Abstract
The circadian clock of the model plant Arabidopsis (Arabidopsis thaliana) is made up of a complex series of interacting feedback loops whereby proteins regulate their own expression across day and night. early bird (ebi) is a circadian mutation that causes the clock to speed up: ebi plants have short circadian periods, early phase of clock gene expression, and are early flowering. We show that EBI associates with ZEITLUPE (ZTL), known to act in the plant clock as a posttranslational mediator of protein degradation. However, EBI is not degraded by its interaction with ZTL. Instead, ZTL counteracts the effect of EBI during the day and increases it at night, modulating the expression of key circadian components. The partnership of EBI with ZTL reveals a novel mechanism involved in controlling the complex transcription-translation feedback loops of the clock. This work highlights the importance of cross talk between the ubiquitination pathway and transcriptional control for regulation of the plant clock.
Most organisms have evolved a molecular timing system known as the circadian clock. The importance of the clock has earlier been demonstrated in studies with clock mutants: those that lacked clocks or whose clocks did not resonate with the environmental cycle showed reduced growth rates and/or loss of fitness (DeCoursey et al., 1997; Ouyang et al., 1998; Green et al., 2002; Dodd et al., 2005).
The circadian clock is typically reset by daily changes in light and temperature. In plants such as Arabidopsis (Arabidopsis thaliana), entraining light signals are perceived through photoreceptors including the phytochromes and cryptochromes (Millar, 2004; Salomé and McClung, 2005; Jones, 2009); what receives the temperature signals and how are not fully understood yet.
Circadian clock mechanisms have been well characterized from animal, fungal, and plant systems (Dunlap, 1999; Harmer, 2009). At their core is an autoregulatory feedback loop whereby proteins repress their own transcription, either directly or indirectly. According to a widely accepted model, the Arabidopsis clock consists of three tightly interlocked feedback loops (Locke et al., 2005, 2006; Zeilinger et al., 2006). The first loop comprises two negative elements, the transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), which both repress expression of the positive factor TIMING OF CAB EXPRESSION1 (TOC1), a member of the five-strong PSEUDORESPONSE REGULATOR (PRR) protein family (Alabadí et al., 2001; Perales and Más, 2007). During the day, CCA1 and LHY are phosphorylated and subsequently targeted for destruction, leading to derepression of TOC1 expression in the afternoon (for review, see Más, 2008).
CCA1 and LHY interact as positive elements in a second loop in which PRR7 and PRR9 feed back to repress the expression of CCA1 and LHY (Eriksson et al., 2003; Ito et al., 2003; Michael et al., 2003; Yamamoto et al., 2003; Farré et al., 2005; Nakamichi et al., 2005, 2010). The third loop was invoked to account for observed rhythms in lhy;cca1 double mutant plants: GIGANTEA (GI) was shown to increase TOC1 transcription in this context, and TOC1 was proposed to feed back as a negative regulator of GI (Locke et al., 2006). GI has been implicated in the control of flowering and circadian clock function, where low levels of GI expression prolonged circadian period and delayed flowering (Fowler et al., 1999; Martin-Tryon et al., 2007).
These loops of temporally coordinated gene expression are synchronized by posttranslational modifications. TOC1 protein is regulated by the F-box protein ZEITLUPE (ZTL), which targets it for degradation via the proteasome by a CULLIN1-containing Skp1-Cullin-F-box protein (SCF) complex at night (Más et al., 2003; Harmon et al., 2008). ztl mutations increase the free-running period (FRP) of the circadian clock and have increased levels of TOC1 protein (Más et al., 2003; Kevei et al., 2006) and lower levels of CCA1 and LHY transcripts and protein (Somers et al., 2004; Baudry et al., 2010). GI associates with ZTL in a light-dependent manner, shaping its expression posttranslationally (Kim et al., 2007). Interaction with ZTL also leads to the degradation of PRR5 by the proteasome (Kiba et al., 2007). The access of ZTL to its substrates TOC1 and PRR5 is regulated by multiple factors, including PRR3, GI, and (blue) light (Kiba et al., 2007; Para et al., 2007; Fujiwara et al., 2008). ZTL thus plays a central role in resetting of the clock in response to dusk; however, all the evidence to date is that ZTL acts posttranslationally to determine protein stability and turnover.
The circadian system of plants is thus based on complex and coordinated interactions between gene expression and protein function. One commonly cited causal factor spurring the evolution of circadian clocks is that matching physiology to environmental rhythms is advantageous, leading to, for example, increased growth (Dodd et al., 2005), survival (Green and Tobin, 2002), and fitness (Ouyang et al., 1998). This model implies that there must be points of connection where the clock and the pathways controlling environmental responses meet, with some clock components also acting in the latter pathways (Legnaioli et al., 2009; Rawat et al., 2009).
Elsewhere, we have shown that early bird (ebi-1) is a novel allele of Arabidopsis NF-X-LIKE2 (AtNFXL2; Ashelford et al., 2011), which, like its homolog AtNFXL1, is a zinc finger protein with putative transcription factor activity and is involved in abiotic stress responses (Lisso et al., 2006). Here, we characterize the circadian phenotypes of the ebi-1 mutant. We provide evidence of its role in circadian regulation and flowering, showing that EBI/AtNFXL2 is capable of regulating the expression of components of the circadian clock. Elevated levels of EBI in protoplasts lead to the repression of CCA1 in the morning and elevate expression levels at night. EBI interacts physically with ZTL, and when ZTL is coexpressed together with EBI in protoplasts, they alter the level of CCA1 RNA. Coexpression of mutated ebi protein and ZTL in protoplasts leads to an expression of TOC1 RNA during the day, when TOC1 is normally not expressed. In accordance with this, we found that ebi-1 plants have an earlier phase and shorter period of TOC1 expression than the wild type. This study thus provides a novel role for ZTL in the plant clock as a regulator, in partnership with EBI, of circadian gene transcription.
RESULTS
ebi-1 Is a New Allele of the Zinc Finger Transcription Factor NFXL2
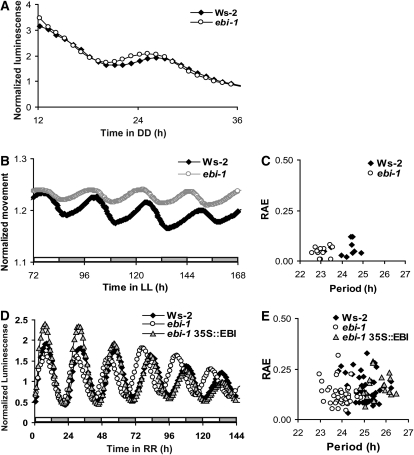
The ebi mutant was isolated from an ethyl methanesulfonate (EMS)-mutagenized population of Wassilewskija (Ws-2) Arabidopsis plants homozygous for the CHLOROPHYLL A/B_BINDING PROTEIN 2 fused to LUCIFERASE (CAB2::LUC+) reporter construct (Kevei et al., 2006). The original ebi-1 plant showed early phase of CAB2::LUC+ relative to wild-type Ws-2 plants (Fig. 1A). Similar to CAB2::LUC+ expression, the FRP of leaf movement rhythms measured in constant white light (25 μmol m−2 s−1) was shorter for ebi-1 than for the wild type (Fig. 1, B and C; Ws-2, mean period = 24.2 h, se = 0.14, n = 11; ebi-1, mean period = 23.1, se = 0.07, n = 12).
Figure 1.
The ebi-1 mutant has an early phase of CAB2::LUC+ expression and a short FRP under constant light conditions. Complementation with CaMV 35S-driven EBI restores the ebi-1 circadian phenotype to the wild type. A, Plants were grown in LD 12:12 for 8 d before transfer to DD at dusk to measure phase. B and D, To measure FRP, plants were entrained in LD 12:12 for 11 or 8 d, then transferred at dawn to constant white light (25 μmol m−2 s−1) for leaf movements of Ws-2 and ebi-1 (B) and to red light (15 μmol m−2 s−1) in order to determine CAB2::LUC+ expression from Ws-2, ebi-1, and ebi-1 carrying CaMV 35S-driven EBI (D). C and E, Relative amplitude error (RAE) of each fitted curve is shown for leaf movement (C) and CAB2::LUC+ (E). Results are representative, and experiments were repeated with similar results (n = 11–40).
