Abstract
Lipolysis is a critical metabolic pathway contributing to energy homeostasis through degradation of triacylglycerides stored in lipid droplets (LDs), releasing fatty acids. Neutral lipid lipases act at the oil/water interface. In mammalian cells, LD surfaces are coated with one or more members of the perilipin protein family, which serve important functions in regulating lipolysis. We investigated mechanisms by which three perilipin proteins control lipolysis by adipocyte triglyceride lipase (ATGL), a key lipase in adipocytes and non-adipose cells. Using a cell culture model, we examined interactions of ATGL and its co-lipase CGI-58 with perilipin 1 (perilipin A), perilipin 2 (adipose differentiation-related protein), and perilipin 5 (LSDP5) using multiple techniques as follows: anisotropy Forster resonance energy transfer, co-immunoprecipitation, [32P]orthophosphate radiolabeling, and measurement of lipolysis. The results show that ATGL interacts with CGI-58 and perilipin 5; the latter is selectively expressed in oxidative tissues. Both proteins independently recruited ATGL to the LD surface, but with opposite effects; interaction of ATGL with CGI-58 increased lipolysis, whereas interaction of ATGL with perilipin 5 decreased lipolysis. In contrast, neither perilipin 1 nor 2 interacted directly with ATGL. Activation of protein kinase A (PKA) increased [32P]orthophosphate incorporation into perilipin 5 by 2-fold, whereas neither ATGL nor CGI-58 was labeled under the incubation conditions. Cells expressing both ectopic perilipin 5 and ATGL showed a 3-fold increase in lipolysis following activation of PKA. Our studies establish perilipin 5 as a novel ATGL partner and provide evidence that the protein composition of perilipins at the LD surface regulates lipolytic activity of ATGL.
Keywords: Adenoviruses, Enzyme Mechanisms, Fluorescence, Fluorescence Resonance Energy Transfer (FRET), Lipase, Lipid Droplet, Lipolysis, Protein/Protein Interactions
Introduction
Hydrolysis of triacylglycerol (TAG)3 stored in the lipid droplet (LD) compartment provides a convenient source of cellular fuel for energy production during conditions such as fasting. However, lipolysis can contribute to the build up of toxic lipid intermediates and/or oxidized lipids in pathological conditions when cellular lipid homeostasis is altered, e.g. with obesity (1). Therefore, TAG hydrolysis must be carefully controlled to meet tissue-specific requirements for energy or lipid substrates in both adipose and non-adipose tissues. A better understanding of the mechanisms by which cells control lipid mobilization is needed to design novel approaches for intervention and prevention of the pathophysiological consequences of obesity.
During the past 10 years, key players in the lipolytic pathway of adipocytes were identified through the study of transgenic mouse models (2–8). Phenotypic analysis of hormone-sensitive lipase null mice suggested the existence of another lipase (7), leading to the identification of adipose triglyceride lipase (ATGL) (6, 9, 10). Characterization of ATGL null mice helped to establish the respective roles of these two lipases in the lipolytic cascade (7). Complete lipolysis requires three enzyme reactions to break down TAG into fatty acids and glycerol. First, ATGL hydrolyzes TAG to produce fatty acid and diacylglycerol; second, hormone-sensitive lipase then acts as a diacylglycerol lipase, and the final step is catalyzed by monoacylglycerol lipase. Studies using RNAi technology in cultured cells have confirmed the role of ATGL in initiating lipolysis (11–14) and generated interest in understanding the regulation of ATGL activity.
TAG hydrolysis requires lipase binding and activation at the LD water/oil interface. As demonstrated previously for hormone-sensitive lipase, ATGL may require interaction with proteins at the surfaces of LDs for optimal catalytic activity. Hence, we hypothesize that both the quality and quantity of proteins at the LD surface regulate ATGL activity.
Among the growing number of LD-associated proteins, members of the perilipin family are likely candidates to regulate ATGL activity. The five members of this family are signature constituents of the LD proteome. The importance of perilipin 1 in the regulation of adipose tissue lipolysis has been well established. Moreover, recent evidence from cell culture models suggests that perilipins 2 and 5 play roles in control of lipolysis in nonadipose tissues (15–17). Perilipin 5 is expressed in oxidative tissues where its expression is regulated by fasting and PPARα (17–19). Our previous work and that of others have shown that perilipin 5 plays an important role in regulating LD accumulation (17–19).
An emerging model suggests that lipolysis is controlled by complex interactions between perilipins, lipases, and additional proteins that modify lipase activity. Members of the perilipin family share scaffolding properties to coordinate lipolysis. Perilipins 1, 2, and 5 interact with CGI-58, a cofactor for ATGL (20–24). Moreover, hormone-sensitive lipase interacts with four members of the perilipin family (25). Most recently, a new protein, G0/G1 switch protein 2, was shown to interact with and negatively regulate ATGL (26). These protein/protein interactions are critical for the regulation of TAG hydrolysis.
