Abstract
An optimized genetic construct for plastid transformation of tobacco (Nicotiana tabacum) for the production of the renewable, biodegradable plastic polyhydroxybutyrate (PHB) was designed using an operon extension strategy. Bacterial genes encoding the PHB pathway enzymes were selected for use in this construct based on their similarity to the codon usage and GC content of the tobacco plastome. Regulatory elements with limited homology to the host plastome yet known to yield high levels of plastidial recombinant protein production were used to enhance the expression of the transgenes. A partial transcriptional unit, containing genes of the PHB pathway and a selectable marker gene encoding spectinomycin resistance, was flanked at the 5′ end by the host plant’s psbA coding sequence and at the 3′ end by the host plant’s 3′ psbA untranslated region. This design allowed insertion of the transgenes into the plastome as an extension of the psbA operon, rendering the addition of a promoter to drive the expression of the transgenes unnecessary. Transformation of the optimized construct into tobacco and subsequent spectinomycin selection of transgenic plants yielded T0 plants that were capable of producing up to 18.8% dry weight PHB in samples of leaf tissue. These plants were fertile and produced viable seed. T1 plants producing up to 17.3% dry weight PHB in samples of leaf tissue and 8.8% dry weight PHB in the total biomass of the plant were also isolated.
Sustainable, efficient, and economical methods for producing high-quality bio-based renewable materials, chemicals, and fuels are necessary to decrease the world’s dependence on petroleum. To gain acceptance, bio-based products must either provide differentiated performance through new properties or be able to compete with their petroleum-derived counterparts on cost. Polyhydroxyalkanoates (PHAs) are a family of biodegradable, renewable plastics that possess properties spanning the range from elastomeric materials to soft films to crystalline materials, allowing their use in a variety of applications currently served by petroleum-based plastics. Differences in monomer unit composition as well as polymer molecular weight form the basis of the wide range of properties that can be obtained with these plastics (Abe and Doi, 2002; Feng et al., 2002; Satkowski et al., 2002; Reddy et al., 2003). PHAs exist in nature within various bacteria, where they serve as a carbon storage material when the microbes are faced with a nutrient limitation that impedes their growth and as a source of energy when more favorable growth conditions return (Madison and Huisman, 1999; Suriyamongkol et al., 2007). PHAs possess excellent stability and shelf life in use coupled with a unique ability to biodegrade in a wide range of environments, including compost, soil, wetlands, marine, and anaerobic digestion facilities (Jendrossek and Handrick, 2002). PHAs can also be converted through simple processes to a range of chemical intermediates. For example, poly-3-hydroxypropionic acid can be converted to acrylic acid using a simple thermal procedure (Zhong and Whitehouse, 2005). Similarly, poly-3-hydroxybutyrate (PHB) can be thermally converted to crotonic acid, a potential renewable platform chemical, which can be readily transformed to a number of commodity chemicals, including propylene by decarboxylation (Peterson and Fischer, 2010) and butanol by hydrogenation (Coons, 2010). The ability to sequester PHAs in bacterial or plant biomass as an inert granular material makes it possible to produce large amounts of a readily convertible polymeric chemical precursor in a biological system where production of the chemical itself might be toxic to the host.
Genetic engineering has allowed the enhancement and/or modification of native microbial pathways for PHA production as well as the transfer of pathways to nonnative producers. Significant progress has been made in efforts to produce PHAs in microbial systems using large-scale industrial fermentations of bacteria, and select compositions of materials produced using this technology are in commercial production (Snell and Peoples, 2009; Coons, 2010). Direct production of PHAs in crop plants is another route for large-scale manufacture of these polymers and could be especially advantageous in energy crops, where a plant by-product, such as biomass or seed oil, could be used for the production of energy (Snell and Peoples, 2009). Most of the efforts to produce PHAs in plants have focused on the homopolymer PHB or the copolymer poly-3-hydroxybutyrate-co-3-hydroxyvalerate, although some effort has been directed toward the production of medium-chain-length PHAs (Suriyamongkol et al., 2007; van Beilen and Poirier, 2008; Snell and Peoples, 2009). To date, the highest levels of PHB have been achieved in plastids (Suriyamongkol et al., 2007; van Beilen and Poirier, 2008; Snell and Peoples, 2009), likely due to the high flux of the PHB pathway substrate acetyl-CoA through this organelle during fatty acid biosynthesis. In the model plant Arabidopsis (Arabidopsis thaliana), PHB has been produced in leaf tissue samples at levels up to 4% of the tissue’s fresh weight (equivalent to approximately 40% dry weight PHB) in plants engineered with nucleus-encoded expression cassettes for plastid-targeted enzymes (Bohmert et al., 2000). In other plants, PHB levels ranging from trace amounts to 7.7% of the dry weight of the leaf or seed sample have been achieved depending on the plant host (Poirier and Gruys, 2002; Suriyamongkol et al., 2007; Snell and Peoples, 2009). PHB synthesis in biomass crops of industrial interest such as switchgrass (Panicum virgatum; Somleva et al., 2008), sugarcane (Saccharum spp. hybrids; Petrasovits et al., 2007; Purnell et al., 2007), and corn stover (leaves and stock of Zea mays; Poirier and Gruys, 2002) have also been reported. Further work, however, is needed to increase levels in these crops for commercial purposes.
One approach to increasing product yield is to increase the expression of the PHB pathway genes in the host plant. Plastid-encoded gene expression has yielded extremely high levels of protein production, with transformants producing 45% (De Cosa et al., 2001) to 70% (Oey et al., 2009) of the transgene-encoded protein per unit of soluble leaf protein and up to 72% (Ruhlman et al., 2010) of the transgene-encoded protein per unit of total leaf protein in tobacco (Nicotiana tabacum). While these expression levels are too high for most metabolic engineering strategies for the production of chemicals, fuels, or materials without creating unwanted stress on the host plant, expression levels can be somewhat controlled by the choice of regulatory elements flanking the transgenes. Plastid gene expression is regulated to a large extent at the posttranscriptional level (Sugita and Sugiura, 1996; Stern et al., 1997; Bruick and Mayfield, 1999; Dubald et al., 2008), and 5′ and 3′ untranslated regions (UTRs) have been shown to impact translational efficiency and mRNA stability, respectively (Eibl et al., 1999). However, these UTR sequences, as well as any other sequences with significant homology to the host’s plastome, must be used with care, since they can cause unwanted rearrangements (Svab and Maliga, 1993; Staub and Maliga, 1994a; Rogalski et al., 2006, 2008; McCabe et al., 2008; Zhou et al., 2008; Gray et al., 2009). Reducing the use of DNA fragments with homology to endogenous sequences and/or replacing them with heterologous sequences can decrease the occurrence of unexpected rearrangements (Nadai et al., 2009) but may yield lower transgene expression levels (Ruhlman et al., 2010).
The first attempts to produce PHB in tobacco with plastid-encoded expression of genes encoding the PHB biosynthetic pathway were successful in producing polymer; however, the success was limited in that polymer levels were very low (Nakashita et al., 2001; Lössl et al., 2003, 2005; Arai et al., 2004). These experiments were performed with plastid transformation vectors that included minimal, if any, optimization of transgene expression cassettes. Efforts included direct placement of a native bacterial operon behind a plastid promoter (Nakashita et al., 2001), optimization of the ribosome-binding sites of PHB genes in a bacterial operon (Arai et al., 2004), placement of a plastid promoter and plastid 5′ UTR in front of a native bacterial operon (Lössl et al., 2003), and creation of a genetic construct where expression of the PHB genes is under the control of an ethanol-inducible expression system (Lössl et al., 2005). Of these strategies, the highest levels of PHB were obtained using a plastidial promoter and 5′ UTR in front of the native bacterial operon. Up to 1.7% dry weight PHB in leaves of tobacco plantlets regenerated from callus was obtained, although subsequent growth of these plantlets for 3 weeks under in vitro conditions reduced the average PHB content of lines to 20 ppm and the lines were sterile (Lössl et al., 2003). Subsequent work reported that high-level, plastid-encoded expression of the phaA gene encoding the 3-ketothiolase enzyme can result in sterile plants (Ruiz and Daniell, 2005). Since little success has been achieved with nucleus-encoded expression systems for PHB genes encoding plastid-targeted enzymes in tobacco (Arai et al., 2001; Bohmert et al., 2002), it was unclear whether the low yields obtained upon plastid-encoded expression of PHB genes were due to problems associated with tobacco as a host system for PHB production, the inability to divert substrate acetyl-CoA from fatty acid biosynthesis pathways, or limitations in the design of the plastid transformation constructs used in the experiments.
In this study, we chose to reexamine plastid-encoded PHB production using tobacco as a host plant to determine if stable production of high levels of polymer could be obtained in soil-grown plants and to explore the potential of plastid-encoded gene expression as a system for reliable engineering of multigene pathways for the production of industrial products. Our efforts focused on the enhancement and stabilization of transgene expression by extending the psbA operon by four transgenes, the three genes encoding the PHB pathway and a selectable marker. Short translational control elements were used to optimize each transgene’s expression. In addition, genes with similar codon usage and GC content to the native tobacco plastome were chosen to further improve expression. This strategy allowed the production of significantly higher levels of PHB in both heterotrophically and autotrophically grown plants compared with previously published results (Nakashita et al., 2001; Lössl et al., 2003, 2005; Arai et al., 2004).
RESULTS
Design of Plastid Transformation Vectors
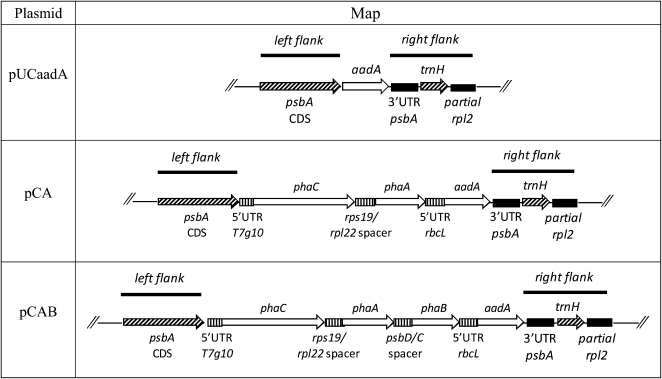
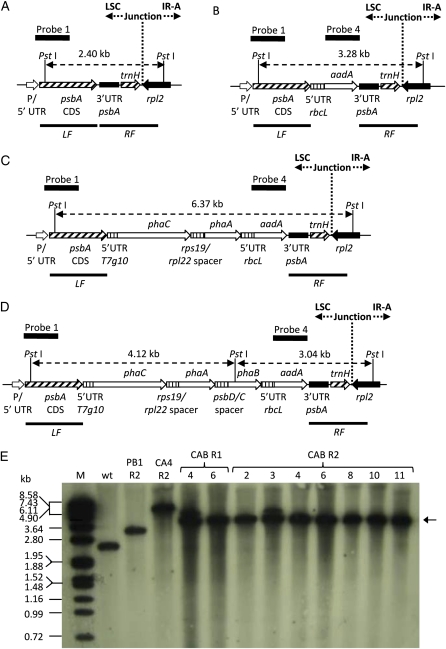
Tobacco was chosen as the host plant for plastid-encoded PHB production since plastid transformation procedures, while available for an increasing number of crops, are considered to be most routine in this plant (Maliga, 2003; Lutz et al., 2007). A set of vectors for plastid transformation was designed to target the insertion of transgenes encoding enzymes of the PHB biosynthetic pathway behind the native psbA promoter and its coding sequence. This so-called operon-extension vector (Herz et al., 2005) reduces the repetition of plastidial sequences that may lead to unwanted rearrangements in the plastome by eliminating the need for a promoter in the inserted DNA that may possess homology to regions in the host’s plastid genome. The tobacco psbA coding sequence was used as a left flank, and the 3′ UTR of psbA, the trnH gene, and a partial sequence of ribosomal protein L2 (rpl2) was used as a right flank within the plastid transformation vectors to promote homologous recombination into the selected site. Genes encoding the Acinetobacter sp. thiolase (phaA) and synthase (phaC; Schembri et al., 1995) and the Bacillus megaterium reductase (phaB; McCool and Cannon, 1999) were chosen from the set of available PHB biosynthetic pathway gene sequences, since their GC content is similar to that of the tobacco plastome and codons with a low frequency of use in the tobacco plastome are either absent or rarely occur. The 5′ UTR of gene 10 from bacteriophage T7 (Kuroda and Maliga, 2001) and short (fewer than 56 nucleotides) spacer elements of plastidial origin (Herz et al., 2005) were used upstream of the individual transgenes to optimize expression. Construct pCAB, containing the PHB pathway genes and the aadA gene conferring resistance to spectinomycin, was prepared using this strategy. Control constructs pUCaadA, containing aadA, and pCA, containing genes encoding synthase and thiolase as well as the aadA gene, were also prepared. The arrangements of transgene expression cassettes and the identity of spacer elements in these transformation vectors are described in detail in Table I.
