three independent experiments, each one containing at least six to eight plants per time point. wt, Wild type. C, Abundance of ProDH1 and ProDH2 transcripts in aerial tissues from 2-week-old plants grown as in B. One representative from three independent experiments is shown. Each experiment involved at least three plants and three leaves from different plants per time point. [See online article for color version of this figure.]
HR Development in the ProDH-Silenced Plants
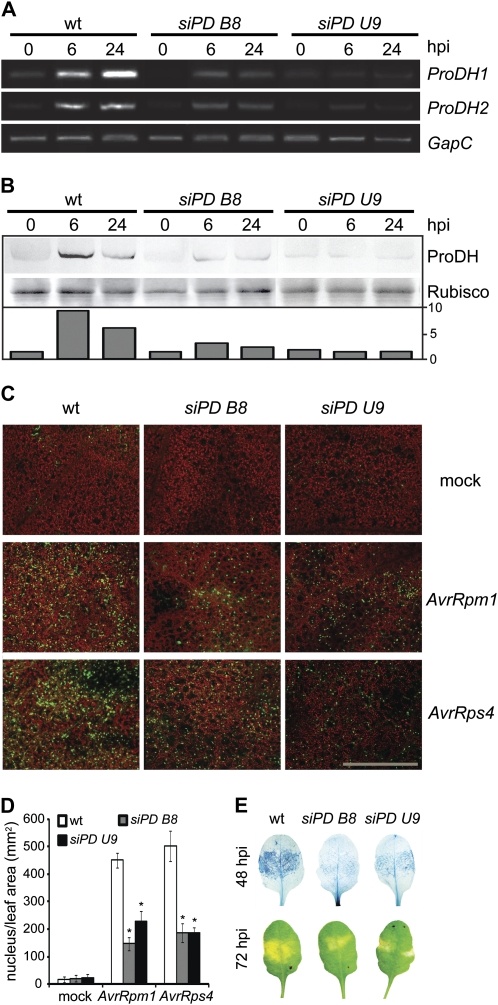
Having determined that siPD B8 and siPD U9 behave as ProDH-deficient plants, we then evaluated how these plants activate HR. In contrast with wild-type tissues, siPD B8 and siPD U9 tissues were unable to accumulate ProDH1 and ProDH2 transcripts in response to Pst-AvrRpm1 at 6 and 24 hpi (Fig. 6A). Furthermore, none of these tissues increased the ProDH content in response to the pathogen (Fig. 6B). Therefore, siPD B8 and siPD U9 plants were analyzed for activation of hypersensitive cell death by Pst-AvrRpm1. At 24 hpi, cell death was detected by SYTOX Green staining and confocal microscopy. This dye enters the cells having the membrane integrity affected and fluoresces upon binding to the nuclear DNA (Truernit and Haseloff, 2008; Fig. 6C). Thus, cell death can be quantified in vivo based on the number of fluorescent nuclei per leaf area. In mock-treated tissues, negligible cell death levels were found (Fig. 6, C and D), while as expected, cell death was notably increased in wild-type tissues treated with Pst-AvrRpm1 (27-fold compared with the mock control). Remarkably, under this condition, the siPD B8 and siPD U9 tissues showed 7- and 10-fold increases in cell death levels with respect to mock controls, respectively. These results indicated 32% (siPD B8) and 50% (siPD U9) reduction in the cell death levels existing in wild-type tissues (Fig. 6D). To verify if these differences were maintained over time, cell death was analyzed at 48 hpi. At this stage, SYTOX Green was detected outside the nuclei (data not shown), thus impairing cell death quantification. Therefore, trypan blue staining was used. We found that at 48 hpi with Pst-AvrRpm1, cell death was still reduced in the transgenic tissues, indicating that the differences observed earlier were not kinetic (Fig. 6E). As expected, trypan blue also detected a reduction of cell death at 24 hpi with Pst-AvrRpm1 in the transgenic plants (data not shown). Moreover, we also observed attenuated HR symptoms in transgenic tissues treated with Pst-AvrRpm1 (Fig. 6E).
Figure 6.
siPD B8 and siPD U9 plants have reduced cell death in response to avirulent bacteria. A and B, Comparative analysis of responses triggered by avirulent pathogens (5 × 106 cfu mL−1) in wild-type (wt) and ProDH-silenced (siPD B8 and siPD U9) plants. ProDH1 and ProDH2 transcript abundance (A) and ProDH levels (B) are quantified in untreated and Pst-AvrRpm1-treated leaves at 6 and 24 hpi, as described in Figure 1, C and D. One representative from three independent experiments is shown, and each experiment included three plants and four leaves from different plants per time point. C, Identification of nonviable cells in Pst-AvrRpm1- or Pst-AvrRps4-challenged leaves isolated at 24 hpi by SYTOX Green staining and laser-scanning confocal microscopy (Truernit and Haseloff, 2008). Green signals indicate probe fluorescence, and red signals indicate chlorophyll autofluorescence. Images are representative of 18 total images. Bar = 400 μm. D, Quantification of stained nucleus per leaf area from the images described in C using ImageJ software (W. Rasband, National Institutes of Health). Values represent means ± se of quantification of 18 images from three biologically independent infection experiments, each one containing three plants per genotype. Significant differences between wild-type and transgenic plants are indicated (* P < 0.01, t test). E, Pst-AvrRpm1-treated leaves were excised at 48 hpi to be stained with trypan blue (top row) or analyzed at 72 hpi for macroscopic HR features (bottom row). One representative from 20 leaves is shown per genotype (three independent infection experiments and four plants each).