The major objective of this study was to investigate the role of three members of the perilipin family, perilipin 1, 2, and 5, in the regulation of ATGL activity at the LD surface, through either modulation of ATGL accessibility to substrate or via direct protein/protein interactions, which may be critical for docking the lipase on LDs. A second objective was to determine whether each of the three perilipins regulates ATGL activity through a common mechanism. In rodent tissues, ATGL is ubiquitously expressed, whereas members of the perilipin family show different patterns of tissue-specific expression (27). Perilipin 1 is expressed most highly in adipose tissue; perilipin 2 is ubiquitous, but most highly expressed in liver; and perilipin 5 is expressed primarily in heart, brown adipose tissue, and skeletal muscle. Heterogeneity in LD perilipin 1, 2, and 5 composition potentially regulates ATGL activity in a tissue-specific manner. Hence, our present studies specifically focused on the ability of these three perilipin proteins to regulate ATGL activity.
In this study, we have used a cell culture system and multiple experimental approaches to reconstitute and investigate the role of perilipin proteins in the regulation of ATGL activity. Our studies provide evidence that ATGL activity is regulated differently by individual proteins of the perilipin family and therefore can be controlled in a tissue-specific manner.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit anti-GFP antibody was from Clontech; mouse anti-GFP antibody was from Covance (Princeton, NJ). Antibodies raised in rabbits against perilipin 1, 2, and 5 and CGI-58 were previously characterized (16, 17, 25, 28, 29). Guinea pig anti-perilipin 5 antibody was from Progen (Heidelberg, Germany). Rabbit anti-ATGL antibody was from Cell Signaling Technology (Danvers, MA). Horseradish peroxidase conjugates of goat anti-rabbit IgG, donkey anti-mouse IgG, and goat anti-guinea pig IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit serum was from Rockland Immunochemicals (Gilbertsville, PA).
Cell Culture
CHO-K1 and CHO Flp-In cells were obtained from ATCC (Manassas, VA) and Invitrogen, respectively. Both types of cells were grown in Ham's F-12 medium supplemented with 10% fetal calf serum, 2 mmol/liter glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2 and 95% humidity. Human skin fibroblasts were from an individual with Chanarin Dorfman syndrome, lacking functional CGI-58 (30); these cells were cultured as described previously (30). Mouse 3T3-L1 cells were obtained from Dr. M. D. Lane (The John Hopkins University, Baltimore) and were grown and differentiated as described previously (31).
The constructs for perilipin 1-YFP, perilipin 5-YFP, and perilipin 2-YFP were introduced into the CHO Flp-In cells according to the manufacturer's instructions and were previously described (25). Stably transfected CHO Flp-In cells were selected 24 h after transfection by adding hygromycin (500 μg/ml) to the growth medium. Wild-type CHO Flp-In cells were grown in the presence of Zeocin (100 μg/ml).
For immunofluorescence studies, CHO-K1 and CHO Flp-In cells were seeded in 35-mm dishes with glass bottoms (MatTek Corp., Ashland, MA) at a density of 2 × 105 cells per dish. The following day, cells were transfected with 1 μg of DNA plasmid containing cDNA coding for the fusion protein of interest per well (co-transfections received 0.5 μg of each DNA plasmid per well) using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. The same day, cells were incubated overnight in growth medium supplemented with 400 μm oleic acid complexed to bovine serum albumin as described previously (25). For immunoprecipitation studies, cells were plated at 2 × 105 cells per well in 6-well plates (Corning Glass, Corning, NY). For lipolysis experiments, cells were plated at 0.5 × 105 cells per well in 24-well plates (Corning Glass).
Molecular Cloning
To generate CFP or YFP fusion vectors, cDNAs of interest, ATGL, CGI-58, perilipin1–3 and 5, were cloned in-frame with monomeric CFP or YFP using pECFP-C1 or pEYFP-C1 plasmids. Names and sequences of oligonucleotide primers used for subcloning and all fluorescent fusion protein constructs are presented in supplemental Table 1. A serine to alanine mutation at position 47 was made in the mouse ATGL cDNA using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), as described previously (32). All constructs were verified by sequencing analysis.
Incubation and Lipolysis of Tissue Pieces
Brown adipose tissues (BAT) and epididymal white adipose tissues were collected from 10 4-week-old C57BL/6 male mice that had been fasted overnight. Tissues were minced finely into 6–10-mg pieces, and a total of 100 mg of each minced tissue was incubated in each well of 6-well dishes for 2 h at 37 °C with 5% CO2. Incubation media were Krebs-Ringer carbonate buffer at pH 7.4, supplemented with 4% fatty acid-free BSA, adenosine deaminase (1 unit/ml), and N6-phenylisopropyladenosine (100 nm) for basal conditions or supplemented with 10 μm isoproterenol for stimulated conditions. Incubation media were collected and assayed for glycerol content, and fat cakes containing lipid droplets were isolated as described previously (33).
