Abstract
Ferrochelatase (FeCH) catalyzes the insertion of Fe2+ into protoporphyrin, forming protoheme. In photosynthetic organisms, FeCH and magnesium chelatase lie at a biosynthetic branch point where partitioning down the heme and chlorophyll (Chl) pathways occurs. Unlike their mammalian, yeast, and other bacterial counterparts, cyanobacterial and algal FeCHs as well as FeCH2 isoform from plants possess a carboxyl-terminal Chl a/b-binding (CAB) domain with a conserved Chl-binding motif. The CAB domain is connected to the FeCH catalytic core by a proline-rich linker sequence (region II). In order to dissect the regulatory, catalytic, and structural roles of the region II and CAB domains, we analyzed a FeCH ΔH347 mutant that retains region II but lacks the CAB domain and compared it with the ΔH324-FeCH mutant that lacks both these domains. We found that the CAB domain is not required for catalytic activity but is essential for dimerization of FeCH; its absence causes aberrant accumulation of Chl-protein complexes under high light accompanied by high levels of the Chl precursor chlorophyllide. Thus, the CAB domain appears to serve mainly a regulatory function, possibly in balancing Chl biosynthesis with the synthesis of cognate apoproteins. Region II is essential for the catalytic function of the plastid-type FeCH enzyme, although the low residual activity of the ΔH324-FeCH is more than sufficient to furnish the cellular demand for heme. We propose that the apparent surplus of FeCH activity in the wild type is critical for cell viability under high light due to a regulatory role of FeCH in the distribution of Chl into apoproteins.
Cyanobacteria, algae, and plants synthesize chlorophyll (Chl), heme, and linear tetrapyrroles such as phycobilins via a common branched pathway. At the beginning of tetrapyrrole biosynthesis, the initial precursor, 5-aminolevulinic acid (ALA), is made from Glu via glutamyl-tRNA and is subsequently converted in several steps to protoporphyrin IX (PPIX), the last common precursor for both Chl and heme biosynthesis. Insertion of Fe2+ into this porphyrin macrocycle by ferrochelatase (FeCH) leads to heme, whereas insertion of Mg2+ by magnesium chelatase leads to magnesium-protoporphyrin IX (MgP), the first biosynthetic intermediate on the “green” Chl branch (for review, see Tanaka and Tanaka, 2007).
Because the levels of heme and Chl vary according to cell development, growth, or light conditions, special regulatory mechanisms have evolved that control heme and Chl formation and coordinate their levels with the synthesis of the corresponding apoproteins (Müller and Eichacker, 1999). Stringent control of this pathway is particularly essential for oxygenic organisms such as plants and cyanobacteria that have to cope with the problem of photooxidation. Chl and its intermediates are readily excited by light and, unquenched, form reactive oxygen species under aerobic conditions. Therefore, for organisms carrying out oxygenic photosynthesis, it is essential to minimize cellular levels of unbound “free” tetrapyrroles.
The mechanisms that control and regulate Chl/heme biosynthesis mostly remain to be elucidated, although it is well established that the total metabolic flow through the pathway is controlled at the point of ALA formation, a rate-limiting step for the whole pathway (for review, see Tanaka and Tanaka, 2007; Masuda and Fujita, 2008). The biosynthetic step controlling ALA formation is most probably that catalyzed by glutamyl-tRNA reductase. This is the first enzyme in the pathway and is modulated by a wide range of regulatory signals, in line with the idea that this is the central controller of total tetrapyrrole flux (Kumar et al., 1996; Tanaka et al., 1996; McCormac et al., 2001). Another key regulatory site is at the branch point between Chl and heme biosynthesis, where distribution of PPIX has to be carefully balanced according to the actual demand for both essential pigments. These regulatory mechanisms that govern the distribution of PPIX remain to be elucidated, although the roles of both chelatases have become apparent (Papenbrock et al., 2000, 2001; Sobotka et al., 2005).
FeCH is an enzyme of special interest regarding the regulation of tetrapyrrole biosynthesis. FeCH activity may regulate the flux down both the heme and Chl branches of the pathway via increased production of heme, which is expected to inhibit the synthesis of ALA at the start of the pathway through feedback control (Weinstein et al., 1993; Vothknecht et al., 1998; Goslings et al., 2004). Differential consumption of the common substrate PPIX by FeCH and magnesium chelatase might also control partitioning at the Chl/heme branch point (for review, see Cornah et al., 2003). Finally, a mechanism whereby the Chl branch exerts control over the heme pathway would provide an even more sophisticated mode of control at this biosynthetic branch point. We have recently demonstrated that FeCH activity influences Chl biosynthesis, as decreased activity of this enzyme was followed by a significant increase in the rate of ALA formation, in the level of Chl precursors, and in the accumulation of Chl-protein complexes (Sobotka et al., 2005, 2008b).
An intriguing feature of FeCH from cyanobacterial and algal sources is the hydrophobic C-terminal Chl a/b-binding (CAB) domain, with a high degree of similarity to the first and third helices of the plant light-harvesting complex II (LHCII) including a Chl-binding motif (Supplemental Fig. S1; Dolganov et al., 1995). Interestingly, the CAB domain is also a part of the FeCH isoform II responsible for the production of most of the heme in chloroplasts of higher plants (Papenbrock et al., 2001). This highly conserved CAB domain is connected to the FeCH catalytic core by a more variable region II, a short, hydrophobic, and Pro-rich sequence (Fig. 1A;Supplemental Fig. S1). This segment is preserved even in a few organisms harboring a plastid-type FeCH enzyme that lacks the CAB domain. This was shown for two cyanobacteria, Synechococcus JA-2-3Ab and JA-2-3B′a (Supplemental Fig. S1; Kilian et al., 2008) and for the green alga Chromera velia (L. Koreny, R. Sobotka, and M. Obornik, unpublished data; Supplemental Fig. S1). The only known plastid-type FeCH lacking both region II and the CAB domain is from the cyanobacterium Gloeobacter, which also lacks thylakoid membranes (Supplemental Fig. S1).
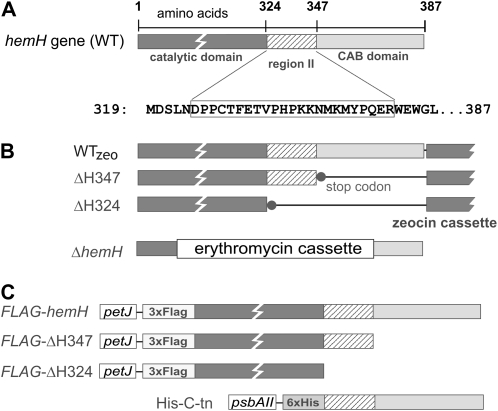
Figure 1.
A, Schematic presentation of the Synechocystis FeCH (coded by the hemH gene) with catalytic and C-terminal CAB domains connected by region II. WT, Wild type. B, Strains with truncated FeCH used in this study; WTzeo and ΔH324 were already described (Sobotka et al., 2008b). The ΔhemH deletion was combined with the FLAG-hemH background (see below) to construct the FLAG-hemH/ΔhemH strain, which expressed the tagged version as the only FeCH enzyme in the cell. C, Constructs used to express full-length and truncated versions of FeCH fused to a protein tag in Synechocystis. 3xFLAG-tagged FeCHs were expressed under the control of the Synechocystis petJ promoter; the His-C-tn strain expressed the 63-residue-long C-terminal fragment of Synechocystis FeCH as a small His6-tagged protein under the control of the Synechocystis psbAII promoter.
Although the functions of both the CAB domain and region II are unknown, we have recently demonstrated that deletion of both of these FeCH features in the ΔH324 strain of the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) dramatically reduces the stability and activity of FeCH and leads to a large accumulation of its substrate, PPIX (Sobotka et al., 2008b; Fig. 1B). In addition, analysis of both full-length and truncated ΔH324 recombinant FeCHs demonstrated that the C-terminal extension comprising region II and the CAB domain is essential for dimerization of the enzyme (Sobotka et al., 2008b).