Table I. Maps of plastid transformation vectors used in this study.
Maps show inserts cloned within bacterial vector pUC19 (Yanisch-Perron et al., 1985). Abbreviations and notes are as follows: left flank of plasmids pUCaadA, pCA, and pCAB contains the complete coding sequence of psbA (D1 protein of PSII), left flank DNA is homologous to the reverse complement of nucleotides 536 to 1,597 of the tobacco plastome (Herz et al., 2005); aadA, gene encoding aminoglycoside 3′-adenyltransferase from E. coli providing spectinomycin/streptomycin resistance (Svab and Maliga, 1993); right flank of plasmids pUCaadA, pCA, and pCAB contains 3′ UTR of psbA, trnH (tRNA-His), and part of rpl2, right flank DNA is homologous to the reverse complement of nucleotides 155,398 to 155,943 and 1 to 530 of the tobacco plastome (Herz et al., 2005); 5′ UTR T7g10, 5′ UTR of gene 10 of bacteriophage T7 (Kuroda and Maliga, 2001), DNA homologous to nucleotides 22,904 to 22,969 of the bacteriophage T7 genome (EMBL accession no. V01146; Dunn and Studier, 1983); phaCAs, gene encoding PHB synthase from Acinetobacter sp., homologous to nucleotides 2,351 to 4,123 of the Acinetobacter sp. PHA biosynthetic gene locus (EMBL accession no. L37761; Schembri et al., 1995); rps19/rpl22 spacer, DNA homologous to nucleotides 86,353 to 86,399 of the tobacco plastome, contains an intergenic region between rps19 and rpl22 (Herz et al., 2005); phaAAs, gene encoding thiolase from Acinetobacter sp., homologous to nucleotides 4,206 to 5,384 of the Acinetobacter sp. PHA biosynthetic gene locus; 5′ UTR rbcL, 15 nucleotides of the 5′ leader sequence of the gene encoding the large subunit of Rubisco, homologous to nucleotides 57,580 to 57,594 of the tobacco plastome (Svab and Maliga, 1993); psbD/C spacer, DNA homologous to nucleotides 35,463 to 35,517 of the tobacco plastome, contains the sequence region upstream of the gene encoding PSII 44-kD protein (psbC), which is overlapped by the coding region of the psbD gene encoding the PSII D2 protein (Herz et al., 2005); phaBBm, gene encoding acetoacetyl-CoA reductase from B. megaterium, homologous to nucleotides 4,758 to 5,501 of the B. megaterium PHA gene cluster (EMBL accession no. AF109909; McCool and Cannon, 1999).
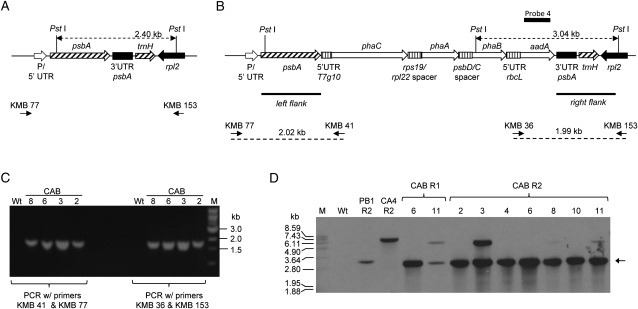
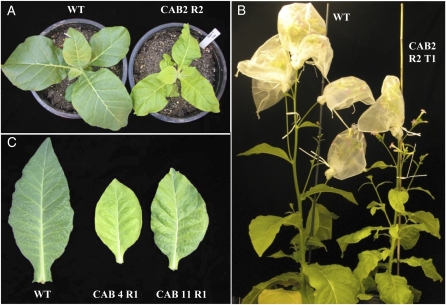
Constructs pCAB, pCA, and pUCaadA were transformed into leaf sections of tobacco cv Petite Havana using particle bombardment, and subsequent selection in the presence of spectinomycin yielded putative transplastomic plants for all three constructs. PCR was used to confirm integration of the pCAB transgenic insert downstream of the psbA coding sequence using primers designed to bind to the host plastidial sequences neighboring either the left or right flank and a sequence within the transgene of interest. Expected bands in PCR designed to detect correct integration were observed in total DNA isolated from candidate transgenic plants grown under in vitro conditions but not in wild-type plants (Fig. 1). Correct integration of DNA into the plastome was also confirmed in Southern-blot experiments with DNA from pCAB and control vector transformants using a probe with homology to a portion of the aadA gene (Fig. 1). In these experiments, the expected 3.04-kb fragment (Fig. 1D) that would be present in plants containing correctly inserted DNA from pCAB into the host plastome was observed. Likewise, 3.3- and 6.4-kb bands indicating correct integration of DNA from the pUCaadA and pCA plastid transformation vectors, respectively, were also observed (Fig. 1D). An additional fragment of 5.4 kb was detected in some of the CAB lines that will be discussed in detail later. Plants from pCAB transformations were found to possess a slightly paler green phenotype and grew slower than wild-type plants (Fig. 2). These phenotypical differences persisted throughout plant development. Four out of five plants transformed with the pCA plasmid possessed a wild-type phenotype. However, one line developed slightly lighter green young leaves when the plant was at a developmental stage in which it possessed between five and 10 leaves. This phenotypic alteration was not detectable during earlier or later developmental stages. Plants generated with the pUCaadA control plasmids possessed a wild-type phenotype.
Figure 1.
Integration of pCAB at the psbA locus. A and B, Diagrams illustrating the genetic arrangement of the wild-type psbA locus (A) and the expected genetic arrangement of the psbA locus after plastid transformation with pCAB (B). Binding sites of PCR primers used to verify correct insertion of the transgenic DNA behind the psbA coding sequence (KMB 77, KMB 41, KMB 36, and KMB 153) are shown in B along with the size of each predicted PCR product. C, Agarose gel electrophoresis of PCR products demonstrating correct integration of the transgenic DNA. M, Marker; Wt, wild-type plant. Samples 2, 3, 6, and 8 are derived from CAB plant lines 2, 3, 6, and 8 in the R2 regeneration cycle (for explanation of regeneration cycles, see Fig. 3). D, Southern blot of PstI-digested genomic DNA from transplastomic and wild-type lines probed with probe 4, a DNA fragment with homology to the aadA gene. Genomic DNA was isolated from the following lines: Wt, the wild type; PB1 R2, line obtained after plastid transformation of pUCaadA in the R2 regeneration cycle; CA 4 R2, line obtained after plastid transformation of pCA in the R2 regeneration cycle; CAB R1 lines 6 and 11, lines obtained after plastid transformation of plasmid pCAB in the R1 regeneration cycle; CAB R2 lines 2, 3, 4, 6, 8, 10, and 11, lines obtained after plastid transformation of pCAB in the R2 regeneration cycle. The arrow shows the expected 3.04-kb band for the expected integration event for CAB lines.
Figure 2.
Phenotype of transplastomic lines obtained after transformation of pCAB. A, Wild-type (left) and transplastomic CAB 2 R2 (right) plants after growth in tissue culture and subsequent transfer to soil. Both plants were grown in the greenhouse in soil for 19 d. The total ages of the wild-type and CAB 2 R2 plants (including tissue culture growth) are 6 and 10 weeks, respectively. Line CAB 2 R2 was obtained after plastid transformation of pCAB, isolation of a regenerant, and one additional cycle of shoot regeneration from an excised leaf (for R2 regeneration cycle description, see Fig. 3). B, Wild-type (left) and transplastomic CAB 2 R2 T1 (right) plants after 63 and 85 d of growth, respectively, in the greenhouse. Line CAB 2 was grown from T1 seed produced from a line obtained after plastid transformation of plasmid pCAB, isolation of a regenerant, and performance of one additional cycle of shoot regeneration from an excised leaf (R2 regeneration cycle). Growth of plants in A and B was staggered so that comparison of plants at similar developmental stages could be recorded. Mesh bags shown in B were used for seed collection. C, Leaves harvested from wild-type and transplastomic CAB plants of equivalent age. Each plant was grown in tissue culture and then transferred to soil and grown for an additional 20 d in a greenhouse. The total age of each plant at the time of harvest was 2 months. Each harvested leaf is leaf 8, where leaf 1 is defined as the oldest leaf of the plant. Lines are as follows: WT, the wild type; CAB 4 R1, line obtained after plastid transformation and isolation of a regenerant (for R1 regeneration cycle description, see Fig. 3); CAB 11 R1, line in the R1 regeneration cycle.
Plants Transformed with Plasmid pCAB Accumulate High Levels of PHB in Leaf Tissue
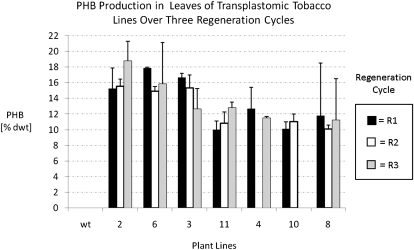
Polymer content in plants transformed with pCAB was measured by gas chromatography-mass spectrometry (GC-MS) procedures after subjecting tissue to a simultaneous extraction and butanolysis procedure (Kourtz et al., 2007). Tissue from seven independent transplastomic CAB lines was analyzed for PHB content over three regeneration cycles using these procedures, and average polymer levels ranging from 10.0% to 18.8% dry weight PHB were observed (Fig. 3). PHB levels in control plants transformed with pUCaadA were similar to the PHB levels detected in wild-type plants (below 0.01% dry weight). Interestingly, plants transformed with the pCA construct possessed PHB levels between 0% and 0.1% dry weight with average PHB levels of 0.02% to 0.03% dry weight. Since these plants do not contain a reductase transgene, this observation suggests that an endogenous enzyme activity present in the plastid is able to reduce acetoacetyl-CoA to R-3-hydroxybutryl-CoA, albeit at a low level.
Figure 3.
