Abstract
The transmembrane adaptor protein Cbp (or PAG1) functions as a suppressor of Src-mediated tumor progression by promoting the inactivation of Src. The expression of Cbp is down-regulated in Src-transformed cells and in various human cancer cells, suggesting a potential role for Cbp as a tumor suppressor. However, the mechanisms underlying the down-regulation of Cbp remain unknown. The present study shows that Cbp expression is down-regulated by epigenetic histone modifications via the MAPK/PI3K pathway. In mouse embryonic fibroblasts, transformation by oncogenic Src and Ras induced a marked down-regulation of Cbp expression. The levels of Cbp expression were inversely correlated with the activity of MEK and Akt, and Cbp down-regulation was suppressed by inhibiting MEK and PI3K. Src transformation did not affect the stability of Cbp mRNA, the transcriptional activity of the cbp promoter, or the DNA methylation status of the cbp promoter CpG islands. However, Cbp expression was restored by treatment with histone deacetylase (HDAC) inhibitors and by siRNA-mediated knockdown of HDAC1/2. Src transformation significantly decreased the acetylation levels of histone H4 and increased the trimethylation levels of histone H3 lysine 27 in the cbp promoter. EGF-induced Cbp down-regulation was also suppressed by inhibiting MEK and HDAC. Furthermore, the inhibition of MEK or HDAC restored Cbp expression in human cancer cells harboring Cbp down-regulation through promoter hypomethylation. These findings suggest that Cbp down-regulation is primarily mediated by epigenetic histone modifications via oncogenic MAPK/PI3K pathways in a subset of cancer cells.
Keywords: Adaptor Proteins, DNA Methylation, Epigenetics, Gene Expression, Histone Deacetylase, MAP Kinases (MAPKs), Ras, Receptor Tyrosine Kinase, Src, Transformation
Introduction
Cbp (Csk-binding protein),2 also known as PAG1, is a transmembrane adaptor protein composed of a short extracellular domain and a longer cytoplasmic domain (1, 2). The cytoplasmic domain of Cbp contains palmitoylation sites that are required for localization to a cholesterol-enriched membrane microdomain and multiple tyrosine residues that can be phosphorylated by Src family kinases (SFKs). Upon phosphorylation by SFKs, Cbp associates with C-terminal Src kinase (Csk), a negative regulator of SFKs, through a specific site (Tyr-317 in humans) and brings it into proximity with membrane-associated SFKs. Csk then phosphorylates the C-terminal negative regulatory tyrosine residue of SFKs, which suppresses their activation. Thus, Cbp acts as a scaffold in the Csk-mediated negative regulation of SFKs. The role of Cbp has been shown in the immune system (2, 3), in cell spreading and migration (4), and in EGF-induced cell transformation (5).
SFKs are membrane-associated non-receptor protein tyrosine kinases that play pivotal roles in regulating various cellular processes including proliferation, differentiation, adhesion, migration, and survival (6). SFKs are comprised of eight members, namely c-Src, Fyn, c-Yes, Lyn, Lck, Hck, c-Fgr, and Blk in mammals. Among these, c-Src, Fyn, and c-Yes are ubiquitously expressed, whereas the others are expressed mainly in the immune system. In a variety of human cancers, protein levels, and/or specific activities of c-Src and c-Yes are frequently up-regulated. Up-regulation of Lyn, Lck, Hck, c-Fgr, and Blk is also observed in some leukemias and lymphomas (6, 7). These observations implicate a role for SFKs in cell transformation, tumorigenesis, and metastasis (8). However, because SFK genes are rarely mutated in human cancer (9, 10), the mechanisms underlying their up-regulation in these cancers remain unclear.
We previously reported that Cbp expression is markedly down-regulated by Src-mediated cell transformation and in some human cancer cells, and that the re-expression of Cbp efficiently suppresses Src transformation and tumorigenesis (11). The same study showed that Cbp suppresses Src function by directly sequestering activated Src in the membrane microdomain (11), and that Cbp can commonly serve as a suppressor for other SFK members (12). In addition, Cbp-deficient fibroblasts are more sensitive to Src transformation than normal cells (11). These findings suggest that the cbp gene serves as a tumor suppressor gene in a subset of cancers, particularly those harboring SFK up-regulation. However, the mechanisms underlying Cbp down-regulation in cancer cells remain unknown.
To elucidate the mechanisms that mediate the down-regulation of Cbp expression, the potential contribution of the oncogenic signaling pathway acting downstream of Src and Ras, was examined based on prior findings showing that Ras- and EGF-mediated transformation can also induce Cbp down-regulation. The present study shows that Cbp is down-regulated by an epigenetic mechanism involving the deacetylation/methylation of histones, but not DNA methylation, in the cbp promoter via the activation of MAPK/PI3K pathways.
EXPERIMENTAL PROCEDURES
Cell Culture
Csk-deficient (Csk−/−) mouse embryonic fibroblasts (MEFs) and wild-type cells (Csk+/+) were kindly donated by Akira Imamoto (13). HT-29 and MCF7 cells were obtained from the American Type Culture Collection (ATCC). A549 cells were a kind gift from Dr. Masuo Yutsudo. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Retroviral-mediated Gene Transfer
All of the gene transfer experiments were carried out using the pCX4 series of retroviral vectors (14). Retroviral vectors carrying wild-type chicken c-Src, v-Src, H-Ras G12V, LA-SDSE MEK, and Myr-Akt were kindly provided by Dr. Tsuyoshi Akagi (Osaka Bioscience Institute, Osaka). The K-Ras G12D vector was kindly provided by Masuo Yutsudo (Osaka University). Wild-type rat Csk was subcloned into the retroviral vector pCX4bleo. Wild-type rat MEK, its constitutively active mutant and a kinase-deficient mutant were subcloned into pCX4puro. A PCR-based procedure was used to generate the MEK mutants. Human HDAC1 cDNA was cloned by PCR and subcloned into a retroviral vector pCX4bsr. The production of retroviral vectors and their infection were performed as described previously (14).
