Abstract
X-linked inhibitor of apoptosis protein (XIAP) overexpression has been found to be associated with malignant cancer progression and aggression in individuals with many types of cancers. However, the molecular basis of XIAP in the regulation of cancer cell biological behavior remains largely unknown. In this study, we found that a deficiency of XIAP expression in human cancer cells by either knock-out or knockdown leads to a marked reduction in β-actin polymerization and cytoskeleton formation. Consistently, cell migration and invasion were also decreased in XIAP-deficient cells compared with parental wild-type cells. Subsequent studies demonstrated that the regulation of cell motility by XIAP depends on its interaction with the Rho GDP dissociation inhibitor (RhoGDI) via the XIAP RING domain. Furthermore, XIAP was found to negatively regulate RhoGDI SUMOylation, which might affect its activity in controlling cell motility. Collectively, our studies provide novel insights into the molecular mechanisms by which XIAP regulates cancer invasion and offer a further theoretical basis for setting XIAP as a potential prognostic marker and specific target for treatment of cancers with metastatic properties.
Keywords: Actin, Cell Migration, Cytoskeleton, Signal Transduction, Tumor Metastases, RhoGDI, XIAP
Introduction
The X-linked inhibitor of apoptosis protein (XIAP)3 is a member of the IAP family that has received substantial attention during the last few years. Biochemical and structural studies have indicated that XIAP has three zinc-binding baculovirus IAP repeat (BIR) domains (BIR1–3), a loop region, and a RING finger (1). The BIR3 domain of XIAP is able to bind and inhibit caspase-9, whereas the BIR2 region binds and inhibits active caspase-3 and caspase-7. The RING domain of XIAP has E3 ligase activity and is able to degrade proteins by linking them to ubiquitin molecules (2–4). More recently, XIAP has been found to be a regulator of the cell cycle through binding the cell cycle regulators MAGED1 and NRAGE and to play an important role in the control of intracellular copper levels through ubiquitin ligase-dependent regulation of the copper-regulating gene MURR1 (5, 6). The ability of XIAP to regulate these pathways, uncoupled with its caspase inhibitory activities, indicates its distinct properties and functions. Based on the finding that XIAP-deficient mice do not display obvious apoptotic phenotypes (7), it was hypothesized that there might be new functions and signaling pathways affected by XIAP, which are probably distinct from those involved in apoptotic caspase cascades.
There is growing evidence showing the correlation between high XIAP overexpression and malignant cancer aggression (8, 9). Comparison of XIAP expression between adjacent malignant tissue and normal tissue invariably demonstrates that XIAP is much more highly expressed in the malignant cancer tissue (10–20). Poorly differentiated carcinomas also display significantly higher levels of XIAP expression than do well differentiated carcinomas (13, 17–20). Moreover, XIAP expression in metastatic specimens is much higher than that in primary cancers (10–12, 14–20). However, the molecular mechanism linking XIAP to cancer metastasis is largely unexplored. Here, we show that XIAP depletion or knockdown in cancer cells leads to a reduction in cell migration and invasion accompanied by decreased actin polymerization and cell motility. Furthermore, we show that the impaired cell migration in XIAP−/− cells can be rescued by knockdown of Rho GDP dissociation inhibitor (RhoGDI) expression, indicating that RhoGDI might be a downstream target of XIAP in the regulation of cancer motility. Finally, our studies show that XIAP forms a complex with RhoGDI through its RING domain and regulates RhoGDI SUMOylation, which might be involved in the modulation of cell migration.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Other Reagents
Plasmids expressing HA-tagged XIAP, ΔRING, ΔBIR, and Δloop and the pEBB empty vector were gifts from Dr. Colin S. Duckett (University of Michigan) (21). The pEGFP-C3/RhoGDI vector expressing GFP-tagged RhoGDI was kindly provided by Dr. Mark R. Philips (New York University School of Medicine) (22). pRNA-U6/siRhoGDI and pEGFP-C3/mRhoGDI (with the RhoGDI gene mutated from 403AAA GGC GTC AAG ATT GAC420 to 403AAG GGA GTA AAA ATC GAT420 to prevent destruction of exogenous mRNA by the corresponding siRNA) were described in a previous study (23). The adenoviruses carrying Dual-His-S-SUMO1/IRES/HA-UBC9 (Ad-SUMO1-WT), Dual-His-S-SUMO1-Q94P/IRES/HA-UBC9 (Ad-SUMO1-Q94P), and Dual-His-S-SUMO1ΔGG/IRES/HA-UBC9 (Ad-SUMO1ΔGG) as well as the control LacZ virus (Ad-LacZ) and all the plasmid constructs were provided by Dr. German Rosas-Acosta (24). The antibodies against XIAP, the phospho-EGF receptor, the EGF receptor, and ubiquitin were purchased from Cell Signaling Technology (Boston, MA). The anti-Arp2, anti-Arp3, anti-GFP, anti-cyclin D1, anti-UBC9, anti-HDAC6, and anti-N-WASP (neuronal Wiskott-Aldrich syndrome protein) antibodies and the agarose-conjugated anti-HA beads were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β-actin and anti-α-tubulin antibodies were obtained from Sigma. The anti-SUMO1 antibody was from Epitomics (Burlingame, CA). The anti-RhoGDI antibody was from Millipore (Billerica, MA). MG132 and cycloheximide were purchased from Calbiochem. EGF was purchased from Invitrogen.
Cell Culture and Transfection
The human colon cancer cell line HCT116 and XIAP−/− HCT116 cells were kind gifts from Dr. Bert Vogelstein (Howard Hughes Medical Institute and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins Medical Institutions) (25). The human bladder cancer cell line T24 was from the laboratory of Dr. Xue-Ru Wu. These cells were maintained at 37 °C in a 5% CO2 incubator in McCoy's 5A medium supplemented with 10% FBS, 2 mm l-glutamine, and 25 μg/ml gentamycin. Human cervix adenocarcinoma HeLa cells were cultured in DMEM with 10% FBS. Cell transfections were performed with Lipofectamine reagent (Invitrogen) or FuGENE® HD transfection reagent (Roche Applied Science) according to the manufacturer's instructions. For stable transfection, cultures were subjected to either hygromycin B (200–400 μg/ml) or G418 drug selection, and cells surviving the antibiotics selection were pooled as stable mass transfectants.
RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen), and cDNAs were synthesized with the ThermoScript RT-PCR system (Invitrogen). Two oligonucleotides (5′-AGA AGG CTG GGG CTC ATT TG-3′ and 5′-AGG GGC CAT CCA CAG TCT TC-3′) were used as the specific primers to amplify human GAPDH cDNA. The human β-actin cDNA fragments were amplified by primers 5′-GCG AGA AGA TGA CCC AGA TCA T-3′ and 5′-GCT CAG GAG GAG CAA TGA TCT T-3′.
