Trench (1969) was the first to characterize the kleptoplastic (i.e. “stolen plastid”) relationship between the sacoglossan mollusc Elysia chlorotica and its algal prey (Vaucheria litorea). In contrast to E. chlorotica, which retains only the plastids of the alga in densely packed digestive tissue (Fig. 1), aquatic invertebrates (e.g. corals, clams, worms, tunicates) and the recently reported spotted salamander (Ambystoma maculatum; work of R. Kerney, reported by Petherick, 2010) owe their photosynthetic capacity to the retention of intact unicellular algae (for review, see Rumpho et al., 2011). Photosynthetic sacoglossans vary in the ability to retain plastids and to maintain their functions. Whereas some of these animals can only utilize transferred photosynthate for several hours before the plastids are degraded, others sustain plastid function for months (for review, see Rumpho et al., 2006, 2011; Händeler et al., 2009; Yamamoto et al., 2009). E. chlorotica exhibits one of the longest time frames for plastid maintenance in the absence of algal food, up to 10 to 12 months in the laboratory (for review, see Rumpho et al., 2011). The obvious question to be asked about this system is how the plastids remain photosynthetically active in the absence of algal nuclei that are presumably required to furnish transcripts for plastid-targeted proteins involved in photosynthesis, signaling, regulation, and protein turnover. This update will compare and contrast past approaches used to understand the basis of plastid maintenance and function with recent work using next-generation sequencing to reconcile what appear to be contradictory outcomes in the observed data.
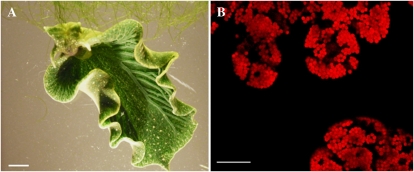
Figure 1.
A, Adult E. chlorotica feeding on its algal prey, the coenocytic heterokont V. litorea. Bar = 5 mm. B, Confocal micrograph showing the red autofluorescent plastids densely packed within the digestive diverticula of E. chlorotica. Once established, these plastids remain photosynthetically active within the adult for several months. Bar = 50 μm.
FUNCTIONAL SEA SLUG KLEPTOPLASTY
The exploitation of photosynthesis by heterotrophic organisms is well documented in aquatic (Trench, 1993; Venn et al., 2008) and terrestrial (Nash, 2008) ecosystems. In these opportunistic relationships, the algal symbiont gains refuge and a stable nutrient source in exchange for supplying the host (e.g. invertebrate, amphibian, plant) with carbon (Trench, 1993; Yellowlees et al., 2008). Symbiotic sacoglossans develop a similar relationship, although exploiting only the plastid captured from specific algal prey (Jensen, 1982; Marin and Ros, 2004; Händeler et al., 2009). This phenomenon was recognized in Elysia species by Kawaguti and Yamasu (1965) and Trench (1969), followed by a description of the ecology (Hinde and Smith, 1974; Jensen, 1986; Clark et al., 1990) and development (Harrigan and Alkon, 1978; West, 1979; West et al., 1984). E. chlorotica has an obligate relationship with Vaucheria species, feeding only on Vaucheria litorea (Fig. 1) or Vaucheria compacta (West et al., 1984). Development is predominantly planktonic, and the deposited eggs and planktonic larval veligers lack plastids. Growth of the veligers occurs by feeding on microalgae in the environment, and metamorphosis occurs after 10 to 21 d in the water column. However, settlement and metamorphosis of the veligers into adult sea slugs require the presence of Vaucheria. Most often, the veligers settle on the algal filaments, ensuring a food supply for recently metamorphosed juveniles (West, 1979). Growth and maturation of E. chlorotica are dependent on feeding on Vaucheria for about 1 week, after which plastids are able to support continued growth of the animal (Rumpho et al., 2011). The mechanisms that allow long-term plastid photosynthetic ability in a heterotrophic host have been studied in detail but remain enigmatic.
HYPOTHESES TO EXPLAIN THE ENIGMA
Retention of Algal Nuclei
A reasonable explanation for long-term plastid maintenance is the presence of algal nuclei within the animal that provide the transcripts needed to support plastid functions. Analyses of adult, green, kleptoplastic sacoglossans using microscopy, PCR, and Southern-blot analysis, however, have failed to substantiate this hypothesis (Graves et al., 1979; Mujer et al., 1996; Green et al., 2000; Rumpho et al., 2001; Mondy and Pierce, 2003; Pierce et al., 2003; Wägele et al., 2011). In addition, molecular markers for nucleus-encoded algal genes (e.g. internal transcribed spacer 1 and spermidine synthase) are not found with PCR using DNA derived from starved animal tissue (Pierce et al., 2007, 2009; Rumpho et al., 2008; Schwartz et al., 2010).