Reduced hypersensitive cell death was also detected in transgenic tissues exposed to the avirulent pathogen Pst-AvrRps4. This pathogen stimulates HR through an ENHANCED DISEASE SUSCEPTIBILITY1-dependent circuit, in contrast to Pst-AvrRpm1, which signals cell death through NDR1 (Aarts et al., 1998). In response to Pst-AvrRps4, both siPD B8 and siPD U9 plants showed 37% of the cell death levels found in wild-type plants (Fig. 6C). However, none of the infection experiments with these avirulent pathogens showed a complete elimination of hypersensitive cell death in the ProDH-silenced plants (data not shown). Therefore, ProDH deficiency reduced the hypersensitive cell death triggered by Pst-AvrRpm1 and Pst-AvrRps4.
Sensitivity to Avirulent Bacteria of ProDH-Silenced Plants
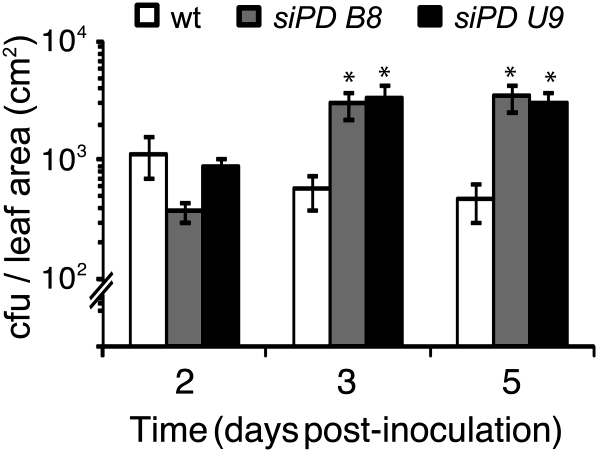
A reduction in hypersensitive cell death may support the proliferation of biotrophic pathogens in planta. To investigate whether this could occur in the ProDH-deficient plants, we compared Pst-AvrRpm1 growth in transgenic and wild-type tissues. At day 2 post inoculation, transgenic and wild-type tissues contained similar bacterial contents. As expected, wild-type leaves were able to maintain and even reduce the bacterial content over the following 3 d, as a consequence of defense activation. In contrast, in siPD B8 and siPD U9 leaves, pathogen proliferation occurred during this period. The bacterial content increased 9- and 4- fold in siPD B8 and siPD U9 leaves, respectively, revealing a low capacity of these tissues to restrict the spread of the pathogen (Fig. 7).
Figure 7.
siPD B8 and siPD U9 plants have enhanced susceptibility to avirulent bacteria. Pst-AvrRpm1 growth in planta is shown. Bacterium was inoculated at 5 × 105 cfu mL−1 to quantify bacterial content at the indicated times. Significant differences between wild-type (wt) and transgenic plants are indicated (* P < 0.01, t test). Data represent means ± se of three independent infection experiments (two sets of six discs isolated from at least three plants per experiment, for a total of 36 discs per time point).