In order to dissect the functional and structural roles of region II and the CAB domain, we have analyzed in this work another FeCH mutant, ΔH347, that retains region II but lacks the CAB domain (Fig. 1B). We have found comparable levels of the FeCH protein and of in vitro activity for the wild type and truncated ΔH347-FeCH, in marked contrast with the very low residual activity of the “fully truncated” ΔH324-FeCH. However, deletion of the CAB domain prevented the ΔH347 strain from growing at higher light intensities and affected the cellular accumulation of tetrapyrroles. In particular, we have found that although the ΔH347 mutant contains a decreased level of PPIX, it specifically accumulates chlorophyllide (Chlide) and fails to reduce cellular levels of Chl-protein complexes under high light. Analysis of the full-length and truncated FeCHs purified from Synechocystis under native conditions demonstrated that whereas the full-length enzyme forms a dimer, the ΔH347-FeCH is active as a monomer. The essential role of the CAB domain for the dimerization of FeCH in vivo was further confirmed by copurification of the full-length FeCHs from the cell using the tagged C-terminal segment of FeCH as bait.
RESULTS
The C-Terminal Domains of FeCH Are Required for the Acclimation of Synechocystis to High Light
To address the proposed role of the FeCH CAB domain in the regulation of tetrapyrrole metabolism and also to elucidate the function of region II, we constructed the Synechocystis mutant ΔH347, which retains the 23-residue region II but lacks the last 40 residues that constitute the putative transmembrane CAB domain (Fig. 1). Another FeCH mutant, ΔH324, prepared for previous work (Sobotka et al., 2008b), was also included in this study. This mutant differs from ΔH347 only by the additional absence of region II at the end of the FeCH catalytic domain (Fig. 1B); thus, a direct comparison of both strains should help to discriminate between the effects of both region II and the CAB domain.
First, we compared the photoautotrophic growth of both mutants and of the control wild-type strain, WTzeo (Fig. 1B), under different light intensities. Under low light (5 μmol photons m−2 s−1), all strains had comparable growth rates and also very similar levels of photosynthetic pigments (Table I; Supplemental Fig. S2). However, at normal light (40 μmol photons m−2 s−1), the ΔH324 mutant grew significantly more slowly, and with a further increase in light intensity to 150 μmol photons m−2 s−1, we observed a rapid loss of photosynthetic pigments. A further increase in light intensity abolished the growth of this strain (Table I; Supplemental Fig. S2). The effect of increased light intensity on the ΔH347 mutant was much less pronounced; its growth was only slightly impaired at normal light, and this strain was able to grow up to 200 μmol photons m−2 s−1 (hereafter high light; Table I). Further increase in light intensity to 250 μmol photons m−2 s−1 also completely inhibited the growth of the ΔH347 mutant, whereas the growth of the control WTzeo strain was not significantly affected (Table I). Interestingly, the ΔH347 mutant did not exhibit any “bleaching” at high light, as we observed for ΔH324; on the contrary, the ΔH347 mutant retained about 40% more Chl per cell than the WTzeo strain after approximately 40 h at high light (Table I; Supplemental Fig. S2). This is noteworthy, as a substantial decrease in Chl level is a typical response of Synechocystis to high light (Hihara et al., 1998; see control WTzeo strain in Table I and Supplemental Fig. S2).
Table I. Growth rate and Chl content of studied Synechocystis strains under different light regimes.
n.g., No growth (strain did not grow under these conditions).
| Strain | Light Intensity (μmol photons m−2 s−1) | |||||||
| 5 | 40 | 200 | 250 | |||||
| Doubling Time | Chl | Doubling Time | Chl | Doubling Time | Chl | Doubling Time | Chl | |
| h | μg mL−1 OD750−1 | h | μg mL−1 OD750−1 | h | μg mL−1 OD750−1 | h | μg mL−1 OD750−1 | |
| WTzeo | 58 | 5.5 ± 0.1 | 13.7 | 5.1 ± 0.2 | 9.8 | 2.6 ± 0.15 | 12.1 | 2.1 ± 0.1 |
| ΔH347 | 61 | 5.5 ± 0.15 | 14.6 | 5.7 ± 0.2 | 21 | 3.6 ± 0.25 | n.g. | n.g. |
| ΔH324 | 56 | 5.8 ± 0.2 | 20.1 | 4.9 ± 0.3 | n.g. | n.g. | n.g. | n.g. |
These results demonstrate that both the CAB and the region II domains are essential for photosynthetic growth of Synechocystis at high light intensities. However, the presence of region II of FeCH substantially improves acclimation to high light, and furthermore, the phenotypes of both FeCH mutants clearly differ under increased light intensities: whereas the ΔH324 strain loses most of its pigments probably due to the destruction of thylakoid membranes, the ΔH347 mutant retains a higher level of Chl, indicating that the viability of ΔH324 and ΔH347 mutants upon a shift to high light is abolished by different processes.
The CAB Domain Is Important for Regulation of the Tetrapyrrole Pathway
A remarkable feature of the ΔH324 strain was the excretion of PPIX into the growth medium at levels high enough to produce a brown color (Sobotka et al., 2008b). As we did not observe this effect in ΔH347, we compared the accumulation of PPIX and Chl intermediates in both mutants grown at normal light levels to elucidate the effect of these two protein truncations on tetrapyrrole metabolism. Surprisingly, we found that levels of tetrapyrroles in these strains are very different: whereas the ΔH324 accumulates high levels of PPIX and also other intermediates in the Chl branch (Fig. 2A; Sobotka et al., 2008b), ΔH347 contains less than 20% of the PPIX found in the control WTzeo strain. On the other hand, levels of magnesium protoporphyrins and protochlorophyllide (PChlide) in the ΔH347 strain are not significantly affected, with the exception of the approximately 2.5-fold increase in the content of the last Chl precursor, Chlide (Fig. 2A).
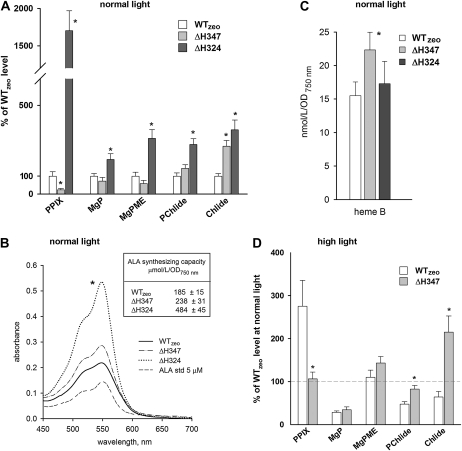
Figure 2.
Analysis of pigments in Synechocystis mutant strains. A, Quantification of the relative contents of PPIX, MgP, magnesium protoporphyrin IX methylester (MgPME), PChlide, and Chlide in Synechocystis cells growing at normal light (40 μmol photons m−2 s−1) and harvested at OD750 = 0.35 to 0.4. Chl precursors were extracted with methanol/0.2% NH4OH and quantified by a combination of HPLC and spectrofluorometry. Values shown represent means ± sd from three independent measurements; asterisks indicate significant differences tested using a paired t test (P = 0.05). B, ALA-synthesizing capacity as determined in a 100-mL cell suspension with OD750 = 0.4 treated for 4 h with levulinic acid to inhibit ALA condensation into porphobilinogen. Representative absorption spectra of developed assays as obtained for each strain together with 5 μm standard of ALA are also presented. C, Quantification of noncovalently bound heme B (protoheme) in cells growing at normal light. Heme was extracted by 90% acetone/2% HCl, separated, and quantified using HPLC. Error bars and the t test of statistical significance are as in A. D, Quantification of the relative contents of Chl precursors in Synechocystis cells growing for approximately 40 h at high light (200 μmol photons m−2 s−1) and harvested at OD750 = 0.35 to 0.4. Obtained values from three measurements were compared with values for the control WTzeo strain growing at normal light. The dashed line indicates 100% in comparison with values at normal light (A). Asterisks indicate statistically significant differences in precursor levels between WTzeo and ΔH347 at high light as tested using a paired t test (P = 0.05).
The observed low PPIX content of the ΔH347 mutant could indicate reduced ALA formation at the beginning of the pathway; however, measurements of ALA-synthesizing capacity revealed only a slightly increased rate of ALA formation in the ΔH347 mutant in comparison with WTzeo (Fig. 2B). This contrasts with a significantly elevated ALA formation in the ΔH324 mutant, which is probably responsible for the accumulation of PPIX in this strain (Fig. 2B; Sobotka et al., 2008b). So, detected changes in tetrapyrrole biosynthesis in ΔH347 do not appear to be caused predominantly by elevated metabolic flow through the whole pathway, as in the case of the ΔH324 mutant, but rather by different events at the heme/Chl branch point between heme and Chl branches influencing the PPIX pool and at the final steps of the pathway resulting in the accumulation of Chlide.