Plastid Genetic Autonomy
Another potential explanation for long-term plastid function in E. chlorotica is the existence of a genetically autonomous V. litorea plastid genome. It is formally possible that this genome has regained, via horizontal gene transfer (HGT), critical genes encoding plastid proteins involved in photosynthesis that have been transferred to the nucleus in other algae and plants. To address this idea, Rumpho et al. (2008) sequenced the plastid genome of V. litorea and found it to be comparable to other algal and plant plastids in terms of gene content and found no evidence for HGT.
Plastid Stability
Although containing a typical algal genome, the Vaucheria plastid exhibits unique physical and biochemical properties that may play a role in the establishment of the sacoglossan symbiosis (Jensen, 1982; Händeler et al., 2009; Wägele et al., 2011). Vaucheria, like many of the algae consumed by sacoglossan molluscs, is filamentous and coenocytic: in essence, a single multinucleate cell. This allows the sea slug to rapidly acquire numerous plastids while feeding but also favors plastids that appear to have a greater longevity than those associated with multicellular algae. Green et al. (2005) investigated the physiology of isolated Vaucheria plastids and found that 30% to 40% (in light versus dark, respectively) remained intact 14 d after isolation from the alga, whereas less than 20% of isolated spinach (Spinacia oleracea) chloroplasts were intact after 24 h. Isolated V. litorea plastids exhibited electron transport activity, CO2-dependent oxygen evolution, and CO2 fixation 72 h post isolation. The isolated plastids were also largely unaffected by osmotic fluctuations, tested up to ±70 mm mannitol. In addition, over a 3-d period, de novo synthesis of plastid proteins in isolated plastids changed minimally in banding patterns and intensity on SDS-PAGE gels, and both Rubisco (large and small subunits) and the PSII D1 protein were synthesized in plastids 3 d post isolation. These data support the unique nature of V. litorea plastids in not requiring σ-factors or other nucleus-encoded regulatory factors for transcription and translation of plastid genes or that the factors are stable for a minimum of 3 d in vitro.
Dual Targeting of Proteins
Dual targeting of animal-derived proteins as an explanation for plastid replenishment has yet to be carefully studied. All of the nucleus-encoded enzymes of the Calvin-Benson carbon reduction cycle have cytosolic counterparts in E. chlorotica except for phosphoribulokinase and sedoheptulose-1,7-bisphosphatase (both subunits of Rubisco are plastid encoded in V. litorea). In addition, transcripts for all of these nucleus-encoded enzymes are present in the partial E. chlorotica transcriptome library discussed in more detail below (Supplemental Table S1). Because these proteins are encoded in the animal genome, they could be coopted for use in the plastid, providing much of the necessary machinery for carbon fixation.
HGT
The most highly cited hypothesis to date, and the focus of this update, is extensive HGT from the algal nucleus to the animal, thereby supporting long-term plastid function (Olendzenski and Gogarten, 2009; Bock, 2010; Boto, 2010; Moran and Jarvik, 2010). The majority of the existing work (Table I; see refs. below) has investigated the presence of genes required for photosynthesis in the nuclear genome and transcriptome of E. chlorotica in both adults and larvae (with and without plastids present, respectively). Until recently, these studies used the candidate gene approach to putatively identify alga-derived genes in the animal. The methods have included PCR (Green et al., 2000; Mondy and Pierce, 2003; Pierce et al., 2003, 2007, 2009; Rumpho et al., 2008, 2009), detection of de novo synthesis of nucleus-encoded plastid proteins using radiolabeling and immunolabeling in combination with specific inhibitors of transcription or translation (Pierce et al., 1996, 2007; Green et al., 2000; Hanten and Pierce, 2001; Rumpho et al., 2001, 2009), northern-blot analysis and genome walking (Rumpho et al., 2008), 14C incorporation and synthesis of chlorophyll in animal tissue (Pierce et al., 2009), and quantitative reverse transcription-PCR of nucleus-encoded algal genes in the animal (Soule, 2009). All of these data support the presence of a few (Rumpho et al., 2008, 2009) to numerous (Pierce et al., 2009; Schwartz et al., 2010) nuclear algal genes or gene fragments in E. chlorotica.