Northern Blotting
Total RNA was extracted from WT and XIAP−/− cells using TRIzol. A probe of β-actin for hybridization was obtained by PCR using primers 5′-CTC GCC TTT GCC GAT CCG CCC-3′ and 5′-AGT GGT ACG GCC AGA GGC GTA C-3′. After purification using a QIAquick PCR purification kit (Qiagen), a 25-ng DNA fragment was labeled with [α-32P]dCTP (PerkinElmer Life Sciences) using a DECAprimeTM II kit (Ambion) following the manufacturer's protocol. The probe was purified using a Sephadex G-25 column, and its radioactivity was detected by liquid scintillation analysis (PerkinElmer Life Sciences). 16 μg of RNA was run on agarose gel (3.7% formaldehyde); after overnight transfer, the membrane was cross-linked using a Stratalinker® 1800 UV system (Stratagene). The membrane was prehybridized, hybridized, and exposed to x-ray film following the protocol recommended for ExpressHybTM hybridization solution (Clontech).
NF-κB-Luciferase Assay
The constructs of the NF-κB-luciferase reporter and pRL-TK were cotransfected into WT or XIAP−/− cells. After starvation with 0.1% FBS-supplemented McCoy's 5A medium for 12 h, cells were treated with or without 60 ng/ml EGF for 12 or 24 h. Luciferase was detected as described in our previous report (26).
Western Blotting
Cell extracts were prepared with cell lysis buffer A (10 mm Tris-HCl (pH 7.4), 1% SDS, and 1 mm Na3VO4). Protein concentrations were determined using a protein quantification assay kit (Bio-Rad). 30 μg of proteins was subjected to a Western blot system as described in our previous reports (27).
Pulse-Chase Assays
A pulse-chase assay was carried out as described in our previous study (28). WT, XIAP−/−, and XIAP−/−(HA-XIAP) cells were incubated with methionine/cysteine-free DMEM (Invitrogen) containing 2% dialyzed fetal calf serum (Invitrogen) for 1 h. [35S]Methionine/cysteine (250 μCi/dish; Tran35S-label, ICN) was added and cultured for 45 min, and cells were harvested in cell lysis buffer B (1% Triton X-100, 150 mm NaCl, 10 mm Tris (pH 7.4), 1 mm EDTA, 1 mm EGTA, 0.2 mm Na3VO4, 0.5% Nonidet P-40, and one Complete protein inhibitor mixture tablet) on ice at the indicated time points. 0.5 mg of total lysate was incubated with anti-β-actin antibody for 2 h at 4 °C. Protein A/G Plus-agarose beads (Santa Cruz Biotechnology) that were precleared with 20 mg/ml BSA for 2 h were incubated with agitation for an additional 2 h at 4 °C. The immunoprecipitated samples were washed with cell lysis buffer B and heated at 100 °C for 5 min. Radiolabeled β-actin proteins were assessed by SDS-PAGE analysis.
F-actin Content Assay
The cells were treated with 50 ng/ml EGF for the indicated time periods, fixed with 3.7% formaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After washing three times with PBS, the cells were blocked in 1% BSA and PBS at room temperature for 20 min and then stained on a rotator with Oregon Green 488-phalloidin (1:40 in 1% BSA and PBS; Molecular Probes) for 30 min. Cells were washed three times again with PBS, and the bound Oregon Green 488-phalloidin was extracted using 100% methanol at 4 °C for 90 min. After extraction, the fluorescence of the methanol extraction solution for each sample was recorded at 465 nm excitation and 535 nm emission and normalized against cell protein concentration (29). The results are expressed as relative F-actin content, where F-actinΔt/F-actin-t0 = (fluorescenceΔt/mg/ml)/(fluorescence-t0/mg/ml). The quantification of polymerized F-actin within cells was also performed by flow cytometry according to Kobayyahsi's method using phallacidin (30). Relative F-actin content was expressed as an F-actin index (mean channel of test cells at specified time/mean channel of buffer sample at 0 min).
Isolation of Cell Fractions
Cells were incubated with or without 50 ng/ml EGF, and the incubations were terminated by the addition of 10 ml of cold PBS. The cells were harvested in HEPES buffer (20 mm HEPES (pH 7.4), 150 mm KCl, 2 mm MgCl2, 4 mm benzamidine, and 1 mm PMSF). After centrifugation, the pellet was resuspended in 0.5 ml of HEPES buffer and forced 15 times through a 25-gauge needle. Intact cells and cell nuclei were pelleted by centrifugation at 4 °C. After centrifugation at 8000 × g for 20 min at 4 °C, the fraction enriched in membrane/cytoskeleton was recovered from the pellet (insoluble fraction), and the supernatant represented predominantly the cytosolic fraction (soluble fraction) (31). The cytosolic, nucleus, membrane/particulate, and cytoskeletal fractions were isolated with the FractionPREPTM cell fractionation system (BioVision) following the manufacturer's protocol.
Immunoprecipitation
For determination of protein-protein interaction and protein ubiquitination and SUMOylation, cells were lysed in cell lysis buffer B on ice. Lysate (0.5 mg) was precleared by incubation with Protein A/G Plus-agarose and then incubated with 2 μg of anti-XIAP, anti-RhoGDI, anti-HA, or anti-GFP antibodies as indicated overnight at 4 °C. Protein A/G Plus-agarose (40 μl) was incubated with agitation for an additional 4 h at 4 °C. The immunoprecipitated samples were washed extensively with cell lysis buffer B and subjected to Western blot assay.
Immunofluorescence Staining and Confocal Microscopy
Cells grown on cover slides were starved for 4 h and treated with EGF (50 ng/ml). The cells were fixed with 3.7% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. The cells were blocked with 1% BSA and PBS for 30 min. Oregon Green 488-conjugated phalloidin was diluted into 5 units/μl, incubated with the cells for 30 min at room temperature, and stained with 0.1 μg/ml DAPI for 1 min. The slides were washed with PBS and mounted with antifade reagent. The cells were observed under a confocal microscope (Leica TCS SP5).
Cell Wound Healing Assay
Wounds were made by sterile pipette tips when the cells reached 80% confluence. Cells were washed with serum-free PBS and then cultured in fresh medium for the time periods indicated. The photographs were taken at the times indicated until the wounds were healed in the control group (32). The wound area was quantified using cell migration analysis software (Muscale LLC, Scottsdale, AZ).
Cell Invasion Assay
A BD BioCoatTM MatrigelTM invasion chamber (BD Biosciences) was used in the invasion assay. Cells were seeded per insert in triplicate in 500 μl of serum-free medium. Inserts were placed in wells containing 500 μl of medium with 5% FBS and 12-O-tetradecanoylphorbol-13-acetate (20 ng/ml). The cells were incubated for 72 h. The same invasion chamber without Matrigel was used as a cell migration control (33). At the end of the culture period, cells on the upper surface of the filters were completely removed by wiping with a cotton swab. The membrane was cut with a sharp knife and placed in a 96-well plate. The invaded/migrated cells were determined using the CellTiter-Glo® luminescent cell viability assay (Promega) according to the manufacturer's instructions. Invasion (%) = (ATP activity of invaded cells/ATP activity of migrated cells) × 100%.