Oxidative Burst in the ProDH-Silenced Plants
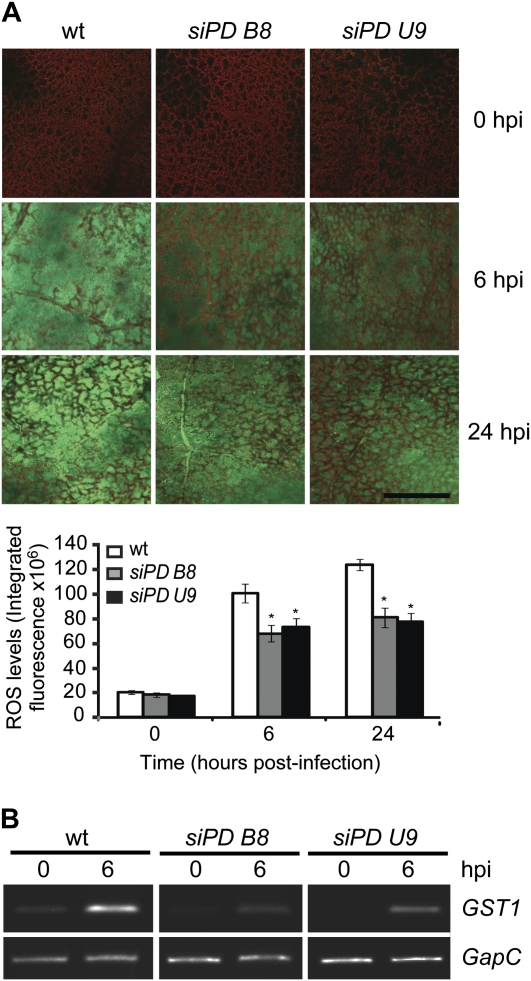
In animals, the activation of ProDH supports ROS accumulation (Donald et al., 2001). Due to the fact that ProDH activation precedes the oxidative burst during HR development (Fig. 4), we examined whether this enzyme affected ROS accumulation under this condition. This aspect was investigated by comparing the ROS content in wild-type and transgenic leaf tissues treated with Pst-AvrRpm1 and excised at 6 and 24 hpi, using untreated leaves as controls. Leaves were stained with 2′,7′- dichlorodihydrofluorescein diacetate (H2DCFDA) and analyzed by confocal microscopy. A similar ROS content was detected in the noninfiltrated tissues of the three plants (Fig. 8A). In contrast, a reduced ROS accumulation was found in the pathogen-challenged tissues of the silenced plants. At 6 hpi, the siPD B8 and siPD U9 tissues achieved only 63% and 68%, respectively, of the fluorescence levels found in wild-type plants (Fig. 8A, bottom). Moreover, reduced ROS levels were observed in these plants at 24 hpi. Consistently, the transgenic plants displayed a lower activation of GST1 (At1g02930; also named GSTF6) compared with wild-type plants (Fig. 8B), with this gene being a sensitive marker of ROS accumulation (Alvarez et al., 1998). Therefore, these results support the idea that ProDH contributed to the generation of the oxidative burst in wild-type HR developing tissues.
Figure 8.
Reduction of the oxidative burst in ProDH-silenced plants. Quantification of ROS in untreated and Pst-AvrRpm1-infiltrated (5 × 106 cfu mL−1) leaf tissues from wild-type (wt) and ProDH-silenced plants (siPD B8 and siPD U9) at 6 and 24 hpi. A, Leaves were stained with H2DCFDA to detect hydrogen peroxide by laser-scanning confocal microscopy. Images were taken with a 10× objective. Green signals indicate probe fluorescence, and red signals indicate chlorophyll autofluorescence. Bar = 400 μm. Images are representative of 16 total images. Green fluorescence values indicated at the bottom were obtained from images analyzed with ImageJ software (W. Rasband, National Institutes of Health). Values represent means ± se of samples from two independent infection experiments, each one containing eight infected leaves isolated from four different plants. Significant differences between wild-type and transgenic plants are indicated (* P < 0.05, t test). B, Abundance of GST1 (also named GSTF6) transcripts of samples from leaves obtained as described in A. One representative of two independent infection experiments (each one including at least three plants and four leaves per time point) is shown.
DISCUSSION
In many different plants, including Arabidopsis, the ProDH activity is sensitive to transcriptional regulation (Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996), with posttranscriptional control of this activity not having been reported (Verbruggen and Hermans, 2008; Szabados and Savouré, 2010). Therefore, the accumulation of ProDH transcripts occurring in Pst-AvrRpm1-treated tissues may be indicative of an increased enzyme activity in these tissues, with the protein accumulating under such conditions (Fig. 1). Unfortunately, our attempts to quantify ProDH activity in leaf tissues have been unsuccessful, apparently due to both the low enzyme activity and the high abundance of components interfering with NADH quantification in these tissues. Nevertheless, we determined ProDH activity in photosynthetic Arabidopsis cultured cells that triggers race-specific defenses through the RPM1 pathway (Cecchini et al., 2009) and reproduces the induction of ProDH1 by SA and the avirulent pathogen that was originally detected in planta (Fig. 4, A and B; see below). The finding that ProDH activity was specifically increased by avirulent bacteria was consistent with the ProDH gene expression patterns observed in vitro and in planta under infection conditions.
The activation of ProDH by Pst-AvrRpm1 is detected from 6 hpi (Figs. 1C, 2A, and 4C). At this initial stage, the enzyme is expected to oxidize the free Pro contained in the challenged tissues. Interestingly, at least during the initial 12 hpi, the induction of ProDH was accompanied by the accumulation of P5CR but not the P5CDH transcripts (Fig. 2A). In Arabidopsis, ProDH, P5CR, and P5CDH are regulated at the transcriptional level, whereas P5CR and P5CDH are also under posttranscriptional control (Verbruggen and Hermans, 2008; Szabados and Savouré, 2010). Therefore, the gene expression patterns described above may suggest stimulation of the Pro-P5C cycle in HR-developing tissues, where ProDH may contribute to generate ROS at the mitochondria (Miller et al., 2009), with the finding of constant Pro and P5C levels until 8 hpi (Fig. 2B) supporting this possibility. Furthermore, the increase in Pro occurring at 12 hpi (Fig. 2B; see Fig. 1A in Fabro et al., 2004), whose source is unknown, is consistent with hyperactivation of the cycle and consequent enhancement of ROS. However, formal studies will be required to determine whether the Pro-P5C cycle contributes to generate mitochondrial ROS in the HR.