Table I. Molecular and biochemical evidence for nucleus-encoded algal genes in E. chlorotica.
| Protein | Method | Gene | Source |
| Fucoxanthin a/c chlorophyll-binding protein | 35S immunolabeling | fcp | Pierce et al. (1996, 2003) |
| Fucoxanthin a/c chlorophyll-binding protein | PCR | fcp | Pierce et al. (2007) |
| Light-harvesting complex-binding protein 1 | 35S immunolabeling | lhcv-1 | Hanten and Pierce (2001) |
| Light-harvesting complex-binding protein 1 | PCR | lhcv-1 | Pierce et al. (2007) |
| Light-harvesting complex-binding protein 2 | PCR | lhcv-2 | Pierce et al. (2007) |
| Light-harvesting complex-binding protein 3 | PCR | lhcv-3 | Pierce et al. (2009) |
| Light-harvesting complex-binding protein 4 | PCR | lhcv-4 | Pierce et al. (2009) |
| Oxygen evolution complex protein | PCR | psbO | Rumpho et al. (2008) |
| Oxygen evolution complex protein | Quantitative PCR | psbO | K. Soule and M.E. Rumpho (unpublished data) |
| Phosphoribulokinase | 35S immunolabeling | prk | Rumpho et al. (2009) |
| Phosphoribulokinase | PCR | prk | Rumpho et al. (2009) |
| Phosphoribulokinase | Quantitative PCR | prk | Soule (2009) |
| Uroporphyrinogen decarboxylase | PCR | uroD | Pierce et al. (2009)a |
| Magnesium chelatase subunit D | PCR | chlD | Pierce et al. (2009)a |
| Magnesium chelatase subunit H | PCR | chlH | Pierce et al. (2009)a |
| Chlorophyll synthase | PCR | chlG | Pierce et al. (2009)a |
| Chlorophyll synthesis | 14C labeling | NDb | Pierce et al. (2009) |
Where identical data were presented in more than one publication, the earliest publication date is noted.
ND, Not determined.
Recently, partial transcriptome data from two kleptoplastic sea slugs, Elysia timida and Plakobranchus ocellatus, were generated by Wägele et al. (2011). Both of these sea slugs feed on multiple chlorophycean algae in nature and retain functional plastids for about 57 and 69 d, respectively (Evertsen et al., 2007). Analysis of 77,648 ESTs from a single P. ocellatus individual and 24,200 ESTs from 15 E. timida individuals showed that 96% to 98% of the expressed genes were metazoan and none were of algal provenance. Based on these data, the authors concluded that HGT plays no role in sustaining long-term plastid function in these sea slugs. Rather, they attributed this unique relationship to the physical characteristics of the algal plastids and the morphology and cellular environment of the animal (physically and biochemically protecting the plastids), based on their ultrastructural, pulse amplitude modulated fluorescence, and phylogenetic analyses (Wägele and Johnsen, 2001; Evertsen et al., 2007; Evertsen and Johnsen, 2009; Händeler et al., 2009).
Using essentially the same approach, we generated partial transcriptome data from E. chlorotica to investigate the extent of HGT within this system (Supplemental Materials and Methods S1; Supplemental Table S1; to download a complete list of the isotigs and singletons produced by GS Assembler, see http://dblab.rutgers.edu/home/downloads/). Unlike the sacoglossans studied by Wägele et al. (2011), cumulative data (summarized above and in Table I) resulting from multiple experimental approaches and investigators over many years have supported HGT in the E. chlorotica/V. litorea system, although the extent of this transfer has been in question: in several key genes (Rumpho et al., 2008, 2009) or in entire metabolic pathways (Pierce et al., 2009; Schwartz et al., 2010). Using 454 pyrosequencing, we generated 148 Mb of cDNA sequence data from starved but actively photosynthesizing adult E. chlorotica (n = 5). From this partial transcriptome, 13,978 assembled unigenes and 99,873 unassembled singletons were analyzed using BLASTx (e-value cutoff ≤ 10−10) to generate putative gene annotations and to assign the taxonomic origin of ESTs. As expected, at least 95% of the predicted proteins had top hits to Metazoa. The putative nonmetazoan top hits returned by BLASTx analysis of the E. chlorotica unigenes (123) and singletons (354) were used as queries in a phylogenomic pipeline specifically designed to identify genes of foreign origin from the host using the methods and database described by Moustafa et al. (2009). This approach identified 20 ESTs of potential foreign origin derived from different prokaryotes, eukaryotes, and viruses (Supplemental Table S2) as well as several plastid-derived transcripts primarily from V. litorea, indicating plastid activity (Supplemental Table S3). None of these 20 ESTs, however, has a direct involvement in photosynthesis. In addition, specific BLASTn searches of both the contig and singleton data for genes previously identified as HGT candidates (lhcv-1, -2, -3, and -4; fcp; psbO; prk; uroD; chlD, -H, and -G; Table I) failed to identify homologs.