Statistical Methods
Student's t test was utilized to determine the significance of differences in actin polymerization, cell migration, and cell invasion among various cell lines and/or treatments. The differences were considered to be significant at p ≤ 0.05.
RESULTS
XIAP Plays an Important Role in β-Actin Protein Expression in HCT116 Cells
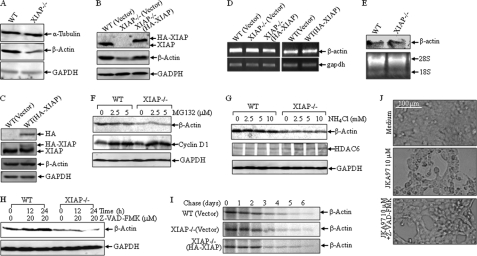
The cytoskeletal system is a dynamic structure in both prokaryotes and eukaryotes and is responsible for maintaining cell shape and enabling cell movement and cell division. There are three main kinds of cytoskeletal filaments in eukaryotic cells, including microfilaments (also known as actin filaments), intermediate filaments, and microtubules. Actin is one of the most highly conserved proteins in all eukaryotic cells and is involved in many important cell processes, such as muscle contraction and cell motility (34, 35). During our experiment utilizing β-actin as a protein loading control for the Western blot assay, we unexpectedly found that β-actin protein expression was dramatically down-regulated in XIAP−/− cells in comparison to that in parental HCT116 cells, whereas α-tubulin and GAPDH expression was comparable between the two types of cells (Fig. 1A). To exclude the possibility of selection bias during the establishment of the knock-out cell line, we transfected the HA-XIAP construct back into XIAP−/− cells, and the stable transfectants were established and identified as shown in Fig. 1B (upper panel). The results showed that reconstituted expression of XIAP in XIAP−/− cells completely restored β-actin protein expression (Fig. 1B), indicating that XIAP plays a role in the regulation of β-actin protein expression. Because the XIAP protein is already overexpressed in HCT116 cells, transfection of the exogenous HA-XIAP construct into WT cells did not render additional effects. Thus, the effect of HA-XIAP expression in HCT116 cells on β-actin expression was marginal (Fig. 1C). Then, we tested β-actin mRNA levels in WT, XIAP−/−, and XIAP−/−(HA-XIAP) cells to further explore the molecular mechanism. Our results showed that β-actin mRNA levels were comparable among the three cell lines (Fig. 1D, upper left panel), which excluded the possibility that XIAP regulates β-actin protein expression at the mRNA level. Consistent with the results of Western blotting, there was no change in β-actin mRNA levels in WT and WT(HA-XIAP) cells (Fig. 1D, upper right panel). Moreover, the results from the Northern blot assay further confirmed that the regulation of β-actin by XIAP did not occur at the mRNA level (Fig. 1E). The ubiquitin-proteasome and autophagy-lysosome pathways are two major routes of protein and organelle clearance in eukaryotic cells (36–38). XIAP has a zinc-binding motif RING domain, which can recruit E2 ubiquitin-conjugating enzymes and catalyze the transfer of ubiquitin onto target proteins for protein degradation by the 26 S proteasome (38). Thus, we employed a 26 S proteasome inhibitor, MG132, to see whether XIAP can regulate β-actin degradation. As shown in Fig. 1F, there was no observable β-actin protein accumulation in either WT or XIAP−/− cells after treatment with MG132, and cyclin D1 accumulation was used as a positive control (39). Similarly, treatment of the cells with ammonium chloride (NH4Cl), a lysosome inhibitor, did not show any observable β-actin accumulation either (Fig. 1G), whereas it showed a positive effect on HDAC6 protein accumulation. It has been reported that XIAP can attenuate the cleavage activity of caspase on actin (1). To test this potential, Z-VAD-fmk, a pan-caspase inhibitor, was used. As shown in Fig. 1H, treatment with Z-VAD-fmk increased β-actin protein levels in HCT116 cells, but not in XIAP−/− cells, suggesting that the down-regulation of β-actin in XIAP−/− cells is not due to caspase-mediated degradation of β-actin protein. The validation of Z-VAD-fmk was achieved by inhibition of the apoptotic response of HCT116 cells exposed to JKA97 (Fig. 1J), an apoptosis inducer, as reported in our previous study (40). Further investigation in a pulse-chase experiment showed that there were no observable differences in β-actin protein degradation rates among the three cell types (Fig. 1I). Taken together, our results demonstrate that XIAP can regulate β-actin protein expression through a mechanism that does not affect mRNA, protein degradation, or proteolysis.
FIGURE 1.
XIAP deficiency results in reduction in β-actin expression in HCT116 cells. A–C, protein expression of β-actin, α-tubulin, HA-XIAP, and GAPDH was determined by Western blotting in various cells as indicated. D and E, β-actin mRNA levels were evaluated by RT-PCR or Northern blotting. F–H, WT and XIAP−/− cells were treated with MG132 (F), NH4Cl (G), or 20 μm Z-VAD-fmk (H) at the indicated concentrations for 24 h (F and G) or for the indicated time periods (H), and β-actin expression was detected by Western blotting. I, the indicated cells were used for pulse-chase assay as described under “Experimental Procedures.” J, WT HCT116 cells were pretreated with Z-VAD-fmk (20 μm) for 60 min and then exposed to JKA97 (10 μm), and changes in cell morphology were photographed under a microscope.
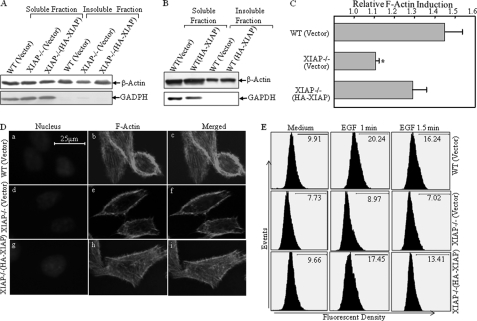
XIAP Deficiency Specifically Decreases the Expression of F-actin, but Not G-actin, and Leads to Reduction in F-actin Polymerization
In eukaryotic cells, actin exists in two forms, a globular monomer called G-actin and a filamentous polymer called F-actin, a linear chain of G-actin subunits. Actin dynamics, or the rapid turnover of actin filaments, plays a central role in numerous cell functions (41). To determine whether XIAP is able to mediate the regulation of actin dynamics, the soluble fraction (cytoplasm) and insoluble fraction (membrane and cytoskeleton) were isolated from WT(Vector), XIAP−/−(Vector), and XIAP−/−(HA-XIAP) cells. As shown in Fig. 2A, G-actin, which exists mostly in soluble fractions, was similar among the three types of cells, whereas F-actin, which is present mainly in insoluble fractions, was much lower in XIAP−/− cells than in parental HCT116(Vector) cells and could be rescued by reconstituted expression of HA-XIAP in XIAP−/− cells, indicating that XIAP may regulate actin polymerization. Consistent with the relatively low expression of HA-XIAP in WT(HA-XIAP) cells, the effect of HA-XIAP on β-actin distribution in the insoluble fraction was also marginal (Fig. 2B). Next, we treated the cells with EGF and then extracted cells for determination of F-actin levels. Induction of actin polymerization by EGF was observed in HCT116 cells, but it was dramatically reduced in XIAP−/− cells, and such reduction was rescued by reconstituted expression of HA-XIAP (Fig. 2C). Consistently, these three types of cells showed differences in cytoskeletal morphology when stained with phalloidin, a dye that specifically binds F-actin. Actin stress fibers in WT HCT116 cells were more abundant than those observed in XIAP−/− cells, whereas the cytoskeleton levels in XIAP−/−(HA-XIAP) and WT cells were comparable (Fig. 2D). Moreover, F-actin polymerization was induced upon EGF treatment by >2-fold at 1 min (20.24% versus 9.91%) in WT cells, whereas only a marginal induction was observed in XIAP−/− cells (8.97% versus 7.73%) (Fig. 2E). Again, reconstituted expression of XIAP in XIAP−/− cells showed remarkable F-actin induction upon EGF treatment (9.96% versus 17.45%) (Fig. 2E). These results demonstrate that XIAP regulates F-actin polymerization and cytoskeleton formation.