We previously observed that in Arabidopsis, Pro synthesis is positively regulated by SA (Fabro et al., 2004). In this work, we report that the Arabidopsis Pro catabolic gene ProDH1 was also induced by this hormone. There, ProDH1 is considered to encode the isoform that contributes most to Pro degradation (Kiyosue et al., 1996; Mani et al., 2002; Nanjo et al., 2003), while the product encoded by ProDH2 has also been recognized as a functional enzyme by recent studies (Funck et al., 2010). Interestingly, ProDH1 and ProDH2 are differentially regulated and encode isoforms suspected to display nonredundant functions (Ribarits et al., 2007; Funck et al., 2010). Consistent with these observations, exogenous SA activated ProDH1 at low concentrations (50 μm) but had no effect on ProDH2 expression even at high doses (2.5 mm; Fig. 3A). Although both ProDH1 and ProDH2 genes responded with similar kinetics to treatment with the pathogen, preliminary results suggest that both isoforms may differentially contribute to defenses (N.M. Cecchini and M.E. Alvarez, unpublished data). It is likely that activation of ProDH1 by Pst-AvrRpm1 occurring at 6 hpi (Fig. 1C) is mediated by SA, as the hormone levels began to increase at this time in Pst-AvrRpm1-treated tissues (Blanco et al., 2009). Moreover, this early activation of ProDH1 appeared to be signaled by both SID2 and NPR1, as infected sid2-2 and npr1-1 plants were impaired in this response (Fig. 3C). Transcription factors belonging to the basic leucine zipper protein (bZIP) family, such as bZIP10, -11, -53, -44, and -2, mediate the transcriptional regulation of ProDH genes in Arabidopsis (Satoh et al., 2004; Weltmeier et al., 2006; Hanson et al., 2008) and may eventually activate the genes in the HR. In addition, AtbZIP10 acts as a positive regulator of the HR antagonized by the negative cell death regulator LSD1 (Kaminaka et al., 2006). At later stages of HR development (24 hpi), a mild activation of ProDH1 was detected in sid2-2 and npr1-1 plants, suggesting that at this time NPR1- and/or SID2-independent pathways may regulate the expression of the gene. To our knowledge, these data provide the first information on the signaling pathway activating the Pro catabolism in infected plants.
In this study, transgenic plants silencing the ProDH1/2 expression were constructed to evaluate the role of ProDH in HR development, with plants having the highest silencing responses displaying the lowest fertility. This phenotype may be due to ProDH deficiency, since the enzyme displays a vital role in reproductive organs (Dashek and Harwood, 1974; Zhang et al., 1982; Hare and Cress, 1997). The siPD B8 and siPD U9 plants selected for our studies were silenced in both ProDH1 and ProDH2 genes, as demonstrated by the response of these plants to Pro or Pst-AvrRpm1 treatment (Figs. 5C and 6A). However, these plants still contained traces of the ProDH transcripts and protein under all analyzed conditions, indicating that silencing was not complete. When exposed to external Pro, both transgenics (Fig. 5) behaved as ProDH-deficient plants (Mani et al., 2002; Nanjo et al., 2003; Funck et al., 2010). In addition, they developed reduced cell death in response to Pst-AvrRpm1 and Pst-AvrRps4 (Fig. 6, C and D), implying that the decrease of ProDH occurring in siPD B8 and siPD U9 plants sufficiently affected the death pathways. However, it is not known whether the residual ProDH activity of these plants was responsible for the remaining cell death levels.
We detected up to a 30% increase in ProDH activity in cultured cells triggering HR (Fig. 4C), a response that was eliminated by incubation with the ProDH inhibitor l-4-thiazolidine carboxylic acid (see “Materials and Methods”). Nevertheless, as mentioned above, the extent of ProDH activation in leaf tissues that develop HR remains to be determined. Although these tissues showed a large increase in the total protein content (Fig. 1D), the ProDH fraction that catalyzes mitochondrial Pro oxidation under these circumstances is not known. It is also unclear whether the in vitro assays used here underestimated the changes in the ProDH activity occurring in vivo, as this catalytic capacity may be modulated by association with the mitochondrial inner membrane or by different electron acceptors under physiological conditions (Elthon and Stewart, 1981). Although our results suggest that reduction, and not necessarily elimination, of ProDH activity is sufficient to alter the normal development of HR, additional studies will be required to determine the activation level required to modulate this defense response.