PHB production in leaf samples of greenhouse-grown transplastomic lines over three regeneration cycles. Regeneration cycles at right are as follows: R1, lines obtained after plastid transformation of pCAB and isolation of regenerant; R2, R1 lines that were subjected to one additional cycle of shoot regeneration from an excised leaf; R3, R2 lines that were subjected to another additional cycle of shoot regeneration from an excised leaf. Samples from the R2 regeneration cycle of line 4 and the R1 regeneration cycle of line 10 were not analyzed. Data points represent averages of two mature to senescent leaf samples harvested from the indicated line. One of these samples was harvested from leaf 4 to 7 (where the oldest leaf of a plant is defined as leaf 1), and the other sample was harvested from leaf 8 to 12. Error bars represent sd. dwt, Dry weight.
Polymer from leaf tissue from the first regeneration cycle (R1) of CAB line 4 (Fig. 3), containing 12.7% dry weight PHB, was found to have a weight-averaged molecular weight (Mw) of 471,000 ± 37,000 using gel-permeation chromatography techniques. The polydispersity index of the polymer, defined as Mw divided by the number-averaged molecular weight (Mn) and an indication of the molecular weight distribution of the polymer sample, was found to be 2.2 ± 0.3.
Granules of PHB Are Localized in Plastids
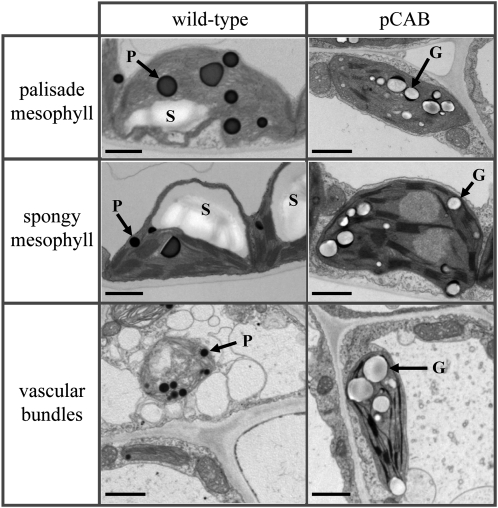
Transmission electron microscopy was used to examine the distribution of polymer granules in palisade and spongy mesophyll cells as well as in vascular tissue (Fig. 4) of CAB transformants. Leaf samples from CAB line 2.1, grown from seeds of plants after two regeneration cycles and containing 6.3% dry weight PHB, were sectioned for these studies. Granular inclusions indicative of PHB formation were found in plastids of all three cell types from the transplastomic line but not in samples obtained from wild-type plants. There was no evidence for PHB granules outside the plastid in any of the CAB lines analyzed. Interestingly, starch granules and plastoglobuli were either significantly reduced or absent within PHB-producing plastids (Fig. 4), and PHB granules often appeared to be colocalized with remnants of the plastoglobuli. This apparent colocalization has also been observed in PHB-producing lines of switchgrass (Somleva et al., 2008).
Figure 4.
PHB accumulation in tobacco leaves. Transmission electron micrographs were obtained from samples of leaf 17 (where leaf 1 is defined as the oldest leaf of a plant) from wild-type plants and CAB line 2.1, a plant obtained from seeds formed from a plant in the R2 regeneration cycle (for definition of the regeneration cycle, see Fig. 3). Cells from the palisade and spongy mesophyll layers as well as from vascular bundles were analyzed. G, PHB granules; P, plastoglobuli; S, starch granules. Bars = 1 μm.
Lines from pCAB Transformations Are Fertile and Produce Progeny with High PHB
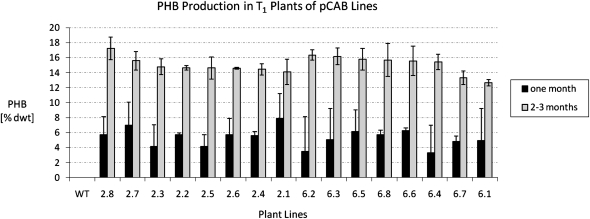
Since previous researchers have obtained sterile lines upon plastid transformation of genes encoding the PHB pathway even though the plants produced low levels of PHB (approximately 20 ppm; Lössl et al., 2003), the fertility of our high-PHB-producing CAB lines was analyzed. All T0 CAB lines were found to be fertile and capable of setting seeds. PHB production in T1 plants was examined by planting T1 seeds obtained from CAB lines 2 and 6 (Fig. 3) and allowing them to grow to maturity in a greenhouse. Leaf material was sampled after 1 and 2 to 3 months of growth. T1 plants were found to produce similar PHB levels as the original T0 parent plants, with average PHB levels in T1 lines 2.1 through 2.8 reaching 15.0% ± 1.0% dry weight and those in T1 lines 6.1 through 6.8 reaching 15.1% ± 1.4% dry weight (Fig. 5).
Figure 5.
PHB production in T1 progeny of CAB lines 2 and 6. Seeds obtained from plants in the R2 regeneration cycle from CAB lines 2 and 6 (Fig. 3) were germinated on tissue culture medium in the presence of spectinomycin as described in “Materials and Methods.” Plantlets were transferred to soil and grown in the greenhouse. Leaf tissue samples were taken after 1 and 2 to 3 months of growth. WT represents the wild-type line. Lines 2.8, 2.7, 2.3, 2.2, 2.5, 2.6, 2.4, and 2.1 are T1 progeny from parental CAB line 2. Lines 6.2, 6.3, 6.5, 6.8, 6.6, 6.4, 6.7, and 6.1 are T1 progeny from parental CAB line 6. Each data point is an average of two different leaf tissue samples, and the error bars represent sd. One of these samples was harvested from leaf 3 to 5 (where the oldest leaf of a plant is defined as leaf 1), and the other sample was harvested from leaf 8 to 13. dwt, Dry weight.
PHB was also found to occur primarily in the leaf tissue of T1 lines, with little produced in the seeds or roots. PHB content in T1 seed samples produced from T0 lines 2, 3, 6, 8, and 11 (Fig. 3) ranged from 0.023% to 0.028% of the seed weight. Root samples from T1 lines CAB 2 and CAB 6 contained PHB ranging from 0.06% to 0.35% dry weight. No PHB was detected in seeds and roots from wild-type plants.
Expression of PHB Pathway Enzymes in T1 Plants
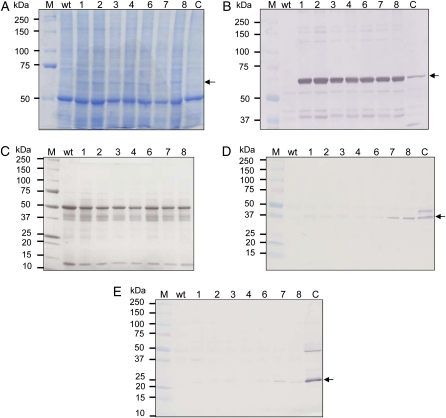
Immunoblot analysis using antibodies prepared against the thiolase, reductase, and synthase proteins was used to confirm the expression of the PHB pathway enzymes in T1 plants. T1 seeds were germinated and grown in tissue culture in the presence of spectinomycin for 46 d, and plantlets were transferred to a greenhouse and grown for an additional 6 d before the third leaf from the bottom of the plant was harvested for analysis. Proteins were separated by SDS-PAGE and detected by Coomassie blue staining. Since PhaC binds to granules of polymer (Gerngross et al., 1993), all SDS-PAGE gel analyses for PhaC were performed using a total protein extract, whereas gels to monitor the expression of thiolase and reductase were examined using soluble protein fractions. Detectable bands at the expected size for PhaC (67.7 kD) were found in all transplastomic lines analyzed on both Coomassie blue-stained gels and western blots probed with polyclonal antibodies raised against the PhaC protein from Ralstonia eutropha (Gerngross et al., 1993), suggesting strong expression of the phaC gene (Fig. 6, A and B). In contrast, expression of phaA and phaB was lower and more variable, and bands for these proteins were only visible on western blots (Fig. 6, D and E) and not on Coomassie blue-stained gels (Fig. 6C). The strong expression of phaC in these plants was likely due to the use of the strong 5′ UTR from gene 10 of bacteriophage T7 to regulate its expression and possibly to its position directly behind the psbA coding sequence in the synthetic operon.
Figure 6.
Expression of transgenes in leaf tissue of T1 plants. Synthase (A and B), thiolase (C and D), and reductase (C and E) enzymes were detected in extracts prepared from leaf tissue of T1 lines obtained from seed of parent line CAB 6 from the R2 regeneration cycle (Fig. 3). A, SDS-PAGE gel of whole SDS-solubilized cell lysates prepared from tobacco leaf tissue. Samples are as follows: M, molecular weight marker; wt, wild-type line; 1, 2, 3, 4, 6, 7, and 8, T1 lines obtained from parent line CAB 6. Lane C contains 268 ng of purified Acinetobacter sp. PHA synthase mixed with 30 μg of whole lysate obtained from a wild-type plant. B, Western blot of whole SDS-solubilized cell lysates probed with polyclonal antibodies prepared against the PHA synthase protein from R. eutropha. For line designations, see A. Lane C contains 268 ng of purified Acinetobacter sp. PHA synthase mixed with 30 μg of whole lysate obtained from a wild-type plant. C, SDS-PAGE gel of soluble protein extracts (15 μg) prepared from tobacco leaf tissue. For line designations, see A. D, Western blot of soluble extracts (15 μg) probed with polyclonal antibodies prepared against the β-ketothiolase protein from Acinetobacter sp. For line designations, see A. Lane C contains 108 ng of purified Acinetobacter sp. β-ketothiolase protein. Antibodies to thiolase were diluted 1:5,000 before use. E, Western blot of soluble extracts (15 μg) probed with polyclonal antibodies prepared against the acetoacetyl-CoA reductase protein from B. megaterium. For line designations, see A. Lane C contains 331 ng of purified Acinetobacter sp. reductase protein. Antibodies to reductase were diluted 1:250 before use. [See online article for color version of this figure.]
T1 Lines Have Delayed Flowering But Are Fertile
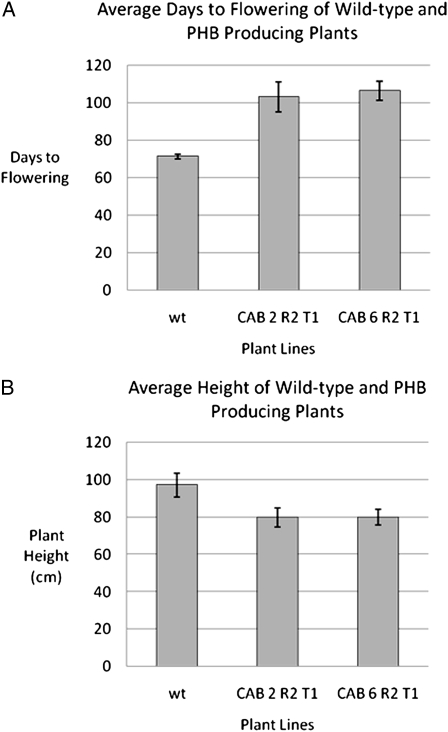
Flowering of T1 lines was found to be delayed such that the T1 lines derived from parental lines 2 and 6 took 44% to 49% longer to flower than wild-type plants (Fig. 7A). The delayed flowering could be in response to the slower growth and delayed development of CAB transgenic lines. The plant height at maturity of T1 lines derived from parental lines 2 and 6 was found, on average, to be approximately 80% of the height of wild-type plants (Fig. 7B). In general, the phenotype of T1 lines was similar to the parental T0 lines. However, in one out of 22 soil-grown T1 plants from line 6, a phenotype substantially different from the parent phenotype was observed. This plant was smaller and bushier and had curly, pointed leaves yet still produced PHB. We are currently investigating the possible reasons for this phenotypic alteration.