siRNA
The siRNA sequences for mouse HDAC1/2/3 genes used are as follows: 5′-GAA CUC UUC UAA CUU CAA A-3′, 5′-UGA CCA ACC AGA ACA CUA A-3′, 5′-UCA AAG AAG AGG UCA AGU U-3′, and 5′-AUA AAC GCA UUG CCU GUG A-3′ (for HDAC1); 5′-CAA AAG UGA UGG AGA UGU A-3′, 5′-ACA GGA GAC UUG AGG GAU A, 5′-CAA UUG GGC UGG AGG ACU A-3′, and 5′-CCA AUG AGU UGC CAU AUA A-3′ (for HDAC2); 5′-GGG AAU GUG UUG AAU AUG U-3′, 5′-CGG CAG ACC UCC UGA CGU A-3′, 5′-AAG UUG AUG UGG AGA UUU A-3′, 5′-GCA CCC GCAU CGA GAA UCA-3′ (for HDAC3) (ON-TARGET plus SMART pool, Thermo). siRNA was introduced with Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen).
Immunochemical Analysis
Cells were lysed in n-octyl-β-d-glucoside (ODG) buffer (25 mm Tris-HCl, pH 7.4, 1 mm EDTA, pH 7.4, 150 mm NaCl, 5% glycerol, 1 mm sodium orthovanadate, 1% Nonidet P-40, 2% ODG, 5 mm β-mercaptoethanol, 50 mm NaF, 1 mm PMSF, 10 mg/ml aprotinin and leupeptin), and immunoblotting was performed as described previously (11). The following antibodies were used: anti-Src pY418 (Invitrogen), anti-K-Ras (Santa Cruz Biotechnology), anti-H-Ras (Calbiochem), anti-MEK (Cell Signaling), anti-ERK (Cell Signaling), anti-ERK pT202/Y204 (Cell Signaling), anti-AKT (Cell Signaling), anti-GSK-3β (Cell Signaling), anti-GSK-3β pS9 (Cell Signaling), and anti-β-tubulin (Santa Cruz Biotechnology). Anti-Cbp antibody was generated as described previously (1).
Real-Time PCR Analysis
Total RNA was prepared using Sepasol (Nacalai Tesque, Kyoto) and reverse transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-Time PCR was performed as described (27). Relative values of the ratio of Cbp mRNA to a control GAPDH mRNA are evaluated in all the experiments. Gene-specific primers for mouse Cbp (ID: Mm00474700_m1), GAPDH (ID: Mm99999915_g1), human Cbp (ID: Hs00179693_m1*) and GAPDH (ID: Hs99999905_m1) were obtained from Applied Biosystems.
Reporter Constructs and Luciferase Assay
The mouse cbp promoter sequence (−3000/+50) and the GAPDH promoter sequence (−1000/+50) were amplified by PCR from the mouse genome extracted from MEFs and then subcloned into the luciferase reporter plasmid PGV-B2 (Toyo Ink, Tokyo). The cbp promoter deletion series was generated by a PCR-based procedure. For luciferase assays, cells were co-transfected with a promoter construct along with the control plasmid pRL-TK (Toyo Ink, Tokyo). Cells were harvested 24h after transfection and luciferase activities were measured using PicaGene DualSeaPansy Luminescence kit (Toyo Ink, Tokyo).
Bisulfite Sequencing
Sodium bisulfite modification of genomic DNA was conducted using the EZ DNA Methylation kit (Zymo Research) according to the manufacturer's instructions. Bisulfite-treated DNA was used as the template for PCR. Primers used for amplification were as follows: 5′-GTT TTA GTT TTT TGT TTT GTA GTT GGT AGT-3′ and 5′-AAA AAA CAA TCA CAA CAC CC-3′ (for mouse); and 5′-AGT TTT GGG TTT ATA AAA TTA GGG TAG-3′ and 5′-TCA ATT AAA AAC CTA CCA AAA AAA A-3′ (for human). Amplified products were subcloned using the TOPO-TA cloning system (Invitrogen). Plasmid DNAs of 10 insert-positive clones were isolated and sequenced.
Chromatin Immunoprecipitation
ChIP assays were performed following the protocol outlined by the manufacturer (Upstate) with minor modifications. Briefly, cells were fixed in culture medium with formaldehyde (final concentration of 1%) for 10 min. After washing with cold PBS, cells were collected by centrifugation and resuspended in SDS lysis buffer (50 mm Tris-HCl (pH 8.1), 10 mm EDTA, 1% SDS, 1 mm PMSF, 10 mg/ml aprotinin and leupeptin) and incubated for 10 min on ice. DNA was sheared into 200–1000bp fragments by sonication. The fragments were immunoprecipitated with 2 μg of antibody against acetyl-histone H3 (Upstate), acetyl-histone H4 (Upstate), histone H3 (Cell Signaling), H3K9me3 (ACTIVE MOTIF), and H3K27me3 (ACTIVE MOTIF). DNA extraction was performed using Wizard SV Gel and PCR Clean-Up System (Promega). The samples were analyzed by Real-Time PCR using SYBR Premix Ex TaqGC (TAKARA BIO, Ohtsu). Primers used for amplification were as follows: 5′-CTG GGA CCC TAA GGC AGT TC-3′ and 5′-AGG GCT GGT GAC CAA AAA CT-3′.