FIGURE 2.
XIAP regulates β-actin polymerization and cytoskeleton formation. A and B, the soluble and insoluble fractions were isolated, and G-actin, F-actin, and GAPDH were detected by Western blotting. C, the induction of F-actin polymerization upon treatment with EGF (50 ng/ml) was evaluated by spectrophotometry in WT, XIAP−/−, and XIAP−/−(HA-XIAP) cells (p < 0.05). D, the indicated cells were stained with Oregon Green 488-conjugated phalloidin, and cytoskeleton and F-actin fibers were observed by confocal microscopy. E, the induction of F-actin polymerization upon treatment with EGF (50 ng/ml) was evaluated by flow cytometry in the various cell lines indicated.
XIAP Promotes Tumor Cell Migration and Invasion
It has been demonstrated that actin polymerization is one of the most essential driving forces for cell motility. To explore the potential role of XIAP in tumor invasion, the wound healing assay was performed in WT(Vector), XIAP−/−(Vector), and XIAP−/−(HA-XIAP) cells. As shown in Fig. 3A, knock-out of XIAP resulted in a great decrease in spontaneous wound healing. The wounded area in WT(Vector) cells was almost covered by migrating cells at day 4, whereas there was still an obvious open area in the XIAP−/−(Vector) cells (Fig. 3A). Consistently, reconstituted expression of XIAP in XIAP−/−(HA-XIAP) cells increased the wound healing rate (Fig. 3A). This finding was further confirmed in human cervix adenocarcinoma HeLa cells and bladder cancer T24 cells using the XIAP knockdown approach. The knockdown efficiency is shown in Fig. 3B. More importantly, knockdown of XIAP expression in HeLa cells (XIAP shRNA) impaired F-actin formation induced by EGF treatment (Fig. 3C) and inhibited the wound healing rate as well (Fig. 3D). A similar result was also reproducible in highly invasive human bladder cancer T24 cells by using XIAP shRNA (Fig. 3E). IAP was reported to regulate cell migration through the NF-κB pathway (42). Thus, we compared the NF-κB activation between WT HCT116 and XIAP−/− cells using the NF-κB-luciferase reporter assay. The results did not show any significant difference in NF-κB transactivation between the two cell lines in the NF-κB promoter-luciferase reporter assay (Fig. 3F), which was further confirmed by the Western blot assay (data not shown). Previous studies have shown that migration of cancer cells is associated with their invasion capability (43). To evaluate this, a Transwell assay was employed to test the role of XIAP in the invasion process. The results showed that knocking out of XIAP inhibited cell invasion compared with that in WT(Vector) cells (1.7% versus 10.0%, p < 0.01), whereas restoration of XIAP expression in XIAP−/− cells rescued the invasion ability to a level similar to that in WT(Vector) cells (10.0% versus 10.1%) (Fig. 3G). Our results demonstrate that XIAP plays a crucial role in the regulation of cancer cell motility.
FIGURE 3.
XIAP is required for cancer cell migration and invasion. A, D, and E, the cell migration behavior was evaluated during a wound healing assay, and images were taken at different time points as indicated. The wound area was quantified using cell migration analysis software, and the quantitative data are shown as indicated. Error bars represent S.D. (n = 3). *, significant difference between/among the indicated cell lines (p < 0.05). Scale bars = 300 μm. B, knockdown of XIAP expression in HeLa and T24 cells was identified by Western blotting. C, the induction of F-actin polymerization upon treatment with EGF (50 ng/ml) was evaluated by spectrophotometry in non-silencing and XIAP shRNA transfectants in HeLa cells. *, significant decrease compared with non-silencing cells (p < 0.05). F, NF-κB-dependent transactivation in both WT and XIAP−/− cells upon treatment with EGF (50 ng/ml) was determined at the indicated times using the NF-κB-luciferase reporter transient transfection assay. G, WT, XIAP−/−, and XIAP−/−(HA-XIAP) cells were used for determination of cancer invasion in the Transwell assay. The results are expressed as a percentage of invasion and are represented as means ± S.D. of the data from three independent experiments. *, significant decrease compared with the WT(Vector) and XIAP−/−(HA-XIAP) groups (p < 0.01).