Figure 7.
Comparison of the average days to flowering (A) and height (B) of wild-type and PHB-producing CAB lines. Plants were grown in the greenhouse, and days to flowering (when first flowers formed) and the height of the plant at seed set were recorded for each line. Each data point is an average of values from eight transplastomic lines or six wild-type plants. Error bars represent sd. Lines are as follows: wt, the wild type; CAB 2 R2 T1 and CAB 6 R2 T1, T1 lines obtained from seed of parental CAB lines 2 and 6 in the R2 regeneration cycle. The PHB value of parental lines 2 and 6 is shown in Figure 3.
Spatial Distribution of Polymer Production in a T1 Line
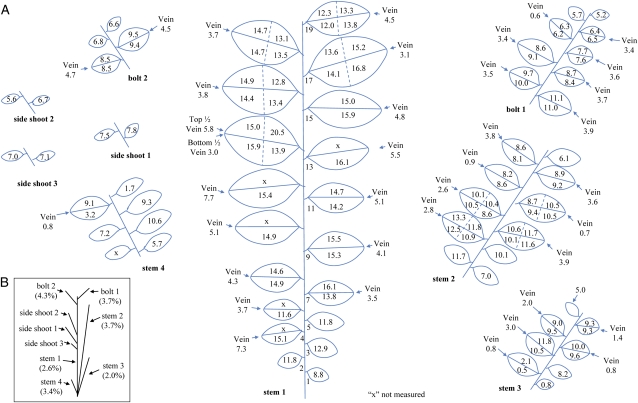
To determine the distribution of PHB accumulation in CAB transplastomic plants, a T1 seed from CAB line 2 from the R2 regeneration cycle (Fig. 3) was germinated and grown in vitro for 7 weeks (after seed imbibition) prior to transfer to soil. The plant was grown to maturity in the greenhouse for an additional 8 weeks. Leaves and stems from this plant were sectioned, and the PHB content and dry weight of each section were measured (Fig. 8). PHB was found to accumulate at significantly higher levels in the leaves than in the stems, with the highest average levels of PHB per leaf reaching approximately 15% to 16% dry weight. Tissue at the tip of the leaf contained higher levels of PHB than the younger base of the leaf in eight out of nine transversally sectioned leaves. In all four stems of the plant analyzed, the leaves at positions in the middle of the stem showed the highest levels of PHB production. In the two bolts, the most basal leaves were the highest PHB producers. The total polymer content of the plant when PHB values of leaf and stem tissue were weight averaged was found to be 8.8% dry weight (Table II). Interestingly, leaf tissue from this CAB plant was a greater percentage of the total plant biomass (71%) than leaf tissue from wild-type plants (54%; Table II).
Figure 8.
Spatial distribution of PHB in a transplastomic tobacco plant grown from a T1 seed of a pCAB transformant. T1 line 2.2 (Fig. 5) was sectioned to determine the spatial distribution of PHB. A, Leaves obtained from the main stem (stem 1), side stems, bolts, and side shoots were sectioned as shown, and the dry weight of each sample was recorded. Numbers within the leaf sections indicate the PHB content (percent dry weight) of each sample as measured by GC-MS. B, Diagram illustrating the positions of stems, bolts, and side shoots in the original plant before sectioning. For purposes of simplicity, the PHB values in stem tissue were recorded as an average value obtained for the whole stem. Leaf slices designated with an “x” were not measured. [See online article for color version of this figure.]
Table II. Comparison of total biomass in a CAB line and a wild-type plant.
| Sample | Biomass of CAB Planta | PHB in CAB Plant | Biomass of Wild-Type Plantb | Mass Ratio CAB to Wild Type |
| g dry wt | % dry wt | g dry wt | ||
| Whole plant | 21.06 | 8.78 | 33.5 ± 3.5 | 0.64 |
| Leaf tissue | 14.98 | 11.18 | 18.4 ± 2.3 | 0.81 |
| Stem tissue | 6.08 | 2.87 | 15.1 ± 1.8 | 0.40 |
Data for CAB is from T1 line 2.2 (Fig. 5).
Data for the wild type is an average of five plants. The weights of inflorescences and seeds were not included in the measurements.
Determination of Homoplasmy
Southern-blot analysis of total DNA isolated from plants grown under in vitro conditions was performed to visually determine the extent of homoplasmy of the lines analyzed. For this experiment, a digoxigenin-labeled DNA probe homologous to a region of the psbA gene (probe 1; Fig. 9) was used and the Southern blot was purposely overexposed so that faint wild-type bands could be detected in DNA samples of transplastomic lines. DNA from a wild-type control plant yielded the expected 2.4-kb fragment of the wild-type plastome when digested with restriction enzyme PstI and probed with the labeled psbA DNA fragment (Fig. 9E, lane wt). DNA from a plant transformed with control plasmid pUCaadA in the R2 regeneration cycle yielded a 3.28-kb fragment, as expected for insertion of the aadA gene (Fig. 9E, lane PB1 R2). There was little if any signal visible at 2.4 kb in the sample lane, suggesting that this plant was near homoplasmy. A plant transformed with control vector pCA in the R2 regeneration cycle yielded a band close to the predicted 6.37-kb size (Fig. 9E, lane CA4 R2). Again, there was little if any signal detectable at 2.4 kb in this sample, suggesting that this plant was near homoplasmy. Plants from transformations of pCAB from either the R1 regeneration cycle (Fig. 9E, lines 4 and 6) or the R2 regeneration cycle (Fig. 9E, lines 2, 3, 4, 6, 8, 10, and 11) yielded a prominent 4.12-kb fragment, as expected for correct integration of the transgenic DNA into the plastome. Little if any of the 2.4-kb fragment expected for the wild-type plastome was observed in these samples, suggesting that these plants had reached near homoplasmy in both the R1 and R2 regeneration cycles. An additional fragment of 5.4 kb was also observed in some of the CAB lines that will be discussed in detail later.
Figure 9.
Genetic analysis of CAB lines. A to D, Diagrams illustrating the genetic arrangement of the wild-type psbA locus (A) and the expected loci after plastid transformation with pUCaadA (B), pCA (C), and pCAB (D). The annealing sites of probe 1 (5′ end of the psbA gene) and probe 4 (aadA gene) as well as the expected size of Southern-blot fragments when genomic DNA is digested with PstI are shown. The junction point of the large single-copy region (LSC) and IR-A of the tobacco plastome (Shinozaki et al., 1986) are shown. Gene abbreviations can be found in Table I. CDS, Coding sequence. E, Southern blot of PstI-digested genomic DNA from transplastomic and wild-type lines probed with probe 1. Genomic DNA was isolated from the following lines: wt, the wild type; PB1 R2, line obtained after plastid transformation of pUCaadA in the R2 regeneration cycle; CA 4 R2, line obtained after plastid transformation of pCA in the R2 regeneration cycle; CAB R1 lines 4 and 6, lines obtained after plastid transformation of plasmid pCAB in the R1 regeneration cycle; CAB R2 lines 2, 3, 4, 6, 8, 10, and 11, lines obtained after plastid transformation of pCAB in the R2 regeneration cycle. The arrow shows the expected 4.12-kb band for the expected integration event for CAB lines. [See online article for color version of this figure.]
While overexposure of Southern blots allows the visualization of faint bands that might occur in samples that are not homoplasmic, detection of a few wild-type plastome copies in a sample dominated by transgenic plastomes is not possible with this technique. Thus, additional screening of select lines judged to be near homoplasmy was performed by screening T1 seeds obtained from self-pollination on medium containing spectinomycin. This procedure allows the detection of residual wild-type copies of the plastome in the parent plant, since these copies should be maternally inherited, producing seeds with spectinomycin-sensitive seedlings. These seedlings are able to germinate in the presence of the antibiotic but are visually distinguishable from spectinomycin-resistant seedlings by their bleached phenotype. The best PHB-producing T0 lines (lines 2, 3, and 6; Fig. 3) were chosen for this analysis, and T1 seeds obtained from self-pollination were germinated on medium both with and without 500 μg mL−1 spectinomycin. While germination rates of lines 2 and 3 were low in this experiment (Table III), subsequent studies with CAB seeds showed that reduced incubation times in the seed sterilization agent can substantially improve germination. Three weeks after seed plating, a portion of the germinating seeds on plates containing spectinomycin possessed mosaic white patches on their cotyledons, suggesting possible sensitivity to the antibiotic and inheritance of some copies of the wild-type plastome. The percentage of seedlings with mosaic cotyledons ranged from 0.6% to 11.6% depending on the line analyzed (Table III). Interestingly, seedlings with mosaic cotyledons did not show any mosaic patterns on their true leaves during growth on medium containing selection agent, and none of the seedlings grown on medium without spectinomycin showed any mosaic patterns (Table III).
Table III. Seed germination tests in the presence and absence of spectinomycin.
| Linea | Medium with Spectinomycin | Medium without Spectinomycin | |||||||
| Seeds Plated | Seeds Germinated | Mosaic Cotyledons | Seeds Plated | Seeds Germinated | Mosaic Cotyledons | ||||
| Total | % | Total | % | Total | % | ||||
| 2 | 11,095 | 2,140 | 19.3 | 49 | 2.3 | 1,783 | 1,219 | 68.4 | 0 |
| 3 | 10,445 | 1,357 | 13 | 158 | 11.6 | 1,790 | 655 | 36.6 | 0 |
| 6 | 7,656 | 4,241 | 55.4 | 28 | 0.6 | 1,823 | 1,366 | 74.9 | 0 |
| Wt | 1,648 | 1,237 | 75.1 | 0b | 0b | 1,369 | 1,025 | 74.9 | 0 |
Lines 2, 3, and 6 refer to CAB lines grown from T1 seed obtained from parental lines 2, 3, and 6 in the R2 regeneration cycle (Fig. 3). Line Wt refers to the wild-type control line.
Cotyledons were completely white/bleached, indicating no spectinomycin resistance.
