Abstract
Members of the C1q/TNF family play important and diverse roles in the immune, endocrine, skeletal, vascular, and sensory systems. Here, we identify and characterize CTRP13, a new and extremely conserved member of the C1q/TNF family. CTRP13 is preferentially expressed by adipose tissue and the brain in mice and predominantly by adipose tissue in humans. Within mouse adipose tissue, CTRP13 is largely expressed by cells of the stromal vascular compartment. Due to sexually dimorphic expression patterns, female mice have higher transcript and circulating CTRP13 levels than males. CTRP13 transcript and circulating levels are elevated in obese male mice, suggesting a potential role in energy metabolism. The insulin-sensitizing drug rosiglitazone also increases the expression of CTRP13 in adipocytes, which correlates with the insulin-sensitizing action of CTRP13. In a heterologous expression system, CTRP13 is secreted as a disulfide-linked oligomeric protein. When co-expressed, CTRP13 forms heteromeric complexes with a closely related family member, CTRP10. This heteromeric association does not involve conserved N-terminal Cys residues. Functional studies using purified recombinant protein demonstrated that CTRP13 is an adipokine that promotes glucose uptake in adipocytes, myotubes, and hepatocytes via activation of the AMPK signaling pathway. CTRP13 also ameliorates lipid-induced insulin resistance in hepatocytes through suppression of the SAPK/JNK stress signaling that impairs the insulin signaling pathway. Further, CTRP13 reduces glucose output in hepatocytes by inhibiting the mRNA expression of gluconeogenic enzymes, glucose-6-phosphatase and the cytosolic form of phosphoenolpyruvate carboxykinase. These results provide the first functional characterization of CTRP13 and establish its importance in glucose homeostasis.
Keywords: Adipose Tissue, AMP-activated Kinase (AMPK), Diabetes, Metabolic Diseases, Metabolism, Metabolic Regulation, Obesity
Introduction
Adipose tissue plays important roles in whole-body energy balance. This specialized tissue serves as a storage depot for triacylglycerol and is the source of many secreted factors, collectively termed adipokines, that directly or indirectly regulate systemic insulin sensitivity and whole-body energy homeostasis (1). The expression of many adipokines is dysregulated in pathologic conditions (e.g. diabetes and obesity), making them important biomarkers and potential drug targets (2). Since the discoveries of leptin (3) and adiponectin (4–6), the list of newly discovered adipokines has significantly expanded, increasing our appreciation for the complexity of intertissue cross-talk mediated by these secreted proteins.
Despite well documented insulin-sensitizing, anti-inflammatory, and anti-atherosclerotic properties, adiponectin knock-out mice display modest phenotypes (7–10). Other factors could probably compensate for the loss of adiponectin (11, 12). Our efforts to discover novel adipokines have led to the identification of a family of secreted proteins designated CTRP1 to -10 (13–15). Both adiponectin and CTRPs4 belong to the C1q/TNF superfamily (16), all members of which contain a signature C-terminal globular domain homologous to the immune complement C1q and whose three-dimensional structures strikingly resemble that of TNF-α (17).
Many CTRPs are expressed by adipose tissue, and most of them circulate in plasma with levels varying according to the genetic backgrounds and metabolic states of mice (14, 15). All CTRPs form trimers as the basic structural unit, and some are further assembled into higher order multimeric structures (14, 15). Physiological functions and mechanisms of action of CTRPs are only beginning to be elucidated. To date, in vitro studies of metabolic regulations (13, 18, 19) using recombinant proteins have been described for CTRP2, CTRP5, and g-CTRP6 (the C-terminal globular domain), whereas in vivo metabolic functions (14, 15, 20) have been demonstrated for CTRP1, CTRP3, and CTRP9. In addition, in vitro studies highlighted the potential cell proliferation/migration and anti-inflammatory roles of CTRP3/CORS-26/cartducin (21, 22). Here, we identify and characterize CTRP13, a novel member of the C1q/TNF family. Our results establish CTRP13 as a novel adipokine with important metabolic functions.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Mouse monoclonal anti-FLAG M2 antibody was obtained from Sigma, and rat monoclonal anti-HA (Clone 3F10) antibody was obtained from Roche Applied Science. Rabbit antibodies that recognize phospho-Akt (Thr-308), phospho-AMPKα (Thr-172), phospho-SAPK/JNK (Thr-183/Tyr-185), Akt, AMPKα, and SAPK/JNK were obtained from Cell Signaling Technology. Rosiglitazone was obtained from Cayman Chemical, TNF-α was from BioSource, phosphatidylinsositol 3-kinase (PI3K) inhibitor (LY294002) was from Cell Signaling Technology, AMPK activator (aminoimidazole carboxamide ribonucleotide; AICAR) was from Calbiochem, and AMPK inhibitor (compound C) was from Calbiochem.
Mice
Leptin-deficient obese (ob/ob) mice on a C57BL/6J background and C57BL/6J lean controls were purchased from Jackson Laboratory. Animal experiments were approved by the Animal Care and Use Committee at the Johns Hopkins University School of Medicine.
Generation of CTRP13-specific Antibody
Rabbit polyclonal anti-CTRP13 antibody directed against the C-terminal globular domain was generated in this study. Sequences that correspond to the globular domain (amino acids 115–255) of CTRP13 were cloned into a bacterial expression vector, pTrcHis (Invitrogen), and verified by DNA sequencing. g-CTRP13 was produced in an Escherichia coli expression system, purified as described for several CTRPs (14), and used as an immunogen for antibody production. Sera from immunized rabbits were collected and tested for their ability to recognize CTRP13 in the supernatants of transfected HEK293T cells.
Cloning of CTRP13
CTRP13 cDNA was cloned from a mouse brain cDNA pool (Clontech) using the primer pair 5′-GGTGATGGTGCTTCTGCTGGTCATC-3′ and 5′-GATTCACTGACGTTAGCCATACG-3′ in a 35-cycle PCR using Platinum Pfx polymerase (Invitrogen) in the presence of 8% DMSO. The PCR product was fractionated in 1% agarose gel, excised, purified, and cloned into the pCR2.1 TOPO vector (Invitrogen). After verification by DNA sequencing, the cDNA insert was excised and cloned into the EcoRI restriction site of a mammalian expression vector, pCDNA3.1 (Invitrogen). The cDNA sequence was deposited in the NCBI GenBankTM data base and assigned accession number EU399230.
cDNA Constructs
The C-terminal FLAG (DYKDDDDK peptide) and HA (YPYDVPDYA peptide) epitope-tagged mouse CTRP13 were generated by PCR and cloned into the pCR2.1 TOPO vector. Epitope-tagged cDNA was excised from the plasmid and cloned into the EcoRI restriction site of pCDNA3 expression vector. All constructs were verified by DNA sequencing. The mammalian expression vectors encoding C-terminal HA-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP9, CTRP10, and CTRP10ΔCys used in this study were described in our previous studies (14, 15).
Site-directed Mutagenesis
A site-directed mutagenesis kit using high fidelity Pfu polymerase from Stratagene was used to mutate Cys-28 and Cys-32 to Ser residues (C28S,C32S mutant) according to the manufacturer's protocol. This construct is designated as CTRP13ΔCys. The following primer pair was used: forward primer, 5′-ACGAGATGCTGGGCACCAGCCGCATGGTCAGCGACCCCTATGGGGGCAC-3′; reverse primer, 5′-GTGCCCCCATAGGGGTCGCTGACCATGCGGCTGGTGCCCAGCATCTCGT-3′. Successful mutagenesis was confirmed by DNA sequencing.
Isolation of Primary Adipocytes and Stromal Vascular Cells from Adipose Tissue
8-week-old male/female obese (ob/ob) mice and their control littermates were used to obtain primary adipocytes and stromal vascular cells (stromal vascular fraction; SVF) from adipose tissue as described previously (14). Total RNAs from primary adipocytes and SVF were isolated with TRIzol reagent (Invitrogen). Potential DNA contaminants were removed by RNase-free DNase I (Ambion).
Quantitative Real-time PCR Analysis of CTRP13 Expression in Mouse and Human Tissues
A quantitative PCR approach was used to screen multiple tissue cDNA panels (Clontech), RNA isolated from adipose tissue of 8-week-old ob/ob mice and lean controls, and RNA isolated from primary adipocytes and SVF for the presence of CTRP13 transcripts. The following PCR primer pairs were used in this study: CTRP13 forward, 5′-AACGCAAGATAAGCAGATGTGTG-3′; CTRP13 reverse, 5′-AAGGAGTATTTGCTTTGGCGG-3′; adipsin forward, 5′-AGCAGTGGGTGCTCAGTGC-3′; adipsin reverse, 5′-CGTCATCCGTCACTCCATCC-3′; resistin forward, 5′-TCTTATGTTGACAGGGACGG-3′; resistin reverse, 5′-CCCTCAGCTTAGACCTGCCC-3′; glucose-6-phosphatase (G6Pase) forward, 5′-CGACTCGCTATCTCCAAGTGA-3′; G6Pase reverse, 5′-GTTGAACCAGTCTCCGACCA-3′; phosphoenolpyruvate carboxykinase (GTP) (cytosolic) (PEPCK-C) forward, 5′-CTGCATAACGGTCTGGACTTC-3′; PEPCK-C reverse, 5′-CAGCAACTGCCCGTACTCC-3′; 18 S rRNA forward, 5′-GCAATTATTCCCCATGAACG-3′; and 18 S rRNA reverse, 5′-GGCCTCACTAAACCATCCAA-3′. The default PCR protocol was used on an Applied Biosystems Prism 7500 sequence detection system. Primary adipocytes and SVF cDNAs were synthesized from 2 μg of total RNA and 200 ng of random hexamers using the Superscript II RNase H-Reverse Transcriptase protocol (Invitrogen). For quantitative PCR, samples were analyzed in 25-μl reactions (10 ng of cDNA, 900 nmol of primer, 12.5 μl of master mix, and water) according to the standard protocol provided for the SyBR® Green PCR Master Mix (Applied Biosystems).