Our results also suggest that ProDH signals hypersensitive cell death by acting downstream of the HR cascade stimulated by RPM1 or RPS4, in a step common to both pathways. As ProDH1 expression is modulated by SID2 and NPR1 (Fig. 3C), the enzyme may work downstream of NPR1. However, it is not known how the enzyme contributes to cell death. Although activation of ProDH without proper stimulation of P5CDH was able to produce P5C accumulation, which might be toxic for plants (Deuschle et al., 2001, 2004), our experiments did not reveal P5C accumulation during the initial 12 hpi (Fig. 2B). Another possibility is that ProDH generates or signals the oxidative stress and thereby stimulates the death process. In agreement with this, ProDH activation paralleled the onset of the oxidative burst in cells developing HR (Fig. 4, C and D), with ProDH-silenced plants reducing the ROS accumulation in response to pathogen (Fig. 8). The homeostasis of mitochondria is strongly modified in infected tissues (Krause and Durner, 2004; Yao and Greenberg, 2006; Mur et al., 2008), and the SA accumulated in these tissues may alter the flux of electrons in the mETC, generating conditions that promote ROS accumulation (Xie and Chen, 2000; Norman et al., 2004). As a flavoprotein, ProDH may charge electrons at different levels of the mETC (Hare and Cress, 1997; Phang et al., 2008), contributing to overload the system and thus promoting an abnormal flow of electrons to the oxygen and the generation of superoxide. Related to this, ProDH supports mitochondrial ROS accumulation in Arabidopsis cells, thereby activating the P5C-Pro cycle (Miller et al., 2009). Remarkably, ProDH seems to promote mitochondrial oxidative burst in animal cells, where the POX gene is a target of p53 (Polyak et al., 1997), with POX overexpression triggering apoptosis by the accumulation of ROS at the mitochondria (Donald et al., 2001; Maxwell and Rivera, 2003). It is interesting that in Thermus thermophilus, ProDH autogenously catalyzes the generation of superoxide in vitro, thus demonstrating its structural flexibility in controlling the exposure of FAD to the solvent (White et al., 2007). However, it is not known whether any of these features are conserved in plant ProDHs. Moreover, it is also unclear how ProDH contributes to the oxidative stress and cell death in the HR and whether the enzyme has a conserved role in the defense systems of plants and animals.
The ProDH-silenced plants showed an enhanced susceptibility to Pst-AvrRpm1 (Fig. 7), suggesting that the enzyme stimulates defenses against avirulent pathogens. Such an effect may be directly associated with the generation of ROS and cell death by the enzyme, as these responses help to restrict pathogen proliferation (Bent and Mackey, 2007). Alternatively, ProDH may have other metabolic implications in infected tissues. First, as photosynthesis is inhibited under this condition, thereby reducing the available energy sources for defense, the activation of ProDH may provide energy at the expense of Pro (Bolton, 2009). In addition, as part of the P5C-Pro cycle, ProDH may shuttle reducing equivalents to mitochondria, thus lowering the cytosolic NADPH levels required for stimulation of the oxidative pentose phosphate pathway (Hare and Cress, 1997; Nemoto and Sasakuma, 2000), a central route for the activation of disease resistance and the generation of secondary defense metabolites (Scharte et al., 2009).
In summary, the functional implication of ProDH in the HR opens a new scenario for the study of the enzyme under biotic stress conditions. The degradation of Pro by ProDH may affect cell death and disease resistance through different mechanisms, including alterations in the cellular redox state and the provision of carbon, energy, and secondary metabolites to infected tissues. Additional studies should help to clarify the mechanisms by which ProDH stimulates defenses.
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Infections
Arabidopsis (Arabidopsis thaliana) wild-type, sid2-2 (Wildermuth et al., 2001), and npr1-1 (Cao et al., 1997) plants from the Col-0 ecotype were used. Plants were grown in soil under 8 h of light/16 h of dark at 22°C. The Arabidopsis (Col-0) photosynthetic cell suspension cultures were grown and maintained as described previously (Cecchini et al., 2009). The Pseudomonas syringae pv tomato DC3000 virulent strain (Pst-vir) and its isogenic avirulent strains carrying the Rpm1 (Pst-AvrRpm1; Ritter and Dangl, 1996) or Rps4 (Pst-AvrRps4; Aarts et al., 1998) gene were grown on King’s B medium supplemented with kanamycin (50 μg mL−1) and rifampicin (100 μg mL−1). Pathogens were inoculated into leaf tissues (Pavet et al., 2005) at 5 × 105 or 5 × 106 colony-forming units (cfu) mL−1 for bacterial growth curves and all other studies, respectively. The bacterial growth curves were determined as reported by Pavet et al. (2005). Inoculation of pathogens on cultured cells (108 cfu mL−1) was performed according to Cecchini et al. (2009).
Gene Expression
Semiquantitative reverse transcription-PCR was performed using 1 μg of total RNA (Pavet et al., 2005), random hexamer primers, and Moloney murine leukemia virus reverse transcriptase (Invitrogen) to synthesize cDNA, with the following primers and number of cycles for the PCRs: ProDH1 (At3g30775) forward (5′-TGATGGAGAAAGCATCAAACGG-3′) and reverse (5′-TCTCCTCTTAAGTTCCATCCTC-3′), 31 cycles; ProDH2 (At5g38710) forward (5′-CGTCGAAGCTGCTAAAACCCT-3′) and reverse (5′-CGTTCGATTCTTGACATCTAAG-3′), 30 cycles; P5CR (At5g14800) forward (5′-GCTCACCCGTCCGAAGTAT-3′) and reverse (5′-AGCTGCAACAGAAACCAGAAT-3′), 30 cycles; P5CDH (At5g62530) forward (5′-ATGTTGGAGCACATGG-3′) and reverse (5′-GTGACGAGTTCGTAGG-3′), 30 cycles; and GapC (At3g04120) forward (5′-CACTTGAAGGGTGGTGCCAAG-3′) and reverse (5′-CCTGTTGTCGCCAACGAAGTC-3′), 25 cycles. The constitutively expressed GapC gene (Kato et al., 2003) was used to control that an equal amount of cDNA was used in each reaction.
