Abstract
The histone chaperone Vps75 presents the remarkable property of stimulating the Rtt109-dependent acetylation of several histone H3 lysine residues within (H3-H4)2 tetramers. To investigate this activation mechanism, we determined x-ray structures of full-length Vps75 in complex with full-length Rtt109 in two crystal forms. Both structures show similar asymmetric assemblies of a Vps75 dimer bound to an Rtt109 monomer. In the Vps75-Rtt109 complexes, the catalytic site of Rtt109 is confined to an enclosed space that can accommodate the N-terminal tail of histone H3 in (H3-H4)2. Investigation of Vps75-Rtt109-(H3-H4)2 and Vps75-(H3-H4)2 complexes by NMR spectroscopy-probed hydrogen/deuterium exchange suggests that Vps75 guides histone H3 in the catalytic enclosure. These findings clarify the basis for the enhanced acetylation of histone H3 tail residues by Vps75-Rtt109.
Keywords: Chaperone Chaperonin, Chromatin Histone Modification, Histone Acetylase, NMR, Protein Structure, X-ray Crystallography, Histone Chaperone, Lysine Acetyltransferase
Introduction
The acetylation of histones at several lysine residues contributes to the regulated disassembly and assembly of nucleosomes, a process essential for DNA replication and repair, and gene transcription. In budding yeast, the lysine acetyltransferase Rtt109 has been implicated in the acetylation of Lys-9, Lys-14, Lys-23, Lys-27, and Lys-56 in newly synthesized histone H3, modifications that are associated with nucleosome assembly during DNA replication and transcription (1–6). The catalytic activity of Rtt109 is regulated by its association with the histone chaperones Asf1 and Vps75. Asf1 is essential for the acetylation of Lys-56 within the globular core of H3-H4 in vivo, whereas Vps75 is dispensable for this enzymatic reaction. In contrast, Vps75 is required for the enhanced acetylation of histone H3 N-terminal tail residues Lys-9, Lys-14, Lys-23, and Lys-27, as well as H3 Lys-56, in vitro (4, 5, 7–12).
Three-dimensional structures of truncated forms of Vps75 and Rtt109 in their free state have been reported, but to date, there is no crystal structure of the Vps75-Rtt109 complex available that could shed light on the 2 orders of magnitude enhanced catalytic activity of Rtt109 when bound to Vps75. To investigate this activation mechanism, we determined the three-dimensional structures of the Vps75-Rtt109 complex with full-length proteins in two different crystal forms. We show that crucial to the tight interaction between Vps75 and Rtt109 are intrinsically disordered regions of the two proteins that become folded upon complex formation, generating an enclosed catalytic area. The enclosure is too small to bind the folded core of an (H3-H4)2 tetramer but can accommodate the N-terminal tail of histone H3 through interaction with Vps75, as probed by NMR spectroscopy. Our studies strongly suggest that Vps75-Rtt109 is primarily a histone H3 tail acetyltransferase, with Vps75 guiding the H3 tail for efficient lysine acetylation by Rtt109.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
Vps75 and Rtt109 were co-expressed in Escherichia coli, and their complex was purified by a combination of affinity and size exclusion chromatography. The protein expression and purification protocols as well as crystallization conditions are reported in the supplemental Experimental Procedures.
Diffraction Data Collection and Processing
Diffraction data for structure determination were collected at 100 K at the 19-ID beamline of the Advanced Photon Source (APS), Argonne National Laboratory. Data were indexed, integrated, and scaled using HKL3000 (13). The space groups for two different crystallization conditions were P6122 and P212121, respectively.
Structure Determination
The Vps75-Rtt109 complex structures were solved by molecular replacement using Phaser (14) with the crystal structures of Rtt109 (Protein Data Bank (PDB) entry 3CZ7) and Vps75 (PDB entry 2ZD7) as starting models. Repeated cycles of model building and refinement were performed with Coot (15) and Refmac5 (16), respectively, followed by further refinement using Phenix (17, 18) with the integration of MolProbity geometry checks (19). Translation libration screw (TLS) motion group definitions were determined using the TLS Motion Determination Server. The two structures were determined to a resolution of 2.7 Å and refined to residual Rwork/Rfree values of 20.6/24.7% and 20.2/22.5% for the P6122 and P212121 crystals, respectively. The x-ray crystallography data collection and refinement statistics are summarized in Table 1.
TABLE 1.