RESULTS
Activation of the ERK MAPK/PI3K Pathway Down-regulates Cbp Expression
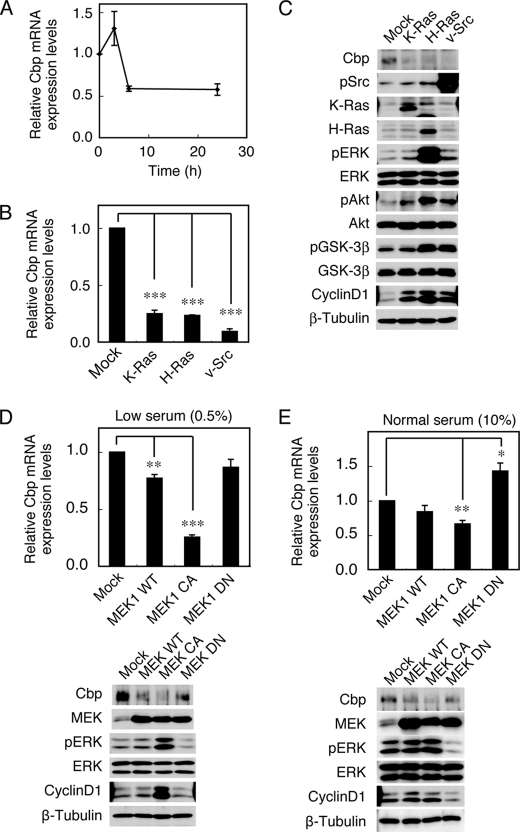
A previous study revealed that Cbp protein and mRNA expression were down-regulated by Src transformation (11). To address the mechanisms underlying Cbp down-regulation, we first examined the effects of serum stimulation on Cbp expression. MEFs were cultured in the absence of serum overnight, followed by stimulation with 10% FBS for 24 h. Although there was a slight up-regulation of Cbp mRNA expression within 3 h after stimulation, Cbp expression was decreased by about 50% within 6 h after stimulation and the reduced levels were sustained up to 24 h (Fig. 1A). This suggests that the expression of Cbp can be regulated through growth-promoting signaling pathways activated by serum. To define the pathways, the effects of introducing K-Ras G12D, H-Ras G12V, and v-Src, which are oncoproteins that activate growth-promoting signals, were examined. MEFs expressing these molecules were cultured under low (0.5%) serum conditions, and the expression levels of Cbp mRNA (Fig. 1B) and protein (Fig. 1C) as well as the activity status of signaling molecules were determined (Fig. 1C). The expression of all these molecules induced a dramatic reduction of Cbp expression. In these cells, there was significant activation of ERK and Akt. The expression of cyclin D1, which reflects ERK activation in the nucleus, was also induced by the expression of these oncogenic molecules. These observations suggest that Cbp down-regulation correlates with the activation of the growth-promoting, oncogenic signaling pathways, namely the ERK MAPK and/or Akt pathways.
FIGURE 1.
Activation of the ERK MAPK pathway down-regulates Cbp expression. A, MEFs were cultured overnight in the absence of serum, followed by stimulation with 10% FBS for 24 h. Expression levels of Cbp mRNA were determined by real-time PCR at the time indicated. Relative values of the ratio of Cbp mRNA to a control GAPDH mRNA are shown. B, MEFs expressing K-Ras G12D, H-Ras G12V or v-Src were cultured under low (0.5%) serum conditions for 16 h, and analyzed for Cbp mRNA expression levels by real-time PCR. C, total cell lysates were immunoblotted with the antibodies indicated. D, MEFs expressing MEK1 WT, MEK1 CA or MEK DN were cultured under low (0.5%) serum conditions for 16 h. Cbp mRNA expression levels were analyzed by real-time PCR (upper), and total cell lysates were immunoblotted with the antibodies indicated (lower). E, MEFs used in D were cultured under normal (10%) serum conditions. Cbp mRNA expression levels were analyzed by real-time PCR (upper), and total cell lysates were immunoblotted with the antibodies indicated (lower). Relative values ± S.D. were obtained from three independent assays. *, p < 0.05; **, p < 0.01; ***, p < 0.001, by Student's t test.
To evaluate the contribution of the ERK MAPK pathway, Cbp expression was examined in MEFs overexpressing wild-type MEK1 (MEK1 WT), a constitutively active mutant with a substitution of Ser-218/222 for Asp (MEK1 CA) or a kinase-deficient mutant with a substitution of Ser-218 for Ala (MEK1 DN) under low serum conditions (Fig. 1D). The overexpression of MEK WT and MEK1 CA significantly down-regulated Cbp expression, while that of MEK1 DN did not. The expression levels of Cbp correlated with the activity status of ERK (Fig. 1D, lower panels). Because the negative effect of MEK1 DN was thought to be due to the low level of basal ERK activity under low serum conditions, the effect of MEK1 DN was re-examined under high (10%) serum conditions (Fig. 1E). Under these conditions, the expression of MEK1 DN successfully suppressed ERK activity and induced an apparent up-regulation of Cbp expression. Overall, the tight correlation between Cbp mRNA expression and ERK MAPK pathway activity suggests that the Cbp down-regulation is at least partially mediated by the ERK MAPK pathway.
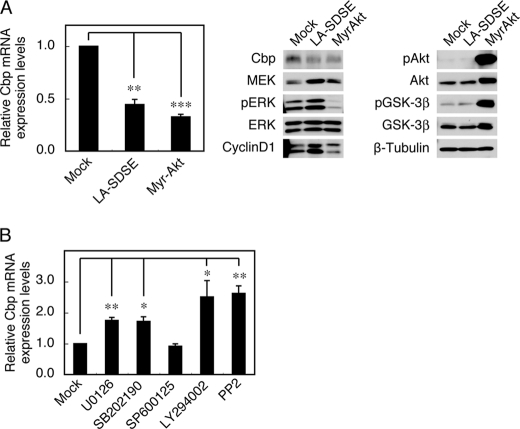
In support of the above possibility, another form of the constitutively active mutant of MEK (LA-SDSE) was able to significantly suppress Cbp expression (Fig. 2A). The potential contribution of the Akt pathway was also examined by introducing a constitutively active Akt (Myr-Akt). In agreement with the activation of Akt, Cbp expression was dramatically down-regulated under these conditions, even though there was no activation of the ERK MAPK pathway (Fig. 2A). We also found that the treatment of MEFs with a MEK inhibitor (U0126) and a p38 MAPK inhibitor (SB202190), but not a JNK inhibitor (SP600125), significantly canceled the repression of Cbp expression (Fig. 2B); this indicates that the p38 MAPK pathway can also participate in Cbp down-regulation. Furthermore, treatment with a PI3K inhibitor (LY294002) effectively restored Cbp expression to a level similar to that gained with a Src inhibitor (PP2). These findings show that Cbp down-regulation can be achieved through multiple signaling pathways that include the ERK MAPK, p38 MAPK, and PI3K-Akt pathways, all of which are located downstream of the Src and Ras oncoproteins.