The low pool of PPIX in ΔH347 could be explained by a rapid “uncontrolled” consumption of PPIX either by magnesium chelatase or by truncated ΔH347-FeCH. The first possibility is not consistent with the almost normal level of MgP in ΔH347. Moreover, we did not observe higher cellular levels of the magnesium chelatase subunits (Fig. 3A). The second possibility, a rapid consumption of PPIX by mutated ΔH347-FeCH, could be reflected in higher levels of cellular heme or phycobilins. To determine levels of heme B (protoheme), total noncovalently bound heme was extracted by acetic acetone as described in “Materials and Methods,” separated by HPLC, and quantified spectroscopically. Indeed, the heme content of ΔH347 was significantly increased (Fig. 2C). Interestingly, the heme content of the ΔH324 mutant was not affected (Fig. 2C), suggesting that the ΔH324-FeCH is able to produce enough of the heme required for normal cellular function, despite a barely detectable in vitro activity of this truncated enzyme (Sobotka et al., 2008b; see below). We did not observe significant changes in phycobilisome levels in any of the strains studied, implying that the accumulation of the light-harvesting apparatus is under tight control and is not influenced by disturbances in tetrapyrrole biosynthesis (Supplemental Fig. S2).
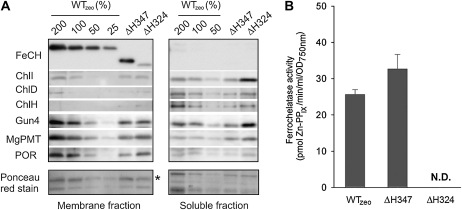
Figure 3.
Accumulation of enzymes of tetrapyrrole biosynthesis, and in vitro activity of FeCH in Synechocystis strains. A, Membrane and soluble protein fractions were prepared as described in “Materials and Methods,” separated by SDS electrophoresis, and blotted to a membrane. Enzymes were detected by specific antibodies. The amount of proteins loaded for 100% of each sample corresponded to 150 μL of cells at OD750 = 1. ChlI, ChlD, and ChlH are subunits of magnesium chelatase (Jensen et al., 1996), and Gun4 is another protein required for the activity of magnesium chelatase (Larkin et al., 2003; Sobotka et al., 2008a). MgPMT, Magnesium-protoporphyrin methyl transferase; POR, light-dependent PChlide oxidoreductase. Below are blotted proteins stained with Ponceau red shown as a loading control; the peripheral subunit of ATPase is marked by an asterisk. B, In vitro FeCH activity in membranes as determined by a continuous spectrofluorometric assay. Activities were monitored as an increase in fluorescence of zinc-PPIX using excitation and emission wavelengths of 420 and 590 nm, respectively. N.D., Not detected.
The enlarged pool of Chlide in the ΔH347 mutant is not easy to explain with regard to the position of FeCH in the pathway and normal or lower levels of other Chl precursors. It should also be noted that the ΔH347 mutation did not significantly alter the accumulation or cell localization of enzymes in the Chl pathway, including the enzyme producing Chlide, PChlide oxidoreductase (Fig. 3A). However, Chlide is the last precursor of the Chl biosynthesis pathway; thus, its accumulation could be related to the failure of ΔH347 to adjust the levels of Chl-protein complexes according to light conditions. Therefore, we compared the levels of Chl precursors in WTzeo and ΔH347 at high light when the growth rate of ΔH347 was retarded (Table I). Whereas the content of Chlide in the control strain decreased when compared with the situation in normal light, the content of Chlide in the ΔH347 remained almost unchanged (Fig. 2D), suggesting that the FeCH CAB domain is required for regulation of the final step(s) of Chl biosynthesis, which could include the attachment of Chl to proteins and Chl turnover (see “Discussion”).
To summarize, it is evident that the two different truncations at the FeCH C terminus have very different consequences for tetrapyrrole metabolism. Elimination of both region II and the CAB domain from FeCH results in a greatly elevated synthesis of PPIX and a consequent increase in the flux down the green branch of tetrapyrrole biosynthesis. In contrast, the ΔH347 strain that retains region II contains only a small PPIX pool but exhibits increased accumulation of Chlide under different light regimes. These results highlight the importance of region II and the CAB domain of FeCH for both its enzymatic and regulatory functions.
Region II Is Critical for Both the Stability and Enzyme Activity of Synechocystis FeCH
We have shown previously that truncation of the whole C-terminal domain of Synechocystis FeCH strongly impairs the stability and activity of the resulting ΔH324 enzyme (Sobotka et al., 2008b). To elucidate the completely different effect of the ΔH347 mutation on tetrapyrrole metabolism, we compared the activity and stability of mutated and full-length FeCHs.
To assess the localization and cellular levels of the ΔH347-FeCH, a whole-cell extract of the ΔH347 mutant was fractionated into cytosolic and membrane fractions, separated by SDS-PAGE, and subjected to immunodetection using anti-FeCH antibody raised against recombinant Synechocystis FeCH. The ΔH347-FeCH was detected only in the membrane fraction despite the lack of the putative transmembrane CAB domain. However, in contrast to the ΔH324 enzyme, both the FeCH levels and in vitro FeCH activities were comparable in membrane fractions from the ΔH347 and WTzeo strains (Fig. 3). As we did not observe any in vitro activity for the ΔH324 enzyme, it is evident that the presence of region II dramatically improves both the stability and the activity of Synechocystis FeCH.
To further determine possible structural functions for both the region II and CAB C-terminal FeCH domains, we analyzed enzymes purified under nondenaturing conditions from Synechocystis using the 3xFLAG tag (FLAG hereafter). To show that the presence of the tag did not interfere with the enzyme function, we deleted the native hemH gene in the FLAG-hemH strain, resulting in a strain expressing just the tagged FeCH version (Fig. 1B). The FLAG-hemH/ΔhemH strain did not display any apparent effects of FeCH deficiency, such as retarded growth or accumulation of PPIX, and the in vitro activity of the FLAG-FeCH in the cell extract corresponded with the enzyme content (data not shown).
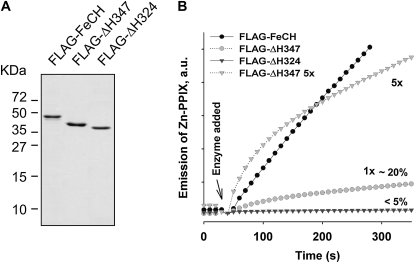
Both full-length and truncated FeCHs were expressed in Synechocystis and purified from cell extracts under native conditions using anti-FLAG resin; the migration of purified proteins on SDS electrophoresis corresponded to theoretical masses of 46.2, 41.7, and 38.9 kD for the FLAG-FeCH, FLAG-ΔH347, and FLAG-ΔH324, respectively (Fig. 4A). Activities of purified FeCHs were compared using an in vitro assay (Fig. 4B). Consistent with previous results on recombinant FeCHs (Sobotka et al., 2008b), deletion of both the region II and the CAB domains dramatically impaired activity of the ΔH324-FeCH (less than 5% of the full-length enzyme activity; Fig. 4B). The specific activity of the FLAG-ΔH347 enzyme was also about 20% of that of the wild-type enzyme. Intriguingly, the rate of incorporation of zinc into PPIX by ΔH347-FeCH rapidly decreases in the first 50 s of the assay, whereas the rate of incorporation of zinc by the full-length tagged enzyme is almost linear for at least 10 min (Fig. 4B). It should be noted that the activity of nontagged ΔH347-FeCH in purified membranes exhibits the same decrease in the rate of zinc-PPIX formation (data not shown). This implies that although the CAB domain is important for functioning of the FeCH, further truncation involving region II affects the FeCH accumulation and activity much more strongly.
Figure 4.
Full-length and truncated FeCH enzymes purified from Synechocystis. A, FLAG-tagged full-length and truncated FeCHs were purified under native conditions on the anti-FLAG affinity gel, separated by SDS electrophoresis, and stained by Coomassie Brilliant Blue; approximately 0.5 μg of protein was loaded in each line. B, In vitro activities of purified FeCH enzymes as determined by continuous spectrofluorometric assay. Activities were monitored as an increase in fluorescence of zinc-PPIX using excitation and emission wavelengths of 420 and 590 nm, respectively; 1x indicates that approximately 0.1 μg of enzyme was assayed. Relative activities of truncated enzymes in comparison with the full-length FeCH (100%) are also indicated.