Gel Filtration Chromatographic Analysis
Supernatant (400 μl) from transfected human embryonic kidney (HEK293T) cells containing FLAG-tagged CTRP13 was loaded onto an AKTA FPLC and fractionated through a 10/30 Superdex 200 column (GE Healthcare) in PBS as described previously for other CTRPs (14, 15). The collected fractions were subjected to immunoblot analysis.
Expression and Purification of Recombinant CTRP13
Full-length recombinant CTRP13, containing a C-terminal FLAG tag, was produced in mammalian HEK293 cells and purified as described for other CTRPs (14). Like adiponectin, all secreted CTRPs form trimers and higher order structures, and most contain posttranslational modifications that include N-linked glycosylation and proline hydroxylation (14, 15). These modifications and the assembly of higher order structures are likely to be functionally important as have been shown for adiponectin (23, 24). Therefore, recombinant CTRP13 was produced in mammalian expression system. Purified proteins were dialyzed against 20 mm Hepes buffer (pH 8.0) containing 135 mm NaCl in a 10 kDa cut-off Slide-A-Lyzer dialysis cassette (Pierce). Protein concentration was determined by the Bradford method (25) using the Coomassie Plus assay kit (Thermo Scientific). Recombinant protein has >95% purity as judged by a Coomassie Blue-stained SDS-polyacrylamide gel.
Glycoprotein Detection
Approximately 50 ng of purified recombinant FLAG-tagged CTRP13 was separated on an SDS-polyacrylamide gel, transferred to PVDF membrane (Bio-Rad), and subjected to the ECL glycoprotein detection protocol according to the manufacturer's instructions (GE Healthcare). Briefly, any carbohydrate moiety on recombinant CTRP13 was oxidized with sodium metaperiodate, and the oxidized sugar aldehyde group was labeled with biotin using biotin-hydrazide (26). The presence of a carbohydrate moiety was then detected using streptavidin conjugated to horseradish peroxidase (HRP) and chemiluminescence substrate (Millipore).
Immunoprecipitation and Immunoblot Analysis
C-terminal HA-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP9, CTRP10, and CTRP13 were transfected in combination with FLAG-tagged CTRP13 into HEK293T cells using Lipofectamine (Invitrogen). An aliquot of the collected supernatants (300 μl) was combined with 500 μl of immunoprecipitation buffer (150 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) and subjected to an immunoprecipitation using the anti-FLAG affinity gel (Sigma). Samples were rotated for 4 h at 4 °C, washed four times with immunoprecipitation buffer, resuspended in 60 μl of NuPAGE LDS Sample buffer (Invitrogen) containing reducing agent (β-mercaptoethanol), heated at 90 °C for 10 min, and separated on 10% BisTris NuPAGE gel (Invitrogen). Proteins from gels were transferred to 0.2-μm Protran BA83 nitrocellulose membranes (Whatman), blocked in 2% nonfat milk for 1 h, and probed with rat anti-HA antibody in the presence of 2% nonfat milk for overnight. Immunoblots were washed three times (10 min each) in PBS containing 0.1% Tween 20 and incubated with sheep anti-rat HRP (Amersham Biosciences) (1:5000) for 1 h. Blots were washed three times (10 min each) in PBS containing 0.1% Tween 20, developed in ECL reagent (Millipore) for 2–5 min, and exposed to Blue XB-1 film (Eastman Kodak Co.). For input controls, aliquots of conditioned medium from transfected cells were analyzed by immunoblot analysis using the anti-FLAG or anti-HA antibody.
Glucose Uptake Assay
3T3-L1 adipocytes or L6 myotubes were differentiated as described (20). On the day of the experiment (day 8 or beyond for 3T3-L1 adipocytes, day 5 or beyond for myotubes), cells were serum-starved in low glucose DMEM for 2 h. Adipocytes or myotubes were treated with vehicle or CTRP13 (5 μg/ml) for 30 min at 37 °C. Cells were then incubated with uptake media containing 0.5 μCi/ml 2-deoxy-d-[1-14C]glucose in Krebs-Ringer-Hepes buffer (25 mm HEPES-NaOH (pH 7.4), 120 mm NaCl, 5 mm KCl, 1.2 mm MgSO4, 1.3 mm CaCl2, 1.3 mm KH2PO4) supplemented with 0.5% BSA for 10 min. Uptake was stopped by aspirating the media, followed by extensive washing with ice-cold PBS buffer. For the glucose uptake assay in clone 9 cells (a non-transformed rat liver cell line; ATCC) (27), cells were cultured in DMEM containing 10% FBS until confluent. Cells were serum-starved overnight and were treated as indicated in the figure legends. Glucose uptake was started by adding 2-deoxy-d-[1-14C]glucose (0.5 μCi/ml) to the medium. After 2 min of incubation, uptake was stopped by aspirating the medium, followed by extensive washing with ice-cold PBS buffer. Cells were lysed with 0.1% Triton in PBS buffer. An aliquot of cell lysate from each sample was used for protein content analysis using a bicinchoninic acid (BCA) assay kit (Pierce). The radioactivity of the cell lysates were counted in Ecoscint® scintillation mixture (National Diagnostics) using a Beckman LS-6000 liquid scintillation counter. The radioactivity in each sample was normalized against the protein content.
Glucose Production and Measurement
Rat H4IIE hepatoma cells were cultured to near confluence in low glucose DMEM containing 10% FBS and antibiotics. Cells were incubated overnight in glucose production buffer (glucose-free DMEM, pH 7.4, supplemented with 20 mm sodium lactate and 2 mm sodium pyruvate without phenol red). The next day, cells were treated with vehicle buffer or CTRP13 (5 μg/ml) for various lengths of time as indicated in the Fig. 8 legend. An aliquot of the medium was taken from each sample for the measurement of glucose concentration using the Amplex Red® glucose assay kit (Invitrogen). Glucose concentration was corrected for cell number variations based on protein content in each sample as measured by the BCA assay. For quantitative real-time PCR analysis of the expression of two key gluconeogenic enzymes, G6Pase and PEPCK-C, H4IIE hepatocytes were cultured to near confluence in low glucose DMEM containing 10% FBS and antibiotics. Cells were then treated with vehicle buffer or CTRP13 (5 μg/ml) for 24 h. Total RNAs were isolated from the treated cells and reverse transcribed into cDNA. Quantitative real-time PCR analyses were performed as described above.
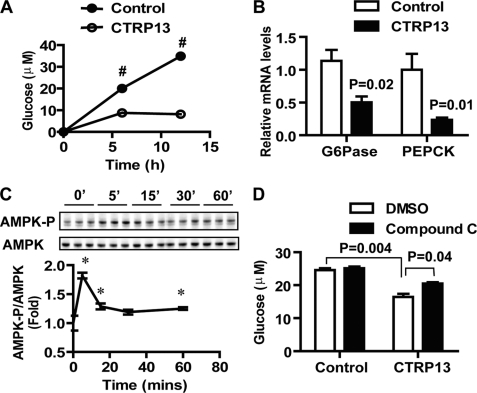
FIGURE 8.
Inhibition of gluconeogenic enzyme gene expression and gluconeogenesis in hepatocytes by CTRP13. A, recombinant CTRP13 (5 μg/ml) inhibits glucose production in H4IIE hepatocytes (n = 3). H4IIE cells were treated with or without CTRP13 in glucose production buffer containing glucose-free and phenol red-free DMEM supplemented with sodium pyruvate and sodium lactate. B, CTRP13 suppresses the expression of two key gluconeogenic enzymes, G6Pase, and PEPCK-C, as revealed by quantitative real-time PCR analysis (n = 6). C, time course of AMPK activation (phosphorylation at Thr-172) in H4IIE hepatocytes induced by CTRP13 (5 μg/ml) (n = 3). D, incubation of H4IIE hepatocytes with compound C (10 μm) partially abolished the suppression of glucose production by CTRP13 (n = 3). All data are expressed as -fold change (normalized to control) and as mean ± S.E. (error bars). *, p < 0.05; #, p < 0.001.
Statistical Analysis
Statistical analyses were performed using Student's t test in Excel. p < 0.05 was considered significant. All quantification results with error bars are expressed as mean ± S.E.
RESULTS
Identification and Analysis of CTRP13 cDNA and Protein
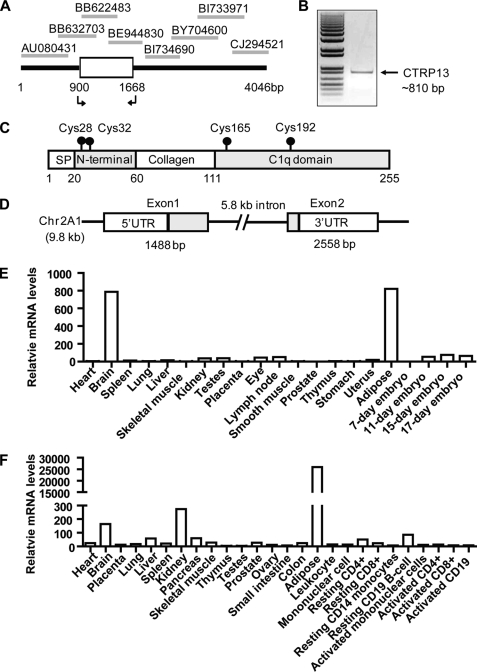
Using the globular C1q domain sequences of adiponectin and CTRPs to query the NCBI GenBankTM data base, we identified a novel member of the C1q/TNF family (Fig. 1). The cDNA is different from the 10 adiponectin paralogs (CTRP1 to -10) we recently identified and characterized (13–15, 28). Because we previously deposited CTRP11 and CTRP12 cDNA sequences in the GenBankTM data base,5 we designated the current cDNA and its encoded protein CTRP13. PCR was used to clone CTRP13 cDNA from mouse brain based on the sequences of overlapping EST clones (Fig. 1, A and B). In searching for proteins that interact with heat shock protein (HSP)-47 through yeast two-hybrid screen, Koide et al. (29) recently described a protein identical to CTRP13, which they designated as C1q-like-3/Gliacolin/K-100. However, no functional information was described for K-100. While the present study was ongoing, Iijima and co-workers (30) characterized in great detail the expression of CTRP13/C1q-like-3 in the central nervous system. The function of CTRP13 in the CNS has not been described. Here, we provide further molecular, biochemical, and functional characterizations of CTRP13 in metabolism. The deduced CTRP13 protein has similar domain structures as other CTRPs (13–15), consisting of a signal peptide, a short N terminus with two highly conserved Cys residues, a collagen domain with 17 Gly-X-Y repeats, and a C-terminal globular domain homologous to the immune complement protein C1q (Fig. 1C). The mouse Ctrp13 gene is 9.8 kb in size, consists of two exons, and is located on chromosome 2A1 (Fig. 1D).