Integrity of the Transplastomic DNA Insertion
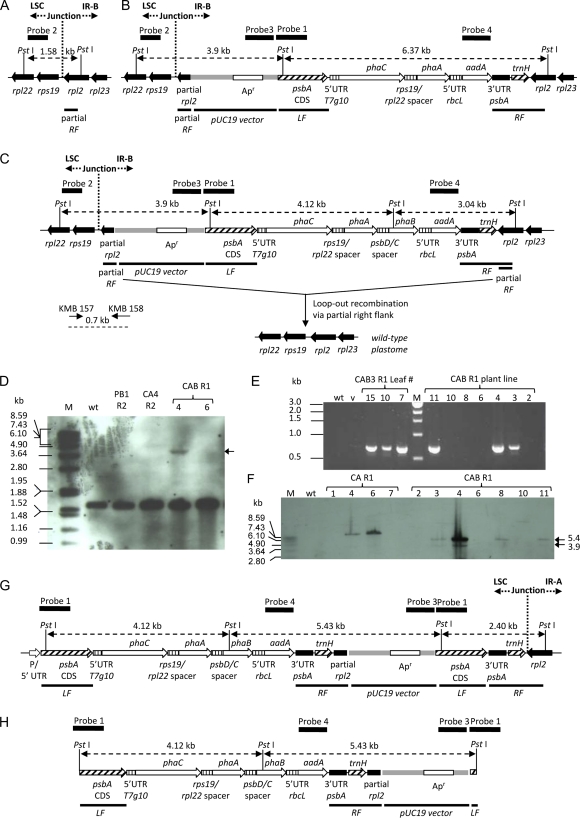
Extensive unwanted recombination of sequences within plastid transformation vectors with the host plastome have been observed previously in our laboratory as well as in other laboratories (Svab and Maliga, 1993; Staub and Maliga, 1994a; Rogalski et al., 2006, 2008; McCabe et al., 2008; Zhou et al., 2008; Gray et al., 2009) when stretches of sequences homologous to the host plastome are present in the plastid transformation vectors. When designing vector pCAB and its corresponding control vectors, care was taken to avoid as many sequences with homology to the tobacco plastome as possible in an attempt to prevent these unwanted recombination events. Despite these efforts, additional bands were observed on Southern blots in some lines using both a labeled aadA fragment (probe 4; Fig. 1D, 5.4-kb band in CAB 11 R1 and CAB 3 R2, and CAB 11 R2) and a labeled psbA fragment (probe 1; Fig. 9, 5.4-kb band in CAB 4 R1 and CAB 3 R2). Southern blotting and PCR experiments designed to determine the source of these unexpected bands led to the realization that a portion of the rpl2 gene included in the right flank could lead to an unwanted additional integration event. The region chosen for the right flank in our plastid transformation vectors spans both the large single-copy region (Shinozaki et al., 1986) and the inverted repeat A region (IR-A; Shinozaki et al., 1986; Staub, 2002; Fig. 9A), such that a portion of the right flank corresponding to a 490-bp fragment of the rpl2 gene is also located in the inverted repeat B region (IR-B; Shinozaki et al., 1986; Fig. 10A). Recombination of only the rpl2 portion of the right flank with rpl2 within IR-B of the host plastome would form the cointegrate (Klaus et al., 2004; Fig. 10C), in which the entire plastid transformation vector is inserted in the plastome. To determine if this unwanted integration event occurred, probe 2 was designed to anneal to the 30S ribosomal protein S19 (rps19) gene downstream of the rpl2 gene such that a 3.9-kb fragment would be detected if a cointegrate (Fig. 10C) had formed at the second rpl2 locus in IR-B. Analysis of total DNA from selected plant lines via Southern-blotting procedures detected an expected 1.58-kb fragment that should be present in all samples, including the wild type, with an intact rpl2 gene (Fig. 10D). However, in some CAB transformants, an additional 3.9-kb fragment expected for unwanted insertion of the transgenes at rpl2 in IR-B was detected (Fig. 10D, lane CAB 4 R1). Since this fragment was faint, it appeared that cointegrates were present in only some of the copies of the plastid genome within the plant. This is not surprising, as cointegrates are thought to be inherently unstable due to the presence of nearby direct repeats of the left and/or right flank sequences. These sequence repeats can promote a loop-out recombination event (Klaus et al., 2004), leading to the wild-type plastome (Fig. 10C).
Figure 10.
Recombination events observed in CA and CAB lines. A, Map of the tobacco wild-type plastome near rpl2 in IR-B. B and C, Maps of partial right flank (RF) cointegrates resulting from recombination of the partial rpl2 gene present in the right flank with rpl2 in IR-B. Cointegrates from recombination of pCA (B) and pCAB (C) are shown. A possible loop-out recombination event leading to a wild-type locus is shown in C for pCAB. D, Southern blot of PstI-digested genomic DNA from transplastomic and wild-type lines probed with probe 2, which binds to the rps19 gene. Lane markings are as follows: M, DIGe molecular weight markers; wt, the wild type; PB1 R2, line obtained after plastid transformation of pUCaadA in the R2 regeneration cycle; CA 4 R2, line obtained after plastid transformation of pCA in the R2 regeneration cycle; CAB R1 lines 4 and 6, lines obtained after plastid transformation of plasmid pCAB in the R1 regeneration cycle. E, Agarose gel electrophoresis of PCR products obtained using primers KMB 157 and KMB 158. Binding sites of these primers as well as the expected size of the PCR product are shown in C. Lane markings are as follows: wt, DNA from the wild-type tobacco line; v, sample of plastid transformation vector pCAB plasmid DNA; CAB 3 R1 Leaf # 15, 10, and 7, leaves 15, 10, and 7 harvested from plant line CAB 3 in the R1 regeneration cycle, where leaf 1 is defined as the oldest leaf of the plant; M, DNA molecular weight marker; CAB R1 11, 10, 8, 6, 4, 3, and 2, CAB lines 11, 10, 8, 6, 4, 3, and 2 in the R1 regeneration cycle. F, Southern blot of PstI-digested genomic DNA from transplastomic and wild-type lines probed with probe 3, which binds to a portion of the pUC19 vector backbone found in plant transformation constructs pUCaadA, pCA, and pCAB. Lane markings are as follows: M, DIGe molecular weight markers; wt, the wild type; CA R1 1, 4, 6, and 7, lines obtained after plastid transformation of pCA in the R1 regeneration cycle; CAB R1 2, 3, 4, 6, 8, 10, and 11, lines obtained after plastid transformation of plasmid pCAB in the R1 regeneration cycle. G, Map of the putative left flank (LF) cointegrate resulting from recombination of pCAB via the left flank into the psbA locus. H, Map of plasmid pCAB. Apr, gene encoding ampicillin resistance; LSC, large single-copy region. Abbreviations of genes located in transformation constructs can be found in Table I. [See online article for color version of this figure.]
PCR experiments using primers KMB 157 and KMB 158 (Fig. 10C) provided confirmation of the formation of the rpl2-mediated cointegrates in IR-B and the presence of vector backbone at the second rpl2 site producing the expected 0.7-kb band in PCR (Fig. 10E). Additional Southern-blotting experiments were performed with probe 3, designed to bind at a portion of the pUC19 vector backbone. A faint 3.9-kb fragment indicating the presence of vector backbone at the rpl2 locus in lines CAB 3 R1, CAB 4 R1, and CAB 11 R1 was observed (Fig. 10F). These same lines had previously tested PCR positive for the presence of vector backbone at the second rpl2 locus (Fig. 10E). A more intense band at 5.4 kb was also detected in CAB lines 3, 4, 8, and 11 in the R1 regeneration cycle (Fig. 10F). Interestingly, the 5.4-kb fragment was previously detected on Southern blots probed with a labeled aadA fragment (probe 4; Fig. 1D, lines CAB 11 R1, CAB 3 R2, and CAB 11 R2) and a labeled psbA fragment (probe 1; Fig. 9E, lines CAB 4 R1 and CAB 3 R2). While the origin of the 5.4-kb band is not certain, bands of this size would be present in a cointegrate formed from integration of the right flank only (Fig. 10G) or in free, unintegrated plasmid (Fig. 10H). Additional experiments were not performed to differentiate between these two possibilities, since this was outside the scope of our investigation. Vector backbone was also observed in some CA lines (Fig. 10, B and F).
Interestingly, CAB lines 2 and 6, our highest PHB-producing lines (Fig. 3) and the lines that appeared to be closest to homoplasmy as determined through T1 seed-plating experiments (Table III), did not contain any vector backbone. In contrast, three of the four lines with vector backbone (Fig. 10F) were determined in separate experiments to be unstable R1 or R2 lines, reverting to the wild type in some tissues, a possible consequence of loop-out recombinations between the duplicated sequences expected in cointegrates (Klaus et al., 2004). Line 3, containing the highest percentage of mosaic cotyledons in the T1 seed-plating experiments and thus likely possessing the most wild-type plastome copies of the three lines analyzed, also contained the vector backbone. These results suggest that screening of plants for “single flank” recombination events and vector backbone insertion should be performed routinely in protocols to identify stable plastid transformants.
DISCUSSION
There is widespread interest among industrialized nations to develop viable technologies for the production of renewable fuels and energy. In the United States, government policy is promoting the production of biofuels with legislation mandating the blending of renewable fuels into gasoline, with timelines for achieving designated quotas (Sissine, 2007). Production of fuels from energy crops, such as perennial grasses and industrial oilseeds, has the potential to help meet these blending quotas. The challenge to the technology sector is to develop robust processes that provide economically viable businesses. The production of value-added coproducts in bioenergy crops makes the production of biofuel and bioenergy more economically attractive by providing an additional revenue stream. PHAs, a family of renewable biodegradable bioplastics, are an ideal coproduct for energy crops, since their material properties allow them to access many of the large markets currently served by petroleum-based plastics, and markets for these materials are scalable with the eventual widespread production of biofuels (Snell and Peoples, 2009). The production of PHAs from renewable resources, as well as their inherent biodegradability, makes these materials particularly attractive to industries that are facing the challenge of becoming more environmentally responsible.
In our quest to continually improve multigene expression systems for the production of industrial products in plants, we evaluated plastid-encoded expression systems for the production of PHB. Plastid transformation systems allow the expression of multiple genes in polycistronic synthetic operons, and the maternal inheritance of plastid DNA in most plants provides a level of, although possibly not complete (Svab and Maliga, 2007; Sheppard et al., 2008), gene containment. Disadvantages of plastid transformation systems include the limited number of plants that can be routinely transformed and the extent of unwanted recombination that can occur if plastid transformation vectors contain sequences that are homologous to the host’s plastome.
To evaluate the robustness of current plastid-encoded multigene expression technologies for industrial purposes, we chose tobacco as the host plant for the production of PHB. Tobacco has historically been a difficult host plant for PHB production, with nucleus-encoded expression of genes fused to sequences encoding plastid targeting signals yielding only 0.3% dry weight PHB (Bohmert et al., 2002). For comparison purposes, similar nucleus-encoded, plastid-targeting strategies have yielded up to approximately 40% dry weight PHB in leaf tissue of Arabidopsis (Bohmert et al., 2000) and up to 5.7% and 3.7% dry weight PHB in corn stover (Poirier and Gruys, 2002) and switchgrass leaves (Somleva et al., 2008), respectively. Previous attempts at plastid-encoded production in tobacco yielded up to approximately 1.7% dry weight PHB in young leaf tissue, a number that dropped to an average of 20 ppm upon subsequent growth of the plants (Lössl et al., 2003).
When we designed and cloned our plastid transformation constructs, there were some suggestions in the literature about unwanted rearrangements occurring between regions of the transgenic insert and the host plastome if sequence homology was present (Svab and Maliga, 1993; Staub and Maliga, 1994a; Eibl et al., 1999). However, researchers still routinely used regulatory elements including promoters and untranslated regions with homology to sequences within the host plastome (Verma and Daniell, 2007), often because these were the only sequences readily available for use. While our experiments were in progress, the magnitude of the recombination issue was realized by several laboratories (Huang et al., 2002; Rogalski et al., 2006; Zhou et al., 2008; Gray et al., 2009), including our own. We had previously constructed several other generations of plastid transformation constructs that contained duplications of endogenous tobacco regulatory sequences and observed extensive recombination of our transgene expression cassettes with the corresponding sites in the tobacco plastome. Therefore, we designed expression cassettes such that minimal sequences with homology to the host plastome were present in the vectors. This task was somewhat challenging, since the majority of regulatory sequences readily available at the time were tobacco sequences. As more sequences with little to no homology to plant plastomes become available, such as the 5′ UTR of gene 10 of bacteriophage T7 (Dunn and Studier, 1983; Kuroda and Maliga, 2001), this task should become more routine.