Sequencing of the ebi-1 genome identified an EMS-induced C-to-T transition in AT5G05660, which resulted in the conversion of a highly conserved Val to Ile in a zinc finger domain (Ashelford et al., 2011). Complementation of ebi-1 with the genomic sequence of AT5G05660 under the control of the cauliflower mosaic virus (CaMV) 35S promoter restored the FRP of ebi-1 to wild-type levels (Fig. 1, D and E), confirming that the correct locus had been identified. This locus had previously been named AtNFXL2 because of its similarity to the mammalian zinc finger transcription factor, NFX1 (Lisso et al., 2006). Homologs of NFXL1 and NFXL2 are conserved across kingdoms (Supplemental Fig. S1; Supplemental Table S1). The Arabidopsis genome encodes two NFX homologs, the other being AtNFXL1 (locus, AT1G10170; Lisso et al., 2006; Asano et al., 2008), which has approximately 30% identity at the amino acid level to EBI/AtNFXL2 (BLASTP pairwise alignment; Altschul et al., 2005). Products of both genes were reported to be regulating the plant’s response to abiotic stress (Lisso et al., 2006).
ebi-1 Shortens FRP by Advancing the Phase of Circadian Gene Expression
Following backcrossing to Ws-2 plants, the behavior of ebi-1/Atnfxl2-2 mutant plants (henceforth and in figures referred to as ebi-1) carrying the CAB2::LUC+ reporter was observed under a range of light conditions. In ebi-1 plants, CAB2::LUC+ rhythms had a short period under higher intensities of continuous red light (RR; Fig. 2A) as well as under continuous white (LL) and blue (BB) monochromatic light compared with the wild type (Ws-2); all data are summarized in Table I. The rhythmicity of ebi-1 plants was robust: although the period was shorter than in Ws-2, variation between individual seedlings was extremely low (se values are given in Table I). Thus, wild-type EBI is required to regulate the FRP of both CAB2::LUC+ and leaf movement. This shortening of the FRP is sufficient to account for the early phase of ebi-1.
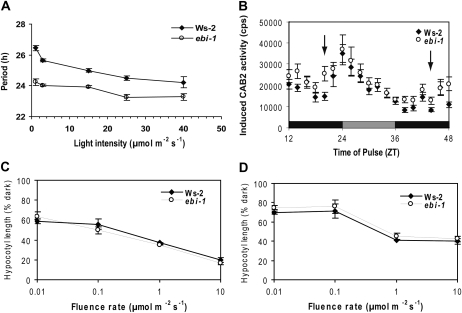
Figure 2.
ebi-1 shortens FRP in response to red light in a similar manner across light intensities, responding specifically during the subjective night. Light-dependent elongation of hypocotyls is not affected. A, Plants were grown in LD 12:12 for 6 to 7 d, when they were transferred to continuous red light at dawn. The panel shows ebi-1 and Ws-2 FRP in response to indicated red light fluency. B, Plants were grown in LD 12:12 for 8 d and then transferred to DD at dusk to measure gating of the acute response of CAB2::LUC+ to 20 min of red light pulses (20 μmol m−2 s−1) at 2-h intervals. Each point is the mean ± se (n = 16). C and D, Hypocotyl length was measured after 4 d under red (C) and blue (D) light of various intensities after an initial 2-h light pulse and dark treatment. Measurements are means ± se of three biological repeats (n = 18–58).
Table I. CAB2::LUC+ period lengths under various light treatments.
Periods of CAB2::LUC+ expression in the wild type, ebi-1, cca1-11;lhy-21, ztl-21, and double and triple mutants (as indicated) were measured under different quality and quantities of light. Rhythm analysis were performed by BRASS and period estimates ± se for plants under constant conditions during 24 to 144 h. Values are from representative experiments and assay conditions, and genotype and number of plants are shown. Art. white, Artificial white light (a mix of 50% blue and 50% red light-emitting diodes).
| Genotype | Irradiance | Wavelength | Period | se | n |
| μmol m−2 s−1 | h | ||||
| Ws-2 | 3 | White | 26.1 | 0.1 | 22 |
| ebi-1 | 3 | White | 25.5 | 0.2 | 19 |
| Ws-2 | 20 | Art. white | 23.4 | 0.1 | 20 |
| ebi-1 | 20 | Art. white | 22.7 | 0.1 | 20 |
| Ws-2 | 3 | Blue | 27.1 | 0.1 | 75 |
| ebi-1 | 3 | Blue | 25.1 | 0.2 | 29 |
| Ws-2 | 15 | Blue | 24.4 | 0.1 | 79 |
| ebi-1 | 15 | Blue | 23.5 | 0.1 | 30 |
| Ws-2 | 3 | Red | 26.5 | 0.2 | 29 |
| ebi-1 | 3 | Red | 24.2 | 0.2 | 30 |
| Ws-2 | 15 | Red | 25.6 | 0.1 | 47 |
| ebi-1 | 15 | Red | 24.0 | 0.1 | 9 |
| Ws-2 | 25 | Red | 25.1 | 0.03 | 100 |
| ebi-1 | 25 | Red | 24.1 | 0.1 | 30 |
| Ws-2 | 40 | Red | 24.5 | 0.1 | 40 |
| ebi-1 | 40 | Red | 23.1 | 0.1 | 43 |
| Ws-2 | 15 | Red | 24.5 | 0.1 | 20 |
| ebi-1 | 15 | Red | 23.5 | 0.1 | 20 |
| cca1-11;lhy-21 | 15 | Red | 18.2 | 0.1 | 12 |
| cca1-11;lhy-21;ebi-1 | 15 | Red | 16.1 | 0.2 | 26 |
| ztl-21 | 15 | Red | 27.1 | 0.2 | 20 |
| ebi-1;ztl-21 | 15 | Red | 26.3 | 0.1 | 41 |
As the phenotypes of ebi-1 resembled those of the canonical clock mutants cca1, lhy, and toc1 (all are short-period, early-flowering mutants), we tested the effect of combining the ebi-1 mutation with cca1 and lhy mutations. In the Ws-2 background, cca1-11, lhy-21, and toc1-21 mutations acted additively, with all double mutants having an FRP shorter than the single mutants; however, circadian rhythmicity is abolished in the triple mutant cca1-11;lhy-21;toc1-21 (Ding et al., 2007). In contrast, the cca1-11;lhy-21;ebi-1 triple mutant retained rhythmic CAB2::LUC+ expression, although its FRP was extremely short (Table I; Supplemental Fig. S2). Thus, the ebi-1 mutation acts in addition to the lhy-21 and cca1-11 mutations with respect to FRP.
ebi-1 Increases the Light Inducibility of the Marker Gene CAB2::LUC+ Only during the Subjective Night
One of the roles of the circadian clock is to regulate its own input (McWatters et al., 2000). For this reason, identical signals elicit different responses depending on the time of day when they are received, a phenomenon known as “gating” (McWatters et al., 2000; Allen et al., 2006). Relative to Ws-2, ebi-1 plants showed an increase in the acute response of the CAB2::LUC+ reporter gene to light during the subjective night, and this response recurred in the next cycle (Fig. 2B); however, no difference in acute response was observed during the subjective day. This indicates that EBI function is required to gate light sensitivity during the night. Consistent with the short-FRP phenotype, peak induction of the acute response occurred in both cycles 2 h earlier than in Ws-2.
The Circadian and Photomorphogenic Functions of EBI Are Separated in the ebi-1 Mutant
The light inhibition of hypocotyl elongation was measured as an assay of hypersensitivity or hyposensitivity to low fluence rates of light. Hypocotyl elongation was measured in Ws-2 and ebi-1 plants grown in constant dark (DD) or in RR or BB, and two-factor (genotype and light intensity) ANOVA was used to determine the effect of light treatment and genotype on hypocotyl growth under each wavelength. Hypocotyls were longer in plants grown at lower light intensities (Fig. 2, C and D); thus, as expected, brighter light inhibited hypocotyl elongation (RR, fluence = 78.7, 4 degrees of freedom, P < 0.001; BB, fluence = 125.1, 4 degrees of freedom, P < 0.001). However, genotype had no effect upon hypocotyl length (RR, fluence = 2.5, 1 degree of freedom, P = 0.15; BB, fluence = 2.8, 1 degree of freedom, P = 0.12). There was no significant interaction between light intensity and genotype, consistent with the conclusion that, as ebi-1 has a response to light indistinguishable from Ws-2 plants, the ebi-1 mutation affects the clock rather than light signaling pathways.
ebi-1 Does Not Affect Red Light Sensing
Wild-type Arabidopsis plants follow Aschoff’s rule for diurnal organisms (Aschoff, 1979): their FRP shortens with increasing light intensity. Ws-2 plants obeyed this rule, as did ebi-1 (Table I). The slopes of the fluence response curve in RR were parallel for Ws-2 and ebi-1, although, consistent with earlier results, the ebi-1 mutants had shorter FRPs than Ws-2 at all red light intensities (Fig. 2A). Overall, genotype and light intensity both had significant effects on period length (two-factor ANOVA; genotype, fluence = 38.2, 1 degree of freedom, P < 0.01; light intensity, fluence = 6.7, 4 degrees of freedom, P < 0.05). No interaction between light intensity and genotype was detected; thus, ebi-1 differed from Ws-2 only in its period length and not in its overall response to light, implying that it does not perceive red light differently from wild-type plants.