ProDH Analysis
For western-blot assays, total proteins were extracted from 50 mg of leaf tissue. Samples were ground in liquid nitrogen, resuspended in 1 mL of ice-cold extraction buffer (50 mm Tris-HCl, pH 6.8, 3% SDS, 30% glycerol, 2% β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail from Sigma-Aldrich), and heated for 5 min at 85°C before being cooled on ice and centrifuged for 10 min at 12,000 rpm at 4°C. Samples (50 μg of protein) were separated on 8% SDS-PAGE gels and transferred to a nitrocellulose membrane. Equal protein loading was confirmed by detection of the Rubisco small subunit in Ponceau S-stained membranes. Membranes were blocked (5% [w/v] nonfat milk in phosphate-buffered saline, 25°C, 1 h) and incubated with anti-ProDH rabbit polyclonal antibodies (1:2,000, 4°C, 16 h). Goat anti-rabbit IgG IRDye 800CW (LI-COR Bioscience) was used as the secondary antibody, and its emission was detected using the Odyssey Infrared Imaging System (LI-COR Bioscience). The ProDH rabbit polyclonal antibodies were prepared according to Harlow and Lane (1988) using the 11-amino acid N-terminal peptide FLMEKASNGSG conserved in ProDH1 and ProDH2 for immunization. These antibodies detected a band of 55 kD in the Arabidopsis total protein extracts (Supplemental Fig. S1). The PDH activity was determined according to Kant et al. (2006). Aliquots (1 mL) of suspension cultures were collected by centrifugation, frozen on liquid nitrogen, and homogenized with a pestle in 4 volumes of extraction buffer (50 mm Tris-HCl, pH 7.4, 7 mm MgCl2, 0.6 m KCl, 3 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 5% [w/v] polyvinylpyrrolidone). Extracts were centrifuged at 16,000g for 10 min, and supernatants were used to determine ProDH activity in vitro by incubation in 0.15 m Na2CO3-HCl buffer (pH 10.3) with 1 mm NAD+ and 20 mm l-Pro to monitor the NAD+ reduction at 340 nm. The specificity of the enzymatic reaction was evaluated by incubation of samples with the ProDH inhibitor l-4-thiazolidine carboxylic acid (Elthon and Stewart, 1984). The ProDH activity was expressed as nmol NADH min−1 mg−1 protein, with the protein content being measured by the Bradford assay.
Pro and P5C Quantification
Pro and P5C were quantified by using the ninhydrin and ortho-aminobenzaldehyde assays (Bates et al., 1973; Miller et al., 2009). Samples were homogenized in 3% (w/v) sulfosalicylic acid and centrifuged for 10 min. The supernatant was divided into two identical aliquots, which were used for Pro and P5C determination. The Pro and P5C values were related to the l-Pro and dl-P5C (Sigma) calibration curves.
ROS Analysis
ROS levels were determined by H2DCFDA (Molecular Probes, Invitrogen) staining. Cell culture aliquots (1 mL) were incubated with the probe (10 μm, pH 7) on multiwell plates to measure fluorescence (520 nm), exactly 5 min after probe addition, using a Fujifilm FLA-3000 Fluorescence and Storage Phosphorimager, as described previously (Cecchini et al., 2009). Leaf samples submerged in H2DCFDA (20 μm, pH 7) were vacuum infiltrated to introduce the dye and then maintained in the dark for 10 min as described by Lorrain et al. (2004). Samples were analyzed in an Olympus FV1000 laser-scanning confocal microscope for parallel detection of the probe (excitation, 488 nm; emission, 505–530 nm) and chlorophyll autofluorescence (excitation, 633; emission, 650 nm). Images were taken using a 10×, 0.3 numerical aperture (UPlanFLN; Olympus) objective and a confocal aperture of 200 μm. Both images for dye and plant autofluorescence were acquired for the same field using a sequential acquisition mode. To quantify ROS, we measured the integrated fluorescence density of the dye from representative images using ImageJ software (W. Rasband, National Institutes of Health).