The CAB Domain Is Necessary for the Dimerization of Synechocystis FeCH in Vivo
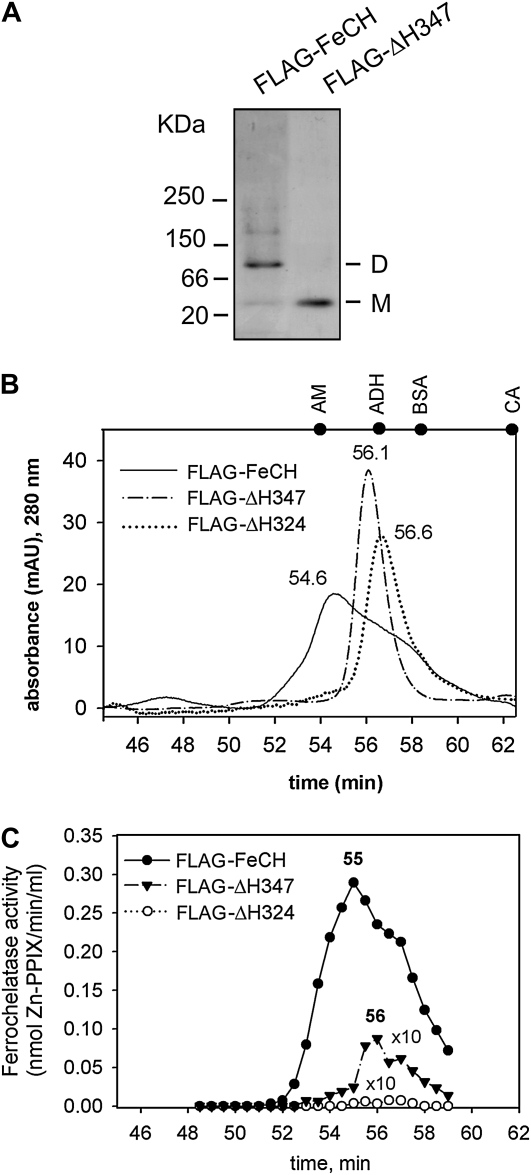
Using recombinant Synechocystis FeCHs, we have previously shown that deletion of the C terminus in ΔH324-FeCH abolishes dimerization of the recombinant enzyme and leads to the loss of its activity (Sobotka et al., 2008b). This led us to the assumption that, in agreement with the situation in mitochondrial FeCH (Grzybowska et al., 2002; Ohgari et al., 2005), the dimer is the active form of the Synechocystis FeCH. Given that the purified ΔH347-FeCH is fairly active and the same enzyme is fully active in membranes (Fig. 3B), we speculated that region II might promote dimerization, resulting in an active enzyme. However, on a native gel, the purified FLAG-ΔH347-FeCH migrated strictly as a monomer, in contrast to the dimeric full-length FLAG-FeCH (Fig. 5A). To facilitate further analysis of enzyme dimerization, all three purified tagged enzymes were separated using size-exclusion chromatography, and the FeCH activity of the column fractions was measured. According to the A280 (Fig. 5B), the full-length FeCH migrates as a broad peak mainly as a dimer but with a significant fraction of monomers and also a small proportion of higher mass aggregates (compare with Fig. 5A), whereas both truncated enzymes migrate in a sharp peak corresponding to monomers with bound detergent molecules (Fig. 5B). Note that it is generally problematic to determine the mass of membrane proteins on gel filtration, as calibration of a column with soluble proteins leads to inaccurate results (Zouni et al., 2005). However, the results in Figure 5 are internally consistent and show that although the mass of the FLAG-ΔH347 protein is only 2.8 kD larger than that of FLAG-ΔH324, its mobility is 0.5 min slower, which corresponds to approximately 25 kD according to our column calibration (Fig. 5B). This could indicate that the loss of region II is accompanied by a significant difference in the conformation of the FeCH protein.
Figure 5.
Aggregation state of purified FeCHs. A, Approximately 0.5 μg of the purified wild-type and truncated FeCH was separated by nondenaturing electrophoresis and stained by Coomassie Brilliant Blue. M and D indicate the positions of FeCH monomer and dimer, respectively. B, Gel filtration of the FLAG-FeCH and truncated FeCH enzymes on the BioSep SEC-S3000 column. Approximately 1 μg of each protein was separated. Positions of standards are shown at the top of the graph: AM = β-amylase (200 kD); ADH = alcohol dehydrogenase (150 kD); BSA = bovine serum albumin (66 kD); CA = carbonic anhydrase (29 kD). C, FeCH activity in 0.5-min fractions as eluted from the gel filtration column during separation of purified FeCH enzymes (described in B). The whole volume of each fraction (75 μL) was assayed immediately after elution in an in vitro FeCH assay. Obtained values for truncated enzymes were multiplied 10-fold to compare activity profiles.
The FeCH activity profile for the collected fractions for the full-length enzyme corresponded well with the protein absorbance, including an obvious shoulder at the position of a predicted monomer (Fig. 5C). This result indicates similar in vitro specific activities for both the dimeric and monomeric forms of the enzyme. The maximum activity of the FLAG-ΔH347-FeCH was measured in the elution peak fraction resembling the monomeric form. Therefore, we expect that the FLAG-ΔH347 was active in the assay in a monomeric form. It should be noted that a substantial portion of the total FLAG-ΔH347 activity was lost during gel filtration, suggesting a lower stability for this truncated enzyme when compared with the full-length FeCH, which was still highly active when eluted from the column (Fig. 5C). As expected, no activity was detected when a similar amount of the FLAG-ΔH324 was analyzed by the same method (Fig. 5C).
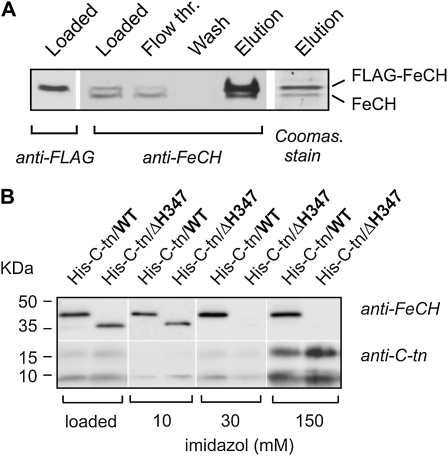
Although previous results strongly support a model where the CAB domain is responsible for FeCH dimerization and the ΔH347-FeCH is therefore a partially active monomer in vivo, it cannot be excluded that dimerization of Synechocystis FeCH is driven by having a high concentration of pure protein. To obtain further evidence that the full-length FeCH physically interacts with another FeCH molecule in vivo, we employed the FLAG-hemH strain expressing both the FLAG-FeCH and nontagged wild-type FeCH. As these two enzyme forms have different mobilities on SDS-PAGE, they can be distinguished by immunodetection using a combination of anti-FeCH and anti-FLAG antibodies (Fig. 6A). After isolation of the FLAG-FeCH on an anti-FLAG affinity gel, the smaller native FeCH was specifically copurified with FLAG-FeCH, and it was the only copurified protein visible on the Coomassie Brilliant Blue-stained gel (Fig. 6A).
Figure 6.
A, Purification of the FLAG-FeCH in a complex with nontagged FeCH. FLAG-FeCH was purified from the FLAG-hemH strain possessing both tagged and nontagged forms of this enzyme; purification was carried out under native conditions on the anti-FLAG affinity gel as described in “Materials and Methods.” Each purification step was separated by SDS electrophoresis and blotted, and both FeCH forms were detected by anti-FeCH antibody; eluted proteins separated by SDS electrophoresis were also stained by Coomassie Brilliant Blue. The position of the FLAG-FeCH was resolved by anti-FLAG antibody. B, Purification of the FeCH C-terminal fragment and its copurification with the full-length FeCH and the ΔH347-FeCH lacking the CAB domain. The 63-residue-long C-terminal fragment of Synechocystis FeCH was expressed as a His-tagged protein (His-C-tn) in the wild type (WT) and in the ΔH347 backgrounds and purified from both strains using Ni2+ affinity chromatography. As the polyclonal anti-FeCH antibody recognized the C-terminal parts of the protein only weakly, the His-C-tn was detected using an antibody raised against a synthetic peptide corresponding to region II of the Synechocystis FeCH (amino acids 333–348). The amount of membrane protein loaded for each sample corresponded to 150 μL of cells at OD750 = 1, 1/50th the volume of each washing step (20 μL from 1 mL), and 1/25th the total elution volume. The imidazole elution is described in “Materials and Methods.”