X-ray crystallography data collection and refinement statistics
One crystal was used for each structure.
| Vps75-Rtt109a | Vps75-Rtt109a | |
|---|---|---|
| Data collection | ||
| Space group | P6122 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 99.4, 99.4, 479.6 | 91.0, 98.1, 171.4 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 |
| Wavelength (Å) | 0.97934 | 0.97901 |
| Resolution (Å) | 50-2.70 (2.75-2.70) | 50-2.70 (2.75-2.70) |
| Rmerge | 0.094 (0.906) | 0.072 (0.904) |
| I/σI | 34.1 (5.3) | 28.4 (2.5) |
| Completeness (%) | 99.4 (99.9) | 99.6 (100.0) |
| Redundancy | 16.8 (17.5) | 8.1 (8.3) |
| Refinement | ||
| Resolution (Å) | 2.7 | 2.7 |
| No. of reflections | 37,636 | 40,782 |
| Rwork/Rfree | 0.206/0.247 | 0.202/0.225 |
| No. of atoms | ||
| Protein | 7273 | 7043 |
| Ligand/ion (SO4) | 10 | 0 |
| Water | 118 | 90 |
| Average B-factors | ||
| Protein | 76.7 | 85.0 |
| Ligand/ion (SO4) | 90.6 | |
| Water | 47.0 | 61.1 |
| r.m.s.b deviations | ||
| Bond lengths (Å) | 0.011 | 0.010 |
| Bond angles (°) | 1.196 | 1.399 |
| Ramachandran | ||
| Most favored (%) | 89.4 | 93.4 |
| Additionally allowed (%) | 9.5 | 6.1 |
| Generously allowed (%) | 1.0 | 0.4 |
| Disallowed (%) | 0.0 | 0.1 |
a Values in parentheses are for highest resolution shell.
b r.m.s., root mean square.
NMR Spectroscopy
NMR spectroscopy measurements were carried out at 22 °C using a Bruker Avance III 700 MHz spectrometer equipped with a 1H, 15N, 13C cryogenic probe. The histone (H3-H4)2 tetramer was reconstituted to include 15N- or 15N/13C-labeled histone H3 and unlabeled histone H4. Details of all NMR experiments are reported in the supplemental Experimental Procedures.
RESULTS AND DISCUSSION
The Vps75-Rtt109 Complex Forms an Asymmetric Assembly with a 2:1 Stoichiometry
To gain insights into the activation mechanism of Rtt109 by Vps75, we undertook a structural characterization of Vps75 in complex with Rtt109 using full-length proteins. Crystals of the Vps75-Rtt109 complex formed in space groups P6122 and P212121, revealing, in each case, an asymmetric assembly composed of two molecules of Vps75 and one molecule of Rtt109 in the asymmetric unit. Both structures were determined to a resolution of 2.7 Å (Fig. 1, A and B, supplemental Fig. S1 and Table 1). The two structures are similar (supplemental Fig. S2A) with a backbone (N, Cα, C′) root mean square deviation of 1.3 Å for the best fit superposition of residues 3–403 of Rtt109, 8–226 of Vps75 (chain A), and 11–219 of Vps75 (chain B). In the P6122 crystal form, a C-terminal α-helix in Rtt109 (residues 412–424) contacts a Vps75 molecule in an adjacent, symmetry-related, 2:1 Vps75-Rtt109 complex (supplemental Fig. S1A). The corresponding region of Rtt109 is disordered in the P212121 crystal (supplemental Fig. S2A). The 2:1 stoichiometry of the Vps75-Rtt109 complex is consistent with equilibrium sedimentation profiles that are best fit to a single component model corresponding to two molecules of Vps75 and one of Rtt109 (20).
FIGURE 1.
Structure of the Vps75-Rtt109 complex. A, stereographic representation of Vps75-Rtt109 complex with Vps75 shown in orange and Rtt109 in blue. B, surface representation of Vps75-Rtt109. The Rtt109 segment (residues 130–175) that undergoes a folding transition upon interaction is encircled in red. C, the electrostatic surface potential of Vps75, calculated using APBS-1.1.0 (28), is plotted on the solvent-accessible surface area with negative and positive areas shown in red and blue, respectively. Rtt109 is shown in graphic representation. D, the electrostatic surface potential on the solvent-accessible surface area of Rtt109, color-coded as in C, is shown. The active site area is encircled in black. Vps75 is depicted in graphic representation. All molecular representations were made using PyMOL (29)).