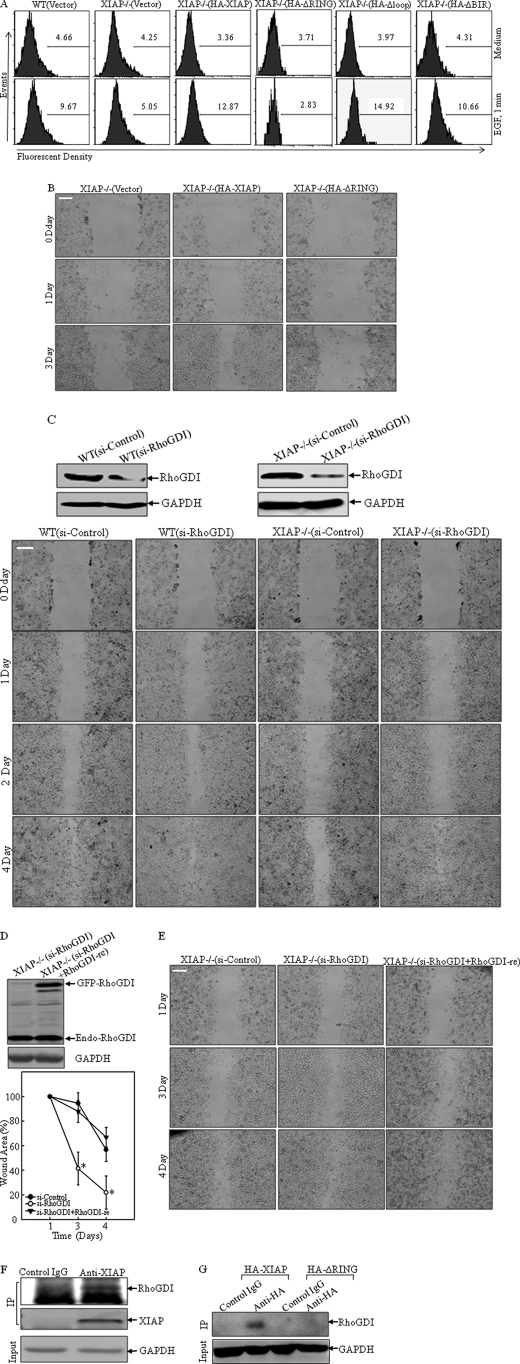
The RING Domain of XIAP Is Crucial for Actin Polymerization, and RhoGDI Is a Putative Downstream Target of XIAP in the Regulation of Cancer Cell Migration
To identify the structure basis of XIAP regulation of actin polymerization, the constructs containing full-length HA-XIAP, deletion of the RING domain (HA-XIAPΔRING), deletion of three BIR domains (HA-XIAPΔBIR), and deletion of the loop region (HA-XIAPΔloop) were transfected into XIAP−/− cells to determine the structure-function relationship in EGF-induced actin polymerization. The results showed that expression of HA-XIAP, HA-XIAPΔBIR, or HA-XIAPΔloop restored actin polymerization in comparison with that in XIAP−/− cells, whereas expression of HA-XIAPΔRING did not show any effect (Fig. 4A). These data demonstrated that the RING domain might be crucial for XIAP-mediated actin polymerization. Consistent with actin polymerization, HA-XIAPΔRING expression was not able to restore cell migration, whereas HA-XIAP did increase cell migration in XIAP−/− cells (Fig. 4B). RhoGDI is a key modulator of F-actin polymerization (44). It interacts with the GTP-bound form of the Rho GTPase and inhibits GTP hydrolysis, therefore allowing RhoGDI to inhibit actin polymerization and cell motility (45). To explore whether RhoGDI acts as an inhibitor of cell migration, an siRNA targeting RhoGDI was transfected into both WT and XIAP−/− cells. Western blot analysis showed that RhoGDI expression was dramatically reduced in both WT(Si-RhoGDI) and XIAP−/−(Si-RhoGDI) cells (Fig. 4C, upper panels). The wound healing assay showed remarkable increase in cell migration in XIAP−/−(Si-RhoGDI) cells compared with XIAP−/−(Si-Control) transfectants (Fig. 4C, lower panels). To further verify this observation, we developed the pEGFP-C3/RhoGDI-re construct, containing a full-length RhoGDI cDNA with numerous mutations in the sequence spanning residues 401–419, the region complementary to the siRNA oligonucleotide targeting RhoGDI. The mutations introduced in pEGFP-C3/RhoGDI-re prevented its destruction by RhoGDI siRNA (23). pEGFP-C3/RhoGDI-re was transfected into XIAP−/−(Si-RhoGDI) cells, and stable transfectants were identified and named XIAP−/−(Si-RhoGDI+RhoGDI-re) (Fig. 4D, right panels). The migration ability of these cells was lower in the wound healing assay compared with XIAP−/−(Si-RhoGDI) cells (Fig. 4D). This finding further suggested that RhoGDI might be an XIAP downstream target for XIAP regulation of cell motility. Therefore, we carried out a co-immunoprecipitation assay utilizing an XIAP-specific antibody to determine whether XIAP can interact with RhoGDI. The results showed that RhoGDI was detected in the co-immunoprecipitated complex (Fig. 4E), suggesting that RhoGDI might interact with endogenous XIAP in the cell. To test whether the RING domain was involved in the XIAP-RhoGDI protein-protein interaction, we utilized XIAP−/−(HA-XIAP) and XIAP−/−(HA-ΔRING) transfectants to carry out co-immunoprecipitation using anti-HA antibody. Our results showed that RhoGDI was detected only in the co-immunocomplex from XIAP−/−(HA-XIAP) cells, whereas there was no detectable RhoGDI in the co-immunocomplex from XIAP−/−(HA-ΔRING) cells (Fig. 4F). These results demonstrate that the RING domain is required by XIAP to interact with RhoGDI and subsequently for its mediation of actin polymerization. Collectively, our results demonstrate that XIAP is able to interact with RhoGDI through its RING domain and that this interaction confers to XIAP the ability to regulate actin polymerization and cell migration.
FIGURE 4.
XIAP regulates cancer cell migration by interacting with RhoGDI. A, the induction of F-actin polymerization upon treatment with EGF (50 ng/ml) was evaluated by flow cytometry as indicated. B–E, cell migration ability was determined by wound healing assays of the indicated transfectants. *, significant difference in the indicated cell lines (p < 0.05). Scale bars = 300 μm. The indicated stable transfectants were identified by Western blotting using anti-RhoGDI antibody (C, upper panels, and D). F–G, lysates from WT cells (F) and XIAP−/−(HA-XIAP) and XIAP−/−(HA-ΔRING) cells (G) were immunoprecipitated (IP) with anti-XIAP (F) or anti-HA (G) antibody, and immunoprecipitates were then subjected to Western blotting with anti-RhoGDI antibody.