FIGURE 1.
Identification of CTRP13 and its tissue expression profile. A, cloning of CTRP13 cDNA from overlapping EST sequences. The arrows indicate forward and reverse primers used to amplify the coding region of CTRP13 cDNA. B, agarose gel electrophoresis showing the PCR product of the cloned CTRP13 cDNA (∼810 bp). Indicated on the left is the 1-kb Plus DNA ladder from Invitrogen. C, the deduced CTRP13 protein domain structure. Four domains are indicated: a signal peptide (SP) for protein secretion, an N-terminal domain with two conserved Cys residues, a collagen domain with 17 Gly-X-Y repeats, and a C-terminal globular domain homologous to the immune complement C1q. The positions of conserved Cys residues found in the mature protein are indicated with a ball and stick. D, Ctrp13 gene structure. The chromosomal position (2A1) and the sizes of intron and exons are indicated. E and F, quantitative real-time PCR analysis of CTRP13 expression in mouse (E) and human (F) tissues. All expression data were normalized to 18 S rRNA level in each sample.
CTRP13 Is Preferentially Expressed by Brain and Adipose Tissue
Quantitative real-time PCR analysis showed that among the 17 adult mouse tissues profiled, the brain and white adipose tissue expressed the highest levels of CTRP13 transcript, with relatively low levels of expression in other tissues (Fig. 1E). In humans, adipose tissue had the highest expression of CTRP13 mRNA, with ∼100-fold or higher enrichment compared with other tissues (Fig. 1F).
CTRP13 Is Highly Conserved throughout Vertebrate Evolution
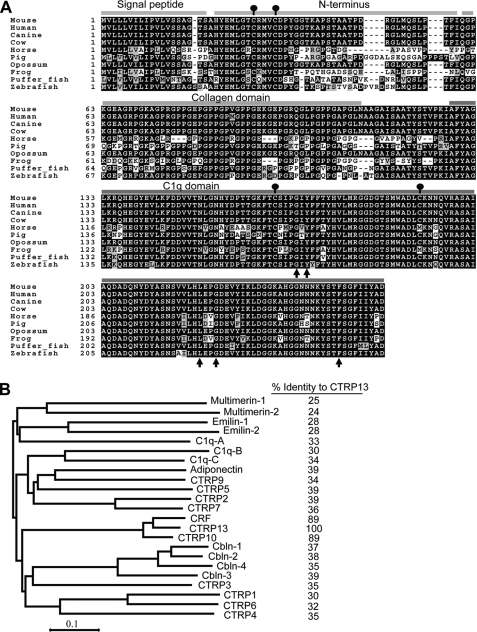
CTRP13 is extremely conserved throughout vertebrate evolution (Fig. 2A). Mouse CTRP13 and its corresponding human, dog, cow, opossum, horse, pig, frog, puffer fish, and zebrafish orthologs share striking degrees of amino acid identity across the full-length protein (Table 1). The protein sequence conservation is even higher when the comparison is restricted to just the C-terminal globular domain (Table 1). For instance, the presumed functional C1q domain of human and zebrafish CTRP13 are 96% identical despite immense evolutionary distance separating these two species. Structure-based alignment between adiponectin, complement C1q, and TNF family members (TNF-α, TNF-β, and CD40L) reveals highly conserved residues important in the packing of the protomer's hydrophobic core (17). These residues are also conserved in CTRP13 (Fig. 2A, arrows). Among the C1q/TNF family members, CTRP13 is most closely related to C1q-related factor (CRF/C1q-like 1) and CTRP10, both of which share 89% amino acid identity to the globular domain of CTRP13 (Fig. 2B). In contrast, adiponectin and CTRP13 share only 39% amino acid identity at their globular domain. The striking conservation of CTRP13 protein sequence throughout vertebrate evolution implies an essential biological function.
FIGURE 2.
Sequence alignment and phylogenetic analysis of CTRP13. A, ClustalW (72) alignment of CTRP13 protein sequences extracted from the NCBI Refseq or the draft genome sequences of mouse (GenBankTM accession number NP_694795), human (NP_001010908), canine (XP_848586), cow (XP_870462.2), horse (XP_001492004), pig (NP_001072144), opossum (XP_001368316), frog (NP_001090380), puffer fish (CAG07104), and zebrafish (CAD61081). Identical amino acids are shaded, and gaps are indicated by a dashed line. The highly conserved Cys residue is indicated by a ball and stick. Four distinct domains of CTRP13 are indicated: signal peptide, N terminus, collagen domain, and globular C1q domain. The arrows indicate the highly conserved residues found in all C1q/TNF superfamily members. Amino acid numbering is indicated on the left. B, phylogenetic analysis of mouse CTRP13 and other C1q/TNF family members based on the ClustalW alignment of the C1q domain sequences. The tree was generated using the MEGA program version 4 (73). The percentage of amino acid identity (of the C1q domain) between each protein and CTRP13 is indicated on the right. GenBankTM accession number for each of the mouse proteins: C1q-A chain (NP_031598), C1q-B chain (NP_033907), C1q-C chain (NP_031600), multimerin-1 (XP_284198), multimerin-2 (NP_878260), emilin-1 (NP_598679), emilin-2 (NP_660140), CTRP1 (NP_064343), CTRP2 (NP_081255), CTRP3 (NP_112150), CTRP4 (NP_080437), CTRP5 (AAY21930), CTRP6 (NP_082607), CTRP7 (NP_780634), CTRP9 (DQ002401), CTRP10 (AAY21934), CRF (NP_035925), CTRP13 (ABY86415), adiponectin (NP_033735), cerebellin-1 (Cbln-1, NP_062600), Cbln-2 (NP_766221), Cbln-3 (NP_062794), and Cbln-4 (NP_783439).
TABLE 1.
Conservation of CTRP13 sequences across vertebrate species
| Amino acid identity to mouse CTRP13 |
||
|---|---|---|
| Full-length | C1q domain | |
| % | % | |
| Mouse (Mus musculus) | 100 | 100 |
| Human (Homo sapiens) | 100 | 100 |
| Dog (Canis familiaris) | 100 | 100 |
| Cow (Bos taurus) | 100 | 100 |
| Opossum (Monodelphis domestica) | 100 | 100 |
| Horse (Equus caballus) | 68 | 83 |
| Pig (Sus scrofa) | 74 | 88 |
| Frog (Xenopus laevis) | 66 | 85 |
| Puffer fish (Tetraodon nigroviridis) | 84 | 96 |
| Zebrafish (Danio rerio) | 88 | 96 |
Sexually Dimorphic Expression Pattern of CTRP13 in Adipose Tissue
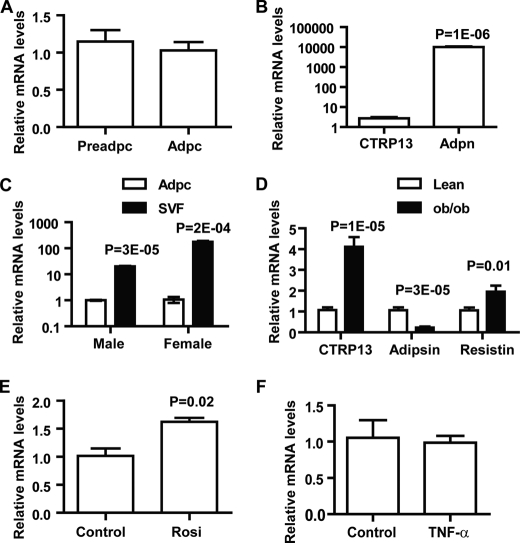
Because CTRP13 is highly expressed in human and mouse adipose tissue, we explored its expression in adipocytes. Using 3T3-L1 cells, we showed that preadipocytes and mature adipocytes have similar levels of CTRP13 transcript (Fig. 3A), indicating that expression of CTRP13 is not significantly modulated by adipocyte differentiation. The expression of CTRP13 transcript is about one-three thousandth of adiponectin, an abundant adipocyte-specific transcript found in mature adipocytes (Fig. 3B); thus, CTRP13 is expressed at relatively low levels in adipocytes. Indeed, comparison between primary adipocytes and the SVF of gonadal fat pads, which contain preadipocytes, fibroblasts, endothelial cells, and occasional immune cells, from male and female mice showed that SVF has significantly higher CTRP13 expression compared with primary adipocytes (Fig. 3C). Interestingly, CTRP13 exhibits pronounced sexually dimorphic expression patterns; females express ∼9-fold higher CTRP13 transcript levels in SVF compared with male mice (Fig. 3C).
FIGURE 3.