To resolve the question of whether the CAB domain alone is sufficient to promote the formation of dimeric FeCH and to investigate whether another part of this enzyme is essential for FeCH-FeCH interaction in vivo, we prepared a Synechocystis strain expressing the FeCH C terminus, consisting of region II and the CAB domain, as a small His6-tagged protein (His-C-tn; Fig. 1C). This artificial protein, lacking the extrinsic catalytic domain, was expressed both in the wild type and the ΔH347 strains and used as in vivo bait for FeCH. If the CAB domain is the only segment required for the FeCH dimerization, the His-C-tn protein would form a stable complex only with the full-length FeCH but not with the truncated ΔH347 enzyme. The His-C-tn protein was purified on a nickel column under native conditions and subjected to gel electrophoresis, and coisolated FeCH was detected by anti-FeCH antibody (Fig. 6B). The full-length FeCH clearly copurified with His-C-tn in contrast to ΔH347-FeCH, confirming in vivo dimerization via the CAB domain (Fig. 6B). Moreover, the His-C-tn protein formed a stable dimer with another His-C-tn molecule, which partly persisted even during separation by denaturing SDS electrophoresis (Fig. 6B). Based on these data, it can be concluded that the CAB domains have affinity for one another and that this domain is strictly required for dimerization of the Synechocystis FeCH in vivo.
DISCUSSION
Cyanobacterial and algal FeCHs as well as the plant FeCH2 isoform possess a unique conserved C-terminal CAB domain with a putative Chl-binding motif (Supplemental Fig. S1). The CAB domain and the FeCH catalytic core are connected by a “linker” (region II), which is quite variable in its length and sequence among organisms (Supplemental Fig. S1). Recently, we have demonstrated that the elimination of both region II and the CAB domain from FeCH in the Synechocystis ΔH324 mutant dramatically impaired the activity and stability of the truncated FeCH (Sobotka et al., 2008b). Given the essential role of dimerization for the function of human and yeast FeCHs (Grzybowska et al., 2002; Ohgari et al., 2005), the low activity of the ΔH324-FeCH was originally attributed to the absence of the CAB domain, yielding a monomeric enzyme (Sobotka et al., 2008b). However, newly discovered thermophilic Synechococcus strains possessing putative monomeric FeCHs lacking the CAB domain but still preserving the short region II (Kilian et al., 2008; Supplemental Fig. S1) stimulated us to mimic these naturally occurring FeCH variants by preparing the ΔH347 strain lacking only the CAB domain. Importantly, elimination of the CAB domain yielded an enzyme with comparable specific activity to the wild-type FeCH (Fig. 3). Direct comparison of ΔH347 with the ΔH324-FeCH lacking both the region II and CAB domains reveals the crucial importance of region II and enabled us, to our knowledge for the first time, to explore the role of the FeCH CAB domain in the regulation of tetrapyrrole biosynthesis.
Our earlier work on the ΔH324 strain (Sobotka et al., 2008b) posed an intriguing question as to why this mutant, possessing a strongly impaired FeCH, does not display any depletion in phycobilins, which are produced by oxidation of heme (Supplemental Fig. S2). Our work here helps to address this issue by demonstrating that the low activity of ΔH324-FeCH is more than sufficient to furnish the cellular demand for heme (Fig. 2C). However, it is still unclear why the wild-type cell maintains FeCH at levels much higher than those actually required by the demands placed on heme biosynthesis. It is noteworthy that under low light or microaerobic conditions, the ΔH324 mutant resembles the wild-type phenotype: it exhibits normal pigmentation and growth rate and does not release PPIX into the growth medium (Table I; Supplemental Fig. S2; R. Sobotka, unpublished data). These data suggest that under low-stress conditions, traces of FeCH activity are sufficient. Nonetheless, at increased light intensity, the growth of the ΔH324 mutant is significantly retarded, and a further increase in light intensity to 150 μmol photons m−2 s−1 caused cell bleaching, probably due to the destruction of thylakoid membranes (Table I; Supplemental Fig. S2). It appears that under such conditions, the high content of FeCH found in the wild type is essential for cell viability to avoid accumulation of the high levels of phototoxic PPIX and other porphyrins found in the ΔH324 mutant.
Based on these data and on our previous results (Sobotka et al., 2005), we expect that the FeCH enzyme is involved in the regulation of tetrapyrrole biosynthesis and that the high content of the enzyme per cell, in relation to the biosynthetic demand for heme, has regulatory implications. Our previous work suggested a regulatory role for FeCH: the cellular FeCH activity increased 3- to 4-fold in Synechocystis mutants impaired in PSII assembly and led to a decrease in metabolic flow through the tetrapyrrole pathway (Sobotka et al., 2005). The cell could balance the distribution of PPIX between both chelatases and could also combat the detrimental effects of a stressful and fluctuating environment using a surplus of “silent” FeCH to dissipate escalating levels of phototoxic PPIX. In the cyanobacterium Thermosynechococcus elongatus, FeCH was found to physically interact with protoporphyrinogen oxidase, the enzyme that produces its substrate PPIX (Masoumi et al., 2008). It is possible that the delivery of PPIX to FeCH is controlled via the formation of such a complex and, if needed, the FeCH quickly interacts with protoporphyrinogen oxidase and channels photosensitizing PPIX into the safer heme metabolite, with the bonus of inhibiting ALA synthesis via a heme feedback loop (see below). In our model, FeCH activity, which includes its access to PPIX, modulates the flow of this metabolite into the heme or Chl branches. The ΔH347 strain shows higher content of noncovalently bound heme but has a low pool of PPIX, although its ALA capacity is comparable to the wild type (Fig. 2). This indicates that the ΔH347 mutant is deficient in the control of PPIX distribution, and we speculate that deletion of the CAB domain facilitates the access of truncated FeCH to PPIX.
According to current models of the regulation of tetrapyrrole biosynthesis, increased consumption of PPIX by FeCH should inhibit the GluTR enzyme via a heme feedback loop (for recent reviews, see Tanaka and Tanaka, 2007; Masuda and Fujita, 2008). However, although an allosteric inhibition of GluTR by heme in plants is supported by convincing evidence, the model of free heme directly interacting with the GluTR and modulating the activity of this enzyme is probably oversimplified. In Chlamydomonas, inhibition of the GluTR activity by heme depends on the presence of an unidentified soluble protein (Srivastava et al., 2005), and heme paradoxically stimulates expression of the gene coding for GluTR (Vasileuskaya et al., 2005). In nonphotosynthetic bacteria, heme controls proteolytic degradation of GluTR, and in the acidophilic bacterium Acidithiobacillus ferrooxidans, heme also regulates the activity of glutamyl-tRNA synthase (Wang et al., 1999; Levicán et al., 2007). The role of heme in the tetrapyrrole signaling network is thus quite complex, and it is not so surprising that the increased protoheme level in the ΔH347 strain did not result in a decreased rate of ALA formation (Fig. 2B). It should be noted that the ΔH324 strain, where ALA synthesis is clearly stimulated, has a practically unchanged protoheme content per cell, so there might be a specific pool of heme used for signaling. However, as almost nothing is known about mechanisms balancing the production of different heme forms (B, A, C) together with linear tetrapyrroles, a regulatory role of a putative heme pool remains speculative.