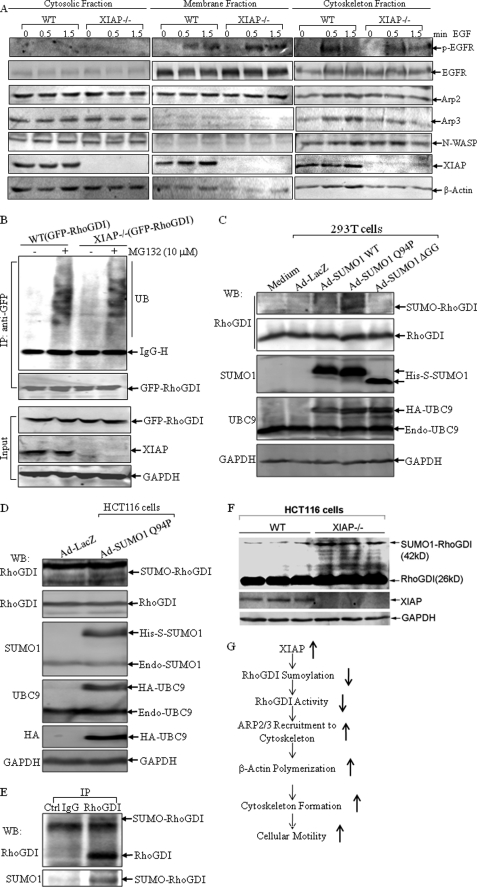
XIAP Mediates Recruitment of the Arp2/3 Complex to the Cytoskeleton and Affects RhoGDI SUMOylation
A family of actin-related proteins (Arp) shares ∼50% homology with actin, and one group of Arp proteins (Arp2/3) is involved in the stimulation of actin assembly. The activated Arp2/3 complex is able to bind to the ends of existing actin filaments and induces actin polymerization (46), and suppression of Arp2/3 activation is a key step in RhoGDI inhibition of actin polymerization (47). To test whether the impairment of actin polymerization induced by EGF in XIAP−/− cells is due to failure to recruit the Arp2/3 complex, the distribution of Arp2/3 protein in cytosolic, membrane, and cytoskeletal fractions was evaluated in EGF-treated WT and XIAP−/− cells. As shown in Fig. 5A, there was a strong recruitment of Arp2/3 in the cytoskeletal fractions of EGF-treated WT HCT116 cells, whereas no obvious change in Arp2/3 translocation was observed in the cytoskeletal fractions of EGF-treated XIAP−/− cells. However, EGF receptor phosphorylation was comparable between WT and XIAP−/− cells, suggesting that the impairment of actin polymerization and Arp2/3 recruitment in EGF-treated XIAP−/− cells occurs downstream of the EGF receptor. The RING domain of XIAP has its E3 ubiquitin ligase activity, which is responsible for regulation of the ubiquitination of several proteins (5). We transfected GFP-tagged RhoGDI into WT and XIAP−/− cells and compared the ubiquitination levels of RhoGDI between the two cells using the anti-GFP antibody to pull down exogenous RhoGDI. As shown in Fig. 5B, although RhoGDI ubiquitination was detected in co-pulldown complexes from both types of cells following MG132 treatment, there were no obvious differences in ubiquitination in WT and XIAP−/− cells, suggesting that RhoGDI ubiquitination is not affected by the E3 ubiquitin ligase activity of XIAP. The post-translational conjugation of the small ubiquitin-like modifier (SUMO) exerts a wide variety of effects on the target proteins, including altered protein conformation, activity, localization, and protein-protein interactions (48). We next tested whether XIAP can alter the activity of RhoGDI by affecting its SUMOylation. However, testing this possibility in vitro was complicated by the unstable and transient nature of this post-translational modification, which is associated with the high abundance and unmatched stability of the de-SUMOylating enzymes. Therefore, two methods were employed to produce substantial increases in overall cellular SUMOylation: transfecting cells with dicistronic expression plasmids for SUMO1 and UBC9 (the E2 SUMO-conjugating enzyme) and transducing cells with recombinant adenoviruses carrying these constructs (24). In addition to the wild-type form of SUMO, two additional forms of SUMO were used in these experiments: SUMO1ΔGG, which carries a C-terminal end deletion of the diglycine motif that renders SUMO non-conjugatable, and SUMO1-Q94P, which contains a single amino acid substitution that prevents deconjugation by the cellular de-SUMOylating enzymes and therefore produces even greater increases in cellular SUMOylation compared with wild-type SUMO. As shown in Fig. 5C for 293T cells and in Fig. 5D for HCT116 cells infected with the recombinant adenoviruses, expression of the Dual-His-S-SUMO1-Q94P/IRES/HA-UBC9 construct (Ad-SUMO1-Q94P) led to the detection of a high molecular weight form of RhoGDI by Western blotting, whereas expression of either the wild-type dicistronic construct, Dual-His-S-SUMO1/IRES/HA-UBC9 (Ad-SUMO1-WT), or the non-conjugatable construct, His-S-SUMO1ΔGG/IRES/HA-UBC9 (Ad-SUMO1ΔGG), did not, indicating that RhoGDI might be modified by SUMOylation. We then used immunoprecipitation to pull down endogenous RhoGDI and detected its SUMOylation with anti-SUMO1 antibody. As shown in Fig. 5E, the anti-RhoGDI antibody not only pull downed the unmodified RhoGDI molecule but also the modified form, indicated by the migrated band, which was also detected by the anti-SUMO1 antibody. Therefore, our results suggested that RhoGDI was post-translationally modified by SUMOylation. Finally, we compared the putative SUMOylation level of RhoGDI in WT and XIAP−/− cells. As shown in Fig. 5F, the retarded migrated band of RhoGDI was stronger in XIAP−/− cells compared with WT cells. Taken together, our studies suggest that XIAP might negatively regulate RhoGDI SUMOylation.
FIGURE 5.
XIAP depletion causes failure to recruit the Arp2/3 complex upon EGF treatment and increased RhoGDI SUMOylation. A, after the WT and XIAP−/− HCT116 cells were treated with EGF (50 ng/ml) for 0.5 or 1.5 min, the cell fractions were isolated and subjected to Western blotting. p-EGFR, phospho-EGF receptor. B, lysates from WT(GFP-RhoGDI) and XIAP−/−(GFP-RhoGDI) cells were co-immunoprecipitated (IP) with anti-GFP antibody, and immunoprecipitates were then subjected to Western blotting. UB, ubiquitin; IgG-H, IgG heavy chain. C and D, 293T and HCT116 cells, respectively, were infected with the indicated viruses for 24 h, and the cells were lysed in 2× SDS sample buffer and subjected to Western blotting (WB). E, lysates from HCT116 cells were immunoprecipitated with anti-RhoGDI antibody, and immunoprecipitates were then subjected to Western blotting. F, WT and XIAP−/− cells were lysed in buffer containing 62.5 mm Tris (pH 6.8) and 2% SDS, boiled for 10 min, and subjected to the Western blot assay. G, the proposed model for XIAP regulation of cancer cell motility.
DISCUSSION
Cancer progression is a multistep process in which cancer cells disperse from a localized primary tumor mass to both invade adnexa and metastasize to distant organs. Clinical studies have shown that the mortality of >90% cancer patients is not due to the primary tumor, but is due to the development of metastasis (49). Therefore, one of the major aims in the cancer research field is to explore and understand the factors involved in cancer metastasis. In this study, we found that either knock-out or knockdown of XIAP decreased insoluble β-actin expression, actin polymerization, cytoskeleton formation, and cell motility. Moreover, reconstituted expression of XIAP in XIAP−/− cells restored all of these biological effects. We have also demonstrated that the cell motility regulated by XIAP depended on its RING domain. Furthermore, we identified RhoGDI as a downstream target of the effects of XIAP on cell motility. The XIAP RING domain was found to form a complex with RhoGDI, which further regulated Arp2/3 recruitment and EGF-induced actin polymerization. Moreover, XIAP seemed to negatively regulate RhoGDI SUMOylation, which could regulate the activity of RhoGDI in controlling cell motility. Collectively, our results demonstrate that XIAP is an essential regulator of cancer cell motility and that this activity is distinct from its reported function as a regulator of caspase cascades.