Regulation and sexually dimorphic expression pattern of CTRP13. A, quantitative real-time PCR analysis of CTRP13 expression in 3T3-L1 preadipocytes (Preadpc) and mature adipocytes (Adpc; day 8) (n = 6). B, comparison between CTRP13 and adiponectin (Adpn) expression levels in mature 3T3-L1 adipocytes (day 8, n = 6). C, expression of CTRP13 in adipocytes (Adpc) and stromal vascular faction (SVF) derived from gonadal fat pads of male and female C57BL/6 mice (8 weeks old) (n = 3). All expression data were normalized to 18 S rRNA and expressed as -fold change over the mRNA levels found in adipocytes. D, expression of CTRP13, adipsin, and resistin in adipose tissue of leptin-deficient obese (ob/ob) mice and age-matched lean controls (n = 10/group; 8 weeks old). E and F, expression of CTRP13 in mature 3T3-L1 adipocytes in response to rosiglitazone (Rosi; 1 μm; n = 6) (E) and TNF-α treatments (2 ng/ml; n = 6) (F). All expression data were normalized to 18 S rRNA or β-actin level in each sample. All data are expressed as mean ± S.E. (error bars).
Expression of CTRP13 in Obesity and Its Up-regulation by Rosiglitazone
Expression of many adipokines is often dysregulated in metabolic diseases. Thus, we explored whether the expression of CTRP13 is differentially regulated under the condition of obesity. In 8-week-old male leptin-deficient ob/ob mice, expression levels of adipsin decreased, whereas resistin increased significantly (Fig. 3D), consistent with previous studies (31, 32). Interestingly, CTRP13 mRNA levels were markedly increased in obese male mice (Fig. 3D). In contrast, no significant up-regulation of CTRP13 expression was observed in female ob/ob mice (data not shown). The observed increase in CTRP13 mRNA levels may simply result from increased fat mass; alternatively, this finding may reflect some functional significance of CTRP13. To distinguish these possibilities, we examined CTRP13 expression in 3T3-L1 adipocytes treated with rosiglitazone, an insulin-sensitizing and anti-diabetic drug, or TNF-α, an inflammatory cytokine known to be up-regulated in obesity leading to insulin resistance (33). CTRP13 expression increased significantly following rosiglitazone treatment (Fig. 3E) but not after TNF-α (Fig. 3F). These results suggest that the observed increase in CTRP13 expression in male ob/ob mice may reflect a compensatory response to insulin resistance found in these hyperphagic mice.
Sex and Nutritional State Affect Circulating CTRP13 Levels
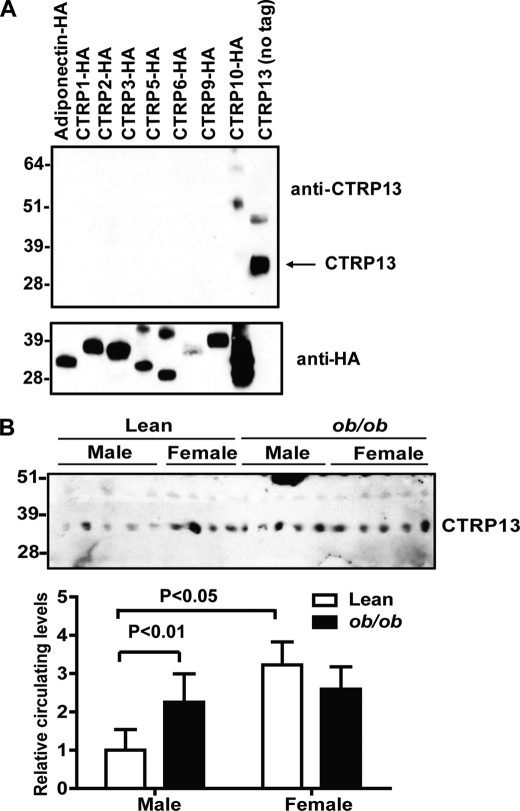
To detect endogenously synthesized CTRP13, we generated rabbit polyclonal antibody specific to CTRP13 (Fig. 4A). We have previously shown that most CTRPs (when specific antibodies are available) circulate in plasma (14, 15). Similarly, here we detected CTRP13 in mouse sera (Fig. 4B). Consistent with the sexually dimorphic expression pattern of CTRP13 mRNA in mouse adipose tissue (Fig. 3C), circulating CTRP13 levels in serum were also higher in lean female mice compared with males (Fig. 3B). Further, as with mRNA levels (Fig. 3D), serum levels of CTRP13 were higher in male ob/ob mice compared with lean controls (Fig. 4B), whereas no difference was observed between lean and obese (ob/ob) female mice.
FIGURE 4.
Circulating levels of CTRP13 in lean and obese mice. A, the specificity of the anti-CTRP13 antibody was demonstrated by an immunoblot analysis (top). The bottom panel indicates the presence of input proteins. B, immunoblot (top) and relative quantification (bottom) of circulating CTRP13 levels in sera obtained from age-matched leptin-deficient obese (ob/ob) mice and lean controls (8 weeks old; n = 5/group). All data are expressed as mean ± S.E. (error bars).
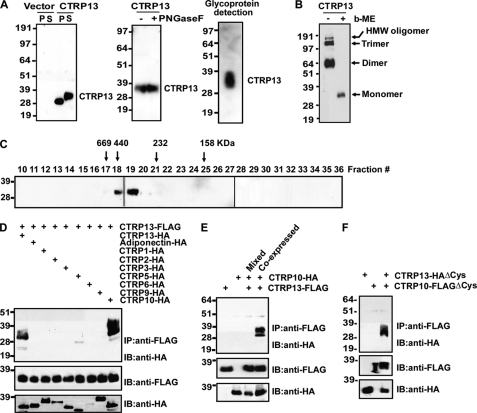
CTRP13 Is a Multimeric Glycoprotein That Forms Homo-oligomers and Heteromeric Complexes with CTRP10
When expressed in mammalian HEK293T cells, CTRP13 was detected in cell lysates and conditioned medium of transfected cells, indicating that CTRP13 is a secreted protein (Fig. 5A, left). The apparent molecular weight of secreted CTRP13 on immunoblot is higher compared with that in the cell lysate, suggesting possible posttranslational modifications of the protein (Fig. 5A). Because most CTRPs contain N-linked glycans (14), we tested if the size shift on immunoblot is due to this modification. Treatment with or without N-glycosidase F did not result in a size shift of CTRP13 on immunoblot (Fig. 5A, middle), indicating the absence of N-linked glycans. However, purified recombinant CTRP13 clearly contains a carbohydrate moiety, as indicated by a glycoprotein detection method that labeled all sugar moieties (Fig. 5A, right). In the presence of reducing agent, CTRP13 migrated as a monomer with an apparent molecular mass of ∼36 kDa on immunoblot (Fig. 5B). In the absence of reducing agent, CTRP13 migrated on immunoblot as a disulfide-linked dimer, trimer, and higher order multimers (Fig. 5B, arrows). The formation of multimeric complexes was confirmed by gel filtration chromatographic analysis, in which recombinant CTRP13 was eluted as a multimer ≥350 kDa in size (Fig. 5C). Importantly, CTRP13 has a larger stoke radius (due to the presence of a rigid collagen triple helical structure in the N terminus) compared with all of the spherical proteins used to calibrate the gel filtration column; thus, the size estimate of CTRP13 appears larger than its actual size. We have previously shown that some CTRPs can form heteromeric complexes with each other (14, 15, 28), representing a potential mechanism to generate an expanded repertoire of functionally distinct ligands with altered function and/or receptor specificity. Similarly, we found that secreted CTRP13 forms heteromeric complexes with the most closely related member, CTRP10, when co-expressed in vitro (Fig. 5D). This physical association is specific; other related family members do not form physical complexes with CTRP13 (Fig. 5D). Interaction between CTRP13 and CTRP10 occurred during biosynthesis; mixing secreted proteins that were expressed separately did not result in co-immunoprecipitation (Fig. 5E). Further, highly conserved Cys residues found in the N terminus of CTRP13 (Cys-28 and Cys-32) or CTRP10 (Cys-29 and Cys-33) are not required for their physical association (Fig. 5F).
FIGURE 5.
CTRP13 is a secreted homo-oligomeric glycoprotein that can also form heteromeric complexes with CTRP10. A, immunoblot analysis of cell pellet (P) and supernatant (S) of HEK293 cells transfected with empty vector or FLAG-tagged CTRP13 (left). Recombinant CTRP13 protein treated with (+) or without (−) peptide N:glycosidase F (PNGaseF) (middle). CTRP13 (50 ng) was subjected to glycoprotein detection (right), indicating the presence of a sugar moiety. B, SDS-PAGE analysis of recombinant CTRP13, carried out in the absence (−) or presence (+) of reducing agent (β-mercaptoethanol; b-ME). C, gel filtration chromatographic analysis of recombinant CTRP13. Fractions 10–36 were analyzed by immunoblot analysis. Arrows, molecular weight markers that correspond to the peak elution fraction of protein standard thyroglobulin, ferritin, catalase, and aldolase, respectively. D, co-expression and immunoprecipitation analyses of epitope-tagged proteins in HEK293 cells. The top panel indicates anti-FLAG pull-down of CTRP13-HA (as positive control) and CTRP10-HA. Input CTRP13-FLAG and HA-tagged proteins are indicated in the middle and bottom panels, respectively. E, conditioned medium containing co-expressed CTRP13 and CTRP10 or a mixture (Mixed) of the two separately expressed proteins was subjected to immunoprecipitation analysis. Top, anti-FLAG pull-down of CTRP10-HA only when both proteins were co-expressed in cells. Input CTRP13-FLAG and HA-tagged proteins are indicated in the middle and bottom panels. F, conditioned media containing N-terminal Cys mutant, CTRP13-FLAGΔCys, and/or CTRP10-HAΔCys were subjected to immunoprecipitation analysis. Top, anti-FLAG pull-down of CTRP10-HAΔCys. Input FLAG- and HA-tagged proteins are indicated in the middle and bottom panels. IP, immunoprecipitation; IB, immunoblot.