FIGURE 2.
Activation of the PI3K pathway down-regulates Cbp expression. A, MEFs expressing LA-SDSE MEK or Myr-Akt were analyzed for Cbp mRNA expression levels by real-time PCR (left), and total cell lysates were immunoblotted with the antibodies indicated (right). B, c-Src-transformed Csk−/− cells were treated with U0126 (10 μm), SB202190 (10 μm), SP600125 (10 μm), LY294002 (20 μm), and PP2 (5 μm), and Cbp mRNA expression levels were analyzed by real-time PCR. Relative values ± S.D. were obtained from three independent assays. *, p < 0.05; **, p < 0.01; ***, p < 0.001; by Student's t test.
EGF Stimulation Down-regulates Cbp Expression through the ERK MAPK Pathway
The ERK MAPK pathway is activated by various extracellular signals, such as growth factors and cytokines, and plays a crucial role in diverse physiological processes. To examine if the activation of the ERK MAPK pathway under physiological conditions would lead to Cbp down-regulation, Cbp mRNA levels were determined in MEFs stimulated with EGF, PDGF, TNF-α, or IL-1β (Fig. 3A). Stimulation with EGF significantly induced the down-regulation of Cbp expression in a dose-dependent manner (Fig. 3B). PDGF and TNF-α had a similar effect but to a lesser extent (Fig. 3A). Kinetic analysis showed that the down-regulation of Cbp mRNA and protein became appreciable 3 h after EGF stimulation and reached a plateau 12 h after stimulation (Fig. 3C). These data demonstrate that Cbp down-regulation is induced by EGF stimulation and that this process occurs at a later phase of cell signaling because EGF-induced activation of the ERK MAPK pathway is terminated within 1 h (data not shown). To confirm the relevance of the ERK MAPK pathway to EGF-induced Cbp down-regulation, the effect of MEK inhibition was examined. MEFs were pretreated with the MEK inhibitor U0126, followed by EGF stimulation for 24 h. Analysis of Cbp mRNA levels showed that the inhibition of MEK robustly reversed EGF-induced Cbp down-regulation.
FIGURE 3.
EGF stimulation down-regulates Cbp mRNA expression via the ERK MAPK pathway. A, MEFs were treated with EGF (100 ng/ml), PDGF (50 ng/ml), TNF-α, and IL-1β (30 ng/ml) for 24 h, and analyzed for Cbp mRNA expression levels by real-time PCR. Relative values ± S.D. were obtained from three independent assays. *, p < 0.05; **, p < 0.01, by Student's t test. B, MEFs were treated with EGF for 24 h and analyzed for Cbp mRNA expression levels by real-time PCR. C, MEFs were treated with EGF (100 ng/ml) for the times indicated and analyzed for Cbp mRNA expression levels by real-time PCR (left), and Cbp protein levels were determined with anti-Cbp (right). D, MEFs treated with or without U0126 (20 μm) for 1 h were stimulated with or without EGF for 24 h and analyzed for Cbp mRNA expression levels by real-time PCR. Total cell lysates were immunoblotted with the antibodies indicated.
Src Activation Does Not Affect Cbp mRNA Stability or cbp Promoter Activity
In a previous study, an experimental system that uses MEFs lacking Csk (Csk−/− cells) was developed for the analysis of c-Src function (13, 15). Csk−/− cells are not morphologically transformed, but exogenous expression of wild-type c-Src can efficiently induce their transformation (16). Because the c-src gene is rarely mutated in human cancer (9, 10), this system can serve as an excellent model to analyze the function of c-Src up-regulation in human cancers. Cbp was also found to be dramatically down-regulated by c-Src transformation in this system (11). This system was therefore used to analyze the mechanism of Cbp down-regulation induced by c-Src up-regulation.
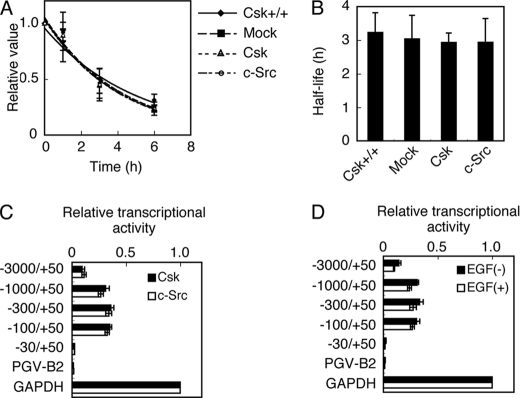
The levels of mRNA can be controlled by the efficiencies of mRNA synthesis and degradation. To examine whether c-Src-induced Cbp down-regulation is caused by a decrease in Cbp mRNA stability, the half-life of Cbp mRNA was analyzed in c-Src-transformed Csk−/− cells (c-Src), nontransformed Csk−/− cells expressing Csk (Csk), and normal MEFs (Csk+/+) (Fig. 4). These cells were treated with actinomycin D to inhibit transcriptional initiation, and Cbp mRNA levels were determined at various time points (Fig. 4A). The half-life of Cbp mRNA in all cell types was ∼3 h (Fig. 4B), indicating that there were no significant differences in Cbp mRNA stability. These results show that c-Src up-regulation does not affect Cbp mRNA stability.