Elevated expression of XIAP has been associated with aggressive malignant cancer behavior and cancer progression and is considered a good predictor of poor clinical outcomes in a number of malignancies (8–20). However, the biological significance of the increased XIAP expression in cancer invasion and metastasis is controversial and still largely unknown. It has been reported that knockdown of XIAP impaired cancer metastasis in hepatocellular carcinoma cells (50), whereas another study suggested that XIAP did not play a role in the regulation of HeLa cell migration (51). cIAP1 and cIAP2 are other important members of the IAP family, whose roles in cancer metastasis are also complicated. Altieri and co-workers (42) have found that knockdown of XIAP as well as cIAP1 and cIAP2 inhibited cell invasion in human breast and prostate adenocarcinoma cells, which is entirely consistent with our findings in human colon cancer HCT116 cells. After completion of this manuscript, there was another report showing that XIAP regulates metastasis in prostate cancer cells through activating the NF-κB pathway (42). Thus, further investigations into the pathological role of XIAP expression in mediation of cancer invasion and metastasis are of significance. Considering the fact that XIAP−/− HCT116 cells contain very few (0–1%) apoptotic cells (25), our results provide clear evidence demonstrating that the requirement for XIAP in the regulation of cancer cell migration is distinct from its well characterized anti-apoptotic function.
Actin polymerization and cytoskeleton formation act as a central intracellular “motor” for cell migration. Our results showed that β-actin expression was significantly down-regulated in XIAP−/− cells and that only F-actin, but not G-actin, was affected, indicating that XIAP can regulate actin polymerization rather than its transcription or protein degradation. Our findings further showed that the defect in actin polymerization in XIAP−/− cells was due to failure to recruit Arp2/3 to the cytoskeleton. By utilizing various XIAP deletions and co-immunoprecipitation, we showed that XIAP formed a complex with RhoGDI through its RING domain and that XIAP-RhoGDI interaction led to the inhibition of RhoGDI function as an inhibitor of Arp2/3 recruitment and actin polymerization. Our results further showed that RhoGDI could be modified by SUMOylation, a newly identified modification of RhoGDI protein. Our findings that knock-out of XIAP led to an increase in RhoGDI SUMOylation, coupled with previous reports that protein SUMOylation can affect protein activity and localization in cells, suggested that XIAP-regulated RhoGDI SUMOylation may be responsible for XIAP-mediated cancer cell motility. Further investigations of this possibility and the detailed molecular mechanisms linking XIAP to RhoGDI SUMOylation and function are among some of the major projects under way in our laboratory. We noted that knockdown of RhoGDI in WT HCT116 cells did not accelerate the cell migration rate, but its effect on XIAP−/− cells was remarkable (Fig. 4C). The explanation for this discrepancy may be that XIAP overexpression in WT HCT116 cancer cells provides a high level of the inhibitory effect on RhoGDI SUMOylation, and consequently, the normal function of RhoGDI in constraining cell migration is very limited or impaired. As a result, knockdown of RhoGDI by siRNA did not show any additional effect on the migration of WT HCT116 cancer cells. On the other hand, in XIAP−/− cells, there was no inhibitory effect of XIAP on RhoGDI SUMOylation, so RhoGDI is a major participant in the negative regulation of cell migration. Therefore, knockdown of RhoGDI shows a marked effect on cell migration.
In summary, the results obtained from this study suggest that XIAP has the function of regulating cell motility via regulation of β-actin polymerization and cytoskeleton formation. We have shown that XIAP regulates actin polymerization and cell motility through its interaction with RhoGDI. Moreover, our study indicates that XIAP negatively regulates RhoGDI SUMOylation, by which XIAP might target and inhibit the RhoGDI biological effect in the regulation of Arp2/3 recruitment and β-actin polymerization. Thus, our study provides new molecular insights into the understanding of XIAP-mediated cancer cell motility as proposed in Fig. 5G. These findings may enable us to explore the potential utilization of XIAP as a target for cancer prevention and therapy.
Acknowledgments
We thank Drs. Colin S. Duckett and Mark R. Philips for generous gifts of plasmids and Dr. Bert Vogelstein for the gift of wild-type and XIAP−/− HCT116 cells.
This work was supported, in whole or in part, by National Institutes of Health Grant CA112557 from NCI and Grants ES012451, ES010344, and ES000260 from NIEHS. This work was also supported by the New York University Urology Center of Excellence.
- XIAP
- X-linked inhibitor of apoptosis protein
- BIR
- baculovirus IAP repeat
- RhoGDI
- Rho GDP dissociation inhibitor
- IRES
- internal ribosome entry site
- Ad
- adenovirus
- Z-VAD-fmk
- benzyloxycarbonyl-Val-Ala-Asp-chloromethyl ketone
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Hunter A. M., LaCasse E. C., Korneluk R. G. (2007) Apoptosis 12, 1543–1568 [DOI] [PubMed] [Google Scholar]
- 2. Schimmer A. D., Dalili S., Batey R. A., Riedl S. J. (2006) Cell Death Differ. 13, 179–188 [DOI] [PubMed] [Google Scholar]
- 3. Morizane Y., Honda R., Fukami K., Yasuda H. (2005) J. Biochem. 137, 125–132 [DOI] [PubMed] [Google Scholar]
- 4. Wilkinson J. C., Wilkinson A. S., Galbán S., Csomos R. A., Duckett C. S. (2008) Mol. Cell. Biol. 28, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burstein E., Ganesh L., Dick R. D., van De Sluis B., Wilkinson J. C., Klomp L. W., Wijmenga C., Brewer G. J., Nabel G. J., Duckett C. S. (2004) EMBO J. 23, 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jordan B. W., Dinev D., LeMellay V., Troppmair J., Gotz R., Wixler L., Sendtner M., Ludwig S., Rapp U. R. (2001) J. Biol. Chem. 276, 39985–39989 [DOI] [PubMed] [Google Scholar]
- 7. Harlin H., Reffey S. B., Duckett C. S., Lindsten T., Thompson C. B. (2001) Mol. Cell. Biol. 21, 3604–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fong W. G., Liston P., Rajcan-Separovic E., St Jean M., Craig C., Korneluk R. G. (2000) Genomics 70, 113–122 [DOI] [PubMed] [Google Scholar]