Temperature cycles can also entrain the circadian system of Arabidopsis. In a previously published microarray data set (Michael et al., 2008), EBI was shown to be rhythmic in DD following temperature entrainment (Supplemental Fig. S3). Ws-2 and ebi-1 seedlings were entrained to warm/cold cycles before transfer to RR or BB for measurement of CAB2::LUC+ rhythms (Table II). As with light entrainment, ebi-1 seedlings showed a shorter FRP of bioluminescence rhythms than Ws-2 plants. These results show that the ebi-1 mutation reduces FRP following temperature and light entrainment but does not alter sensitivity to red or blue light.
Table II. Period lengths after temperature entrainment.
Periods of CAB2::LUC+ expression in the wild type and ebi-1 were measured following temperature entrainment under 12 h, 22°C/12 h, 12°C for 7 d when transferred to a constant ambient temperature of 22°C and hourly imaging in red or blue light of 3 and 15 μmol m−2 s−1. Rhythm analyses were performed by BRASS and period estimates ± se for plants under constant conditions during 24 to 144 h. Values are from representative experiments and assay conditions, and genotype and number of plants are shown.
| Genotype | Irradiance | Wavelength | Period | se | n |
| μmol m−2 s−1 | h | ||||
| Ws-2 | 3 | Red | 25.5 | 0.3 | 15 |
| ebi-1 | 3 | Red | 24.5 | 0.2 | 17 |
| Ws-2 | 15 | Blue | 23.6 | 0.1 | 55 |
| ebi-1 | 15 | Blue | 22.1 | 0.1 | 32 |
ebi-1 Alters the Expression of Core Clock Components
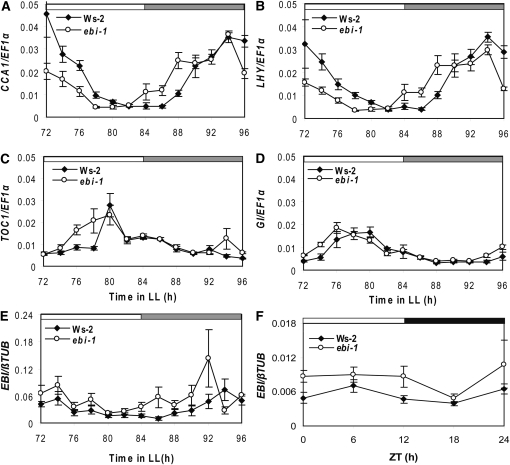
We measured the expression of core clock components after 3 d in white LL following entrainment to light/dark conditions of 12/12 h (LD 12:12). This regime was chosen to reflect the initial experimental regime that had shown ebi-1 had a short period and early phase (Fig. 1; Table I). Expression of the morning clock components CCA1 and LHY had an early phase in free-running ebi-1 plants, relative to expression in Ws-2 (Fig. 3, A and B), consistent with the observation of early phase and short period for CAB2::LUC+ and leaf movement rhythms (Fig. 1). Expression of evening clock genes TOC1 and GI was also advanced in ebi-1 (Fig. 3, C and D). TOC1 showed a second, smaller peak before subjective dawn in ebi-1 plants but not in the wild type. These results suggest that EBI is required for the correct functioning of the feedback loop between CCA1, LHY, and TOC1.
Figure 3.
ebi-1 mutation leads to altered expression of core clock components. A, CCA1. B, LHY. C, TOC1. D, GI. Seedlings were grown in LD 12:12 for 6 d and then transferred to constant light for 3 d before sampling started at ZT72. E and F, EBI expression in LL (E) or LD 12:12 cycles (F). Measurements are means ± se of three (A–C and F) or means ± range of two (D and E) biological repeats.
Quantitative real-time reverse transcription (RT)-PCR was performed to determine whether EBI was rhythmic in LD and LL. The expression of EBI expression was slightly increased in ebi-1 seedlings compared with the wild type under LL as well as LD 12:12 conditions (Fig. 3, E and F). In each case, EBI expression was weakly rhythmic, albeit with different phases in LL and LD conditions. Similarly, EBI appeared slightly rhythmic under cycles of light and temperature (Supplemental Fig. S3).
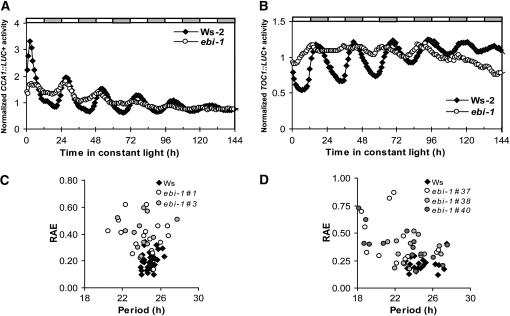
We examined the function of the feedback loop between CCA1 and TOC1 in more detail by analysis of CCA1::LUC+ and TOC1::LUC+ reporters in plants free running in constant artificial white light (equal amounts of red and blue light) after entrainment to LD 12:12. As with the quantitative RT-PCR, the waveform of CCA1::LUC+ expression showed a short-FRP, early-phase pattern (compare Figs. 3A and 4A). However, in two independently transformed lines, CCA1::LUC+ expression showed low amplitude and shorter period in ebi-1 plants relative to the wild type (Fig. 4, A and C; Ws-2, mean period = 24.9 h, se = 0.1, n = 38; ebi-1, mean period = 24.2, se = 0.4, n = 30; average relative amplitude error for Ws-2 = 0.2 and for ebi-1 = 0.4) . TOC1::LUC+ expression was also weakly rhythmic with a shorter period in ebi-1, as shown for three independently transformed transgenic lines (Fig. 4, B and D; Ws-2, mean period = 24.9 h, se = 0.3, n = 19; ebi-1, mean period = 23.0, se = 0.4, n = 40; average relative amplitude error for Ws-2 = 0.3 and for ebi-1 = 0.4) . These results confirm that the ebi-1 mutation affects the function of the feedback loop.
Figure 4.
ebi-1 mutation leads to altered regulation of CCA1 and TOC1. Plants were entrained in LD12:12 cycles for 6 d before being transferred to constant artificial white light (RR + BB; 20 μmol m−2 s−1). A, CCA1::LUC+ expression of T3 plants from three independent transgenic lines. B, TOC1::LUC+ expression of T2 plants from two independent transgenic lines. C, Relative amplitude error (RAE) for CCA1::LUC+ lines. D, Relative amplitude error for TOC1::LUC+ lines. Each experiment was repeated two times with similar results (n = 12–20 per line).
The ebi-1 Mutant Is Early Flowering
Arabidopsis is a facultative long-day plant, flowering earlier in long days than in short days (Thomas and Vince Prue, 1997). Controlling this photoperiodic response is one of the major functions of the clock. Clock mutants often have alterations in physiological responses, such as hypocotyl elongation (Dowson-Day and Millar, 1999) and flowering time (Zagotta et al., 1996), possibly because of the altered phase of gene expression relative to the external day/night cycle (Roden et al., 2002; Yanovsky and Kay, 2002). In constant white light, leaf movement rhythms of ebi-1 had an FRP length 1 h shorter than the wild type. We measured the time to flowering of Ws-2 and ebi-1 mutants in long-day (LD 16:8), intermediate-day (LD 12:12), and short-day (LD 9:15) photoperiods (Table III). Although ebi-1 retained the ability to distinguish between long and short photoperiods, ebi-1 was early flowering under each daylength relative to the wild type (Table III). Thus, ebi-1 alters but does not abolish the perception of photoperiod.
Table III. Flowering time.