As we already discussed, the ΔH324 dies at high light probably due to massive accumulation of PPIX and also other porphyrins. The growth of the ΔH347 mutant is also impaired by high light intensities, although it does not accumulate any tetrapyrroles other than Chlide (Table I); interestingly, at the same time, this strain contains approximately 40% more Chl per cell than WTzeo. We expect that the failure of ΔH347 to grow at high light is not caused primarily by a high concentration of phototoxic tetrapyrroles but due to an inability of this strain to control the synthesis and turnover of Chl-protein complexes. A similar “stay-green” phenotype was already described in the Synechocystis pmgA mutant, which had lost the ability to regulate photosystem stoichiometry, particularly to decrease the level of PSI relative to PSII upon a switch to high light (Hihara et al., 1998). Such an adjustment of the PSI/PSII ratio was shown to be essential for the survival of Synechocystis when grown at 300 μmol photons m−2 s−1 (Sonoike et al., 2001), and since PSI contains most of the Chl in the cell, the process of high-light acclimation results in a remarkable decrease in total Chl content per cell (see WTzeo in Table I). Interestingly, the down-regulation of PSI was reported to be determined primarily by limited availability of Chl for the synthesis of PSI subunits (Muramatsu et al., 2009); thus, this process can be affected by an elevated level of Chlide, the last Chl precursor. On the other hand, the regulation of the PSI/PSII ratio cannot be accomplished simply by a total restriction of Chl formation but rather by Chl redirection, as, in contrast to PSI, the synthesis of PSII subunits is accelerated under high-light stress due to a faster turnover of PSII (Nowaczyk et al., 2006). We expect that a sophisticated mechanism exists ensuring the distribution of Chl to Chl-binding proteins under changing environmental conditions and that such an apparatus has to recycle Chlide originating from degraded Chl-protein complexes via Chl turnover (Vavilin and Vermaas, 2007).
The presence of a Chl-binding motif in the FeCH CAB domain led us to the speculation that this domain is involved in the mechanism proposed above to balance Chl biosynthesis, Chl turnover, and the synthesis of particular Chl-protein complexes. Using the FLAG-tagged FeCH and its C terminus as bait, we have provided clear evidence that the CAB domain mediates a specific interaction between FeCH molecules in vivo. In the FeCH dimer, the interacting CAB domains should resemble the “cross helix” structure found in the LHCII complex of higher plants; the Chl-binding motif ExxNGR is preserved in the CAB domain (Supplemental Fig. S1; Liu et al., 2004). Our data thus support the possibility of Chl(ide) interacting with the CAB domain and may form an (alternative) route for the transfer of Chl(ide) to photosystem apoproteins. In this context, it is interesting that both cyanobacteria and plants contain high/early light-inducible proteins (ELIP/HLIP), which are structurally very similar to the FeCH CAB domain, including a conserved Chl-binding motif (Dolganov et al., 1995). These proteins, which quickly accumulate at high levels upon a switch to high light, might form part of a dynamic network of interacting CAB domains in the membrane that perhaps transiently form a complex with the FeCH CAB domain, yielding a monomeric enzyme that is virtually fully active but localized adjacent to different protein complexes, with different affinity to Chl or with different access to PPIX. Indeed, both ELIP and HLIP were found to regulate Chl biosynthesis (Xu et al., 2002; Tzvetkova-Chevolleau et al., 2007) and to physically interact with Chl-binding proteins (Promnares et al., 2006).
The CAB domain apparently has an important role also in the stability of Synechocystis FeCH; in contrast to the comparable membrane-associated catalytic activities of the wild type and the ΔH347-FeCH, the specific activity of the purified FLAG-ΔH347-FeCH was much lower (Fig. 4). This low specific activity is probably caused by lower stability of the enzyme, as the purified FLAG-ΔH347 enzyme lost its activity much faster than the full-length enzyme, which made it difficult for example to measure the activity of FLAG-ΔH347-FeCH separated by gel filtration. It is probable that one of the reasons for higher stability of the full-length FeCH is its dimerization.
The critical role of region II for the activity of Synechocystis FeCH is noteworthy, regarding its rather low sequence similarity among different groups of organisms and its position at the end of the extrinsic catalytic domain of the enzyme. The only common features perceptible from amino acid alignments are the rather hydrophobic nature of this sequence and a higher frequency of Pro residues (Supplemental Fig. S1). When compared with the structure of the spinach (Spinacia oleracea) LHCII (Liu et al., 2004), the FeCH CAB domain best corresponds to the third helix and the Pro-rich region II can be aligned with the stromal loop connecting the second and third helices (Supplemental Fig. S1). Indeed, this short sequence in LHCII contains five Pro residues that bend this region to form a curled but relatively flat structure on the membrane surface (Supplemental Fig. S3). So, the FeCH Pro-rich domain could form a similar structure, although we cannot account for the importance of this domain for enzyme activity/stability. A clue could be the unusual kinetics of the ΔH347-FeCH in our assay (Fig. 4B). The rapid decline in the initial rate of chelation is specific only for the ΔH347 enzyme, as the activity of recombinant ΔH324, although very weak, exhibits a linear rate of zinc-PPIX formation (R. Sobotka, unpublished data). Also, this effect is not caused by low stability of the ΔH347 enzyme in the assay, as the purified FLAG-ΔH347-FeCH can be incubated in assay buffer without zinc for at least 15 min with no decrease in activity (data not shown). The observed kinetics could be related to zinc uptake by FeCH, which is known to be quite complex, as zinc, even at low concentrations, inhibits the release of product from the enzyme (Davidson et al., 2009). To clarify this question, a detail enzymological study is required.
We conclude that this study, based on a combined analysis of several Synechocystis strains and purified FeCHs, has demonstrated both regulatory and structural roles for the FeCH CAB domain and, unexpectedly, revealed a critical role of region II for the catalytic function of the plastid-type FeCH enzyme. In addition, as the mitochondrial FeCH is known to be active only as a homodimer (Grzybowska et al., 2002; Ohgari et al., 2005), the Synechocystis FeCH can be described as a new type of dimeric FeCH, since the deletion of the C-terminal CAB domain, responsible for dimerization, did not abolish the catalytic activity of the resulting monomeric enzyme.
MATERIALS AND METHODS
Growth Conditions
If not stated otherwise, Synechocystis strains were grown photoautotrophically in liquid BG-11 medium (Rippka et al., 1979) supplemented by 10 mm TES at 30°C and 40 μmol photons m−2 s−1 (normal light) on the rotary shaker. To induce the expression of tagged FeCHs from the petJ promoter (see below), FLAG-hemH/ΔhemH, FLAG-ΔH347, and FLAG-ΔH324 strains were grown in BG-11 medium lacking copper in the trace metal mix.
Construction of Synechocystis Mutants
Construction of the WTzeo strain and the ΔH324 mutant has been described by Sobotka et al. (2008b). Essentially the same approach was adopted for the preparation of the ΔH347 mutant, with a stop codon in the hemH gene at amino acid position 347 (for all oligonucleotides used for strain preparation, see Supplemental Table S1).
To obtain the FLAG-hemH strain expressing 3xFLAG-tagged FeCH (FLAG-FeCH), the coding sequence of the hemH gene (locus slr0839) encoding the FeCH was subcloned into the pDrive vector (Qiagen). Subsequently, two complementary oligonucleotides encoding the 3xFLAG peptide (Sigma-Aldrich) were hybridized, thus creating NdeI-compatible overhangs, and then ligated into the NdeI site of the pDrive-hemH construct, thereby eliminating the second NdeI site. Using NdeI and BglII restriction enzymes (sites were introduced by the primers; Supplemental Table S1), a DNA fragment encoding the FLAG-FeCH was excised and then ligated into pSK9 vector for chromosomal integration under the control of the copper-dependent petJ promoter (Tous et al., 2001). The plasmid was transformed into Synechocystis wild type, and transformants were selected on BG-11 agar plates containing 7 μg mL–1 chloramphenicol.
In order to inactivate the entire FeCH gene and thus obtain a strain expressing the FLAG-FeCH as the only FeCH enzyme in the cell (FLAG-hemH/ΔhemH strain), the original hemH gene in the FLAG-hemH strain was replaced by an erythromycin resistance cassette using PCR-mediated insertion (Lee et al., 2004; Sobotka et al., 2008b). To prepare the FLAG-ΔH347 and FLAG-ΔH324 strains expressing truncated FeCHs, modified hemH genes containing stop codons were amplified from the ΔH347 and ΔH324 mutants and cloned into the pSK9 plasmid as described above.
The Synechocystis His-C-tn strain expresses the 63-residue-long C-terminal fragment of Synechocystis FeCH as a His6-tagged protein under the control of the psbAII promoter. To prepare this strain, the hemH gene fragment was amplified by PCR using gene-specific primers with artificial restriction sites for NdeI and BglII and containing six His codons (CAC) in the forward primer (Supplemental Table S1). After restriction, the PCR fragment was cloned into NdeI and BamHI sites of the pSBA plasmid containing the upstream and downstream regions of the Synechocystis psbAII gene (Lagarde et al., 2000). The plasmid was transformed into the Synechocystis psbAII-KS strain as described previously (Lagarde et al., 2000).