- 9. Yang L., Cao Z., Yan H., Wood W. C. (2003) Cancer Res. 63, 6815–6824 [PubMed] [Google Scholar]
- 10. Burstein D. E., Idrees M. T., Li G., Wu M., Kalir T. (2008) Ann. Diagn. Pathol. 12, 85–89 [DOI] [PubMed] [Google Scholar]
- 11. Kleinberg L., Lie A. K., Flørenes V. A., Nesland J. M., Davidson B. (2007) Hum. Pathol. 38, 986–994 [DOI] [PubMed] [Google Scholar]
- 12. Kleinberg L., Flørenes V. A., Silins I., Haug K., Trope C. G., Nesland J. M., Davidson B. (2007) Cancer 109, 228–238 [DOI] [PubMed] [Google Scholar]
- 13. Akyurek N., Ren Y., Rassidakis G. Z., Schlette E. J., Medeiros L. J. (2006) Cancer 107, 1844–1851 [DOI] [PubMed] [Google Scholar]
- 14. Nemoto T., Kitagawa M., Hasegawa M., Ikeda S., Akashi T., Takizawa T., Hirokawa K., Koike M. (2004) Exp. Mol. Pathol. 76, 253–259 [DOI] [PubMed] [Google Scholar]
- 15. Li B., Zhang Y. D., Tian B. N. (2007) Nan Fang Yi Ke Da Xue Xue Bao 27, 1746–1748 [PubMed] [Google Scholar]
- 16. Nagi C., Xiao G. Q., Li G., Genden E., Burstein D. E. (2007) Ann. Diagn. Pathol. 11, 402–406 [DOI] [PubMed] [Google Scholar]
- 17. Li M., Song T., Yin Z. F., Na Y. Q. (2007) Chin. Med. J. 120, 469–473 [PubMed] [Google Scholar]
- 18. Mizutani Y., Nakanishi H., Li Y. N., Matsubara H., Yamamoto K., Sato N., Shiraishi T., Nakamura T., Mikami K., Okihara K., Takaha N., Ukimura O., Kawauchi A., Nonomura N., Bonavida B., Miki T. (2007) Int. J. Oncol. 30, 919–925 [PubMed] [Google Scholar]
- 19. Kluger H. M., McCarthy M. M., Alvero A. B., Sznol M., Ariyan S., Camp R. L., Rimm D. L., Mor G. (2007) J. Transl. Med. 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gordon G. J., Mani M., Mukhopadhyay L., Dong L., Edenfield H. R., Glickman J. N., Yeap B. Y., Sugarbaker D. J., Bueno R. (2007) J. Pathol. 211, 447–454 [DOI] [PubMed] [Google Scholar]
- 21. Lewis J., Burstein E., Reffey S. B., Bratton S. B., Roberts A. B., Duckett C. S. (2004) J. Biol. Chem. 279, 9023–9029 [DOI] [PubMed] [Google Scholar]
- 22. Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. (2001) J. Cell Biol. 152, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang B., Zhang Y., Dagher M. C., Shacter E. (2005) Cancer Res. 65, 6054–6062 [DOI] [PubMed] [Google Scholar]
- 24. Pal S., Rosas J. M., Rosas-Acosta G. (2010) J. Virol. Methods 163, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cummins J. M., Kohli M., Rago C., Kinzler K. W., Vogelstein B., Bunz F. (2004) Cancer Res. 64, 3006–3008 [DOI] [PubMed] [Google Scholar]
- 26. Song L., Li J., Ye J., Yu G., Ding J., Zhang D., Ouyang W., Dong Z., Kim S. O., Huang C. (2007) Mol. Cell. Biol. 27, 2713–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song L., Li J., Zhang D., Liu Z. G., Ye J., Zhan Q., Shen H. M., Whiteman M., Huang C. (2006) J. Cell Biol. 175, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang D., Li J., Costa M., Gao J., Huang C. (2010) Cancer Res. 70, 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taniuchi S., Kinoshita Y., Yamamoto A., Fujiwara T., Hattori K., Hasui M., Kobayashi Y. (1999) Pediatr. Int. 41, 37–41 [DOI] [PubMed] [Google Scholar]
- 30. Chan A. Y., Raft S., Bailly M., Wyckoff J. B., Segall J. E., Condeelis J. S. (1998) J. Cell Sci. 111, 199–211 [DOI] [PubMed] [Google Scholar]
- 31. Diakonova M., Payrastre B., van Velzen A. G., Hage W. J., van Bergen en Henegouwen P. M., Boonstra J., Cremers F. F., Humbel B. M. (1995) J. Cell Sci. 108, 2499–2509 [DOI] [PubMed] [Google Scholar]
- 32. Shan D., Chen L., Njardarson J. T., Gaul C., Ma X., Danishefsky S. J., Huang X. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3772–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rouleau C., Roy A., St Martin T., Dufault M. R., Boutin P., Liu D., Zhang M., Puorro-Radzwill K., Rulli L., Reczek D., Bagley R., Byrne A., Weber W., Roberts B., Klinger K., Brondyk W., Nacht M., Madden S., Burrier R., Shankara S., Teicher B. A. (2006) Mol. Cancer Ther. 5, 219–229 [DOI] [PubMed] [Google Scholar]
- 34. Yamazaki D., Kurisu S., Takenawa T. (2005) Cancer Sci. 96, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ananthakrishnan R., Ehrlicher A. (2007) Int. J. Biol. Sci. 3, 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 37. Lee H. K., Marzella L. (1994) Int. Rev. Exp. Pathol. 35, 39–147 [PubMed] [Google Scholar]
- 38. Vaux D. L., Silke J. (2005) Nat. Rev. Mol. Cell Biol. 6, 287–297 [DOI] [PubMed] [Google Scholar]
- 39. Ouyang W., Zhang D., Li J., Verma U. N., Costa M., Huang C. (2009) J. Cell. Physiol. 218, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo W., Liu J., Li J., Zhang D., Liu M., Addo J. K., Patil S., Zhang L., Yu J., Buolamwini J. K., Chen J., Huang C. (2008) J. Biol. Chem. 283, 8624–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Staiger C. J., Blanchoin L. (2006) Curr. Opin. Plant Biol. 9, 554–562 [DOI] [PubMed] [Google Scholar]
- 42. Mehrotra S., Languino L. R., Raskett C. M., Mercurio A. M., Dohi T., Altieri D. C. (2010) Cancer Cell 17, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaguchi H., Wyckoff J., Condeelis J. (2005) Curr. Opin. Cell Biol. 17, 559–564 [DOI] [PubMed] [Google Scholar]
- 44. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 45. DerMardirossian C., Bokoch G. M. (2005) Trends Cell Biol. 15, 356–363 [DOI] [PubMed] [Google Scholar]
- 46. Disanza A., Steffen A., Hertzog M., Frittoli E., Rottner K., Scita G. (2005) Cell. Mol. Life Sci. 62, 955–970 [DOI] [PubMed] [Google Scholar]
- 47. Zhang B. (2006) Drug Resist. Updates 9, 134–141 [DOI] [PubMed] [Google Scholar]
- 48. Rosas-Acosta G., Russell W. K., Deyrieux A., Russell D. H., Wilson V. G. (2005) Mol. Cell. Proteomics 4, 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Entschladen F., Drell T. L., 4th, Lang K., Joseph J., Zaenker K. S. (2004) Lancet Oncol. 5, 254–258 [DOI] [PubMed] [Google Scholar]
- 50. Shi Y. H., Ding W. X., Zhou J., He J. Y., Xu Y., Gambotto A. A., Rabinowich H., Fan J., Yin X. M. (2008) Hepatology 48, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dogan T., Harms G. S., Hekman M., Karreman C., Oberoi T. K., Alnemri E. S., Rapp U. R., Rajalingam K. (2008) Nat. Cell Biol. 10, 1447–1455 [DOI] [PubMed] [Google Scholar]