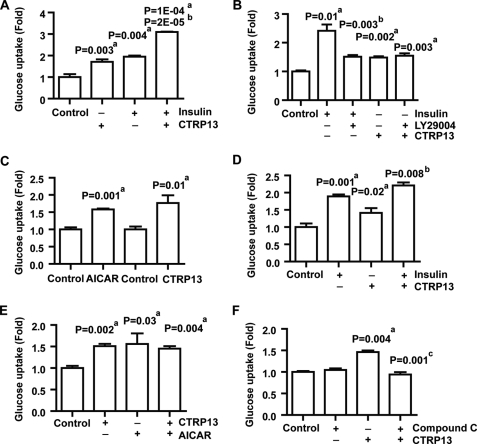
CTRP13 Increases Glucose Uptake in Adipocytes, Myotubes, and Hepatocytes
CTRP13 expression in adipocytes was increased by the insulin-sensitizing drug rosiglitazone (Fig. 3E), suggesting that CTRP13 may contribute to enhancing insulin signaling or function. Thus, we explored the role of CTRP13 in glucose uptake in adipocytes. Treatment with insulin significantly increased glucose uptake in adipocytes as expected (Fig. 6A). Importantly, recombinant CTRP13 treatment also resulted in a significant increase in glucose uptake, to an extent similar to that of insulin (Fig. 6A). Interestingly, treatment with a combination of CTRP13 and insulin resulted in further enhancement of glucose uptake in adipocytes, an effect greater than insulin or CTRP13 treatment alone, suggesting that both work in an additive manner (Fig. 6A). The ability of CTRP13 to further enhance glucose uptake in adipocytes in the presence of a nearly saturating dose of insulin (10 nm) suggests that CTRP13 works through a signaling pathway distinct from that of insulin signaling. Because insulin robustly activates the phosphatidylinositol 3-kinases (PI3K) pathway downstream of the insulin receptor to exert its biological effects (34), we tested whether CTRP13 increases glucose uptake in adipocytes in a PI3K-independent manner using a PI3K-specific inhibitor, LY294002 (35). Co-treatment with LY294002 suppressed insulin-induced glucose uptake in adipocytes as expected (Fig. 6B). In contrast, LY294002 was ineffective in suppressing glucose uptake induced by CTRP13 (Fig. 6B), indicating that CTRP13 activates a signaling pathway independent of PI3K and protein kinase B/Akt. Because GLUT4 is an insulin-responsive glucose transporter, we explored whether other glucose transporters mediate the effect of glucose uptake induced by CTRP13. To address this question, we utilized clone 9 liver cells, a non-transformed rat liver cell line that expresses only GLUT1 but not other glucose transporters (36). As a positive control, AICAR, an AMPK activator (37), significantly increased glucose uptake in clone 9 liver cells (Fig. 6C). Similarly, CTRP13 also increased glucose uptake in clone 9 liver cells to the same extent as AICAR (Fig. 6C). These results indicate that CTRP13 increases glucose uptake through the GLUT1 transporter, at least in clone 9 liver cells. To test whether CTRP13 has any effect in glucose uptake in muscle cells, rat L6 myotubes were treated with CTRP13, insulin, or both. Treatment with insulin or CTRP13 alone resulted in increased glucose uptake in myotubes (Fig. 6D). Only modest enhancement of glucose uptake was observed when cells were treated with a combination of insulin and CTRP13 (Fig. 6D). Because AMPK activation has been linked to increased glucose uptake in myotubes and is PI3K-independent, we examined whether CTRP13 induces glucose uptake in myotubes through the AMPK signaling pathway. L6 myotubes treated with CTRP13 or AICAR exhibited increased glucose uptake to the same extent (Fig. 6E). However, there was no further enhancement of glucose uptake when cells were treated with a combination of CTRP13 and AICAR, suggesting that they possibly target the same signaling pathway. Indeed, incubation of L6 myotubes with compound C, an AMPK inhibitor, abolished glucose uptake induced by CTRP13 (Fig. 6F). These results confirmed the activation of the AMPK signaling pathway by CTRP13 to mediate glucose uptake in myotubes.
FIGURE 6.
CTRP13 increases glucose uptake in adipocytes, myotubes, and liver cells. A, 2-deoxy-d-[1-14C]glucose (2-DG) uptake in 3T3-L1 adipocytes treated with vehicle control, CTRP13 (5 μg/ml; 30 min), and/or insulin (10 nm; 15 min). B, 2-deoxy-d-[1-14C]glucose uptake in adipocytes treated with vehicle control, insulin (10 nm; 15 min), or CTRP13 (5 μg/ml; 30 min) in the presence or absence of PI3K inhibitor, LY29004 (25 μm; 1 h). C, 2-deoxy-d-[1-14C]glucose uptake in clone 9 liver cells treated with vehicle control, AICAR (2 mm; 1 h), or CTRP13 (5 μg/ml; 1 h). D, 2-deoxy-d-[1-14C]glucose uptake in L6 myotubes treated with vehicle control, insulin (100 nm, 15 min), and/or CTRP13 (5 μg/ml; 30 min). E, 2-deoxy-d-[1-14C]glucose uptake in L6 myotubes treated with vehicle control, AICAR (2 mm, 1 h), and/or CTRP13 (5 μg/ml; 30 min). F, 2-deoxy-d-[1-14C]glucose uptake in L6 myotubes treated with vehicle control or CTRP13 in the presence or absence of an AMPK inhibitor, compound C (10 μm; 1 h). Statistical analysis was as follows: compared with vehicle control (a); compared with insulin treatment (b); compared with CTRP13 treatment (c). All data are expressed as -fold change (normalized to control) and as mean ± S.E. (error bars) (n = 3 in all experiments).
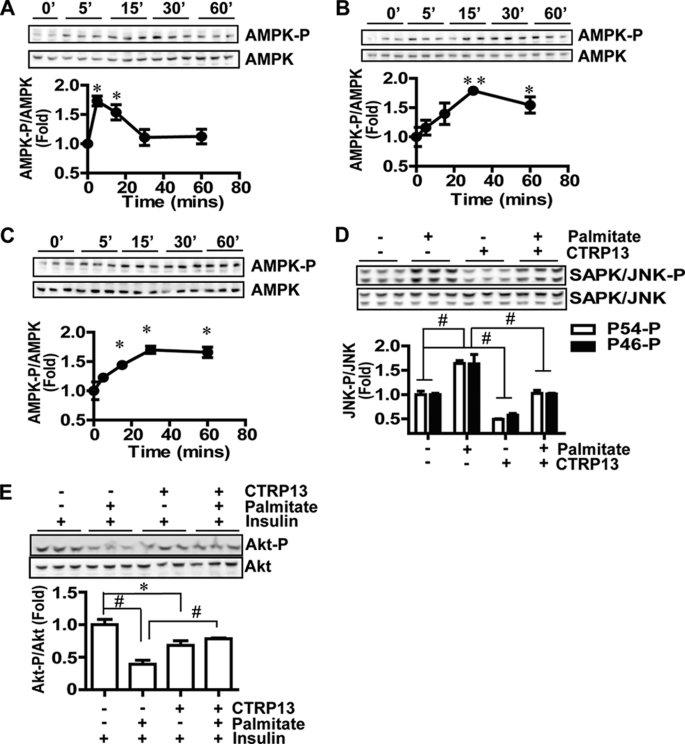
CTRP13 Activates the AMPK Signaling Pathway and Ameliorates Lipid-induced Insulin Resistance
Consistent with our glucose uptake studies (Fig. 6), recombinant CTRP13 significantly activated the AMPK signaling in 3T3-L1 adipocytes (Fig. 7A), L6 myotubes (Fig. 7B), and clone 9 liver cells (Fig. 7C). In all cases, AMPK activation was rapid, reaching maximum phosphorylation at or before 30 min, probably indicating a direct signaling activation by CTRP13. AMPK activation is known to enhance insulin sensitivity (38). Also, the anti-diabetic drug, rosiglitazone, increased CTRP13 expression in adipocytes (Fig. 3E), thus suggesting a possible role for CTRP13 in insulin sensitization. Therefore, we explored whether CTRP13 can decrease insulin resistance associated with obesity. To address this question, we utilized a free fatty acid-induced insulin resistance model in rat H4IIE hepatocytes, a cell type known to be susceptible to lipid-induced insulin resistance (39). Consistent with previous studies, treatment with palmitate, a saturated 16-carbon fatty acid, strongly activates cellular stress signaling, indicated by activation of the SAPK/JNK pathway (Fig. 7D), which is believed to be a mechanism by which free fatty acids induce insulin resistance in cells (39). Treatment with CTRP13 reduced base-line stress signaling and potently suppressed palmitate-induced SAPK/JNK activation in H4IIE hepatocytes (Fig. 7D). As a consequence of insulin resistance, palmitate-treated cells showed a significant reduction in insulin-stimulated phosphorylation of Akt (Fig. 7E). However, CTRP13 relieved palmitate-induced insulin resistance and enhanced insulin-stimulated phosphorylation of Akt in palmitate-treated cells (Fig. 7E). Together, these results indicate that CTRP13 activates the AMPK signaling pathway in multiple cell types and ameliorates free fatty acid-induced insulin resistance at the level of Akt by suppressing the SAPK/JNK stress signaling pathway that interferes with insulin signaling.
FIGURE 7.
Activation of the AMPK and suppression of the SAPK/JNK signaling pathway by CTRP13. A, time course of AMPK activation in 3T3-L1 adipocytes induced by recombinant CTRP13 (5 μg/ml). AMPK-P, AMPKα phosphorylated at Thr-172. B, time course of AMPK activation in L6 myotubes induced by CTRP13 (5 μg/ml). C, time course of AMPK activation in clone 9 liver cells induced by CTRP13 (5 μg/ml). D, suppression of SAPK/JNK stress signaling (phosphorylation at Thr-183/Tyr-185) by CTRP13 (5 μg/ml) in control (BSA) or palmitate (300 μm)-treated H4IIE hepatocytes. H4IIE cells were treated with or without palmitate in 2% free fatty acid-free BSA in the absence or presence of CTRP13 for 5 h. E, enhanced insulin signaling (Akt phosphorylation at Thr-308) by CTRP13 in palmitate (300 μm)-treated H4IIE hepatocytes in response to insulin (10 nm). All data are expressed as -fold change (normalized to control) and as mean ± S.E. (error bars) (n = 3 in all experiments). *, p < 0.05; **, p < 0.01; #, p < 0.001.