One molecule of Rtt109 and a homodimer of Vps75 assemble through three distinct interfacing areas (detailed below), generating a cylinder-shaped enclosure whose interior consists of the active site-containing surface of Rtt109 and of a largely electronegative surface created by the two Vps75 molecules (Fig. 1, C and D). The long axis of the cavity runs parallel to a channel in Rtt109 thought to accommodate Rtt109 acetylation substrates. From previously reported structures of free Rtt109, this channel holds the acetyl-CoA binding cavity (20–22).
The most noticeable difference between free and Vps75-bound Rtt109 is the disorder-to-order transition of a large segment of Rtt109 (residues 130–175) upon interaction (supplemental Fig. S2, B and C). In the absence of Vps75, this Rtt109 segment is sensitive to proteolysis, indicating that it is unfolded (data not shown) (22). Indeed, all three reported structures of Rtt109 were determined after this disordered region was deleted (20–22) (supplemental Fig. S2B). Nonetheless, this Rtt109 region is essential for association with Vps75 as its deletion prevents complex formation. From the complex structures, this segment constitutes the main binding motif of the three interaction interfaces (interface 1), adopting a stable conformation via extensive hydrophobic and charge-charge interactions with the two Vps75 molecules (Figs. 1 and 2). In particular, residues 144–155 within this Rtt109 segment form an amphipathic α-helix, with the hydrophobic side interacting with a complementary hydrophobic surface formed by the N-terminal end of the long α-helix of Vps75 chain B (residues 11–23) and by an α-helix of Vps75 chain A (residues 212–220) (Fig. 2A). Two bidentate salt bridges involving Rtt109 Arg-149 and Arg-154 and Vps75 Glu-23 (chain B) and Asp-222 (chain A), respectively, complement the hydrophobic contacts (Fig. 2A).
FIGURE 2.
Details of the Vps75-Rtt109 interaction interface 1. A, an amphipathic α-helix of Rtt109 (blue) folds upon interaction with chains A (orange) and B (red) of Vps75. The left shows the complex structure, and the right shows the details of this interaction in stereographic representation. B, additional details of the interaction from a different angle of the complex. Amino acid side chains in the Rtt109 hydrophobic cavity surrounding Vps75 Leu-223 are highlighted. Details of the Vps75-Rtt109 interaction interfaces 2 and 3 are presented in supplemental Fig. S3.
Several Rtt109 amino acids (residues 124–143 and 158–177) preceding and following the amphipathic α-helix fold into a globular motif that surrounds Vps75 Leu-223 (chain A). The motif is stabilized by a hydrophobic core made up of Rtt109 Tyr-130, Ile-133, Pro-135, Leu-137, Leu-163, Phe-171, Leu-175, and Tyr-176 (Fig. 2B). Leu-223 nicely illustrates the asymmetric nature of the Vps75-Rtt109 complex as this residue from Vps75 chain A is totally buried at the intermolecular interface, whereas in chain B, it is fully accessible to solvent. Further stabilizing the complex, Rtt109 Arg-128 participates in salt bridges with Vps75 Asp-225 and Glu-226 (chain A) (Fig. 2B). The Vps75 amino acids 226–229 (chain A) form a β-turn anchored to Rtt109 by a salt bridge between Vps75 Glu-229 and Rtt109 Arg-355 (not shown).
The other two interfaces between Rtt109 and Vps75, one on each side of the catalytic enclosure, are shown in Fig. 1, C and D and supplemental Fig. S3. Both combine hydrophobic and electrostatic interactions with an overall negative charge for Rtt109 and positive charge for the Vps75 molecules. One of these interfaces (interface 2) involves α-helices from Rtt109 and Vps75 chain A. Hydrophobic contacts are formed by Rtt109 Ala-360, Tyr-364, Phe-386, and Leu-389 and Vps75 Val 66, Ala-74, and Phe-77. In addition, Rtt109 Glu-374, Glu-378, and Arg-390 form salt bridges with Vps75 Arg-173, Arg-73, and Asp-81, respectively (supplemental Fig. S3A).
The other interface (interface 3) includes regions of the two Vps75 chains. A mainly hydrophobic area of Rtt109 composed of His-30, Thr-34, Pro-35, Val-279, Tyr-280, Leu-284, Ile-294, His-295, Leu-303, and Leu-304 contacts the Vps75 (chain A) segment made up of residues Val-248, Asp-249, Pro-250, Leu-251, Leu-252, and Ser-253 (supplemental Fig. S3B). These residues of Vps75 are missing in two of the previously determined structures of free Vps75 (5, 20) and located at the crystal packing interface in the third structure (23). Adjacent to the hydrophobic area, negatively charged side chains of Rtt109 are proximal to a positively charged region of Vps75 (chain B). There is also a salt bridge between Rtt109 Asp-301 and Vps75 Lys-177 (supplemental Fig. S3B).