Flowering response was measured as number of leaves at bolting, scored as stem at least 1 cm, for the wild type, ebi-1, cca1-11;lhy-21, ztl-21, and double and triple mutants (as indicated). Values are from representative experiments and assay conditions, and genotype and number of plants are shown.
| Genotype | Growth Conditions(Light:Dark) | No. of Leaves | se | n |
| Ws-2 | 16 h:8 h/23°C:18°C | 11.9 | 0.2 | 30 |
| ebi-1 | 16 h:8 h/23°C:18°C | 10.7 | 0.2 | 30 |
| Ws-2 | 9 h:15 h/23°C:18°C | 36.4 | 0.6 | 29 |
| ebi-1 | 9 h:15 h/23°C:18°C | 28.2 | 0.9 | 28 |
| Ws-2 | 16 h:8 h/22°C:22°C | 8.9 | 0.2 | 20 |
| ebi-1 | 16 h:8 h/22°C:22°C | 7.0 | 0.2 | 20 |
| cca1-11;lhy-21 | 16 h:8 h/22°C:22°C | 7.2 | 0.2 | 17 |
| cca1-11;lhy-21;ebi-1 | 16 h:8 h/22°C:22°C | 6.6 | 0.2 | 12 |
| Ws-2 | 12 h:12 h/22°C:22°C | 14.1 | 0.4 | 30 |
| ebi-1 | 12 h:12 h/22°C:22°C | 8.2 | 0.2 | 30 |
| cca1-11;lhy-21 | 12 h:12 h/22°C:22°C | 7.4 | 0.1 | 29 |
| cca1-11;lhy-21;ebi-1 | 12 h:12 h/22°C:22°C | 7.7 | 0.2 | 10 |
In LD 12:12 cycles, which induce a moderate short-day response, ebi-1 plants were early flowering (8.2 leaves) compared with the wild type (14.1 leaves; Table III). However the triple mutant cca1-11;lhy-21;ebi-1 flowered at a similar stage as the cca1-11;lhy-21 double mutant, reminiscent of the cca1-11;lhy-21;toc1-21 triple mutant (Ding et al., 2007).
EBI Interacts with ZTL
Since the ebi-1 mutation affected gating of light input during the night (Fig. 2B) and the expression of EBI peaked at or before dawn (Fig. 3; Supplemental Fig. S3), the interaction between EBI and the nighttime clock-associated proteins GI, TOC1, and ZTL was investigated. Myc-tagged EBI was coexpressed in protoplasts with hemagglutinin (HA)-tagged GI, TOC1, or ZTL.
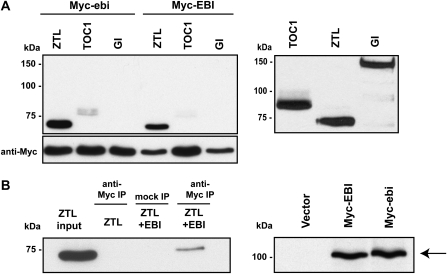
Immunoprecipitation using anti-Myc followed by probing with HA antibody did not reveal any interaction between GI and EBI proteins, and only a weak association between TOC1 and EBI was apparent. However, there was a strong association between ZTL and EBI; the mutated ebi protein also interacted with ZTL and weakly with TOC1 (Fig. 5A). Independently, the interaction of EBI with ZTL was shown by the yeast two-hybrid system (Supplemental Fig. S4). Thus, EBI physically interacts with ZTL, and this interaction is apparently not affected by the ebi-1 mutation. We could not detect HA-ZTL protein if Myc-EBI was not expressed or when HA-ZTL and Myc-EBI were coexpressed, but the complex was mock precipitated with normal mouse IgG beads (Fig. 5B). Expression of EBI and ebi proteins was confirmed by probing blots with anti-Myc antibody (Fig. 5B).
Figure 5.
EBI and ebi proteins interact with ZTL following cotransfection in protoplasts. A, Myc-EBI interacts strongly with HA-ZTL, weakly with HA-TOC1, and not with HA-GI protein (top left panel). Myc-EBI and Myc-ebi expression following immunoprecipitation with anti-Myc antibody and detected on gel blots with anti-Myc antibodies (bottom left panel). HA-ZTL, HA-TOC1, and HA-GI detected by anti-HA antibodies (right panel). B, The interaction with HA-ZTL is specific (left panel). To test specificity of the coimmunoprecipitation reaction, HA-ZTL was expressed either alone or together with Myc-EBI, and proteins were immunoprecipitated either with anti-Myc antibody or with normal mouse IgG (mock IP). HA-ZTL protein was detected by blotting immunocomplexes with an anti-HA antibody. Myc-EBI and Myc-ebi protein expression is shown compared with an empty vector control following detection with an anti-Myc antibody (right panel). The experiment was repeated two times with similar results.
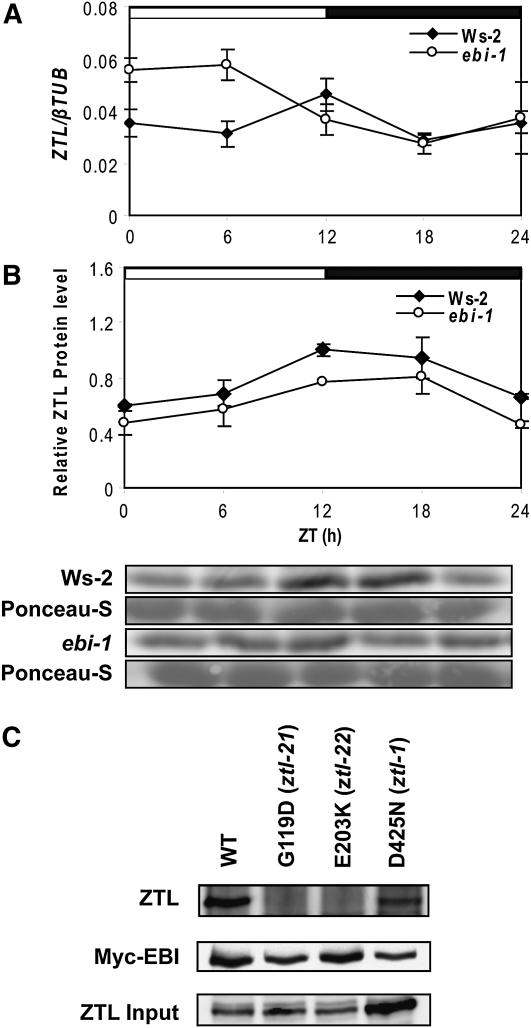
Because the results indicated an interaction between ZTL and EBI, we measured ZTL transcript and protein levels in wild-type and ebi-1 seedlings (Fig. 6, A and B). Consistent with previous reports (Somers et al., 2000; Kim et al., 2003b), ZTL transcript did not appear rhythmic under these conditions, but the protein was rhythmic in light/dark cycles, with a peak at the end of the subjective day. The level of ZTL transcript was slightly increased during the light period in ebi-1 relative to the wild type, while ZTL protein appeared slightly lower in ebi-1 compared with the wild type.
Figure 6.
Levels of ZTL transcript but not protein are altered in the ebi-1 mutant under LD 12:12. A, ZTL transcript. B, ZTL protein. Results are means ± se of three biological replicates, and representative immunoblots are shown. C, Mutations in ZTL LOV domain and F-box regions reduce its affinity for EBI. G119D, ztl-21 LOV domain mutation; E203K, ztl-22 F-box mutation; D425N, ztl-1 Kelch repeat mutation. Anti-ZTL antibody was used to detect ZTL protein bound to Myc-EBI following immunoprecipitation with anti-Myc (top panel). To visualize EBI, the blot was reprobed with anti-Myc antibody (middle panel). Expression level of ZTL proteins used for each immunoprecipitation reaction (Input) was tested in parallel transfections and anti-ZTL immunoblot (bottom panel). WT, Wild type.
Mutations in the N Terminus of ZTL Interfere with Binding to EBI
TOC1 and GI interact with ZTL via the LIGHT, OXYGEN, OR VOLTAGE (LOV) domain (Kim et al., 2007). We tested which region(s) of ZTL interacted with EBI by coexpressing Myc-tagged EBI protein with HA-tagged ZTL constructs carrying mutations in the Kelch repeat (D425N: ztl-1 mutation), LOV domain (G119D: ztl-21 mutation), and F-box (E203K: ztl-22 mutation) regions (all mutant 35S::ZTL constructs were previously described by Kim et al. [2007]). Wild-type HA-tagged ZTL protein was included as a positive control, and ZTL antibodies were used to detect levels of EBI-bound ZTL following anti-Myc precipitation (Fig. 5C, top panel). Levels of Myc-EBI following precipitation and input of ZTL were used as controls (Fig. 5C, middle and bottom panels).
A strong interaction was found between EBI and ztl-1 proteins, but no interactions between EBI and either ztl-21 or ztl-22 were found (Fig. 5C). It is still unknown how these mutations affect ZTL protein structure, but the ztl-21 mutation abolishes ZTL-GI interactions (Kim et al., 2007). Thus, both LOV and F-box domain mutations interfere with ZTL binding to EBI, suggesting that it may compete with GI and/or TOC1 for access to the LOV domain.