In all of these transcriptome studies, no evidence exists to indicate that nucleus-encoded algal genes are expressed in photosynthetic sacoglossans. Wägele et al. (2011) noted the high expression level of photosynthetic genes in the transcriptome of the source alga Acetabularia, comparable to that observed in the model green plant Arabidopsis (Arabidopsis thaliana), and yet none of the “top 50” highly expressed nuclear transcripts for plastid protein were found in the two sacoglossans used in their study. Although data are not publicly available from the V. litorea transcriptome to conduct such a comparative analysis (Pierce et al., 2009), one can assume that a similar hierarchy of expression patterns would be observed in this alga. In the partial transcriptome data presented here from E. chlorotica, we also did not find evidence of expression of any of the photosynthetic top 50 nucleus-encoded algal genes. However, it is important to note that expression patterns of a putatively transferred gene in the animal may not mirror what is observed in the alga, and to assume similar expression patterns in the foreign environment is highly speculative. Not only might the copy number of the genes be markedly different, but the expression levels and modes of regulation of expression in the animal are unknown. Recent studies employing quantitative reverse transcription-PCR support the expression of both prk and psbO in starved E. chlorotica. However, the expression levels of these genes in the animal were markedly lower, as much as 525 times lower for prk (Soule, 2009) and 63 times lower for psbO (K. Soule and M.E. Rumpho, unpublished data), compared with expression levels measured in V. litorea under identical (2 h post illumination) conditions. Differential expression of both prk and psbO in the animal was observed over a diurnal cycle, but again these patterns were markedly different from that observed in V. litorea. Similar patterns were also observed with the plastid-encoded transcripts rbcL and psaA. These studies provide evidence of the expression of nucleus-encoded plastid-targeted proteins in E. chlorotica starved for several months, but more importantly, they suggest that levels and patterns of expression of foreign genes may not mirror those in the alga (e.g. Acetabularia or Vaucheria). Thus, although Wägele et al. (2011) provide compelling arguments to suggest that nucleus-encoded algal transcripts could not have been missed in their transcriptome libraries, we feel that more exhaustive sequencing may be required to adequately address this issue.
Of interest in both studies is the finding of plastid gene expression, indicating that this genome is actively transcribed in the animals. The 46 plastid ESTs identified included 19 unique genes in E. chlorotica, some of which were represented multiple times (Supplemental Table S3). Together with the data from Wägele et al. (2011), these data suggest that nucleus-encoded transcription factors or regulatory molecules are not necessarily required for plastid gene expression in these taxa.
FUTURE DIRECTIONS
Alternative hypotheses to HGT need to be considered to explain plastid function in sacoglossans. It is apparent that plastid stability is essential for the success of these symbioses, from initial uptake to establishment of kleptoplasty (Green et al., 2005; Wägele et al., 2011). The extent to which HGT is occurring is less clear and warrants further exploration to resolve the conflicting results to date. Wägele et al. (2011) conclude that HGT does not explain plastid longevity and function in the two sacoglossans investigated in their study. A similar conclusion cannot yet be drawn for E. chlorotica due to the longevity of this functional association and multiple lines of evidence that indicate that nuclear algal genes have been transferred. Therefore, either these sacoglossan species (E. chlorotica versus E. timida and P. ocellatus) have evolved unique mechanisms to obtain and sustain photosynthetic abilities, one utilizing gene transfer and the others not, or additional explanations are required to reconcile current data.
Finally, reconciliation of these data will only occur through the investigation of new mechanisms that could be working synergistically with limited HGT and plastid stability toward plastid function. This could include investigating whether (1) the animal cells transiently use proteins and transcripts in an opportunistic fashion when available, (2) transcripts are integrated into extrachromosomal elements, which may restrict detection due to extremely low copy number and expression levels, (3) the unique stability of the plastids translates to unusual protein turnover rates of essential plastid- and nucleus-encoded photosynthetic proteins in the animal versus alga and/or protease activity, and (4) the regulation of expression of foreign genes in the animal impacts traditional detection approaches.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of Metazoa-derived unigenes.