A synthetic operon extending from the endogenous copy of the psbA coding sequence with transcription initiated at the endogenous psbA promoter was designed to test PHB production. The necessity of a promoter within our transgene insert, and thus the sequence homology that would be associated with the additional promoter, was therefore avoided. So called “operon-extension vectors” that allow the placement of transgenes downstream of an endogenous promoter and coding sequence have been previously described using the reporter gene uidA encoding GUS and the selectable marker aadA providing resistance to spectinomycin (Herz et al., 2005). To our knowledge, our efforts are the first time this strategy has been used for the expression of four transgenes. Transformation of the synthetic operon into tobacco and subsequent selection for transformants yielded plants that contained significantly higher PHB levels in both in vitro- and greenhouse-grown plants than has previously been reported. In greenhouse-grown plants, we observed on average 10% to 18% dry weight PHB in tissue harvested from portions of leaves of transgenic lines (Fig. 3). The maximum levels reported by previous researchers in greenhouse-grown plants were ppm (approximately 0.0008% dry weight PHB; Nakashita et al., 2001), 60 ppm (approximately 0.006% dry weight PHB; Arai et al., 2004), and 1,031 ppm (approximately 0.1% dry weight PHB; Lössl et al., 2003). While the reasons for the vast difference in PHB accumulation levels between our experiments and those previously reported are not definitively known, the design of our transformation vector for enhanced expression of each transgene and limited homology to the host plastome is a probable reason. Although our lines were paler green and smaller than wild-type plants, they were fertile and produced viable seed. Transgenes remained stably integrated into the next generation (T1 generation), and similar levels of PHB were found to accumulate in the T0 and T1 generations. Previous researchers reported loss of flowers and male sterility with PHB production at 0.1% or less dry weight in soil-grown plants (Lössl et al., 2003).
The unexpected occurrence of low levels of PHB in plants transformed with the pCA construct lacking the acetoacetyl-CoA reductase transgene suggests the presence of an endogenous enzyme capable of converting acetoacetyl-CoA to R-3-hydroxybutyrl-CoA. While it is currently unknown which endogenous activity in the plastids is capable of catalyzing this reaction, one possible candidate is 3-ketoacyl-acyl carrier protein reductase, an enzyme in the fatty acid biosynthetic pathway. Bacterial equivalents of this enzyme have been used to produce PHAs containing 3-hydroxybutrate monomers in recombinant bacterial hosts (Park et al., 2005; Nomura et al., 2008), and the presence of this enzyme in plant plastids makes it a likely candidate for the low levels of PHB observed in plants transformed with the pCA construct.
PHB was found to accumulate at minimal levels in seeds and roots of our plants, likely due to the transcription of the synthetic operon from the endogenous psbA promoter combined with different translational control elements. Although the psbA promoter is known to produce mRNA in all tissues, its 5′ UTR regulates the accumulation of the psbA-encoded D1 protein in photosynthetic tissue in response to light (Staub and Maliga, 1993, 1994b). Low translational activity in nonphotosynthetic plastids initiated from the translational control elements used in our expression constructs might be the cause for the low PHB accumulation observed in nongreen tissues.
PHB accumulated to higher levels in leaves than in stems of the transplastomic tobacco plants. The highest PHB levels were found in leaves located in the middle of the stem, not in the oldest basal leaves of the plant, possibly because the oldest basal leaves developed and matured while the plant was still small and had not reached its full “production capacity.” Consistent with this interpretation, basal leaves on bolts of the plant that formed when the plant was more mature contained higher levels of PHB than leaves in the middle of the bolt. The PHB distribution pattern in the tobacco plant analyzed in this study is similar to what has previously been observed in PHB-producing tobacco (Lössl et al., 2003), switchgrass (Somleva et al., 2008), and sugarcane (Purnell et al., 2007), where maximum PHB accumulation was observed in fully developed mature leaf tissues and older basal sections of leaves in general possessed more PHB than the younger tips.
While it is unknown why our PHB-producing plants contained a higher percentage of their total biomass in leaf tissue than the corresponding wild-type plants (Table II), this phenotypical difference could be a physiological response to reduced or impaired photosynthesis. Previous researchers have shown that plants stressed by low-light conditions, as well as variegated plants, compensate for the reduced photosynthetic efficiency of leaves by producing more leaf area per unit of biomass (Lee, 1986; Sadof and Raupp, 1991; Yang and Sadof, 1995). This same response has also been observed in tobacco transformed with a construct designed to reduce levels of the small subunit of Rubisco via antisense oligonucleotides (Quick et al., 1991; Stitt and Schulze, 1994). Additional studies involving detailed analyses of the photosynthetic capacity of the PHB-producing transplastomic tobacco lines would need to be performed to corroborate this theory.
Despite our efforts to prevent recombination, we did observe some unexpected events. In addition to the expected insertion of our transgenes downstream of the psbA coding sequence, we observed the unexpected integration of the entire pCAB plastid transformation vector, including the pUC19 vector backbone, into the second rpl2 gene that is not adjacent to psbA. While this was a low-frequency event, the plant lines that possessed this recombination event also contained additional vector backbone in an unknown location and were not our best lines in terms of PHB production or homoplasmy. Future efforts using the psbA locus as the insertion site for plastid transformation experiments can prevent this unwanted recombination event by removing the rpl2 region from the right flank, yielding a considerably shorter right flank sequence for homologous recombination. Alternatively, the unwanted presence of vector backbone can easily be screened for via Southern blotting or PCR, allowing the affected plants to be identified and discarded from the pool of transgenics. Since single recombination events initiated by homology between transgene and endogenous host plastome sequences are known to happen frequently, this kind of vector backbone screening has become a routine task in our laboratory.
One of the T1 plants produced during this study was smaller than the other T1 plants, with a bushy phenotype. The cause of this altered phenotype is currently unknown but could be due to unexpected random insertion of the construct into the nuclear genome (tagged mutation), a spontaneous nuclear mutation, or an additional yet uncharacterized plastidial recombination event(s). Additional studies are needed to determine the cause of this phenotype.
The polymer produced in CAB lines was found to be a high-molecular weight molecule with an Mw of 471,000. This value is lower than what is typically observed in a native bacterial PHB producer such as Cupriavidus necator (formerly Alcaligenes eutrophus; Kawaguchi and Doi, 1992). This lower molecular weight may be due to the high levels of PHA synthase observed in our plant lines via western blotting. It has been previously shown that the molecular weight of PHB can be controlled in some bacterial (Sim et al., 1997) and in vitro (Gerngross and Martin, 1995) systems with PHA synthase, such that the Mw of the polymer produced is inversely proportional to synthase activity.
CONCLUSION
Plastid transformation is clearly a promising technique for expressing genes necessary for producing fuels, chemicals, and materials in plants. In this study, we evaluated the utility of plastid transformation in engineering a robust system for expression of a multigene biosynthetic pathway for the production of the bioplastic PHB. The low levels of PHB as well as the plant sterility obtained by previous researchers had provided some concern regarding the utility of this technique for stable high-level production of industrial products in plants. In this study, we have shown that plastid-encoded expression of the PHB multigene pathway can produce high levels of PHB in tobacco without affecting its fertility and that the engineered pathway is stably transferred to the next generation. We have also shown that tobacco has the capability as a host system to produce high levels of PHB, up to an average of 15% to 18% dry weight PHB in samples of leaf tissue and 8.8% in the total biomass of the plant, despite earlier efforts with nuclear and plastid transformation that resulted in the production of only low levels in greenhouse-grown plants (Nakashita et al., 2001; Bohmert et al., 2002; Lössl et al., 2003; Arai et al., 2004). Duplication of this effort in the high-yielding bioenergy crops that are under consideration for the production of biofuels will require the development and/or optimization of reliable, efficient plastid transformation procedures for these crops.
MATERIALS AND METHODS
Plastid Transformation Vectors
The plastome of tobacco (Nicotiana tabacum) contains 23 codons with a low frequency of use (less than 10 in 1,000; http://www.kazusa.or.jp/codon/). The presence of these codons in PHB pathway genes from various natural PHA producers as well as the overall GC content of the genes was compared with data available for the tobacco plastome. Genes from Acinetobacter sp. (Schembri et al., 1995) and Bacillus megaterium (McCool and Cannon, 1999) were found to be most similar to the codon usage (i.e. avoids the use of codons with a low frequency of use [less than 10 in 1,000]) and GC content (less than 50%) of the tobacco plastome sequence and were chosen for use in plastid transformation vectors.
Detailed descriptions of the plastid transformation vectors used in this study as well as references to pertinent DNA sequences are available in Table I. Plasmid pJKD1425 (Schembri et al., 1995) was used as the source of the Acinetobacter sp. PHA operon. Plasmid pGM10 (McCool and Cannon, 1999) was used as the source of PHB genes from B. megaterium.
Plant Material and Transformation
Seeds of tobacco (Nicotiana tabacum ‘Petite Havana SR1’) were obtained from Lehle Seeds. Plants in tissue culture were grown (16-h light period, 20–30 μmol photons m−2 s−1, 23°C; 8-h dark period, 20°C) on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 2% (w/v) Suc. Plastid transformation was performed using a PDS 1000 system (Bio-Rad) and 0.6-μm gold particles as described previously (Svab et al., 1990; Daniell, 1997). Selection of transplastomic lines was performed on Murashige and Skoog/Suc medium supplemented with 500 mg L−1 spectinomycin. Once transferred to soil, plants were grown in growth chambers (16-h light period, 40–80 μmol photons m−2 s−1, 23°C; 8-h dark period, 20°C) or in a greenhouse with supplemental lighting (16-h light period, minimum 150 μmol photons m−2 s−1, 23°C–25°C; 8-h dark period, 20°C–22°C).
Genetic Analysis
Total DNA was isolated from in vitro- or greenhouse-derived tobacco leaves using the DNeasy Kit (Qiagen). PCR analysis of plants was performed using 10 to 12 ng of total DNA with the PCR Supermix Kit (Invitrogen). Oligonucleotides for the detection of transgenes included the following: KMB 41 (5′-TTGAGCTGCGCCAAAGCCTC-3′), KMB 77 (5′-CTTGTGCTAGAACTTTAGCTCG-3′), KMB 153 (5′-CCACCCATGTGGTACTTCATTCTACG-3′), and KMB 36 (5′-GAGTTGTAGGGAGGCAACCATGGCAG-3′). Binding sites of PCR primers designed to detect the PHB transgene integration are shown in Figure 1. Oligonucleotides designed to detect the presence of a vector backbone-containing cointegrate at the second rpl2 locus in IR-B are KMB 157 (5′-GACCGATCATTGTGGGTATAATGG-3′) and KMB 158/M13R (5′-CAGGAAACAGCTATGAC-3′).