Cell Death Analysis
In the cell culture system, cell death was analyzed by Evans blue staining by determining a cell death value of 100% with heat-killed cells (65°C, 15 min) from 6-d-old untreated cultures (Cecchini et al., 2009). In leaf tissues, dying cells were visualized by lactophenol trypan blue (Pavet et al., 2005) or SYTOX Green (Molecular Probes, Invitrogen; Truernit and Haseloff, 2008) staining. The second assay analyzed cell death in vivo. Leaves were vacuum infiltrated with SYTOX Green (0.2 μm, pH 7) and maintained in the dark for 20 min before being washed with distilled water and observed with an Olympus FV1000 laser-scanning confocal microscope. Images were taken using a 10×, 0.3 numerical aperture (UPlanFLN; Olympus) objective and a confocal aperture of 200 μm. Both images for dye (excitation, 488 nm; emission, 505–530 nm) and plant autofluorescence (excitation, 633; emission, 650 nm) were acquired for the same field using a sequential acquisition mode. The number of nuclei per leaf area (mm2) was quantified using ImageJ software (W. Rasband, National Institutes of Health).
ProDH-Silenced Plants
The pENTR/D-TOPO (Gateway system; Invitrogen) vector was used to clone a 192-bp cDNA from the ProDH1 gene, having 76% identity with ProDH2, which was synthesized by PCR (forward primer, 5′-CACCATTTTTGCGCCGGTGAAGAT-3′; reverse primer, 5′-CTGAGCTAAAGTGAGATGTTGGTA-3′). This silencing cassette was transferred to the binary vector pK7GWIWG2D(II) (Karimi et al., 2002), and then the construct integrity was confirmed by sequencing. Binary vectors were electroporated to Agrobacterium tumefaciens strain GV3101 pMP90 and used for transformation of Arabidopsis (Col-0) plants by the floral dip technique (Clough and Bent, 1998). Primary transformed (T0) seedlings were selected on Murashige and Skoog agar plates supplemented with kanamycin (50 μg mL−1) and evaluated for segregation of the silencing cassette. Two representative T3 plants, named siPD B8 and siPD U9, carrying either a single copy of the cassette in homozygosis or multiple copies of this construct, respectively, were characterized in detail. The phenotypes of both plants in response to pathogens or exogenous Pro were also present in their parental lines as well as in other T3 plants obtained in these transformation experiments (data not shown).
Responses of Plants to Exogenous Pro
Seeds were germinated on GM synthetic medium (0.5× Murashige and Skoog salts, 1× Gamborg vitamins, 1% Suc, and 1.6% agar) supplemented with different Pro concentrations, which were then used in the studies. Plant growth on supplemented medium was evaluated at the age of 3 weeks, and then Pro and ProDH protein levels were determined at the age of 2 weeks, using aerial tissues from six to eight plantlets for each measurement. Tissues were washed with distilled water prior to being used for Pro quantification or western-blot assays.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Anti-ProDH antibody validation.
Acknowledgments
We thank Dr. László Szabados for critical reading of the manuscript. N.M.C. and M.I.M. are CONICET fellows. M.E.A. is a senior Career Investigator of CONICET.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME. (2000) Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol Biol 44: 429–442 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C. (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Roberts JK, Mitchell HJ, Zhang R, Lawrence GJ, Ellis JG, Pryor TJ. (2002) A plant gene up-regulated at rust infection sites. Plant Physiol 129: 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. (1973) Rapid determination of free proline for water stress studies. Plant Soil 39: 205–207 [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Boggess SF, Koeppe DE. (1978) Oxidation of proline by plant mitochondria. Plant Physiol 62: 22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense: fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bonner CA, Williams DS, Aldrich HC, Jensen RA. (1996) Antagonism by L-glutamine of toxicity and growth inhibition caused by other amino acids in suspension cultures of Nicotiana silvestris. Plant Sci 113: 43–58 [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cecchini NM, Monteoliva MI, Blanco F, Holuigue L, Alvarez ME. (2009) Features of basal and race-specific defences in photosynthetic Arabidopsis thaliana suspension cultured cells. Mol Plant Pathol 10: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dashek W, Harwood H. (1974) Proline, hydroxyproline and lily pollen tube elongation. Ann Bot (Lond) 38: 947–959 [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. (2004) The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16: 3413–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Hellmann H, Däschner K, Binder S, Frommer WB. (2001) A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J 27: 345–356 [DOI] [PubMed] [Google Scholar]
- Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. (2001) Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res 61: 1810–1815 [PubMed] [Google Scholar]
- Elthon TE, Stewart CR. (1981) Submitochondrial location and electron transport characteristics of enzymes involved in proline oxidation. Plant Physiol 67: 780–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Stewart CR. (1984) Effects of the proline analog l-thiazolidine-4-carboxylic acid on proline metabolism. Plant Physiol 74: 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro G, Kovács I, Pavet V, Szabados L, Alvarez ME. (2004) Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol Plant Microbe Interact 17: 343–350 [DOI] [PubMed] [Google Scholar]
- Funck D, Eckard S, Müller G. (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Stadelhofer B, Koch W. (2008) Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol 8: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn CH, Phang JM. (1983) Transfer of reducing equivalents into mitochondria by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch Biochem Biophys 225: 95–101 [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949 [DOI] [PubMed] [Google Scholar]
- Hare P, Cress W. (1997) Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul 21: 79–102 [Google Scholar]