FIGURE 4.
Src activation does not affect the Cbp mRNA stability or cbp promoter activity mediated by transcription factors. A, Csk+/+ cells and Csk−/− cells expressing Csk or c-Src were treated with actinomycin D (5 μg/ml) for the times indicated and Cbp mRNA expression levels were analyzed by real-time PCR. Relative values ± S.D. were obtained from three independent assays. B, half-life of Cbp mRNA. Average half-lives ± S.D. were obtained from three independent assays. C, Csk−/− cells expressing Csk or c-Src were co-transfected with reporter plasmids containing the fragments of the cbp promoter indicated, and analyzed for luciferase activity 24 h after transfection. D, MEFs transfected with the reporter plasmids were treated with or without EGF and analyzed for luciferase activity 24 h after transfection. The normalized values ± S.D. were obtained from three independent assays.
To examine whether c-Src-induced Cbp down-regulation is caused by a decrease in the efficiency of mRNA transcription regulated by transcription factors, the effects of c-Src transformation on cbp promoter activity were analyzed. Luciferase reporter assays were performed using the promoter region (3 kbp) upstream of the transcriptional initiation site of the cbp gene and its deletion constructs. Results showed that promoter activity was present in the region between −100 and +50 bp, but that there were no significant differences in promoter activity between c-Src-transformed and nontransformed cells (Fig. 4C). Similar results were obtained with EGF-stimulated and unstimulated MEFs (Fig. 4D). These results suggest that the activation of c-Src and EGF signaling does not affect the activity of transcription factors that regulate the promoter activity of the cbp gene.
Cbp Down-regulation Is Mediated by Histone Modifications
The contribution of epigenetic modifications, such as DNA methylation, histone deacetylation, and histone methylation, to Cbp down-regulation was examined. These mechanisms affect DNA transcription by remodeling chromatin structure (17, 18). The c-Src-transformed Csk−/− cells (c-Src) and nontransformed Csk−-expressing cells (Csk) were treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-Aza-dC) and/or the histone deacetylase (HDAC) inhibitor trichostatin A (TSA), and Cbp mRNA levels were determined (Fig. 5A). TSA treatment restored Cbp mRNA levels in both cell types, and the effect was greatly enhanced in c-Src transformed cells, suggesting the involvement of histone deacetylation in Cbp down-regulation. Essentially the same results were obtained when the cells were treated with another HDAC inhibitor, Scriptaid (data not shown). To examine the contribution of histone deacetylation, the effects of the siRNA-mediated knockdown of HDAC family members, HDAC1/2/3, were examined (Fig. 5B). Knockdown of HDAC1, but not HDAC2 or HDAC3, caused significant up-regulation of Cbp expression; this indicates that HDAC1 is responsible for Cbp down-regulation. However, there was a synergistic effect when HDAC1 siRNA was used in combination with HDAC2 siRNA, which suggest that there is functional redundancy between HDAC1 and HDAC2. Conversely, overexpression of HDAC1 induced the repression of Cbp expression, and Cbp up-regulation induced by HDAC1/2 siRNA was abrogated by the expression of siRNA-resistant human HDAC1 (Fig. 5C). These findings strongly suggest that HDAC1/2-mediated histone deacetylation is involved in Cbp down-regulation.
FIGURE 5.
Down-regulation of Cbp expression is mediated by epigenetic histone modifications. A, Csk−/− cells expressing Csk or c-Src were treated with 5-Aza-dC (300 nm for 72 h) and/or TSA (33 nm for 24 h) and analyzed for Cbp mRNA levels by real-time PCR. B, c-Src-transformed Csk−/− cells were transfected with combinations of siRNAs for HDACs. Expression levels of HDACs were analyzed by RT-PCR (upper panels) and Cbp mRNA expression levels were analyzed by real-time PCR (lower graph). C, c-Src-transformed Csk−/− cells were transfected with or without human HDAC1 (hHDAC1), followed by transfection with or without siRNAs for HDAC1 and 2. Expression levels of HDACs were analyzed by RT-PCR (upper panels) and Cbp mRNA expression levels were analyzed by real-time PCR (lower graph). D, Csk−/− cells expressing Csk or c-Src were analyzed for histone acetylation levels of the cbp promoter by ChIP assay with anti-acetyl-histone H3 and anti-acetyl-histone H4 (left). The efficiency of the ChIP assay was confirmed with anti-histone H3 (right). Average rates ± S.D. were obtained from five independent assays. **, p < 0.01; by Student's t test. E, Csk−/− cells expressing Csk or c-Src were analyzed for histone methylation levels of the cbp promoter by ChIP assay with anti-H3K9me3 and anti-H3K27me3. F, MEFs treated with 5-Aza-dC and/or TSA for 4 h were stimulated with or without EGF and analyzed for Cbp mRNA levels 24 h after stimulation. Relative values ± S.D. were obtained from three independent assays.
In c-Src-transformed cells, treatment with 5-Aza-dC failed to restore Cbp mRNA expression, while combination with TSA caused a slight additive effect (Fig. 5A). This suggests a potential involvement of DNA methylation in c-Src-induced Cbp down-regulation. To evaluate this possibility, the methylation status of the CpG islands in the cbp promoter was examined by bisulfite sequencing. The CpG islands were equally hypomethylated in these cell types (data not shown). These results suggest that histone deaceylation is the major, if not the only, cause of Cbp down-regulation in c-Src-transformed MEFs.