Like ebi-1 but unlike other ztl mutants, ztl-21 altered circadian FRP but not light sensitivity (Kevei et al., 2006). Since the ztl-21 mutation abolished the interaction between EBI and ZTL, we assayed the FRP of the double mutant ebi-1;ztl-21 and found it was longer than that of Ws-2 but shorter than that of ztl-21 plants (Table I). Thus, ebi-1 caused period shortening in a ztl-21 background. This suggests that EBI and ZTL separately have opposite effects on a common target, as the ebi-1 mutation mitigated the effect of ztl-21 when EBI could not interact with ZTL due to the mutation ztl-21.
ZTL Does Not Degrade EBI
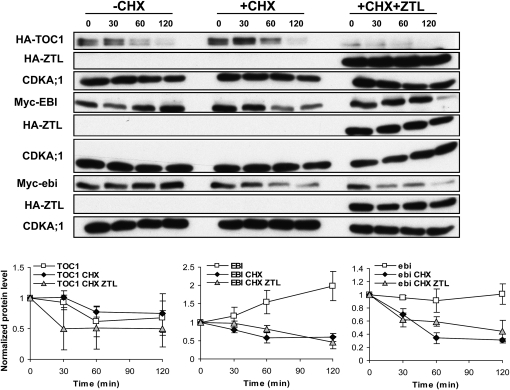
Given that ZTL is the substrate-binding subunit of an E3 ubiquitin ligase complex that targets TOC1 for degradation, we investigated whether ZTL also caused degradation of EBI in the protoplast system. TOC1 or EBI protein was coexpressed with ZTL in protoplasts, then further protein synthesis was blocked by the addition of cycloheximide. In accordance with previous studies (Más et al., 2003; Kim et al., 2007), TOC1 protein disappeared within 30 min when ZTL was coexpressed (Fig. 7). In contrast, the level of EBI was unchanged in the presence of ZTL compared with expression of EBI alone (Fig. 7). However, the level of EBI decreased slowly over time, suggesting that it is eventually degraded in a ZTL-independent manner. We repeated this experiment with the mutated version of ebi protein with similar results (Fig. 7). Thus, ZTL does not facilitate the degradation of EBI.
Figure 7.
ZTL degrades TOC1 but not EBI. HA-TOC1, Myc-EBI, or Myc-ebi was coexpressed with HA-ZTL in Arabidopsis protoplasts. Protein synthesis was blocked 18 h posttransfection by the addition of 100 μm cycloheximide (CHX). TOC1 and ZTL proteins were detected with anti-HA antibody, and EBI and ebi were detected with anti-Myc antibody. Equal protein loading was revealed by probing the blot with anti-CYCLIN-DEPENDENT KINASE A;1 (CDKA;1, PSTAIRE) antibody. The experiment was repeated twice with similar results, and a representative data set is shown. The bottom panels show the quantification of HA-TOC1, Myc-EBI, and Myc-ebi in the absence or presence of CHX with or without added HA-ZTL. Mean protein levels ± se are shown from three experiments where the amount of each protein at time zero (0) in each treatment was arbitrarily set to 1.
ZTL Opposes the Effects of EBI on Clock Components during the Day
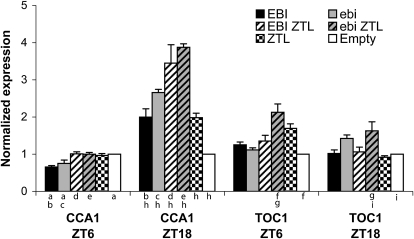
Since EBI resembles a mammalian transcription factor (Lisso et al., 2006), we investigated whether constitutive expression of EBI or ebi was capable of altering the expression of clock components. We transfected protoplasts with Myc-tagged wild-type or mutated EBI protein in the presence or absence of transfected HA-tagged ZTL and measured the expression of CCA1 and TOC1 from their endogenous promoters at zeitgeber time (ZT) ZT 6 (middle of the light period) and ZT 18 (middle of the dark period). The expression of transcript following transfection with empty vector was set at an arbitrary level of 1 (Fig. 8).
Figure 8.
Both EBI and ebi act on CCA1 expression, but only mutant ebi alters TOC1 induction. ZTL counteracts the effect of EBI during the day and increases it at night. Endogenous gene expression is shown following constitutive expression of EBI or ebi with and without ZTL in Arabidopsis protoplasts. Expression of CCA1 and TOC1 (relative to βTUB) was normalized to expression of cells receiving an empty vector control at ZT 6 to 7 (light) or ZT 18 to 19 (dark) under LD 12:12 cycles with 25 μmol m−2 s−1 white light received during the light period. Significance was tested using ANOVA (P < 0.001) followed by Student-Newman-Keuls test for differences between the categories (n = 3–6). Letters show which samples were significantly different (P < 0.05) from other samples. Results are representative, and experiments were repeated with similar results.
At ZT 6, transfection with either wild-type EBI or the mutated form, ebi, reduced CCA1 expression (Fig. 8); similarly, transfection with ZTL plus either form of EBI abolished this effect. However, at ZT 18, transfection of either EBI or ZTL increased levels of CCA1, and simultaneous transfection with both proteins had a greater effect than either alone. Transfection with the mutated form ebi, either alone or in the presence of ZTL, also increased the level of CCA1. There was thus a considerable time-of-day effect upon CCA1 induction. These results suggest that EBI induces CCA1 during the night and represses it during the day. However, as similar effects were observed for EBI and ebi, it is unlikely that the short FRP of ebi-1 is due to direct misregulation of CCA1 expression by the mutated protein.
There was no time-of-day effect of transfections of wild-type EBI protein on TOC1 expression, regardless of whether ZTL was cotransfected. However, ebi plus ZTL induced greater TOC1 expression at ZT 6 than at ZT 18 (Fig. 8). Expression of TOC1 was also higher at ZT 6 in protoplasts transfected with ebi alone. Given the apparently high levels of TOC1::LUC+ expression observed in ebi-1 plants (Fig. 4) and the early phased expression of TOC1 rhythm (Fig. 3), it is possible that increased expression of TOC1 during the daytime may be the primary cause of the short-period ebi-1 phenotype.
DISCUSSION
EBI Regulates Period Length
We have shown that ebi-1 is a mutant of Arabidopsis that displays a short-FRP, early-phase phenotype of the circadian clock. Our data indicate that the likely cause of this phenotype is misregulation of the feedback loop between CCA1, LHY, and TOC1 that lies at the heart of the plant clockwork. Although ebi-1 appears to have high levels of TOC1, this is not reflected in an increase in CCA1 or LHY expression, implying that EBI acts at about the point where transcriptional activation of CCA1/LHY by TOC1 protein takes place and regulates the FRP. We have demonstrated that EBI can bind ZTL (part of the SCF complex) directly and that this binding alters the activation of clock components regulated by EBI and ZTL, including CCA1 and TOC1. Thus, EBI acts on the circadian clock in partnership with ZTL, presumably in a time-dependent manner, since the level of ZTL protein fluctuates rhythmically in planta.
EBI together with its family member AtNFXL1 represents a phylogenetically conserved group of proteins with homologs represented across phyla (Supplemental Fig. S1). Both ebi-1 (this paper) and ebi-2, an independent T-DNA insertion allele in Columbia (Ashelford et al., 2011), had short-FRP, early-phase rhythms under a variety of entrainment conditions, and EBI is thus revealed as a novel regulator of clock speed. So far, no clock phenotype related to NFX-like genes has been reported from other phyla, although the Drosophila melanogaster gene shuttle craft, a homolog of AtNFXL1 (Supplemental Fig. S1), is contained within a quantitative trait locus affecting locomotion, a trait under clock control in this species (Jordan et al., 2006).
The ebi-1 mutation was originally identified in a screen for altered CAB2::LUC+ expression as an early-phase mutant, a characteristic shared by the Columbia allele ebi-2 (Ashelford et al., 2011). The ebi-1 mutation shortened the FRP of the clock across a range of backgrounds; it acted additively to the cca1-11 and lhy-21 mutations and also decreased the FRP of ztl-21 mutants. Although the timing of EBI protein in planta is not known, our data on the circadian behavior of the ebi-1 mutant predicts that it acts on the clock at night.