- Harlow E, Lane D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Haudecoeur E, Planamente S, Cirou A, Tannières M, Shelp BJ, Moréra S, Faure D. (2009) Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci USA 106: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. (2000) Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol 122: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH, Cavalieri AJ. (1979) Proline oxidase and water stress-induced proline accumulation in spinach leaves. Plant Physiol 63: 531–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, III, Merkle T, Schäfer E, Harter K, et al. (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25: 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S. (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421–426 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. (1996) A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 8: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Durner J. (2004) Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol Plant Microbe Interact 17: 131–139 [DOI] [PubMed] [Google Scholar]
- Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, Oberley LW, Phang JM. (2005) MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis 26: 1335–1342 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balagué C, Roby D. (2004) Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16: 2217–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S, Majerus V, Kinet JM. (1999) NaCl effects on proline metabolism in rice (Oryza sativa) seedlings. Physiol Plant 105: 405–458 [Google Scholar]
- Mani S, Van De Cotte B, Van Montagu M, Verbruggen N. (2002) Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol 128: 73–83 [PMC free article] [PubMed] [Google Scholar]
- Maxwell SA, Davis GE. (2000) Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA 97: 13009–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell SA, Rivera A. (2003) Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. J Biol Chem 278: 9784–9789 [DOI] [PubMed] [Google Scholar]
- Meon S, Fisher JM, Wallace HR. (1978) Changes in free proline following infection of plants with either Meloidogyne javanica or Agrobacterium tumefaciens. Physiol Plant Pathol 12: 251–256 [Google Scholar]
- Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A. (2009) Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem 284: 26482–26492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell HJ, Ayliffe MA, Rashid KY, Pryor AJ. (2006) A rust-inducible gene from flax (fis1) is involved in proline catabolism. Planta 223: 213–222 [DOI] [PubMed] [Google Scholar]
- Mohanty SK, Sridhar R. (1982) Physiology of rice tungro virus disease: proline accumulation due to infection. Physiol Plant 56: 89–93 [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Fujita M, Seki M, Kato T, Tabata S, Shinozaki K. (2003) Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol 44: 541–548 [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Sasakuma T. (2000) Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.). Plant Sci 158: 53–60 [DOI] [PubMed] [Google Scholar]
- Norman C, Howell KA, Millar AH, Whelan JM, Day DA. (2004) Salicylic acid is an uncoupler and inhibitor of mitochondrial electron transport. Plant Physiol 134: 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH. (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139: 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Lu Q, Verma DP. (1996) Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet 253: 334–341 [DOI] [PubMed] [Google Scholar]
- Phang JM, Pandhare J, Zabirnyk O, Liu Y. (2008) PPARgamma and proline oxidase in cancer. PPAR Res 2008: 542694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. (1997) A model for p53-induced apoptosis. Nature 389: 300–305 [DOI] [PubMed] [Google Scholar]
- Radwan DE, Fayez KA, Mahmoud SY, Hamad A, Lu G. (2007) Physiological and metabolic changes of Cucurbita pepo leaves in response to zucchini yellow mosaic virus (ZYMV) infection and salicylic acid treatments. Plant Physiol Biochem 45: 480–489 [DOI] [PubMed] [Google Scholar]
- Ribarits A, Abdullaev A, Tashpulatov A, Richter A, Heberle-Bors E, Touraev A. (2007) Two tobacco proline dehydrogenases are differentially regulated and play a role in early plant development. Planta 225: 1313–1324 [DOI] [PubMed] [Google Scholar]
- Ritter C, Dangl JL. (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P, et al. (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh R, Fujita Y, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. (2004) A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol 45: 309–317 [DOI] [PubMed] [Google Scholar]
- Scharte J, Schön H, Tjaden Z, Weis E, von Schaewen A. (2009) Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc Natl Acad Sci USA 106: 8061–8066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant Microbe Interact 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Skubatz H, Meeuse BJD, Bendich AJ. (1989) Oxidation of proline and glutamate by mitochondria of the inflorescence of voodoo lily (Sauromatum guttatum). Plant Physiol 91: 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakman EC. (1915) Relation between Puccina graminis and plants highly resistant to its attack. J Agric Res 4: 193–199 [Google Scholar]
- Szabados L, Savouré A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Truernit E, Haseloff J. (2008) A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. (2008) Proline accumulation in plants: a review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hua XJ, May M, Van Montagu M. (1996) Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc Natl Acad Sci USA 93: 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. (2008) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W. (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TA, Krishnan N, Becker DF, Tanner JJ. (2007) Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem 282: 14316–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xie Z, Chen Z. (2000) Harpin-induced hypersensitive cell death is associated with altered mitochondrial functions in tobacco cells. Mol Plant Microbe Interact 13: 183–190 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT. (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HQ, Croes A, Linskens H. (1982) Protein synthesis in germinating pollen of Petunia: role of proline. Planta 154: 199–203 [DOI] [PubMed] [Google Scholar]