Fractionation of Synechocystis Cells
A total of 50 mL of cells (optical density at 750 nm [OD750] ∼ 0.5) was washed and resuspended in buffer A containing 20 mm HEPES, pH 7.4, 10 mm MgCl2, 5 mm CaCl2, and 20% glycerol. The cell suspension was mixed with glass beads and broken in a Mini-BeadBeater-16 (BioSpec), and the resulting homogenate was centrifuged at 50,000g for 30 min at 4°C. The supernatant containing soluble proteins was transferred to a new tube. Pelleted membranes were washed two times in buffer A, resuspended, and solubilized in the same buffer containing 1% dodecyl-β-maltoside, and unbroken cells and unsolubilized material were discarded by centrifugation.
Western Blot and Immunoblotting
Unless otherwise stated, proteins were denatured by 2% SDS and 1% dithiothreitol for 30 min at room temperature and separated on a denaturing 12% SDS-polyacrylamide gel. Proteins separated on the gel were transferred onto a polyvinylidene difluoride membrane. The membrane was incubated with specific primary antibodies and then with secondary antibody conjugated to horseradish peroxidase (Sigma). The anti-FeCH was raised in rabbits against recombinant Synechocystis glutathione S-transferase-FeCH expressed in Escherichia coli. The anti-FeCH C-terminal antibody (anti-C-tn) was raised in rabbits against residues 332 to 347 of the Synechocystis FeCH. The antibodies against Synechocystis magnesium chelatase subunits, Gun4 and Magnesium-protoporphyrin methyl transferase, were raised in rabbits using recombinant proteins prepared in E. coli.
Affinity Purification of 3xFLAG-Tagged FeCHs from Synechocystis
Two liters of FLAG-hemH/ΔhemH and FLAG-ΔH347 cells and 4 L of FLAG-ΔH324 cells harvested at OD750 = 0.7 to 0.8 were washed and resuspended in buffer A (see above) containing protease inhibitors (complete protease inhibitor cocktail; Roche). The cell suspension was mixed with glass beads and broken in a Mini-BeadBeater-16 using six 1-min cycles. Membranes were pelleted by centrifugation at 50,000g for 20 min at 4°C, washed in buffer A, and resuspended in 10 mL of the same buffer containing 1% dodecyl-β-maltoside. Membranes were solubilized by gentle mixing for 30 min at 10°C, and unbroken cells were discarded by centrifugation. Proteins were loaded onto a column containing approximately 100 μL of anti-FLAG M2 affinity gel (Sigma) and then washed with 15 mL of buffer A containing 0.04% dodecyl-β-maltoside (buffer A-DDM). The FLAG-tagged FeCH was eluted into buffer A-DDM by incubation of the M2 affinity gel with 3xFLAG peptide (100 μg mL−1) for 30 min.
Affinity Purification of the His-C-tn Protein from Synechocystis
For purification of the C-terminal fragment of Synechocystis FeCH (His-C-tn protein), membranes from 250 mL of cells (OD750 ∼ 0.7–0.8) were washed and resuspended in buffer B (20 mm HEPES, pH 7.7, 10 mm MgCl2, 5 mm CaCl2, 0.1 m NaCl, and 10% glycerol) containing protease inhibitor (complete protease inhibitor cocktail; Roche). Cells were broken and membrane proteins were prepared in the same way as already described for the purification of FLAG-FeCHs. Proteins were loaded onto a column containing approximately 75 μL of His-select resin (Sigma) charged with Ni2+ and preequilibrated with buffer B containing 0.04% dodecyl-β-maltoside (buffer B-DDM). To remove any loosely bound contaminants, the column was first washed with 8 mL of buffer B-DDM and then successively with 1 mL of buffer B-DDM containing 10 mm, 20 mm, and 30 mm imidazole. The His-C-tn protein was eluted with buffer B-DDM containing 150 mm imidazole.
FeCH Activity Assay
FeCH activity was monitored spectrofluorometrically at 35°C by directly recording the rate of zinc-PPIX formation using a Spectronic Unicam series 2 spectrofluorometer. The reaction mixture (1.5 mL final volume) contained 100 mm Tris-HCl (pH 8.0), 0.03% Tween 80, 5 μm PPIX, and 1 μm ZnSO4. The measurement of FeCH activity in the cell membrane fraction was initiated by adding proteins in amounts corresponding to 0.3 mL of cells at OD750 = 1. This value was calculated from the Chl content in the analyzed sample and Chl level per OD750 of the particular strain.
Quantification of Chl and Chl Precursors
For Chl quantification, pigments were extracted from cell pellets (5 mL, OD750 ∼ 0.4) with 100% methanol, and Chl content was measured spectrophotometrically (Porra et al., 1989).
For quantitative determination of Chl precursors in the cells, 75 mL of culture at OD750 = 0.35 to 0.4 was filtered through a 4-μm cellulose filter to remove all precipitated pigments in growth medium and harvested. Pigments were extracted by 1 mL of methanol/0.2% NH4OH using a Mini-BeadBeater-16 with two breaking cycles. Subsequently, the sample was centrifuged and the supernatant containing extracted pigments was collected. The pellet was then extracted again by 0.3 mL of methanol/0.2% NH4OH with one breaking cycle, and the combined supernatants were mixed with 150 μL of 1 m NaCl. This solution was extracted two times by 400 μL of hexane to remove Chl and β-carotene, concentrated to 750 μL on a vacuum evaporator, and extracted two times by 400 μL of petroleum ether (boiling range, 45°C–60°C) to remove zeaxanthin. Remaining solution was evaporated to dry on a vacuum evaporator. Pigments were than resuspended in 140 μL of methanol/0.2% NH4OH and mixed with 60 μL of water, and precipitated myxoxanthophylls were discarded by centrifugation. Samples were immediately separated by HPLC (Agilent-1200) on a reverse phase column (Nova-Pak C18, 4 μm particle size, 3.9 × 150 mm; Waters) using 30% methanol in 0.5 m ammonium acetate and 100% methanol as solvents A and B, respectively. Porphyrins were eluted with a linear gradient of solvent B (65%–75% in 30 min) at a flow rate of 0.9 mL min−1 at 40°C. HPLC fractions containing MgP (retention time of approximately 6.5 min), Chlide (approximately 8.5 min), PChlide (approximately 11.5 min), magnesium protoporphyrin IX methylester (approximately 13 min), and PPIX (approximately 15 min) were collected, and concentrations of the corresponding compounds were determined fluorometrically using a Spectronic Unicam series 2 spectrofluorometer.
Quantification of Hemes and Determination of ALA-Synthesizing Capacity
For quantification of heme B (protoheme), a sample of cell debris, already extracted by methanol/0.2% NH4OH for quantification of Chl precursors, was extracted further by 1 mL of acetone:water:HCl (90:8:2). After centrifugation, the solution was evaporated to dry on a vacuum evaporator, resuspended in 100 μL of acetone, and immediately separated by HPLC on a Nova-Pak C18 column (4-μm particle size, 3.9 × 150 mm; Waters) using a linear gradient from 50% to 80% solvent B (40% acetone in methanol) in solvent A (30% methanol in 0.5 m ammonium acetate) in 30 min at a flow rate of 1 mL min−1 at 40°C. Heme B was detected by a diode array detector (Agilent-1200) and quantified using an authentic hemin standard (Sigma).
To determine ALA-synthesizing capacity, a total of 100 mL of Synechocystis cells at OD750 = 0.4 was supplemented with 5 mm Glc and 15 mm levulinic acid/KOH, pH 7.5, to enhance ALA formation but inhibit its condensation into porphobilinogen. After 4 h, cells were harvested, resuspended in 0.3 mL of double-distilled water, and mixed with 20 μL of 50% TCA. Precipitated proteins were discarded by centrifugation, and the supernatant was adjusted to pH 6.7 by 90 μL of 0.5 m Na3PO4. The supernatant was then mixed with 12 μL of ethylacetoacetate, boiled for 15 min at 100°C, cooled on ice, and cleared by centrifugation. A total of 450 μL of modified Ehrlich’s reagent was added to the supernatant, and the content of ALA was determined by A553 (Mauzerall and Granick, 1956) on a Shimadzu 2000 spectrophotometer.