In summary, parts of Rtt109 and Vps75 forming the tripartite interface undergo disorder-to-order folding transitions upon complex formation. Although both Vps75 chains contribute interactions with Rtt109, chain A accounts for ∼73% of the total Vps75-Rtt109 interface area of ∼2300 Å3 (supplemental Fig. S4).
The Vps75-Rtt109 Complex Recognizes the N-terminal Tail of Histone H3
The cylindrical enclosure shaped by Vps75 and Rtt109 has dimensions (∼35 × 30 Å) that preclude full insertion of an (H3-H4)2 histone tetramer (Fig. 1). The regions of Rtt109 (residues 130–175) and Vps75 chain A (residues 248–253) that contribute to interfaces 1 and 3 partly close off one side of this cavity. Thus, the cavity likely accommodates only a partial insertion of histone tetramer, even if there are unrevealed conformational rearrangements upon binding. Because crystallographic studies have demonstrated that Asf1 associates with an H3-H4 heterodimer, blocking the formation of (H3-H4)2 tetramer (24, 25), we reasoned that Vps75-Rtt109 might also disrupt (H3-H4)2 by accommodating the H3-H4 dimer into the catalytic enclosure.
To test this possibility and gain information on the interaction interface of full-length Vps75-Rtt109 complex and Vps75 alone with (H3-H4)2 in solution, we devised an NMR spectroscopy-based hydrogen/deuterium (H/D)3 exchange protection assay (26, 27). Briefly, the (H3-H4)2 tetramer was prepared with 15N- and 15N/13C-labeled histone H3 and unlabeled histone H4 in H2O. The protonation state of amide nitrogen atoms in histone H3 was then probed after exchange into D2O in the absence and presence of unlabeled binding partners Vps75-Rtt109 or Vps75. Amide hydrogen atoms buried in the protein core or close to the binding interface are protected from exchange and could be identified after the 1H-15N heteronuclear single quantum coherence NMR signals of histone H3 were assigned. The procedure, which involves dissociating the complexes by denaturation combined with H/D exchange quenching, is detailed in the supplemental Experimental Procedures.
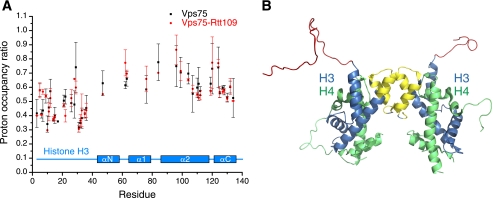
Results of the H/D protection assays are summarized in Fig. 3. In the presence of Vps75-Rtt109, there is preferentially reduced H/D exchange for amino acids at the N terminus of histone H3, a region corresponding to the flexible tail of H3 bearing 4 lysine residues known to be acetylated by Vps75-Rtt109. Residues in the last α-helix of H3, as well as the C terminus of the penultimate α-helix and the linker between these two helices, are also protected from H/D exchange, albeit to a lower extent than the tail residues (Fig. 3A). The N-terminal tail and C-terminal part of H3 lie on the same side of the histone tetramer and are close in space (Fig. 3B).
FIGURE 3.
Interaction of Vps75-Rtt109 with (H3-H4)2 probed using NMR spectroscopy-based hydrogen/deuterium exchange measurements. A, per amino acid ratio of histone H3 amide proton occupancies after exchange in D2O for 10 min in the absence and presence of Vps75 (black squares) or Vps75-Rtt109 (red dots). Analysis was possible for 42 histone H3 amino acids whose signals are resolved in the NMR spectra and could be assigned. Shown are the average ratios and error bars for standard deviations from triplicate experiments. B, residues protected from solvent exchange by Vps75-Rtt109 are color-coded on the surface of the (H3-H4)2 tetramer (from PDB identification code 1EQZ), with the N-terminal tail of H3 in red and residues from the two C-terminal α-helices and interhelical region in yellow (corresponding to a region where H/D exchange ratio values are less than 0.6 for 10 residues).
Our results show that the interaction interface of Vps75-Rtt109 with histone H3, within the context of (H3-H4)2, is limited to the flexible H3 tail and an adjacent region of H3 in the tetramer, in agreement with the limiting volume of the catalytic enclosure. The crystal structures of free Vps75 show a negatively charged concave surface that could in principle cover a larger area of the histone tetramer than does Vps75-Rtt109. Remarkably, however, the H/D exchange data reveal the binding interface in Vps75-(H3-H4)2 alone is also restricted to the N-terminal tail and C-terminal residues of histone H3 (Fig. 3). Although the H/D exchange protection assays do not probe the contact surface with histone H4, the limited change in H3 protection by Vps75-Rtt109 or Vps75 lends no support for the disruption of histone tetramers by these proteins.