For Southern-blot analysis, 2.5 to 7.5 μg of total DNA was digested with the indicated restriction enzymes and blotted onto positively charged nylon membranes (Roche Molecular Biochemicals). Digoxigenin-labeled hybridization probes for the detection of genetic elements were prepared with the DIG Probe Synthesis Kit (Roche Molecular Biochemicals) using the following oligonucleotides: for probe 4/aadA, KMB 85 (5′-GGCAGAAGCGGTGATCGCCGAAGTATCGACT-3′) and KMB 86 (5′-GCCGACTACCTTGGTGATCTCGCCTTTCACGTAGTGGAC-3′); for probe 1/psbA, KMB 96 (5′-CTTCTGTAACTGGATAACTAGCACTG-3′) and KMB 97 (5′-GTTACCAAGGAACCATGCATAGCACTG-3′); for probe 2/rpl22,rps19, large single copy region (Shinozaki et al., 1986), KMB 177 (5′-GGCCGCGAATTTGATTAATTACTCTTCG-3′) and KMB 178 (5′-CCGGGCATCTACCATTATACCCACAATG-3′); and for probe 3/pUC19 vector backbone, KMB 123 (5′-CCGGGAGCTGCATGTGTCAGAGG-3′) and KMB 181 (5′-CATTCTGAGAATAGTGTATGCGGC-3′). Hybridization signals were detected with alkaline phosphatase-conjugated anti-digoxigenin antibody and chemiluminescence detection (CDP-Star; Roche Molecular Biochemicals). The Dige marker VII (Roche Molecular Biochemicals) was used as the DNA Mr marker for all southern blots.
Intein-Mediated Purification of Proteins
Purified protein samples of Acinetobacter sp. β-ketothiolase (PhaAAs) and B. megaterium acetoacetyl-CoA reductase (PhaBBm) were generated using the IMPACT-CN Protein Fusion and Purification System (New England Biolabs). The coding sequence of each gene was amplified by PCR and cloned separately into vector pCR-BluntII-TOPO (Invitrogen). Primers used for coding sequence amplification are as follows: for phaA, KMB 18 (5′-CTCGGATCCCATATGAAAGATGTTGTGATTGTTGCAG-3′) and KMB 19 (5′-TGGAATTCCCGGGGTCACGTTCAACTGCAAGTGCAACACCC-3′); for phaB, KMB 17 (5′-CTCGGATCCCATATGACAACATTACAAGGTAAAGTAG-3′) and KMB 20 (5′-TGGAATTCCCGGGCATGTATAAGCCGCCGTTAATGTTTAACTG-3′). The resulting plasmids were digested with NdeI and SmaI, and fragments encoding PhaAAs (1.1 kb) and PhaBBm (0.75 kb) were ligated into the C-terminal intein fusion vector pTYB2 (New England Biolabs). Plasmids pTYB2A2 and pTYB2B19 were isolated encoding thiolase and reductase proteins, respectively, fused at the C terminus to the intein/chitin-binding domain. Plasmids were separately transformed into Escherichia coli strain ER2566 (New England Biolabs) and grown at 37°C in 1 L of Luria-Bertani medium supplemented with 100 μg mL−1 ampicillin. Cell cultures were grown to midlog phase prior to induction with 0.3 mm isopropyl-β-d-1-thiogalactopyranoside for 20 h at 16°C. Cells were lysed by sonication, and soluble extracts were purified using the IMPACT Protein Purification System (New England Biolabs) according to the manufacturer’s instructions. Purified B. megaterium reductase samples possessed proteins that migrated at the expected relative Mr values on SDS-PAGE gels. N-terminal sequencing (Macromolecular Structure Facility, Department of Biochemistry, Michigan State University, East Lansing) confirmed the isolation of the expected reductase protein. SDS-PAGE analysis of Acinetobacter sp. β-ketothiolase obtained from purified protein preparations of E. coli strain ER2566/pTYB2A2 revealed two major bands (40.6 and 55 kD), both possessing an N-terminal amino acid sequence in the first five amino acids identical to β-ketothiolase from Acinetobacter sp. The band with the expected Mr for Acinetobacter sp. thiolase (40.6 kD) was chosen for the production of polyclonal antibodies.
Production of Antibodies
Samples of Acinetobacter sp. thiolase and B. megaterium reductase proteins purified with the IMPACT system were loaded onto SDS-PAGE gels, stained with Coomassie Brilliant Blue, and bands corresponding to each protein were excised for use as antigen. Polyclonal antibodies were produced by Fitzgerald Industries International in specific pathogen-free (Pasturella) New Zealand White rabbits. The IgG fraction was purified from the crude antibody mixture by protein A affinity chromatography.
The production of polyclonal antibodies to the Ralstonia eutropha synthase has been described previously (Gerngross et al., 1993). Aliquots of these antibodies were further purified with an acetone powder prepared from crude wild-type tobacco protein extracts prior to use and were found to cross-react sufficiently with the Acinetobacter sp. synthase to allow its detection.
Protein Analysis
Crude cell lysates were prepared in 125 μL of cell extraction buffer (100 mm Tris-HCl, pH 6.8, 10 mm EDTA, 4 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride) by grinding 100 to 150 mg of tobacco tissue in liquid nitrogen. Insoluble proteins were removed by centrifugation. The resulting crude soluble extracts were diluted 1:5 in cell extraction buffer, and protein concentrations were measured using the Bradford assay (Bradford, 1976) with reagent purchased from Bio-Rad using bovine serum albumin as a standard. Samples of soluble protein extracts were diluted in 3× SDS sample buffer (New England Biolabs), boiled, and loaded onto Novex precast Tris-Gly gels (Invitrogen).
Total protein extracts for analysis of PHB synthase were prepared in cell extraction buffer supplemented with 4% SDS to solubilize granule-bound PHB synthase. Samples were diluted with 3× SDS sample buffer (New England Biolabs) prior to gel loading. To standardize amounts of SDS-containing total protein extracts loaded in each well of SDS-PAGE gels, an initial gel was run with an equal amount of lysate in each lane and stained with Coomassie Brilliant Blue. Loading volumes for subsequent SDS-PAGE gels were adjusted based on comparative differences in the band intensity of lanes.
For western blots, proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad) using a Bio-Rad Mini-Trans Blot cell. Transfer buffer consisted of 20% methanol, 25 mm Tris base, and 192 mm Gly. Blots were blocked in Tris-buffered saline buffer (20 mm Tris-HCl, pH 7.5, 500 mm NaCl) supplemented with 0.05% Tween 20 and 3% blotting-grade nonfat dry milk (Bio-Rad). Protein detection by immunoblotting was performed using goat anti-rabbit alkaline phosphatase-linked secondary antibodies in Tris-buffered saline supplemented with 0.05% Tween 20 and 1% blotting-grade nonfat dry milk. Protein bands were visualized using the alkaline phosphatase conjugate substrate kit and 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium color development reagents (Bio-Rad).
PHB Analysis
PHB concentrations in plant tissue were measured by GC-MS as described previously (Kourtz et al., 2007) using 30 to 150 mg of lyophilized leaf material.
Polymer Extraction and Molecular Weight Determination
For molecular weight determination, plant tissue was prepared for analysis as follows. A plant from line 4 obtained from transformation with plasmid pCAB after one regeneration cycle (Fig. 3) was grown in a greenhouse with supplemental lighting for approximately 4 months prior to Mw analysis. Samples of plant tissue (approximately 450 mg dry weight) were heated in chloroform (1 mL per 17 mg of dry tissue) at 61°C for 4 h. The CAB plant tissue sample was obtained from a plant that previously had been determined to contain an average content of 12.7% dry weight PHB in a mixed sample of leaves 6 and 11. Cooled samples were triple filtered with 2-μm Teflon filters. The Mw of the extracted polymer was determined by gel-permeation chromatography in chloroform calibrated against monodisperse polystyrene standards with a Waters 2414 Refractive Index detector and three miniMix B 10-μm columns in series (Polymer Laboratories). The polydispersity index, equivalent to Mw/Mn, was calculated from Mw and Mn values determined in gel-permeation chromatography experiments. Data reported for Mw, Mn, and polydispersity index of CAB line 4 are average values obtained from two tissue samples from the same plant.
Transmission Electron Microscopy
Leaf samples were prepared for analysis by transmission electron microscopy by fixing in 2% paraformaldehyde, 2% glutaraldehyde, 4% Suc, 1 mm CaCl2, and 2 mm MgCl2 in 50 mm sodium cacodylate buffer, pH 7.2. One-centimeter-square leaf pieces were cut from the midblade area and cut into strips 0.5 to 1.0 mm wide while submerged in the fixative. The fixative was vacuum infiltrated into the leaf tissue at approximately 70 kPa for several cycles until most pieces sank. The fixation was conducted for 2 h at room temperature. Tissue was rinsed in three changes of 50 mm sodium cacodylate buffer containing 4% Suc and postfixed in the same buffer with 1% osmium tetroxide for 8 h at 4°C. Tissue was rinsed in several changes of distilled water and dehydrated in acetone by 10% increments to 100% acetone and gradually infiltrated (1:3, 1:2, 1:1, 2:1, and 3:1, 100%) with Ellis low-viscosity epoxy resin formulation (Ellis, 2006), an update of the Spurr’s resin mixture (Spurr, 1969). The samples received three changes of 100% resin at 2-h intervals and were embedded in the same and polymerized for 16 h at 70°C. Sections were cut at 60-nm thickness, mounted on copper grids, and stained for 20 min at room temperature with uranyl acetate (uranyl acetate solution was saturated at 4°C in 50% ethanol) and for 3 min in lead citrate (2.5 mg mL−1 in 0.1 n NaOH). Sections were observed at 80 kV in a JEOL JEM-100S transmission electron microscope and photographed with a CCD camera (Scientific Instruments and Applications, Inc.; model 7C). The images were adjusted for brightness and contrast, and occasionally γ, to best represent the image details of interest.
Growth of T1 Plants
T1 seeds of CAB lines 2 and 6 from the R2 regeneration cycle (Fig. 3) were germinated on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 2% (w/v) Suc. The medium for transgenic lines was supplemented with 500 μg mL−1 spectinomycin. Wild-type plants were grown in tissue culture medium without spectinomycin. Plants in tissue culture were grown with a 16-h light period (20–30 μmol photons m−2 s−1, 23°C) and an 8-h dark period at 20°C. Three weeks after seed imbibition, germinated seedlings were transferred to tissue culture vessels and maintained on the medium described above. Six weeks after seed imbibition, six wild-type plants and eight plants of CAB lines 2 and 6 were transferred to a greenhouse with supplemental lighting (16-h light period, minimum 150 μmol photons m−2 s−1, 23°C–25°C; 8-h dark period, 20°C–22°C). The onset of the formation of inflorescences was monitored. Days until flowering were calculated from the day of seed imbibition until opening of the first flower of the first inflorescence. Plant height was measured at the end of the plant’s life cycle.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers V01146, L37761, and AF109909.
Acknowledgments
We thank Renate Ruszczyk (Metabolix) for assistance with plant sampling and data analysis and Mirel Sharxhi and Muna Ray (Metabolix) for analytical help during the course of this study. Transmission electron microscopy was performed by Dale Callaham at the Central Microscopy Facility of the University of Massachusetts in Amherst.