To further confirm that c-Src activation induces histone deacetylation in the cbp promoter, the acetylation status of histones H3 and H4 in the cbp promoter was examined by the ChIP assay (Fig. 5D). A significant decrease in the acetylation of histone H4 was observed in c-Src-transformed cells, whereas the acetylation of histone H3 was only moderately decreased by c-Src transformation. These results suggest that c-Src activation promotes the deacetylation of histone H4 and, to a lesser extent, histone H3 associated with the cbp promoter, which may be associated with the suppression of Cbp mRNA transcription.
Histone methylation is also known to be involved in the epigenetic regulation of gene expression (18, 19). The trimethylation status of histone H3 lysines 27 (H3K27me3) and 9 (H3K9me3), which mark transcriptionally silent chromatin, was examined in the cbp promoter by ChIP assay. Although there was no dramatic change in H3K9me3, the level of H3K27me3 was significantly increased in c-Src-transformed cells (Fig. 5E). This suggests that polycomb repressive complex 2 (PRC2)-mediated gene silencing (18) may also be activated in these cells.
To assess whether EGF-induced down-regulation of Cbp expression is also controlled by epigenetic mechanisms, MEFs treated with 5-Aza-dC and/or TSA were stimulated with or without EGF, and Cbp mRNA levels in these cells were assessed (Fig. 5F). Treatment with 5-Aza-dC did not affect Cbp mRNA down-regulation, whereas treatment with TSA restored Cbp expression. Combination treatment with 5-Aza-dC and TSA did not show any additive effect. These results demonstrate that histone deacetylation is also involved in EGF-induced Cbp down-regulation.
Cbp Down-regulation Is Mediated by Histone Modifications in Cancer Cells
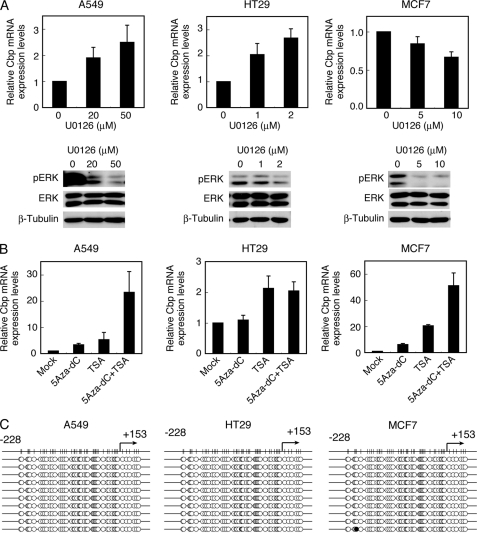
The above results suggest that c-Src-induced deacetylation and methylation of histones associated with the cbp promoter results in the down-regulation of Cbp mRNA transcription. In a previous study, Cbp mRNA was shown to be down-regulated in several cancer cell lines and tumor tissues (11). To examine whether the above mentioned epigenetic mechanisms are indeed functional in human cancer cells, the effects of the inhibition of the ERK MAPK pathway were tested in the colon cancer cell line HT29 and the lung cancer cell line A549, both of which harbor c-Src up-regulation and substantial down-regulation of Cbp (11) (Fig. 6A). The breast cancer cell line MCF7, which has Cbp down-regulation but has no c-Src up-regulation (11), was also studied (Fig. 6A). Treatment with U0126 restored the expression of Cbp in A549 and HT29 cells in a concentration-dependent manner, although a higher concentration of U0126 was required for A549 cells. These results indicate that Cbp down-regulation is mediated by the ERK MAPK pathway in some cancer cells as well. However, treatment with U0126 suppressed Cbp expression in MCF7 cells, which suggests that the contribution of the ERK MAPK pathway is cell context-dependent and potentially dependent on the patterns of activated oncoproteins.
FIGURE 6.
Cbp is down-regulated by epigenetic histone modifications in human cancer cells. A, A549, HT29, and MCF7 cells were treated with U0126 at the concentrations indicated for 24 h and analyzed for Cbp mRNA levels by real-time PCR, and total cell lysates were immunoblotted with the antibodies indicated. B, A549, HT29, and MCF7 cells were treated with 5-Aza-dC (5 μm for 72 h) and/or TSA (300 nm for 24 h) and analyzed for Cbp mRNA levels by real-time PCR. Relative values ± S.D. were obtained from three independent assays. C, A549, HT29, and MCF7 cells were analyzed for cbp promoter methylation status by bisulfite sequencing according to the method described under “Experimental Procedures.” The methylation status of ten clones obtained from each cell line is shown. Methylated and non-methylated cytosines are indicated by closed and open circles, respectively.
Treatment of these cancer cells with 5-Aza-dC and/or TSA also exhibited cell type-dependent effects (Fig. 6B). In A549 cells, treatment with 5-Aza-dC or TSA restored Cbp expression and the combination of 5-Aza-dC and TSA induced the synergistic up-regulation of Cbp expression. In HT29 cells, TSA treatment restored Cbp expression, but 5-Aza-dC treatment did not affect Cbp expression. MCF7 cells showed a response similar to that of A549 cells. The positive effects of 5-Aza-dC in A549 and MCF7 cells suggest that DNA methylation might contribute to Cbp down-regulation in these cells. However, the CpG islands in the promoter region of the cbp gene were equally hypomethylated in these cell types (Fig. 6C), which indicates that the effects of treatment with 5-Aza-Dc might be elicited by an indirect mechanism. These findings suggest that, although signaling pathways may vary among cancer types, histone modification is one of the major causes of Cbp down-regulation in some cancer cells.