We propose that EBI promotes the expression of CCA1 at night but not by day and that this activational effect is enhanced by interaction with ZTL. Several pieces of evidence support this: during the night, changes in transcriptional activation after transfection were greater following coexpression of ZTL and EBI, ZTL protein immunoprecipitated with EBI, and the gating of the acute response to light was altered during subjective night but not subjective day. As CAB2 is a direct target of CCA1 (Wang et al., 1997; Wang and Tobin, 1998), a plausible explanation for the early phase of CAB2 is the early rise and fall of CCA1 in ebi-1: advancing the phase of CCA1 results in early expression of CCA1-regulated genes, including CAB2; hence, the increased induction of CAB2 during subjective night. However, both mutant and wild-type forms of EBI had similar effects upon CCA1 expression, so the ebi-1 mutation does not alter this function directly. Induction of TOC1 expression at ZT 6 by expressed ebi was greater than that by EBI, and TOC1 expression was high during subjective day in the ebi-1 mutant (Fig. 3C), with an early phase and short period (Figs. 3C and 4, B and D). We conclude, therefore, that the mutation acts by allowing TOC1 to increase during the day and hence speed up the central loop.
When the clock is free running in constant light, peak expression of EBI occurs at or around subjective dawn, as des the expression of CCA1 and LHY. The observation of rhythmicity in the cca1-11;lhy-21;ebi-1 triple mutant (this paper) means that EBI, like CCA1 and LHY, is not essential for rhythmic behavior. Rather, its function is to build a delay into the clockwork such that the feedback loops run with circadian (i.e. approximately 24-h) periods. Therefore, although none of these genes is essential for rhythmicity per se, all are required if the periodicity of the plant is to approximate the 24-h solar day. Their products thus act together to convert the basic oscillator into a circadian clock.
EBI Acts in Partnership with ZTL
Expression of ZTL was high in the morning in ebi-1 seedlings. Overexpression of ZTL has previously been associated with a shortening of the FRP (Somers et al., 2004). However, there was little or no difference in the steady-state level of ZTL protein; thus, the ebi-1 mutation does not affect the translational and posttranslational regulation of ZTL in planta. Therefore, ebi-1 seedlings must retain mechanisms for regulating ZTL levels at the posttranscriptional and/or translational level, and the increase in ZTL observed in ebi-1 during the daytime can be dismissed as the explanation for the short-FRP/early-phase phenotype. Our data show the ebi-1 mutation may act by altering the time of transcription of clock components. A change to this EBI function may be responsible for the specific timing defect of ebi-1, which is associated with a premature rise in the expression of CCA1, LHY, TOC1, and GI (Fig. 3).
ZTL establishes periodicity of the clock by regulating the proteasome-dependent degradation of TOC1 and PRR5 (Más et al., 2003; Kiba et al., 2007). In this context, it would be interesting to determine whether EBI alters the effect of ZTL on these proteins. GI is known to bind the LOV domain of ZTL; the ztl-21 mutation abolishes both the GI-ZTL and TOC1-ZTL interactions and specifically affects the circadian clock (Kevei et al., 2006; Kim et al., 2007). Interaction with GI stabilizes ZTL and is required for the circadian rhythm of ZTL protein (Kim et al., 2007). Our results suggest that EBI, together with ZTL, affects the coordination of morning and evening clock loops in order to synchronize the timing of CCA1 and TOC1 expression. The mutated form retained the ability to complex with ZTL (Fig. 5) and induce or repress CCA1 in a time-of-day-dependent manner but differed from the wild type in its effects on TOC1 expression (Fig. 8). As the ebi-1 mutation caused a premature rise in TOC1 expression in the presence of ZTL, it appears that the short FRP results from a failure properly to repress TOC1 during the day.
The EBI protein possesses a RING finger and zinc finger domain and putative nuclear localization domains (Lisso et al., 2006), indicating that it is capable of binding to both DNA and proteins. We investigated both its ability to form complexes with other clock proteins and its effects on clock gene expression. The association of EBI protein with ZTL was weakened by mutations in the N-terminal region of ZTL, specifically in the F-box and LOV domain, suggesting that this part might interact with EBI. In contrast to ZTL-dependent TOC1 degradation, association with ZTL did not lead to the degradation of EBI. Therefore, EBI is not a ZTL substrate in the manner of TOC1 or PRR5 (Más et al., 2003; Kiba et al., 2007) and the short phenotype of ebi-1 is not likely to be due to enhanced turnover of the protein.
ZTL is an F-box-containing protein that is part of an SCF E3 ubiquitin ligase complex that targets TOC1 for ubiquitination and degradation via the proteasome (Somers et al., 2004). There is evidence from mammalian systems that the ubiquitination pathway has the additional role, beyond its effect on protein degradation, of enhancing transcription (e.g. Skp2; Kim et al., 2003a) via proteolysis by the ubiquitin-proteasome system (Lipford et al., 2005). Transfection with ZTL alone increased the expression of CCA1 at ZT 18 and TOC1 at ZT 6. Therefore, we cannot exclude the possibility that similar mechanisms operate in plants.
ZTL is principally present in the cytosol (Kim et al., 2007); thus, interaction with the rhythmic ZTL may define the timing of EBI action. ebi-1 led to shortening of the FRP of ztl-21 plants. The ztl-21 mutation disrupts physical interaction between EBI and ZTL (Fig. 6) and so implies that, in a ztl-21 background, ZTL cannot counteract ebi’s effect upon CCA1 and TOC1. Interaction with EBI provides another mechanism by which ZTL sets the period of the clock beyond its well-known posttranslational function. It underlines the importance of ZTL as a regulator of circadian period.
MATERIALS AND METHODS
Plant Material and Transgenic Lines
ebi-1 was isolated from a screen of EMS-mutagenized M2 Arabidopsis (Arabidopsis thaliana) Ws-2 plants (Kevei et al., 2006) by its early phase of CAB2::LUC+ expression in DD. Full-genome sequencing revealed a single-nucleotide mutation in AT5G05660 (Ashelford et al., 2011).
The original ebi-1 plant was backcrossed to the cognate wild type, and the early-phase mutation was subsequently observed to be inherited in a codominant fashion. Complementation of the ebi-1 mutation was obtained by using the Myc-tagged CaMV 35S:EBI coding sequence in pRT104 plasmid (as described below), the construct was digested with SbfI (New England Biolabs), and the resulting fragment was ligated into PstI-digested and dephosphorylated promoterless binary vector pGREEN0229. The resulting plasmid was introduced into ebi-1 using Agrobacterium tumefaciens-mediated floral dipping (Clough and Bent, 1998), selected by spraying with 5.75% BASTA (Hoechst Schering AgrEvo) twice, and confirmed by PCR. All experiments were performed using the third or fourth backcrossed line.
ebi-1 (fourth backcross) was introduced into clock mutant backgrounds by crossing with the ztl-21 and cca1-11;lhy-21 mutants (Kevei et al., 2006; Ding et al., 2007). cca1-11;lhy-21 double mutants were identified using PCR primers for CCA1 and LHY in combination with T-DNA primers (JL202/JL270) as described previously (Hall et al., 2003).
CCA1::LUC+ and TOC1::LUC+ constructs have been described previously (Doyle et al., 2002; McWatters et al., 2007). ebi-1 plants without the CAB2::LUC+ reporter (obtained from a cross with wild-type Ws-2) were transformed with CCA1::LUC+ or TOC1::LUC+ reporters using floral dipping as described above.
Genotyping of plants at the EBI and ZTL loci was performed by PCR and derived cleaved-amplified polymorphic sequence mapping (Neff et al., 2002). For EBI, primers were EBI-F (5′-GGACACCTATGGTAGCCACAACCAAGAT-3′) and EBI-R (5′-ATGCTTAATTGCGGGAAACA-3′). The product was digested with MboI; the ebi1-1 sequence remained undigested. For ZTL, primers were ZTL21-F (5′-AAGAAAATGTATTGATGAGGGCATTGAATTTCTAG-3′) and ZTL21-R (5′-TCCAAGGTCAATGTCTGTCTCG-3′). The product was digested with XbaI, which cleaved the ztl-21 but not the wild-type sequence. CAB2::LUC+ rhythms were measured using the homozygous ztl-21;ebi-1 double mutant.
All plants except those in the hypocotyl elongation and flowering time assays (described below) were grown on plates containing 1× Murashige and Skoog salts (Duchefa Biochemie) supplemented with 3% Suc and 0.8% agar (w/v). Unless stated elsewhere, all molecular biology reagents and primers were obtained from Invitrogen. All experiments were repeated at least twice with similar results.
Hypocotyl Measurement
Seeds were stratified and treated essentially as described by Fankhauser and Casal (2004); they were grown on 1× Murashige and Skoog salts and 0.8% agar without added Suc. Imbibed seeds were given a 2-h light treatment of 100 μmol m−2 s−1 before being returned to DD for 24 h prior to light treatment with calibrated levels of blue (474 nm) and red (660 nm) light-emitting diodes (MD Electronics) or kept dark during 4 d. Plates were scanned and hypocotyl lengths measured by image analysis using Metamorph 6.3 (Molecular Devices). Results represent means ± se of three biological replicates each with at least 18 hypocotyls.