CTRP13 Suppresses the Expression of Gluconeogenic Enzymes and Gluconeogenesis
Because CTRP13 ameliorates lipid-induced insulin resistance in H4IIE hepatocytes, we tested whether it also modulates other biological processes in these cells relevant to glucose metabolism. As shown in Fig. 8A, H4IIE hepatocytes treated with recombinant CTRP13 showed a marked reduction in de novo glucose production over time relative to vehicle control-treated cells. Consistent with decreased glucose output, the expression of two key gluconeogenic enzymes, G6Pase and PEPCK-C, were also markedly suppressed by CTRP13 (Fig. 8B). Similar to adipocytes, L6 myotubes, and clone 9 liver cells (Fig. 7), H4IIE cells also demonstrated rapidly activated AMPK signaling in response to CTRP13 treatment (Fig. 8C). Because AMPK activation suppresses hepatic glucose output (40–42), we explored whether AMPK mediates the suppression of gluconeogenesis by CTRP13. Compound C, an AMPK inhibitor, did not affect glucose output when given to H4IIE cells alone (Fig. 8D). However, compound C partially abolished the ability of CTRP13 to inhibit glucose production (Fig. 8D), indicating that AMPK mediates, at least in part, the effect of CTRP13 on gluconeogenesis.
DISCUSSION
CTRP13 is one of the most conserved members of the C1q/TNF family characterized to date. The striking conservation, with 96% amino acid identity in the presumed functional C1q domain of CTRP13 between zebrafish and humans, implies an essential and highly conserved function of CTRP13. In contrast, the widely studied insulin-sensitizing adipokine, adiponectin, shares only 56% amino acid identity at the globular domain with its zebrafish ortholog. Unlike the intensely studied adiponectin, little is known about the function, regulation, and mechanism of action of CTRPs. Table 2 shows the similarities and differences among CTRPs based on recent studies (13–15, 18, 19, 22, 43–45).
TABLE 2.
Comparison of CTRPs
Information was compiled from recent studies (13–15, 18, 19, 22, 43–45). Studies on CTRP4, CTRP11, and CTRP12 have not been published and are not included here. CTRP8 is absent in mice and rats (28) and therefore is not included here. HMW, high molecular weight; ND, not determined; g-CTRP6, truncated globular domain of CTRP6.
| CTRP1 | CTRP2 | CTRP3 | CTRP5 | CTRP6 | CTRP7 | CTRP9 | CTRP10 | CTRP13 | |
|---|---|---|---|---|---|---|---|---|---|
| Primary site of expression | Adipose, placenta | Adipose | Adipose, kidney, testis, stomach | Eye, Testis, adipose | Placenta | Adipose, lung | Adipose | Eye, brain, placenta | Adipose, brain |
| Serum levels in ob/ob mice | ND | ND | Up-regulated | Up-regulated | ND | ND | Up-regulated | ND | Up-regulated |
| Sexual dimorphism in circulating levels | No | No | No | Yes | No | ND | Yes | nd | Yes |
| Rosiglitazone's effect on adipocytes | ND | ND | ND | ND | ND | ND | ND | ND | Up-regulated |
| Forms trimers | Yes | Yes | Yes | Yes | Yes | ND | Yes | Yes | No |
| Forms hexamers | No | No | Yes | Yes | Yes | ND | Yes | Yes | No |
| Forms HMW oligomers | No | No | Yes | Yes | Yes | ND | No | Yes | Yes |
| Forms hetero-oligomers with: | CTRP6 | Adiponectin CTRP7 | None | None | CTRP1 | CTRP2 | Adiponectin | CTRP13 | CTRP10 |
| Functional studies using bacteria-produced protein | No | Yes | No | Yes | Yes (g-CTRP6) | No | No | No | No |
| Functional studies using protein expressed in mammalian cells | Yes | No | Yes | Yes | No | No | Yes | No | Yes |
| Functional studies using protein expressed in Sf9 insect cells | Yes | No | Yes | No | No | No | No | No | No |
| Glucose uptake in myotubes | ND | ND | No | Yes | ND | ND | ND | ND | Yes |
| Fatty acid oxidation in myotubes | ND | Yes | No | Yes | Yes | ND | ND | ND | ND |
| Suppression of gluconeogenesis | ND | ND | Yes | ND | ND | ND | ND | ND | Yes |
| Akt activation | Yes | No | Yes | ND | ND | ND | Yes | ND | No |
| AMPK activation | No | Yes | No | Yes | Yes | ND | Yes | ND | Yes |
| Suppression of lipid-induced JNK signaling | ND | ND | ND | ND | ND | ND | ND | ND | Yes |
| Lower blood glucose in mice | Yes | ND | Yes | No | ND | ND | Yes | ND | ND |
| Antagonize LPS action | ND | ND | Yes | ND | ND | ND | ND | ND | ND |
| Prevent collagen-induced platelet thrombosis in vivo | Yes | ND | ND | ND | ND | ND | ND | ND | ND |
| Increase aldosterone production | Yes | ND | ND | ND | ND | ND | ND | ND | ND |
Of the characterized CTRPs, CTRP13 has the most pronounced sexually dimorphic expression pattern. The higher CTRP13 mRNA and circulating levels in female mice suggest that sex hormones may play a role in regulating the expression of CTRP13 in vivo, as has been reported for adiponectin (46). Aside from CTRP13, CTRP5 and CTRP9 also exhibit sexual dimorphism, with female mice having higher circulating levels of the proteins (14, 15). The functional significance of a sexually dimorphic expression pattern for CTRP13 is unclear. However, males are much more susceptible to the development of obesity and diabetes than are females (47, 48). Our functional studies demonstrate that CTRP13 promotes glucose uptake, suppresses de novo glucose production, and ameliorates lipid-induced insulin resistance; therefore, higher circulating levels of CTRP13 in female mice may partly account for sex differences seen in the development of metabolic abnormalities associated with obesity. Whether a sexually dimorphic CTRP13 expression pattern also exists in humans remains to be determined.
CTRP13 is a secreted multimeric protein, a characteristic shared by all C1q/TNF family proteins that have been biochemically characterized (14–16, 49–57). All CTRPs with a C1q domain form trimers as their basic structural unit (14, 15). In addition, some CTRPs are further assembled into hexamers and higher order structures, similar to the octadecamers found in classical complement C1q (58) and adiponectin (59, 60). As has been shown for adiponectin, different oligomeric forms appear to bind to different receptors (61, 62) and further activate distinct signal transduction pathways (50). Analysis using gel filtration chromatography indicated that CTRP13 forms oligomeric complexes of ≥350 kDa, suggesting an octameric-like or higher order structure. A higher resolution analytical method, such as sedimentation equilibrium centrifugation analysis (59), is needed to determine the number of subunits that comprise the oligomeric form of CTRP13. The presence of the carbohydrate moiety and the formation of disulfide-linked higher order structure underscore the importance of using recombinant protein produced in the mammalian expression system for functional study. Recombinant protein produced in bacteria is unlikely to contain functionally important posttranslational modifications.
CTRP13 forms heteromeric complexes in vitro with a closely related family member, CTRP10. This interaction has been independently confirmed by Iijima et al. (30). Heteromeric complex formations appear to be a characteristic feature of the C1q domain-containing proteins (14, 15, 28, 30, 58, 63, 64) and represent a potential mechanism to generate functionally distinct protein complexes. In most cases, only closely related family members form heteromeric complexes. For instance, CTRP13 and CTRP10 share 89% amino acid identity at the C1q domain while sharing only <40% sequence identity with other CTRPs. Hence, CTRP13 specifically forms heteromeric complexes with CTRP10 but not with other more distantly related CTRPs. Once formed, the heteromeric complex is stable and does not interchange subunits. Whether such an association also occurs in vivo awaits future investigation.
The relevance of CTRP13 to in vivo energy metabolism is indicated by the fact that obese male mice have higher mRNA and circulating levels of CTRP13 compared with lean controls. The beneficial metabolic effects of CTRP13 could partially compensate for the development of insulin resistance. Consistent with this notion, the anti-diabetic drug, rosiglitazone, increased CTRP13 expression in vitro. Recent studies indicate that the insulin-sensitizing effect of thiazolidinediones (e.g. rosiglitazone and pioglitazone) is mediated in part by adiponectin; the full pharmacologic effect of rosiglitazone in lowering blood glucose is blunted in adiponectin knock-out mice (10, 65). Further, depending on the dose used, there appears to be an adiponectin-dependent and -independent mechanism by which pioglitazone exerts its effect in vivo (65). CTRP13 may be one of the molecular components mediating the adiponectin-independent, insulin-sensitizing effect of pioglitazone.
In obesity, ectopic deposition of fatty acids in non-adipose tissues, such as liver and muscle, has been proposed as a mechanism underlying insulin resistance in these tissues (67). Elevated intracellular free fatty acids and their derivatives (e.g. ceramide) activate stress signaling pathways, such as the SAPK/JNK pathway (67). Activation of SAPK/JNK impairs signal transduction downstream of the insulin receptor (39, 68). Importantly, we show that CTRP13 can ameliorate insulin resistance by lowering basal levels of SAPK/JNK signaling in H4IIE hepatocytes and further suppresses this stress signaling pathway induced by saturated fatty acids. This novel aspect of CTRP13 function, not previously described for any other CTRPs, underscores the potential of CTRP13 to improve insulin sensitivity in disease conditions where insulin resistance is present.