Supplemental Table S2. ESTs of putative nonmetazoan origin.
Supplemental Table S3. ESTs of plastid origin.
Supplemental Materials and Methods S1. Library preparation and bioinformatic analysis.
References
- Bock R. (2010) The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci 15: 11–22 [DOI] [PubMed] [Google Scholar]
- Boto L. (2010) Horizontal gene transfer in evolution: facts and challenges. Proc Biol Sci 277: 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KB, Jensen KR, Stirts HM. (1990) Survey for functional kleptoplasty among west Atlantic Ascoglossa (=Sacoglossa) (Mollusca, Opisthobranchia). Veliger 33: 339–345 [Google Scholar]
- Evertsen J, Burghardt I, Johnsen G, Wägele H. (2007) Retention of functional chloroplasts in some sacoglossans from the Indo-Pacific and Mediterranean. Mar Biol 151: 2159–2166 [Google Scholar]
- Evertsen J, Johnsen G. (2009) In vivo and in vitro differences in chloroplast functionality in the two north Atlantic sacoglossans (Gastropoda, Opisthobranchia) Placida dendritica and Elysia viridis. Mar Biol 156: 847–859 [Google Scholar]
- Graves DA, Gibson MA, Bleakney JS. (1979) Digestive diverticula of Alderia modesta and Elysia chlorotica (Opisthobranchia, Sacoglossa). Veliger 21: 415–422 [Google Scholar]
- Green BJ, Fox TC, Rumpho ME. (2005) Stability of isolated algal chloroplasts that participate in a unique mollusc/kleptoplast association. Symbiosis 40: 31–40 [Google Scholar]
- Green BJ, Li WY, Manhart JR, Fox TC, Summer EJ, Kennedy RA, Pierce SK, Rumpho ME. (2000) Mollusc-algal chloroplast endosymbiosis: photosynthesis, thylakoid protein maintenance, and chloroplast gene expression continue for many months in the absence of the algal nucleus. Plant Physiol 124: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händeler K, Grzymbowski YP, Krug PJ, Wägele H. (2009) Functional chloroplasts in metazoan cells: a unique evolutionary strategy in animal life. Front Zool 6: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanten JJ, Pierce SK. (2001) Synthesis of several light-harvesting complex I polypeptides is blocked by cycloheximide in symbiotic chloroplasts in the sea slug, Elysia chlorotica (Gould): a case for horizontal gene transfer between alga and animal? Biol Bull 201: 34–44 [DOI] [PubMed] [Google Scholar]
- Harrigan JF, Alkon DL. (1978) Laboratory cultivation of Haminoea solitaria (Say, 1822) and Elysia chlorotica (Gould, 1870). Veliger 21: 299–305 [Google Scholar]
- Hinde R, Smith DC. (1974) Chloroplast symbiosis and the extent to which it occurs in Sacoglossa (Gastropoda, Mollusca). Biol J Linn Soc Lond 6: 349–356 [Google Scholar]
- Jensen KR. (1982) Chemoreception as a factor in food location of Elysia cauze Marcus (Opisthobranchia, Ascoglossa). Mar Behav Physiol 8: 205–218 [Google Scholar]
- Jensen KR. (1986) Observations on copulation in two species of Elysia from Florida (Opisthobranchia, Ascoglossa). Ophelia 25: 25–32 [Google Scholar]
- Kawaguti S, Yamasu T. (1965) Electron microscopy on the symbiosis between an elysioid gastropod and chloroplasts from a green alga. Biol J Okayama Univ 2: 57–64 [Google Scholar]
- Marin A, Ros J. (2004) Chemical defenses in Sacoglossan Opisthobranchs: taxonomic trends and evolutive implications. Sci Mar 68: 227–241 [Google Scholar]
- Mondy WL, Pierce SK. (2003) Apoptotic-like morphology is associated with annual synchronized death in kleptoplastic sea slugs (Elysia chlorotica). J Invertebr Biol 122: 126–137 [Google Scholar]
- Moran NA, Jarvik T. (2010) Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328: 624–627 [DOI] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324: 1724–1726 [DOI] [PubMed] [Google Scholar]
- Mujer CV, Andrews DL, Manhart JR, Pierce SK, Rumpho ME. (1996) Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc Natl Acad Sci USA 93: 12333–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T. (2008) Lichen Biology, Ed 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Olendzenski L, Gogarten JP. (2009) Evolution of genes and organisms the tree/web of life in light of horizontal gene transfer. Witzany G, , Natural Genetic Engineering and Natural Genome Editing, Vol 1178. Blackwell Publishing, Oxford, 137–145 [DOI] [PubMed] [Google Scholar]