Native Electrophoresis and Size-Exclusion Chromatography
Nondenaturing PAGE was performed with the precast 4% to 16% NativePAGE Bis-Tris Gel (Invitrogen) at 10 V cm−1 at 4°C using the XCell SureLock cell (Invitrogen). Cathode buffer contains 0.25 mm Tricine, 7.5 mm Bis-Tris-HCl, pH 7.0, 0.05% sodium deoxycholate, and 0.02% dodecyl-β-maltoside; anode buffer contains 0.25 mm Bis-Tris-HCl, pH 7.0. Approximately 0.25 μg of protein in buffer A-DDM was loaded for each sample, and after electrophoresis, the gel was stained with Coomassie Brilliant Blue.
Gel filtration chromatography was carried out on the BioSep SEC-S3000 300- × 7.80-mm column (Phenomenex) connected to a photodiode array detector (Agilent-1200). The column was equilibrated with buffer A containing 0.1% dodecyl-β-maltoside. The flow rate was 0.15 mL min−1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment among the C-terminal ends of FeCH proteins from evolutionarily distant Chl-producing organisms.
Supplemental Figure S2. Whole-cell absorbance spectra of Synechocystis strains grown under different light regimes.
Supplemental Figure S3. Crystal structure of the spinach LHCII with the highlighted stromal loop connecting the second and third helices.
Supplemental Table S1. Oligonucleotides used for the construction of Synechocystis mutant strains.
Acknowledgments
We thank Eva Prachova for her technical assistance and Ulf Dühring for preparation of the pSK9-FLAG-hemH construct. The vector pSK9 was a kind gift of Prof. S. Zinchenko.
References
- Cornah JE, Terry MJ, Smith AG. (2003) Green or red: what stops the traffic in the tetrapyrrole pathway?. Trends Plant Sci 8: 224–230 [DOI] [PubMed] [Google Scholar]
- Davidson RE, Chesters CJ, Reid JD. (2009) Metal ion selectivity and substrate inhibition in the metal ion chelation catalyzed by human ferrochelatase. J Biol Chem 284: 33795–33799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganov NA, Bhaya D, Grossman AR. (1995) Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc Natl Acad Sci USA 92: 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K. (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J 40: 957–967 [DOI] [PubMed] [Google Scholar]
- Grzybowska E, Góra M, Plochocka D, Rytka J. (2002) Saccharomyces cerevisiae ferrochelatase forms a homodimer. Arch Biochem Biophys 398: 170–178 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Sonoike K, Ikeuchi M. (1998) A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis species PCC 6803 in response to high light. Plant Physiol 117: 1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Henningsen KW, Hunter CN. (1996) Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J Biol Chem 271: 16662–16667 [DOI] [PubMed] [Google Scholar]
- Kilian O, Steunou AS, Grossman AR, Bhaya D. (2008) A novel two domain-fusion protein in cyanobacteria with similarity to the CAB/ELIP/HLIP superfamily: evolutionary implications and regulation. Mol Plant 1: 155–166 [DOI] [PubMed] [Google Scholar]
- Kumar AM, Csankovszki G, Söll D. (1996) A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol Biol 30: 419–426 [DOI] [PubMed] [Google Scholar]
- Lagarde D, Beuf L, Vermaas W. (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 66: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee J, Lee HJ, Shin MK, Ryu WS. (2004) Versatile PCR-mediated insertion or deletion mutagenesis. Biotechniques 36: 398–400 [DOI] [PubMed] [Google Scholar]
- Levicán G, Katz A, de Armas M, Núñez H, Orellana O. (2007) Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci USA 104: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Masoumi A, Heinemann IU, Rohde M, Koch M, Jahn M, Jahn D. (2008) Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus. Microbiology 154: 3707–3714 [DOI] [PubMed] [Google Scholar]
- Masuda T, Fujita Y. (2008) Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci 7: 1131–1149 [DOI] [PubMed] [Google Scholar]
- Mauzerall D, Granick S. (1956) The occurrence and determination of δ-amino-levulinic acid and porphobilinogen in urine. J Biol Chem 219: 435–446 [PubMed] [Google Scholar]
- McCormac AC, Fischer A, Kumar AM, Söll D, Terry MJ. (2001) Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J 25: 549–561 [DOI] [PubMed] [Google Scholar]
- Müller B, Eichacker LA. (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11: 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Sonoike K, Hihara Y. (2009) Mechanism of downregulation of photosystem I content under high-light conditions in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 155: 989–996 [DOI] [PubMed] [Google Scholar]
- Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, Rögner M. (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18: 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgari Y, Sawamoto M, Yamamoto M, Kohno H, Taketani S. (2005) Ferrochelatase consisting of wild-type and mutated subunits from patients with a dominant-inherited disease, erythropoietic protoporphyria, is an active but unstable dimer. Hum Mol Genet 14: 327–334 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mishra S, Mock HP, Kruse E, Schmidt EK, Petersmann A, Braun HP, Grimm B. (2001) Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J 28: 41–50 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mock HP, Tanaka R, Kruse E, Grimm B. (2000) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Promnares K, Komenda J, Bumba L, Nebesarova J, Vacha F, Tichy M. (2006) Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. J Biol Chem 281: 32705–32713 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herman M, Stanier RY. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Sobotka R, Dühring U, Komenda J, Peter E, Gardian Z, Tichy M, Grimm B, Wilde A. (2008a) Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J Biol Chem 283: 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotka R, Komenda J, Bumba L, Tichy M. (2005) Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J Biol Chem 280: 31595–31602 [DOI] [PubMed] [Google Scholar]
- Sobotka R, McLean S, Zuberova M, Hunter CN, Tichy M. (2008b) The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J Bacteriol 190: 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K, Hihara Y, Ikeuchi M. (2001) Physiological significance of the regulation of photosystem stoichiometry upon high light acclimation of Synechocystis sp. PCC 6803. Plant Cell Physiol 42: 379–384 [DOI] [PubMed] [Google Scholar]
- Srivastava A, Lake V, Nogaj LA, Mayer SM, Willows RD, Beale SI. (2005) The Chlamydomonas reinhardtii gtr gene encoding the tetrapyrrole biosynthetic enzyme glutamyl-trna reductase: structure of the gene and properties of the expressed enzyme. Plant Mol Biol 58: 643–658 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Yoshida K, Nakayashiki T, Masuda T, Tsuji H, Inokuchi H, Tanaka A. (1996) Differential expression of two hemA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol 110: 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tous C, Vega-Palas MA, Vioque A. (2001) Conditional expression of RNase P in the cyanobacterium Synechocystis sp. PCC6803 allows detection of precursor RNAs: insight in the in vivo maturation pathway of transfer and other stable RNAs. J Biol Chem 276: 29059–29066 [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Franck F, Alawady AE, Dall’Osto L, Carrière F, Bassi R, Grimm B, Nussaume L, Havaux M. (2007) The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J 50: 795–809 [DOI] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF. (2005) Mg-protoporphyrin IX and heme control HEMA, the gene encoding the first specific step of tetrapyrrole biosynthesis, in Chlamydomonas reinhardtii. Eukaryot Cell 4: 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavilin D, Vermaas W. (2007) Continuous chlorophyll degradation accompanied by chlorophyllide and phytol reutilization for chlorophyll synthesis in Synechocystis sp. PCC 6803. Biochim Biophys Acta 1767: 920–929 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Kannangara CG, von Wettstein D. (1998) Barley glutamyl tRNAGlu reductase: mutations affecting haem inhibition and enzyme activity. Phytochemistry 47: 513–519 [DOI] [PubMed] [Google Scholar]
- Wang L, Elliott M, Elliott T. (1999) Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol 181: 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JD, Howell RW, Leverette RD, Grooms SY, Brignola PS, Mayer SM, Beale SI. (1993) Heme inhibition of δ-aminolevulinic acid synthesis is enhanced by glutathione in cell-free extracts of Chlorella. Plant Physiol 101: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Funk C, Vermaas W. (2002) Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 49: 149–160 [DOI] [PubMed] [Google Scholar]
- Zouni A, Kern J, Frank J, Hellweg T, Behlke J, Saenger W, Irrgang KD. (2005) Size determination of cyanobacterial and higher plant photosystem II by gel permeation chromatography, light scattering, and ultracentrifugation. Biochemistry 44: 4572–4581 [DOI] [PubMed] [Google Scholar]