References
- Abe H, Doi Y. (2002) Molecular and material design of biodegradable poly(hydroxyalkanoate)s. Doi Y, Steinbuchel A, , Biopolymers, Vol 3b. Polyesters II: Properties and Chemical Synthesis. Wiley-VCH, Weinheim, Germany, pp 105–132 [Google Scholar]
- Arai Y, Nakashita H, Doi Y, Yamaguchi I. (2001) Plastid targeting of polyhydroxybutyrate biosynthetic pathway in tobacco. Plant Biotechnol J 18: 289–293 [Google Scholar]
- Arai Y, Shikanai T, Doi Y, Yoshida S, Yamaguchi I, Nakashita H. (2004) Production of polyhydroxybutyrate by polycistronic expression of bacterial genes in tobacco plastid. Plant Cell Physiol 45: 1176–1184 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey RN, Willmitzer L. (2000) Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta 211: 841–845 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Steinbüchel A, Tischendorf G, Willmitzer L. (2002) Constitutive expression of the β-ketothiolase gene in transgenic plants: a major obstacle for obtaining polyhydroxybutyrate-producing plants. Plant Physiol 128: 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bruick RK, Mayfield SP. (1999) Light-activated translation of chloroplast mRNAs. Trends Plant Sci 4: 190–195 [DOI] [PubMed] [Google Scholar]
- Coons R. (2010) Industrial biotechnology turning process engineering into profits. Chem Week November 15 [Google Scholar]
- Daniell H. (1997) Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Tuan R, , Methods in Molecular Biology, Vol 62 Humana Press, Totowa, NJ, pp 463–489 [DOI] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee S-B, Miller M, Daniell H. (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubald M, Tissot G, Dufourmantel N, Goutorbe F. (2008) The engineering of recombinant plastids in higher plants. Kumar A, Sopory SK, , Recent Advances in Plant Biotechnology and Its Applications. JK International, New Delhi, pp 36–59 [Google Scholar]
- Dunn JJ, Studier FW. (1983) Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol 166: 477–535 [DOI] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop H-U. (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19: 333–345 [DOI] [PubMed] [Google Scholar]
- Ellis EA. (2006) Solutions to the problem of substitution of ERL 4221 for vinyl cyclohexene dioxide in Spurr low viscosity embedding formulations. Microscopy Today 14: 32–33 [Google Scholar]
- Feng L, Watanabe T, Wang Y, Kichise T, Fukuchi T, Chen G-Q, Doi Y, Inoue Y. (2002) Studies on comonomer compositional distribution of bacterial poly(3-hydroxybutyrate-co-3-hydroxyhexanoate)s and thermal characteristics of their factions. Biomacromolecules 3: 1071–1077 [DOI] [PubMed] [Google Scholar]
- Gerngross TU, Martin DP. (1995) Enzyme-catalyzed synthesis of poly[(R)-(−)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc Natl Acad Sci USA 92: 6279–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerngross TU, Reilly P, Stubbe J, Sinskey AJ, Peoples OP. (1993) Immunocytochemical analysis of poly-beta-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J Bacteriol 175: 5289–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray BN, Ahner BA, Hanson MR. (2009) Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res 18: 559–572 [DOI] [PubMed] [Google Scholar]
- Herz S, Füssl M, Steiger S, Koop H-U. (2005) Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res 14: 969–982 [DOI] [PubMed] [Google Scholar]
- Huang FC, Klaus SM, Herz S, Zou Z, Koop HU, Golds TJ. (2002) Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol Genet Genomics 268: 19–27 [DOI] [PubMed] [Google Scholar]
- Jendrossek D, Handrick R. (2002) Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56: 403–432 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Doi Y. (1992) Kinetics and mechanism of synthesis and degradation of poly(3-hydroxybutyrate) in Alcaligenes eutrophus. Macromolecules 25: 2324–2329 [Google Scholar]
- Klaus SM, Huang FC, Golds TJ, Koop HU. (2004) Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nat Biotechnol 22: 225–229 [DOI] [PubMed] [Google Scholar]
- Kourtz L, Dillon K, Daughtry S, Peoples OP, Snell KD. (2007) Chemically inducible expression of the PHB biosynthetic pathway in Arabidopsis. Transgenic Res 16: 759–769 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2001) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol 125: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. (1986) Unusual strategies of light absorption in rain-forest herbs. Givnish T, , On the Economy of Plant Form and Function. Cambridge University Press, New York, pp 105–131 [Google Scholar]
- Lössl A, Bohmert K, Harloff H, Eibl C, Mühlbauer S, Koop H-U. (2005) Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol 46: 1462–1471 [DOI] [PubMed] [Google Scholar]
- Lössl A, Eibl C, Harloff HJ, Jung C, Koop H-U. (2003) Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep 21: 891–899 [DOI] [PubMed] [Google Scholar]
- Lutz KA, Azhagiri AK, Tungsuchat-Huang T, Maliga P. (2007) A guide to choosing vectors for transformation of the plastid genome of higher plants. Plant Physiol 145: 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. (1999) Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63: 21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. (2003) Progress towards commercialization of plastid transformation technology. Trends Biotechnol 21: 20–28 [DOI] [PubMed] [Google Scholar]
- McCabe MS, Klaas M, Gonzalez-Rabade N, Poage M, Badillo-Corona JA, Zhou F, Karcher D, Bock R, Gray JC, Dix PJ. (2008) Plastid transformation of high-biomass tobacco variety Maryland Mammoth for production of human immunodeficiency virus type 1 (HIV-1) p24 antigen. Plant Biotechnol J 6: 914–929 [DOI] [PubMed] [Google Scholar]
- McCool GJ, Cannon MC. (1999) Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol 181: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nadai M, Bally J, Vitel M, Job C, Tissot G, Botterman J, Dubald M. (2009) High-level expression of active human alpha1-antitrypsin in transgenic tobacco chloroplasts. Transgenic Res 18: 173–183 [DOI] [PubMed] [Google Scholar]
- Nakashita H, Arai Y, Shikanai T, Doi Y, Yamaguchi I. (2001) Introduction of bacterial metabolism into higher plants by polycistronic transgene expression. Biosci Biotechnol Biochem 65: 1688–1691 [DOI] [PubMed] [Google Scholar]
- Nomura CT, Tanaka T, Eguen TE, Appah AS, Matsumoto K, Taguchi S, Ortiz CL, Doi Y. (2008) FabG mediates polyhydroxyalkanoate production from both related and nonrelated carbon sources in recombinant Escherichia coli LS5218. Biotechnol Prog 24: 342–351 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445 [DOI] [PubMed] [Google Scholar]
- Park SJ, Choi J-I, Lee SY. (2005) Engineering of Escherichia coli fatty acid metabolism for the production of polyhydroxyalkanoates. Enzyme Microb Technol 36: 579–588 [Google Scholar]
- Peterson AA, Fischer CR, inventors September 9, 2010. Conversion of natural products including cellulose to hydrocarbons, hydrogen and/or other related compounds. US Patent Application No. 2010/0228067 [Google Scholar]
- Petrasovits LA, Purnell MP, Nielsen LK, Brumbley SM. (2007) Production of polyhydroxybutyrate in sugarcane. Plant Biotechnol J 5: 162–172 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Gruys KJ. (2002) Production of polyhydroxyalkanoates in transgenic plants. Doi Y Steinbüchel A, , Biopolymers, Vol 3a. Polyesters I: Biological Systems and Biotechnological Production. Wiley-VCH, Weinheim, Germany, pp 401–435 [Google Scholar]
- Purnell MP, Petrasovits LA, Nielsen LK, Brumbley SM. (2007) Spatio-temporal characterization of polyhydroxybutyrate accumulation in sugarcane. Plant Biotechnol J 5: 173–184 [DOI] [PubMed] [Google Scholar]
- Quick WP, Schurr U, Fichtner K, Schulze ED, Rodermel SR, Bogorad L, Stitt M. (1991) The impact of decreased Rubisco on photosynthesis, growth, allocation and storage in tobacco plants which have been transformed with antisense rbcS. Plant J 1: 51–58 [Google Scholar]
- Reddy CSK, Ghai R, Rashmi, Kalia VC. (2003) Polyhydroxyalkanoates: an overview. Bioresour Technol 87: 137–146 [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R. (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R. (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Verma D, Samson N, Daniell H. (2010) The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol 152: 2088–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. (2005) Engineering cytoplasmic male sterility via the chloroplast genome by expression of β-ketothiolase. Plant Physiol 138: 1232–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadof CS, Raupp MJ. (1991) Effect of variegation in Euonymus japonica var. aureus on two phloem feeding insects, Unaspis euonymi (Homoptera: Diaspididae) and Aphis fabae (Homoptera: Aphididae). Environ Entomol 20: 83–89 [Google Scholar]
- Satkowski M, Melik D, Autran J-P, Green P, Noda I, Schechtman L. (2002) Physical and processing properties of polyhydroxyalkanoate copolymers. Doi Y Steinbuchel A, , Biopolymers, Vol 3b. Polyesters II: Properties and Chemical Synthesis. Wiley-VCH, Weinheim, Germany, pp 231–263 [Google Scholar]
- Schembri MA, Bayly RC, Davies JK. (1995) Phosphate concentration regulates transcription of the Acinetobacter polyhydroxyalkanoic acid biosynthetic genes. J Bacteriol 177: 4501–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, Ayliffe MA, Blatch L, Day A, Delaney SK, Khairul-Fahmy N, Li Y, Madesis P, Pryor AJ, Timmis JN. (2008) Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol 148: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SJ, Snell KD, Hogan SA, Stubbe J, Rha C, Sinskey AJ. (1997) PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat Biotechnol 15: 63–67 [DOI] [PubMed] [Google Scholar]
- Sissine F. (2007) Energy Independence and Security Act of 2007: a summary of major provisions. A CRS Report for Congress. http://energy.senate.gov/public/_files/RL342941.pdf (February 25, 2011)
- Snell KD, Peoples OP. (2009) PHA bioplastic: a value-added coproduct for biomass biorefineries. Biofuel Bioprod Bior 3: 456–467 [Google Scholar]
- Somleva MN, Snell KD, Beaulieu JJ, Peoples OP, Garrison BR, Patterson NA. (2008) Production of polyhydroxybutyrate in switchgrass, a value-added co-product in an important lignocellulosic biomass crop. Plant Biotechnol J 6: 663–678 [DOI] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Staub JM. (2002) Expression of recombinant proteins via the plastid genome. Vinci VA, Parekh SR, , Handbook of Industrial Cell Culture: Mammalian, Microbial, and Plant Cells. Humana Press, Totowa, NJ, pp 259–278 [Google Scholar]
- Staub JM, Maliga P. (1993) Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J 12: 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. (1994a) Extrachromosomal elements in tobacco plastids. Proc Natl Acad Sci USA 91: 7468–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Maliga P. (1994b) Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J 6: 547–553 [DOI] [PubMed] [Google Scholar]
- Stern DS, Higgs DC, Yang J. (1997) Transcription and translation in chloroplasts. Trends Plant Sci 2: 308–315 [Google Scholar]
- Stitt M, Schulze D. (1994) Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology. Plant Cell Environ 17: 465–487 [Google Scholar]
- Sugita M, Sugiura M. (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Suriyamongkol P, Weselake R, Narine S, Moloney M, Shah S. (2007) Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants: a review. Biotechnol Adv 25: 148–175 [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. (1990) Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA 87: 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. (2007) Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA 104: 7003–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Poirier Y. (2008) Production of renewable polymers from crop plants. Plant J 54: 684–701 [DOI] [PubMed] [Google Scholar]
- Verma D, Daniell H. (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sadof C. (1995) Variegation in Coleus blumei and the life history of Citrus mealybug (Homoptera: Pseudococcidae). Environ Entomol 24: 1650–1655 [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119 [DOI] [PubMed] [Google Scholar]
- Zhong L, Whitehouse RS, inventors May 24, 2005. Methods of making intermediates from polyhydroxyalkanoates. US Patent No. 6,897,338 [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AM, Maloney AP, Kavanagh TA, Gray JC, Bock R. (2008) High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6: 897–913 [DOI] [PubMed] [Google Scholar]