We have shown recently that hepatocytes are the major target cells of CTRP3 action in vitro and in vivo (20). Mechanistically, CTRP3 regulates hepatic glucose output via activation of the Akt signaling. The Akt signaling pathway, downstream of PI3K, is known to mediate the inhibition of gluconeogenesis by insulin (34) through regulation of the Forkhead box O (FoxO1) transcription factor (69). Phosphorylation of FoxO1 by Akt leads to its nuclear exclusion, thereby turning off gluconeogenic gene transcription regulated by FoxO1. In contrast, CTRP13 suppresses gluconeogenesis in hepatocytes via activation of the AMPK signaling pathway. The AMPK signaling pathway, activated in response to elevated cellular AMP/ATP ratio or by upstream kinases (e.g. LKB1), has been shown to inhibit gluconeogenesis through phosphorylation and nuclear exclusion of CREB-regulated transcription factor coactivator 2 (CRTC2) (70). Although CTRP13 and CTRP3 differ in tissue expression profiles and share only limited amino acid identity (35%) at the presumed functional C-terminal globular domain, they share similar function in regulating de novo glucose synthesis. However, they do so by activating two distinct signaling pathways.
Liver is a major target tissue of CTRP3 in mice (20). Consistent with this, CTRP3 failed to elicit metabolic changes in myotubes and adipocytes in vitro. In contrast, CTRP13 activates the AMPK signaling pathway to promote glucose uptake in myotubes and adipocytes, suggesting that CTRP13 may regulate glucose metabolism in skeletal muscle and adipose tissue in addition to liver. Hepatic glucose output and glucose disposal in peripheral tissues are the two major mechanisms employed by mammals to control blood glucose levels. Therefore, activating distinct signaling pathways and targeting different metabolic tissues may represent important functional differences between CTRP3 and CTRP13. Although the receptor(s) for CTRP3 and CTRP13 have not been identified, it is likely that they interact with distinct receptors to activate the Akt and AMPK signaling pathway in cells, respectively.
Besides CTRP13, recent studies show that CTRP5 and CTRP6 can also activate the AMPK signaling pathway to promote glucose uptake and fatty acid oxidation (18, 19). Although bacterially produced recombinant GST-CTRP5 fusion protein can activate the AMPK signaling pathway in mouse myotubes (18), its prominent expression in the eye suggests a primary role in the visual system (14, 45). Indeed, mutation in the CTRP5 gene has been found to cause late onset macular degeneration in humans (45, 71). The missense mutation (S163R) identified in the globular C1q domain of human CTRP5 causes the protein to form abnormal high molecular weight aggregate (71). Aside from the defect in the eye, no metabolic abnormality has been reported for patients with a CTRP5 mutation (45, 71). Also, consistent with its role in the visual system, injection of recombinant CTRP5, produced in mammalian cells and thus possessing all of the proper posttranslational modifications (e.g. proline hydroxylation within the collagen domain) and disulfide-linked higher order oligomeric structures failed to elicit any changes in blood glucose levels in mice (14). It is not known whether GST-CTRP5 fusion protein produced in bacteria can be correctly assembled into proper trimers and higher order oligomers. As such, the physiological relevance of CTRP5 in regulating glucose metabolism in vivo remains to be fully established.
Bacterially produced g-CTRP6, consisting of just the C-terminal globular domain, can also activate the AMPK signaling pathway to enhance fatty acid oxidation in mouse myotubes (19). Because the full-length protein does not express well in bacteria (19), it remains to be determined whether the full-length protein has the same function as the truncated g-CTRP6. When expressed in mammalian HEK293 cells, secreted CTRP6 contains N-linked glycans. It forms trimer as its basic structural unit, and the trimeric protein can be further assembled into presumed hexamers and high molecular weight oligomers via intermolecular disulfide bonds (14). It is not known whether g-CTRP6 produced in bacteria can be properly assembled into correct higher order structure. Moreover, CTRP6 mRNA is primarily expressed in mouse placenta (14), suggesting a possible role in reproductive biology. Thus, the physiological significance of g-CTRP6 in regulating fatty acid metabolism in skeletal muscle remains to be fully established. In contrast to CTRP5 and CTRP6, CTRP13 is primarily expressed in adipose tissue and the brain, and the expression of CTRP13 mRNA and protein is modulated by the metabolic state of the animals.
In sum, we provide evidence that CTRP13 is a novel and highly conserved adipokine that promotes glucose uptake and suppresses gluconeogenesis. This secreted multimeric protein improves insulin action in hepatocytes by suppressing the basal and lipid-induced stress signaling that interferes with insulin signaling. Future studies aimed at elucidating the in vivo action of CTRP13 in normal and pathophysiologic conditions will probably yield novel insights into its function and mechanisms of action. Such knowledge may provide new avenues to treat metabolic diseases associated with diabetes.
Acknowledgment
We thank Dr. Peter Pedersen (Department of Biological Chemistry, Johns Hopkins University School of Medicine) for providing the rat clone 9 liver cell line.
This work was supported, in whole or in part, by National Institutes of Health Grant DK084171 (to G. W. W.). This work was also supported in part by American Heart Association Grant SDG2260721 and Baltimore Diabetes Research and Training Center Grant P60DK079637.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) EU399230.
G. W. Wong, unpublished results.
- CTRP
- C1q/TNF-related protein
- AICAR
- aminoimidazole carboxamide ribonucleotide
- AMPK
- AMP-activated kinase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SVF
- stromal vascular fraction
- G6Pase
- glucose-6-phosphatase
- PEPCK-C
- phosphoenolpyruvate carboxykinase (GTP) (cytosolic).
REFERENCES
- 1. Scherer P. E. (2006) Diabetes 55, 1537–1545 [DOI] [PubMed] [Google Scholar]
- 2. Shetty S., Kusminski C. M., Scherer P. E. (2009) Trends Pharmacol. Sci. 30, 234–239 [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J. M. (1994) Nature 372, 425–432 [DOI] [PubMed] [Google Scholar]
- 4. Scherer P. E., Williams S., Fogliano M., Baldini G., Lodish H. F. (1995) J. Biol. Chem. 270, 26746–26749 [DOI] [PubMed] [Google Scholar]
- 5. Hu E., Liang P., Spiegelman B. M. (1996) J. Biol. Chem. 271, 10697–10703 [DOI] [PubMed] [Google Scholar]
- 6. Maeda K., Okubo K., Shimomura I., Funahashi T., Matsuzawa Y., Matsubara K. (1996) Biochem. Biophys. Res. Commun. 221, 286–289 [DOI] [PubMed] [Google Scholar]
- 7. Ma K., Cabrero A., Saha P. K., Kojima H., Li L., Chang B. H., Paul A., Chan L. (2002) J. Biol. Chem. 277, 34658–34661 [DOI] [PubMed] [Google Scholar]
- 8. Maeda N., Shimomura I., Kishida K., Nishizawa H., Matsuda M., Nagaretani H., Furuyama N., Kondo H., Takahashi M., Arita Y., Komuro R., Ouchi N., Kihara S., Tochino Y., Okutomi K., Horie M., Takeda S., Aoyama T., Funahashi T., Matsuzawa Y. (2002) Nat. Med. 8, 731–737 [DOI] [PubMed] [Google Scholar]
- 9. Kubota N., Terauchi Y., Yamauchi T., Kubota T., Moroi M., Matsui J., Eto K., Yamashita T., Kamon J., Satoh H., Yano W., Froguel P., Nagai R., Kimura S., Kadowaki T., Noda T. (2002) J. Biol. Chem. 277, 25863–25866 [DOI] [PubMed] [Google Scholar]
- 10. Nawrocki A. R., Rajala M. W., Tomas E., Pajvani U. B., Saha A. K., Trumbauer M. E., Pang Z., Chen A. S., Ruderman N. B., Chen H., Rossetti L., Scherer P. E. (2006) J. Biol. Chem. 281, 2654–2660 [DOI] [PubMed] [Google Scholar]
- 11. Yano W., Kubota N., Itoh S., Kubota T., Awazawa M., Moroi M., Sugi K., Takamoto I., Ogata H., Tokuyama K., Noda T., Terauchi Y., Ueki K., Kadowaki T. (2008) Endocr. J. 55, 515–522 [DOI] [PubMed] [Google Scholar]
- 12. Davis K. E., Scherer P. E. (2008) Biochem. J. 416, e7–e9 [DOI] [PubMed] [Google Scholar]
- 13. Wong G. W., Wang J., Hug C., Tsao T. S., Lodish H. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong G. W., Krawczyk S. A., Kitidis-Mitrokostas C., Revett T., Gimeno R., Lodish H. F. (2008) Biochem. J. 416, 161–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong G. W., Krawczyk S. A., Kitidis-Mitrokostas C., Ge G., Spooner E., Hug C., Gimeno R., Lodish H. F. (2009) FASEB J. 23, 241–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kishore U., Gaboriaud C., Waters P., Shrive A. K., Greenhough T. J., Reid K. B., Sim R. B., Arlaud G. J. (2004) Trends Immunol. 25, 551–561 [DOI] [PubMed] [Google Scholar]