DISCUSSION
A wide range of genes has been shown to be up-regulated or down-regulated in cancer cells. Some of these down-regulated genes are tumor suppressors. Epigenetic modifications are involved in the down-regulation of some of these tumor suppressors (17). The main epigenetic modifications in mammals are DNA methylation and histone modifications (acetylation, methylation, phosphorylation, etc.). A number of studies show that tumor suppressors are transcriptionally silenced through hypermethylation of promoter CpG islands (20, 21) and/or modifications of promoter-associated histones (17). The present study showed that Cbp, which functions as a suppressor of Src-mediated tumor progression, is down-regulated by an epigenetic mechanism involving deacetylation/methylation of histones, but not DNA methylation, via the activation of the oncogenic MAPK/PI3K pathways (Fig. 7). Although the mechanisms by which these pathways regulate histone deacetylases, e.g. HDAC1, or histone methylase complexes, e.g. PRC2 (18), remain unknown, understanding the molecular link between these pathways and the regulation of histone modification would provide new insights into the molecular basis of cancer-related alterations of gene expression through chromatin remodeling.
FIGURE 7.
A schematic model of the mechanisms of Cbp down-regulation. Activation of Src, EGFR, or Ras induces activation of the ERK MAPK and/or PI3K pathway. Although the underlying mechanisms are unknown, HDAC1/2 and PRC2 complex are activated and promote deacetylation and trimethylation of histones, respectively, resulting in suppressing the cbp gene transcription.
The genes down-regulated by an oncogenic form of Src, v-Src, have been studied extensively. The neuroretina specific gene QR1 is transcriptionally down-regulated by a v-Src-responsive element (22). The Von Hippel-Lindau tumor-suppressor gene (vhl) and 3Y1 tumor suppressor gene (tsg) are also down-regulated by v-Src (23). In these cases, v-Src up-regulates the expression of DNA methyltransferase 1, resulting in the hypermethylation of the vhl and tsg promoters. The scaffolding protein SSeCKS, which functions as a suppressor of tumorigenesis and metastasis, is down-regulated by v-Src and oncogenic Ras (24). v-Src down-regulates SSeCKS expression by facilitating the recruitment of HDAC1 to its promoter, which results in a decrease in the acetylation of histones H3 and H4. In the present study, Src transformation was shown to down-regulate Cbp by promoting the deacetylation of histone H4 in the cbp promoter through HDAC1/2. In this context, the mechanism of Cbp down-regulation appears similar to that of SSeCKS down-regulation. In the case of SSeCKS, however, v-Src can inhibit the expression of SSeCKS by recruiting Sp1/Sp3 to the SSeCKS promoter, while the activity of the cbp promoter itself is not affected by the activation of c-Src or EGF signaling. Moreover, activation of Raf, which is a component of the MAPK pathway, does not decrease SSeCKS expression, which suggests that the activation of the MAPK pathway is not sufficient for SSeCKS down-regulation. Although there are some inconsistencies, SSeCKS and Cbp may share a common regulatory mechanism mediated by Src-induced histone deacetylation in their promoter regions. A comparative analysis of the mechanisms involved in Cbp and SSeCKS down-regulation would help elucidate the molecular mechanisms associated with the Src-mediated regulation of particular sets of tumor suppressor genes.
The present study showed that the activation of the ERK MAPK pathway contributes to Cbp down-regulation. This is consistent with observations that Cbp is down-regulated in cells transformed by various oncogenes, including c-Src, v-Src, H-Ras, and K-Ras, all of which activate the ERK MAPK pathway. We also observed that Cbp was down-regulated in various human cancer cells harboring c-Src up-regulation and Ras mutations (11). Moreover, growth factors and cytokines that activate the ERK MAPK pathway, such as EGF, PDGF, and TNFα, caused Cbp down-regulation. Up-regulation and/or hyperactivation of such growth factors and cytokines and their receptors are frequently detected in cancers as well as in other cellular processes such as cell growth, inflammation, and development, which implies that these events might also be associated with Cbp down-regulation. These findings suggest that the ERK MAPK pathway contributes to Cbp down-regulation even under physiological conditions. We showed that the PI3K and p38 MAPK pathways, which are also activated by various cytokines, were likely to participate in Cbp down-regulation. Since these pathways are known to affect gene transcription, common factors downstream of these oncogenic pathways might activate critical molecules that induce the epigenetic down-regulation of Cbp. Alternatively, these pathways might be differentially utilized in different cell types. These observations suggest that Cbp expression can be regulated through multiple pathways activated not only by pathological processes, such as cancer progression and chronic inflammation, and by normal physiological cellular events, such as cell growth and differentiation/development. Therefore, analysis of Cbp down-regulation mechanisms would shed new light on the physiological functions of Cbp in regulating signaling pathways, particularly the c-Src-MAPK/PI3K pathway.
The down-regulation of Cbp, a suppressor of Src-mediated transformation, by c-Src activation is seemingly contradictory. However, the present results showed that full Cbp down-regulation requires a longer time (>12 h) than MAPK pathway activation (<1 h). This phenomenon could be due to the amount of time required for stable histone modification-mediated transcriptionally silencing. Therefore, during acute cell stimulation, Cbp is able to function as a suppressor of transiently activated c-Src; however, when Cbp is down-regulated epigenetically under chronic conditions, c-Src can no longer be suppressed by Cbp and exhibits constitutive activity. This suggests that the epigenetic silencing of Cbp would in turn create a positive feedback mechanism for c-Src: hyperactivation of c-Src-related pathways promotes chronic cellular events that induce epigenetic down-regulation of Cbp, resulting in the further activation of c-Src. Thus, it is likely that Cbp down-regulation occurs only under chronic conditions, such as cancer, inflammation, and development, which accompany a global shift in gene expression patterns via the epigenetic mechanisms.