Tagged Protein Constructs
Epitope-tagged versions of proteins were produced in pRT104 plasmids (Fülöp et al., 2005); expression is driven by the constitutive CaMV 35S promoter. EBI and ebi proteins were expressed with a Myc epitope; GI, TOC1, and ZTL proteins carried a HA epitope.
EBI
EBI and ebi-1 gene sequences were obtained by amplification from Ws-2 and ebi-1 cDNA with the primers EBIFL-F (5′-CCGGAATTCATGGCCGGAACCGCCA-3′) and EBIFL-R (5′-CCGCTCGAGGATTCGAGGGTATCTTCTAGACT-3′). Primers included EcoRI or XhoI restriction sites (boldface) to facilitate subsequent cloning into pRT104-3xMyc (Fülöp et al., 2005). Transformants were obtained following heat shock transformation of XLI MRF′ cells (Stratagene). Correct constructs were identified by restriction mapping and derived cleaved-amplified polymorphic sequence analysis (as described above) and confirmed by sequencing.
GI
GI cDNA was obtained from RNA extracted from Ws-2 plants using the primers GI-F (5′-CTTTTGCGAATTCATGGCTAGTTCATCTTCATCTGAGAGA-3′), incorporating an EcoRI restriction site, and GI-R (5′-TTTGCGCTCGAGTTAGCGGCCGCATTGGGACAAGGATATAGTACAGCC-3′), incorporating a NotI restriction site. Amplified cDNA was first cloned into pMALc2x-cHIS (New England Biolabs), from which GI was recovered as an EcoRI-NotI fragment. This was ligated into pRT104-3xHA (Fülöp et al., 2005), previously digested with EcoRI and XbaI, to obtain pRT104-3xHA-GI.
TOC1
TOC1 cDNA was amplified using TOC1-F (5′-TTGGCTCGAGGAATTCATGGATTTGAACGGTGAGTGTAAAGG-3′), incorporating an EcoRI restriction site, and TOC1-R (5′-TTCTGAGCTCCTACTCGAGAGTTCCCAAAG-CATCATCCTG-3′), incorporating a SacI restriction site. Amplified cDNA was cloned into pGADT7 (Clontech Laboratories) to create pGADT7-TOC1. This was digested with EcoRI and XhoI, and the TOC1 fragment was subsequently ligated into pRT104-3xHA, previously digested with EcoRI and SalI, to produce pRT104-3xHA-TOC1.
ZTL
ZTL cDNA was amplified with ZTL-F (5′-TGGACTCGAGGGATCCGTATGGAGTGGGACAGTGGTTC-3′), incorporating a BamHI restriction site, and ZTL-R (5′-TTTCCCGGGTTACTCGAGATTCGTGAGATAGCTCGCTAGTGAT-3′), incorporating a SmaI site. The product was digested with BamHI and SmaI to obtain the ZTL fragment, which was ligated into pGADT7 to create pGADT7-ZTL. This was digested with EcoRI and XhoI to release TOC1 that was subsequently ligated into pRT104-3xHA, previously digested with EcoRI and SalI, to obtain pRT104-3xHA-ZTL.
Transformed Escherichia coli was obtained and confirmed as above. Constructs were used for transient transformation in Arabidopsis protoplasts (Fülöp et al., 2005) and immunoprecipitation studies (see below).
Luminescence Assays
For LUC activity measurements, plants were entrained for 6 to 7 d with LD 12:12 cycles under white light of 100 μmol m−2 s−1 (TLD 32W/840; Philips Electronics) and 22°C in a growth cabinet (Percival Scientific) or under constant monochromatic light-emitting diodes (MD Electronics) and temperature cycles of 12 h of 22°C/12 h of 12°C for 7 d and were prepared for imaging (as described by Millar et al., 1992) before they were transferred to a constant ambient temperature of 22°C and hourly imaging.
Luminescence levels were measured by low-light video imaging using a Hamamatsu ORCA-II-ERG 1024 cooled CCD camera system and Wasabi imaging software (Hamamatsu Photonics). Data analysis was performed using Metamorph 6.3 image-analysis software (Molecular Devices) and the macro suite Biological Rhythms Analysis Software System (BRASS; available at http://www.amillar.org). The fast Fourier-transformed nonlinear least-squares analysis function was used to estimate circadian parameters, such as rhythmicity, phase of peak expression, and period length, as described previously (Millar et al., 1995; Plautz et al., 1997; Locke et al., 2005). Rhythmic robustness was evaluated using relative amplitude error; values less than 0.6 were considered an indication of robust rhythmicity. If data were normalized, it was as the quotient of the absolute data point over the mean of the entire data set.
The gating experiment was carried out using a Packard Topcount according to the protocol described by McWatters et al. (2000). Plants were given a 20-min red light (660 nm; 20 μmol m−2 s−1) pulse, after which the acute response of CAB2::LUC+ activity was measured and compared with an unpulsed control.
Leaf Movement Assays
Plants were entrained in LD 12:12 at 22°C and 120 μmol m−2 s−1 white light for 11 d before they were transferred to constant 25 μmol m−2 s−1 white light. Leaf movements were monitored and periods estimated using the best-curve fit for each leaf position from the emerging first leaves following 72 h in constant conditions (Doyle et al., 2002).
Flowering Time Assays
For flowering time measurements, seeds were stratified (4°C for 3 d) and sown in a 1:3 mix of vermiculite and peat in a controlled-growth room under long-day conditions (LD 16:8, 23°C:18°C), intermediate-day conditions (LD 12:12, 22°C:22°C), or short-day conditions (LD 9:15, 23°C:18°C), all with light fluence of 100 to 120 μmol m−2 s−1. Rosette and cauline leaf numbers were counted when a 1-cm bolt was present. Data are representative of two independent experiments, each containing 10 to 30 plants that gave very similar results.
Quantitative Real-Time RT-PCR
Seedlings were grown for 6 d in LD 12:12, 22°C:22°C (as for LUC assays above) and then in constant white fluorescent light (100 μmol m−2 s−1) for 3 d, when samples were harvested at the indicated times starting 72 h after the last dawn. Samples were collected and immediately frozen in liquid nitrogen. Seedlings for the LD time course were treated in the same way but collected under an ongoing light/dark cycle.
RNA was extracted, and the resulting RNA was purified by an RNeasy Plant Mini Kit (Qiagen), including DNase treatment as described in the manufacturer’s protocol. One microgram of RNA was used as a template for cDNA synthesis using iScript cDNA Synthesis Kit procedures (Bio-Rad Laboratories). Real-time PCR was performed with a MyIQ, ICycler, or CFX96 Real-Time PCR Detection System (all Bio-Rad Laboratories) using iQ SYBR Green Supermix (Bio-Rad Laboratories).
The efficiency of amplification was assessed relative to β-TUBULIN (βTUB) or ELONGATION FACTOR1α (EF1α) expression. Each experiment was repeated at least two times with independent biological material. Expression levels were calculated relative to the reference gene using a comparative threshold cycle method (Livak and Schmittgen, 2001). The results are means of two to three independent experiments, each with three technical repeats, and are expressed relative to the mean of the wild-type series after standardization to the βTUB or EF1α control.
Primers for βTUB and EF1α controls were as published previously (Czechowski et al., 2004; Knight et al., 2008, respectively). Gene-specific primer sequences were as follows: CCA1-F, 5′-TCTGTGTCTGACGAGGGTCGAATT-3′; CCA1-R, 5′-ACTTTGCGGCAATACCTCTCTGG-3′; EBI-F, 5′-TGCGAGAATATGCTTAATTGC-3′; EBI-R, 5′-CCACAACATCACAAGACAAG-3′; LHY-F, 5′-CAACAGCAACAACAATGCAACTAC-3′; LHY-R, 5′-AGAGAGCCTGAAACGCTATACGA-3′; TOC1-F, 5′-ATCTTCGCAGAATCCCTGTGATA-3′; TOC1-R, 5′-GCACCTAGCTTCAAGCACTTTACA-3′; ZTL-F, 5′-TGACGAGGTTGTGTCTATGA-3′; ZTL-R, 5′-AGCACCAGGAACAGTCTCTA-3′; GI-F, 5′-CTGTCTTTCTCCGTTGTTTC-3′; GI-R, 5′-ATCAACAACCTGTCTCCATC-3′.