- 17. Shapiro L., Scherer P. E. (1998) Curr. Biol. 8, 335–338 [DOI] [PubMed] [Google Scholar]
- 18. Park S. Y., Choi J. H., Ryu H. S., Pak Y. K., Park K. S., Lee H. K., Lee W. (2009) J. Biol. Chem. 284, 27780–27789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee W., Kim M. J., Park E. J., Choi Y. J., Park S. Y. (2010) FEBS Lett. 584, 968–972 [DOI] [PubMed] [Google Scholar]
- 20. Peterson J. M., Wei Z., Wong G. W. (2010) J. Biol. Chem. 285, 39691–39701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akiyama H., Furukawa S., Wakisaka S., Maeda T. (2007) Mol. Cell Biochem. 304, 243–248 [DOI] [PubMed] [Google Scholar]
- 22. Kopp A., Bala M., Buechler C., Falk W., Gross P., Neumeier M., Schölmerich J., Schäffler A. (2010) Endocrinology 151, 5267–5278 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Lam K. S., Chan L., Chan K. W., Lam J. B., Lam M. C., Hoo R. C., Mak W. W., Cooper G. J., Xu A. (2006) J. Biol. Chem. 281, 16391–16400 [DOI] [PubMed] [Google Scholar]
- 24. Richards A. A., Stephens T., Charlton H. K., Jones A., Macdonald G. A., Prins J. B., Whitehead J. P. (2006) Mol. Endocrinol. 20, 1673–1687 [DOI] [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Gershoni J. M., Bayer E. A., Wilchek M. (1985) Anal. Biochem. 146, 59–63 [DOI] [PubMed] [Google Scholar]
- 27. Weinstein I. B., Orenstein J. M., Gebert R., Kaighn M. E., Stadler U. C. (1975) Cancer Res. 35, 253–263 [PubMed] [Google Scholar]
- 28. Peterson J. M., Wei Z., Wong G. W. (2009) Biochem. Biophys. Res. Commun. 388, 360–365 [DOI] [PubMed] [Google Scholar]
- 29. Koide T., Aso A., Yorihuzi T., Nagata K. (2000) J. Biol. Chem. 275, 27957–27963 [DOI] [PubMed] [Google Scholar]
- 30. Iijima T., Miura E., Watanabe M., Yuzaki M. (2010) Eur. J. Neurosci. 31, 1606–1615 [DOI] [PubMed] [Google Scholar]
- 31. Flier J. S., Cook K. S., Usher P., Spiegelman B. M. (1987) Science 237, 405–408 [DOI] [PubMed] [Google Scholar]
- 32. Steppan C. M., Bailey S. T., Bhat S., Brown E. J., Banerjee R. R., Wright C. M., Patel H. R., Ahima R. S., Lazar M. A. (2001) Nature 409, 307–312 [DOI] [PubMed] [Google Scholar]
- 33. Hotamisligil G. S., Shargill N. S., Spiegelman B. M. (1993) Science 259, 87–91 [DOI] [PubMed] [Google Scholar]
- 34. Saltiel A. R., Kahn C. R. (2001) Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]
- 35. Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. (1994) J. Biol. Chem. 269, 5241–5248 [PubMed] [Google Scholar]
- 36. Shetty M., Loeb J. N., Vikstrom K., Ismail-Beigi F. (1993) J. Biol. Chem. 268, 17225–17232 [PubMed] [Google Scholar]
- 37. Sullivan J. E., Brocklehurst K. J., Marley A. E., Carey F., Carling D., Beri R. K. (1994) FEBS Lett. 353, 33–36 [DOI] [PubMed] [Google Scholar]
- 38. Kahn B. B., Alquier T., Carling D., Hardie D. G. (2005) Cell Metab. 1, 15–25 [DOI] [PubMed] [Google Scholar]
- 39. Nakamura S., Takamura T., Matsuzawa-Nagata N., Takayama H., Misu H., Noda H., Nabemoto S., Kurita S., Ota T., Ando H., Miyamoto K., Kaneko S. (2009) J. Biol. Chem. 284, 14809–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iglesias M. A., Ye J. M., Frangioudakis G., Saha A. K., Tomas E., Ruderman N. B., Cooney G. J., Kraegen E. W. (2002) Diabetes 51, 2886–2894 [DOI] [PubMed] [Google Scholar]
- 41. Lochhead P. A., Salt I. P., Walker K. S., Hardie D. G., Sutherland C. (2000) Diabetes 49, 896–903 [DOI] [PubMed] [Google Scholar]
- 42. Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., Eto K., Akanuma Y., Froguel P., Foufelle F., Ferre P., Carling D., Kimura S., Nagai R., Kahn B. B., Kadowaki T. (2002) Nat. Med. 8, 1288–1295 [DOI] [PubMed] [Google Scholar]
- 43. Lasser G., Guchhait P., Ellsworth J. L., Sheppard P., Lewis K., Bishop P., Cruz M. A., Lopez J. A., Fruebis J. (2006) Blood 107, 423–430 [DOI] [PubMed] [Google Scholar]
- 44. Jeon J. H., Kim K. Y., Kim J. H., Baek A., Cho H., Lee Y. H., Kim J. W., Kim D., Han S. H., Lim J. S., Kim K. I., Yoon do Y., Kim S. H., Oh G. T., Kim E., Yang Y. (2008) FASEB J. 22, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 45. Ayyagari R., Mandal M. N., Karoukis A. J., Chen L., McLaren N. C., Lichter M., Wong D. T., Hitchcock P. F., Caruso R. C., Moroi S. E., Maumenee I. H., Sieving P. A. (2005) Invest. Ophthalmol. Vis. Sci. 46, 3363–3371 [DOI] [PubMed] [Google Scholar]
- 46. Combs T. P., Berg A. H., Rajala M. W., Klebanov S., Iyengar P., Jimenez-Chillaron J. C., Patti M. E., Klein S. L., Weinstein R. S., Scherer P. E. (2003) Diabetes 52, 268–276 [DOI] [PubMed] [Google Scholar]
- 47. Gale E. A., Gillespie K. M. (2001) Diabetologia 44, 3–15 [DOI] [PubMed] [Google Scholar]
- 48. Ding E. L., Song Y., Malik V. S., Liu S. (2006) JAMA 295, 1288–1299 [DOI] [PubMed] [Google Scholar]
- 49. Pajvani U. B., Du X., Combs T. P., Berg A. H., Rajala M. W., Schulthess T., Engel J., Brownlee M., Scherer P. E. (2003) J. Biol. Chem. 278, 9073–9085 [DOI] [PubMed] [Google Scholar]
- 50. Tsao T. S., Tomas E., Murrey H. E., Hug C., Lee D. H., Ruderman N. B., Heuser J. E., Lodish H. F. (2003) J. Biol. Chem. 278, 50810–50817 [DOI] [PubMed] [Google Scholar]
- 51. Waki H., Yamauchi T., Kamon J., Ito Y., Uchida S., Kita S., Hara K., Hada Y., Vasseur F., Froguel P., Kimura S., Nagai R., Kadowaki T. (2003) J. Biol. Chem. 278, 40352–40363 [DOI] [PubMed] [Google Scholar]
- 52. Colombatti A., Doliana R., Bot S., Canton A., Mongiat M., Mungiguerra G., Paron-Cilli S., Spessotto P. (2000) Matrix Biol. 19, 289–301 [DOI] [PubMed] [Google Scholar]
- 53. Hayward C. P., Warkentin T. E., Horsewood P., Kelton J. G. (1991) Blood 77, 2556–2560 [PubMed] [Google Scholar]
- 54. Bao D., Pang Z., Morgan J. I. (2005) J. Neurochem. 95, 618–629 [DOI] [PubMed] [Google Scholar]
- 55. Kvansakul M., Bogin O., Hohenester E., Yayon A. (2003) Matrix Biol. 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 56. Mongiat M., Mungiguerra G., Bot S., Mucignat M. T., Giacomello E., Doliana R., Colombatti A. (2000) J. Biol. Chem. 275, 25471–25480 [DOI] [PubMed] [Google Scholar]
- 57. Bogin O., Kvansakul M., Rom E., Singer J., Yayon A., Hohenester E. (2002) Structure 10, 165–173 [DOI] [PubMed] [Google Scholar]
- 58. Reid K. B., Porter R. R. (1976) Biochem. J. 155, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suzuki S., Wilson-Kubalek E. M., Wert D., Tsao T. S., Lee D. H. (2007) FEBS Lett. 581, 809–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Briggs D. B., Jones C. M., Mashalidis E. H., Nuñez M., Hausrath A. C., Wysocki V. H., Tsao T. S. (2009) Biochemistry 48, 12345–12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M., Murakami K., Ohteki T., Uchida S., Takekawa S., Waki H., Tsuno N. H., Shibata Y., Terauchi Y., Froguel P., Tobe K., Koyasu S., Taira K., Kitamura T., Shimizu T., Nagai R., Kadowaki T. (2003) Nature 423, 762–769 [DOI] [PubMed] [Google Scholar]
- 62. Hug C., Wang J., Ahmad N. S., Bogan J. S., Tsao T. S., Lodish H. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10308–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iijima T., Miura E., Matsuda K., Kamekawa Y., Watanabe M., Yuzaki M. (2007) Eur. J. Neurosci. 25, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 64. Illidge C., Kielty C., Shuttleworth A. (2001) Int. J. Biochem. Cell Biol. 33, 521–529 [DOI] [PubMed] [Google Scholar]
- 65. Kubota N., Terauchi Y., Kubota T., Kumagai H., Itoh S., Satoh H., Yano W., Ogata H., Tokuyama K., Takamoto I., Mineyama T., Ishikawa M., Moroi M., Sugi K., Yamauchi T., Ueki K., Tobe K., Noda T., Nagai R., Kadowaki T. (2006) J. Biol. Chem. 281, 8748–8755 [DOI] [PubMed] [Google Scholar]
- 66. Deleted in proof.
- 67. Samuel V. T., Petersen K. F., Shulman G. I. (2010) Lancet 375, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002) Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- 69. Gross D. N., van den Heuvel A. P., Birnbaum M. J. (2008) Oncogene 27, 2320–2336 [DOI] [PubMed] [Google Scholar]
- 70. Cheng A., Saltiel A. R. (2006) BioEssays 28, 231–234 [DOI] [PubMed] [Google Scholar]
- 71. Hayward C., Shu X., Cideciyan A. V., Lennon A., Barran P., Zareparsi S., Sawyer L., Hendry G., Dhillon B., Milam A. H., Luthert P. J., Swaroop A., Hastie N. D., Jacobson S. G., Wright A. F. (2003) Hum. Mol. Genet. 12, 2657–2667 [DOI] [PubMed] [Google Scholar]
- 72. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 73. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]