Treatment of A549 and MCF7 cells with TSA or 5-Aza-dC up-regulated Cbp mRNA expression, and treatment with both inhibitors showed synergistic effects. This result suggests that the activity of the cbp promoter in these cells can be down-regulated not only by histone modification but also by DNA methylation. However, analysis of DNA methylation showed that the cbp promoter CpG islands were hypomethylated in these cells. Cancer cells contain localized hypermethylation at CpG islands (25), which is associated with the transcriptional inactivation of various cancer-related genes besides tumor suppressor genes (20, 21, 26). It is thus likely that 5-Aza-dC may target other genes that are indirectly involved in repressing Cbp expression, although the possibility that other distal elements of the cbp gene might be regulated by DNA methylation could not be excluded. In contrast, the treatment of HT29 cells with 5-Aza-dC did not affect Cbp expression, which indicates that the cbp promoter is down-regulated predominantly by histone modification in HT29 cells. Overall, these findings suggest that the cbp gene is not directly silenced by DNA methylation and that histone modification is the primary mechanism for suppressing Cbp expression at least in a subset of cancer cells.
In conclusion, the present study demonstrated the involvement of epigenetic histone modifications, activated through multiple oncogenic pathways, in the down-regulation of the tumor suppressor Cbp. These findings suggest that elements involved in the aberrant positive feedback loop created by Cbp down-regulation, which consists of c-Src-related oncogenic signaling pathways and histone modifiers, could serve as potential targets for the development of anticancer therapeutics against human cancers harboring c-Src up-regulation.
Acknowledgments
We thank Dr. A. Imamoto, Dr. T. Akagi, and Dr. M. Yutsudo for generous gifts of reagents.
This study was supported in part by the Uehara Foundation and a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
- Cbp
- Csk-binding protein
- TSA
- trichostatin A
- 5-Aza-dC
- 5-aza-2′-deoxycytidine
- HDAC
- histone deacetylase
- Csk
- C-terminal Src kinase
- SFK
- Src family kinases.
REFERENCES
- 1. Kawabuchi M., Satomi Y., Takao T., Shimonishi Y., Nada S., Nagai K., Tarakhovsky A., Okada M. (2000) Nature 404, 999–1003 [DOI] [PubMed] [Google Scholar]
- 2. Brdicka T., Pavlistová D., Leo A., Bruyns E., Korinek V., Angelisová P., Scherer J., Shevchenko A., Hilgert I., Cerný J., Drbal K., Kuramitsu Y., Kornacker B., Horejsi V., Schraven B. (2000) J. Exp. Med. 191, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davidson D., Bakinowski M., Thomas M., Horejsi V., Veillette A. (2003) Mol. Biol. Cell 23, 2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shima T., Nada S., Okada M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14897–14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang L., Feng X., Zhou W., Knyazev P., Ullrich A., Chen Z. (2006) Oncogene 25, 5495–5506 [DOI] [PubMed] [Google Scholar]
- 6. Parsons S. J., Parsons J. T. (2004) Oncogene 23, 7906–7909 [DOI] [PubMed] [Google Scholar]
- 7. Hu Y., Liu Y., Pelletier S., Buchdunger E., Warmuth M., Fabbro D., Hallek M., Van Etten R. A., Li S. (2004) Nat. Genet. 36, 453–461 [DOI] [PubMed] [Google Scholar]
- 8. Summy J., Gallick G. (2003) Cancer Metastasis Rev. 22, 337–358 [DOI] [PubMed] [Google Scholar]
- 9. Irby R. B., Mao W., Coppola D., Kang J., Loubeau J. M., Trudeau W., Karl R., Fujita D. J., Jove R., Yeatman T. J. (1999) Nat. Genet. 21, 187–190 [DOI] [PubMed] [Google Scholar]
- 10. Irby R. B., Yeatman T. J. (2000) Oncogene 19, 5636–5642 [DOI] [PubMed] [Google Scholar]
- 11. Oneyama C., Hikita T., Enya K., Dobenecker M. W., Saito K., Nada S., Tarakhovsky A., Okada M. (2008) Mol. Cell 30, 426–436 [DOI] [PubMed] [Google Scholar]
- 12. Oneyama C., Iino T., Saito K., Suzuki K., Ogawa A., Okada M. (2009) Mol. Cell. Biol. 29, 6462–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imamoto A., Soriano P. (1993) Cell 73, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 14. Akagi T., Sasai K., Hanafusa H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nada S., Yagi T., Takeda H., Tokunaga T., Nakagawa H., Ikawa Y., Okada M., Aizawa S. (1993) Cell 73, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 16. Oneyama C., Hikita T., Nada S., Okada M. (2008) Genes to Cells 13, 1–12 [DOI] [PubMed] [Google Scholar]
- 17. Sharma S., Kelly T. K., Jones P. A. (2010) Carcinogenesis 31, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Margueron R., Reinberg D. (2011) Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 20. Baylin S. (2001) Dev. Biol. 106, 85–87; discussion 143–160 [PubMed] [Google Scholar]
- 21. Yan P. S., Chen C. M., Shi H., Rahmatpanah F., Wei S. H., Caldwell C. W., Huang T. H. (2001) Cancer Res. 61, 8375–8380 [PubMed] [Google Scholar]
- 22. Pierani A., Pouponnot C., Calothy G. (1993) Mol. Cell Biol. 13, 3401–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sung J., Turner J., McCarthy S., Enkemann S., Li C. G., Yan P., Huang T., Yeatman T. J. (2005) Carcinogenesis 26, 487–494 [DOI] [PubMed] [Google Scholar]
- 24. Bu Y., Gelman I. H. (2007) J. Biol. Chem. 282, 26725–26739 [DOI] [PubMed] [Google Scholar]
- 25. Goelz S. E., Vogelstein B., Hamilton S. R., Feinberg A. P. (1985) Science 228, 187–190 [DOI] [PubMed] [Google Scholar]
- 26. Momparler R. L., Bovenzi V. (2000) J. Cell. Physiol. 183, 145–154 [DOI] [PubMed] [Google Scholar]
- 27. Yagi R., Waguri S., Sumikawa Y., Nada S., Oneyama C., Itami S., Schmedt C., Uchiyama Y., Okada M. (2007) EMBO J. 26, 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]