Transient Protoplast Expression Assays
Arabidopsis Columbia cell cultures were grown at constant LD 12:12, 23°C:23°C, with light intensity of 25 μmol m−2 s−1. Protoplasts were transformed with Myc-EBI or Myc-ebi constructs together with, if appropriate, HA-ZTL, HA-GI, or HA-TOC1 constructs at ZT 12 (Meskiene et al., 2003). Following transformation, cells were cultivated at 23°C under constant light and rotation until 18 h posttransfection, when cells were harvested for protein extraction. All data represent means of three or more independent experiments.
For testing protein interactions, cotransfected cells were harvested as above and suspended in lysis buffer containing 25 mm Tris-HCl, pH 7.8, 10 mm MgCl2, 75 mm NaCl, 5 mm EGTA, 60 mm β-glycerophosphate, 1 mm dithiothreitol, 10% glycerol, 0.2% Igepal CA-630, and 1× Protein Inhibitor Cocktail (Sigma-Aldrich). The cell suspension was frozen in liquid nitrogen and then thawed on ice and centrifuged. Supernatants were precleared by mixing with 10 μL of Protein G-Sepharose beads for 1 h at 4°C before adding 1.5 μL of anti-Myc antibody (9E10; Covance) and incubating reactions for 2 h at 4°C on a rotating wheel. Immunocomplexes were captured on 10 μL of Protein G-Sepharose beads, washed three times in 25 mm sodium phosphate, 150 mm NaCl, 5% glycerol, and 0.2% Igepal CA-630 buffer and then eluted by boiling with 35 μL of SDS sample buffer. The presence of ZTL, TOC1, or GI protein in the immunocomplex was assessed by probing blots with anti-HA antibody (3F10; Roche Diagnostics). Finally, blots were stripped and incubated with anti-Myc antibody to confirm the presence of EBI or ebi in the protein complex.
Clock gene expression studies in protoplasts were performed following transient transformation with Myc-EBI, Myc-ebi, or an empty vector control with or without HA-ZTL, similar to Uemukai et al. (2005). The protoplasts were kept under entrained conditions (LD 12:12, 23°C:23°C) and harvested at ZT 6 or 7 and light intensity of 25 μmol m−2 s−1 or at ZT 18 to 19 under green safelight. Then, 2.5 μg of each DNA was transfected in a volume of 15 μL in each experiment, and three to four biological replicates of each condition were performed, each containing three technical replicates. Real-time quantitative RT-PCR was performed to measure levels of CCA1 and TOC1 transcripts using the primers described above. The biological replicates gave similar results.
Western Blotting
To determine levels of ZTL protein in planta, pools of 8-d-old seedlings were sampled at 3-h intervals and frozen immediately in liquid nitrogen. Approximately 100 mg of ground plant material was used for protein extraction with 200 μL of extraction buffer containing 25 mm Tris-HCl, pH 8, 75 mm NaCl, 10% glycerol, 0.2% Tween 20, 1 mm dithiothreitol, and 0.001% Protein Inhibitor Cocktail (Sigma-Aldrich). Samples were sonicated and then centrifuged at 10,000g for 3 min, and the supernatant was transferred to a new tube. Protein concentrations were measured using a Bradford assay and a SpectraMax Spectrophotometer (Molecular Devices).
Then, 30 μg of sample was loaded onto an 8% SDS-polyacrylamide gel. After the gel was run, the proteins were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore). To measure protein loading levels, membranes were stained with Ponceau S solution (0.1% Ponceau S in 5% acetic acid) and scanned. Membranes were blotted with ZTL affinity-purified antibody in 1:666 dilution (Kim et al., 2003a, 2007). Secondary antibody (GE Healthcare ECL Anti-Rabbit IgG) was applied in 1:5,000 dilution, and the protein signal was detected using enhanced chemiluminescence (GE Healthcare) and a FUJIFILM LAS-3000 Luminescent Image Analyzer. Protein levels were normalized to the Ponceau S stain for the same sample using MetaMorph Imaging Software (Molecular Devices).
For studies of posttranslational regulation of EBI and ebi, protoplasts were cotransfected with EBI or ebi and EBI or ebi plus ZTL constructs. After 18 h, further protein synthesis was arrested by the addition of 100 μm cycloheximide. Samples were collected and proteins extracted at the indicated times prior to western blotting. Levels of EBI, TOC1, and ZTL proteins were determined using anti-Myc and anti-HA antibodies.
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL/DDBJ data libraries under the following accession numbers: CCA1 (AT2G46830), EBI/AtNFXL2 (AT5G05660), EF1α (AT5G60390) GI (AT1G22770), LHY (AT1G01060), TOC1 (AT5G61380), βTUB4 (AT5G44340), and ZTL (AT5G57360).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of NFX-like genes.
Supplemental Figure S2. CAB2::LUC+ expression in red light.
Supplemental Figure S3. EBI expression data from the Bio-Array Resource for Plant Functional Genomics.
Supplemental Figure S4. Yeast two-hybrid interaction between EBI and ZTL.
Supplemental Table S1. List of NFX-like genes.
Acknowledgments
We are grateful to the anonymous reviewers who provided critical and helpful comments in order to refine the findings of this paper. We are thankful to Seth Davis for the gift of the TOC1::LUC+ construct.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Allen T, Koustenis A, Theodorou G, Somers DE, Kay SA, Whitelam GC, Devlin PF. (2006) Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18: 2506–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272: 5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Masuda D, Yasuda M, Nakashita H, Kudo T, Kimura M, Yamaguchi K, Nishiuchi T. (2008) AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J 53: 450–464 [DOI] [PubMed] [Google Scholar]
- Aschoff J. (1979) Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z Tierpsychol 49: 225–249 [DOI] [PubMed] [Google Scholar]
- Ashelford K, Eriksson ME, Allen CM, D'Amore L, Johansson M, Gould P, Kay S, Millar AJ, Hall N, Hall A. (2011) Full genome re-sequencing reveals a novel circadian clock mutation in Arabidopsis. Genome Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- DeCoursey PJ, Krulas JR, Mele G, Holley DC. (1997) Circadian performance of suprachiasmatic nuclei (SCN)-lesioned antelope ground squirrels in a desert enclosure. Physiol Behav 62: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Ding Z, Doyle MR, Amasino RM, Davis SJ. (2007) A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM. (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Dunlap JC. (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ. (2003) Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218: 159–162 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Casal JJ. (2004) Phenotypic characterization of a photomorphogenic mutant. Plant J 39: 747–760 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han LQ, Suh SS, Salomé PA, McClung CR, Somers DE. (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fülöp K, Pettkó-Szandtner A, Magyar Z, Miskolczi P, Kondorosi E, Dudits D, Bakó L. (2005) The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J 42: 810–820 [DOI] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM. (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. (2002) The role of CCA1 and LHY in the plant circadian clock. Dev Cell 2: 516–518 [DOI] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al. (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM. (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Jones MA. (2009) Entrainment of the Arabidopsis circadian clock. J Plant Biol 52: 202–209 [Google Scholar]
- Jordan KW, Morgan TJ, Mackay TFC. (2006) Quantitative trait loci for locomotor behavior in Drosophila melanogaster. Genetics 174: 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Hall A, Kozma-Bognár L, Kim WY, Eriksson ME, Tóth R, Hanano S, Fehér B, Southern MM, et al. (2006) Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. (2003a) Skp2 regulates Myc protein stability and activity. Mol Cell 11: 1177–1188 [DOI] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han LQ, David K, Putterill J, Nam HG, Somers DE. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kim WY, Geng R, Somers DE. (2003b) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Thomson AJW, McWatters HG. (2008) Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol 148: 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Más P. (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Smith GT, Chi Y, Deshaies RJ. (2005) A putative stimulatory role for activator turnover in gene expression. Nature 438: 113–116 [DOI] [PubMed] [Google Scholar]
- Lisso J, Altmann T, Müssig C. (2006) The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Lett 580: 4851–4856 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Millar AJ, Turner MS. (2005) Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol 234: 383–393 [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL. (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P. (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18: 273–281 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ. (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ. (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H. (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen RK, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ. (2004) Input signals to the plant circadian clock. J Exp Bot 55: 277–283 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA. (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H. (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P. (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. (1997) Independent photoreceptive circadian clocks throughout Drosophila. Science 278: 1632–1635 [DOI] [PubMed] [Google Scholar]
- Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng YF, Andersson CR, Zhao YD, Ljung K, Harmer SL. (2009) REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106: 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden LC, Song HR, Jackson S, Morris K, Carre IA. (2002) Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proc Natl Acad Sci USA 99: 13313–13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince Prue D. (1997) Photoperiodism in Plants, Ed 2. Academic Press, San Diego [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, de Uemukai S, Kato K, Kondorosi E, Murray JAH, Ito M, Shinmyo A, Sekine M. (2005) Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol Biol 57: 83–100 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T. (2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ., III (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]