The similar H/D exchange protection in H3 by Vps75-Rtt109 and Vps75 implies an important Vps75 function in orienting histone H3. Vps75 can be seen as contributing to the presentation of (H3-H4)2 to Rtt109, resulting in efficient catalysis. The histone guidance effect of Vps75 likely increases the frequency of Rtt109/(H3-H4)2 encounters that are productive for acetyl transfer, therefore providing an explanation for the ∼100–250-fold enhanced catalytic rate constant (kcat) of Rtt109 and virtually unchanged Km in the presence of Vps75 (5, 11, 12). We note also that the confinement of the Rtt109 catalytic site to a small volume in the Vps75-Rtt109 complex may contribute to the increased enzyme efficiency by increasing the formation rate of enzyme-substrate complexes.
Conclusion
The structural and binding data reported here strongly suggest that Vps75-Rtt109 is foremost a histone H3 tail binding and acetylation complex. This interpretation is consistent with the main target of Vps75-Rtt109 being Lys-9 in the H3 tail in vitro and in vivo (5, 9) and with the observation that Vps75-Rtt109 cannot efficiently acetylate Lys-56 in the globular core of (H3-H4)2 tetramers (7). The efficient acetylation of Lys-56 by Vps75-Rtt109 necessitates the chaperone Asf1 (7). However, unlike Vps75, which forms a tight complex with Rtt109 (KD = ∼10 nm) (11) and co-purifies with Rtt109 from yeast cell extracts, Asf1 and Rtt109 do not co-purify, demonstrating the much lower affinity of the latter complex (7). We propose that Asf1, by dissociating (H3-H4)2 into dimers, allows partial dimer insertion in the catalytic enclosure of Vps75-Rtt109, leading to efficient acetylation of Lys-56. More work is needed to test this hypothesis.
Supplementary Material
Acknowledgments
We acknowledge the use of beamline 19-ID at the APS, Argonne National Laboratory. The APS is operated by UChicago Argonne, LLC, for the United States Department of Energy under Contract DE-AC02-06CH11357. We are grateful to Younchang Kim and Rongguang Zhang at the APS for expert assistance with x-ray data collection. We thank Karolin Luger and Aaron Hieb for the advice on histone purification. We thank Maria Victoria Botuyan for careful reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA132878 (to G. M.) and GM81838 (to Z. Z.).
The atomic coordinates and structure factors (codes 3Q66 and 3Q68) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S4.
- H/D
- hydrogen/deuterium.
REFERENCES
- 1. Ransom M., Dennehey B. K., Tyler J. K. (2010) Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 3. Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berndsen C. E., Tsubota T., Lindner S. E., Lee S., Holton J. M., Kaufman P. D., Keck J. L., Denu J. M. (2008) Nat. Struct. Mol. Biol. 15, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 7. Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 28587–28596 [DOI] [PubMed] [Google Scholar]
- 8. Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 14158–14164 [DOI] [PubMed] [Google Scholar]
- 9. Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. (2008) Mol. Cell. Biol. 28, 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgess R. J., Zhou H., Han J., Zhang Z. (2010) Mol. Cell 37, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albaugh B. N., Kolonko E. M., Denu J. M. (2010) Biochemistry 49, 6375–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolonko E. M., Albaugh B. N., Lindner S. E., Chen Y., Satyshur K. A., Arnold K. M., Kaufman P. D., Keck J. L., Denu J. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20275–20280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 14. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 18. Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) J. Synchrotron Radiat. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 19. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang Y., Meeth K., Jiang E., Luo C., Marmorstein R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin C., Yuan Y. A. (2008) Structure 16, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 22. Stavropoulos P., Nagy V., Blobel G., Hoelz A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12236–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park Y. J., Sudhoff K. B., Andrews A. J., Stargell L. A., Luger K. (2008) Nat. Struct. Mol. Biol. 15, 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K. (2006) Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007) Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 26. Dyson H. J., Kostic M., Liu J., Martinez-Yamout M. A. (2008) FEBS Lett. 582, 1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato H., Gruschus J., Ghirlando R., Tjandra N., Bai Y. (2009) J. Am. Chem. Soc. 131, 15104–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.