- Petherick A. (2010) A solar salamander. NATNEWS 10.1038/news.2010.384 (February 1, 2011) [Google Scholar]
- Pierce S, Biron R, Rumpho M. (1996) Endosymbiotic chloroplasts in molluscan cells contain proteins synthesized after plastid capture. J Exp Biol 199: 2323–2330 [DOI] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE, Hanten JJ, Boerner SL, Schwartz JA. (2007) Transfer, integration and expression of functional nuclear genes between multicellular species. Symbiosis 43: 57–64 [Google Scholar]
- Pierce SK, Curtis NE, Schwartz JA. (2009) Chlorophyll a synthesis by an animal using transferred algal nuclear genes. Symbiosis 49: 121–131 [Google Scholar]
- Pierce SK, Massey SE, Hanten JJ, Curtis NE. (2003) Horizontal transfer of functional nuclear genes between multicellular organisms. Biol Bull 204: 237–240 [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Dastoor FP, Manhart JR, Lee J. (2006) The kleptoplast. Wise RR, Hoober JK, , Advances in Photosynthesis and Respiration: The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, 451–473 [Google Scholar]
- Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. (2011) The making of a photosynthetic animal. J Exp Biol 214: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Pochareddy S, Worful JM, Summer EJ, Bhattacharya D, Pelletreau KN, Tyler MS, Lee J, Manhart JR, Soule KM. (2009) Molecular characterization of the Calvin cycle enzyme phosphoribulokinase in the stramenopile alga Vaucheria litorea and the plastid hosting mollusc Elysia chlorotica. Mol Plant 2: 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Green BJ, Fox TC, Manhart JR. (2001) Mollusc/algal chloroplast symbiosis: how can isolated chloroplasts continue to function for months in the cytosol of a sea slug in the absence of an algal nucleus? Zoology (Jena) 104: 303–312 [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. (2008) Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci USA 105: 17867–17871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JA, Curtis NE, Pierce SK. (2010) Using algal transcriptome sequences to identify transferred genes in the sea slug, Elysia chlorotica. Evol Biol 37: 29–37 [Google Scholar]
- Soule K. (2009) Light regulated photosynthetic gene expression and enzyme activity in the heterokont alga Vaucheria litorea and its symbiotic partner the sacoglossan mollusc Elysia chlorotica. MS thesis University of Maine, Orono: [DOI] [PubMed] [Google Scholar]
- Trench RK. (1969) Chloroplasts as functional endosymbionts in the mollusc Tradachia crispata (Bergh) (Opisthobrancha, Sacoglossa). Nature 222: 1071–1072 [Google Scholar]
- Trench RK. (1993) Microalgal-invetebrate symbiosis: a review. Endocytobiosis Cell Res 9: 135–175 [Google Scholar]
- Venn AA, Loram JE, Douglas AE. (2008) Photosynthetic symbioses in animals. J Exp Bot 59: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Wägele H, Deusch O, Händeler K, Martin R, Schmitt V, Christa G, Pinzger B, Gould SB, Dagan T, Klussmann-Kolb A, et al. (2011) Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol 28: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele H, Johnsen G. (2001) Observations on the histology and photosynthetic performance of “solar-powered” opisthobranchs (Mollusca, Gastropoda, Opisthobranchia) containing symbiotic chloroplasts or zooxanthellae. Org Divers Evol 1: 193–210 [Google Scholar]
- West HH. (1979) Chloroplast symbiosis and development of the ascoglossan opisthobranch Elysia chlorotica. PhD thesis Northeastern University, Boston [Google Scholar]
- West HH, Harrigan JF, Pierce SK. (1984) Hybridization of two populations of a marine opisthobranch with different developmental patterns. Veliger 26: 199–206 [Google Scholar]
- Yamamoto Y, Yusa Y, Yamamoto S, Hirano Y, Hirano Y, Motomura T, Tanemur T, Obakata J. (2009) Identification of photosynthetic sacoglossans from Japan. Endocytobiosis Cell Res 19: 112–119 [Google Scholar]
- Yellowlees D, Rees TA, Leggat W. (2008) Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ 31: 679–694 [DOI] [PubMed] [Google Scholar]