Confocal Imaging
Confocal imaging of live cells was performed at 37 °C and 5% CO2 using a Zeiss LSM510 microscope equipped with an S-M incubator (Carl Zeiss MicroImaging, Inc.) and controlled by a CTI temperature regulator along with humidification and an objective heater. Emitted light was passed through bandpass filters for collection of CFP (470–510 nm) and YFP (530–550 nm).
Co-immunoprecipitations and Immunoblot Analysis
Antibodies used for immunoprecipitations were purified with the Melon gel IgG spin purification kit (Pierce). Rabbit serum was used as the negative control. Protein/protein interactions were investigated using extracts from cells co-transfected with the appropriate pair of protein fusion constructs (25). Briefly, cells were lysed by extrusion through a syringe in a buffer containing 150 mm NaCl, 1% Triton X-100, 60 mm n-octyl glucoside (Sigma), 10 mm Tris, pH 8.0, supplemented with protease inhibitors, and incubated for 15 min on ice (25). Cell lysates were pre-cleared with 30 μl of immobilized protein A/G-agarose (Pierce) for 15 min on a rotator at room temperature and then centrifuged at 14,000 × g at 4 °C for 30 min. 250 μl of the pre-cleared supernatant was incubated with the precipitating antibodies at room temperature for 2 h on a rotator. 50 μl of immobilized protein A/G-agarose was then added, and suspensions were incubated at room temperature for 2 h on a rotator. Protein complexes were precipitated by centrifugation at 2300 × g at 4 °C for 5 min, washed three times with cold PBS, and finally mixed with 60 μl of Laemmli sample buffer (Bio-Rad) containing β-mercaptoethanol. 20 μl of the pre-cleared supernatant and 30 μl of the immunoprecipitated proteins were separated on 4–12% polyacrylamide NuPAGE gels (Invitrogen), transferred to nitrocellulose membranes, and probed with the specific antibodies indicated, followed by a corresponding horseradish peroxidase-conjugated secondary antibody. The immunoblot signals were detected with SuperSignal chemiluminescence reagents (Pierce). In some experiments, AML12 cells transduced with perilipin 5-YFP adenovirus or differentiated 3T3-L1 adipocytes were incubated with 100 μCi/ml [32P]orthophosphate (PerkinElmer Life Sciences) for 2 h prior to exposure to forskolin (20 μm) and IBMX (0.5 mm) for 30 min. The incubations were performed at 37 °C under an atmosphere of 5% CO2.
Adenovirus
To generate adenovirus to drive the expression of perilipin 5 and ATGL, the perilipin 5-YFP cDNA fragment was excised from perilipin 5-YFP (see supplemental Table 1) (25) with AgeI (blunted) and KpnI, and the ATGL-CFP cDNA fragment was excised from ATGL-CFP (see supplemental Table 1) with NheI and HindIII. Subsequently, perilipin 5-YFP cDNA and ATGL-CFP cDNA were subcloned into a modified pAdTrack-CMV shuttle vector (34) lacking the GFP cassette. The PmeI-digested vector was used for transformation into AdEasy BJ5183 cells. Correct recombination of the resulting viral vector was confirmed by restriction enzyme digestions. Finally, the PacI-digested viral DNA was transfected into human embryonic kidney 293 cells for virus production and amplification (34).
Cellular Lipolysis
Assessment of the effects of ectopic proteins on lipolysis was performed (25). Briefly, CHO-FlpIn cells constitutively expressing perilipin 1-YFP or perilipin 5-YFP or perilipin 2-YFP (25) were seeded into a 24-well plate at a density of 0.5 × 105 cells per well and then infected with adenovirus driving the expression of ATGL-CFP. After 48 h, cells were incubated overnight with growth medium supplemented with 400 μm oleic acid complexed to 0.4% bovine serum albumin to promote triacylglycerol deposition. [3H]Oleic acid (2 × 106 dpm/well) was included as a tracer. Supplemental fatty acids were then removed, and re-esterification of fatty acids during subsequent incubations was prevented by inclusion of 5 μm Triacsin C (Biomol, Plymouth Meeting, PA), an inhibitor of acyl co-enzyme A synthetase, in the medium (16, 25, 29). To stimulate PKA, cAMP levels in cells were elevated by the addition of 20 μm forskolin. Lipolysis was determined by measuring radioactivity released into media (16, 25, 29). Similar experiments were performed using mouse AML12 cells (25). Quadruplicate wells were tested for each condition. Efficiency of transduction with Ad-ATGL-CFP was checked by immunoblotting.
Statistical Analysis
Statistical significance was tested using either one-way analysis of variance or a two-tailed Student's t test (GraphPad Software, Inc.).
RESULTS
Localization of ATGL to LDs Is Affected by the Perilipin Protein Composition of the LD
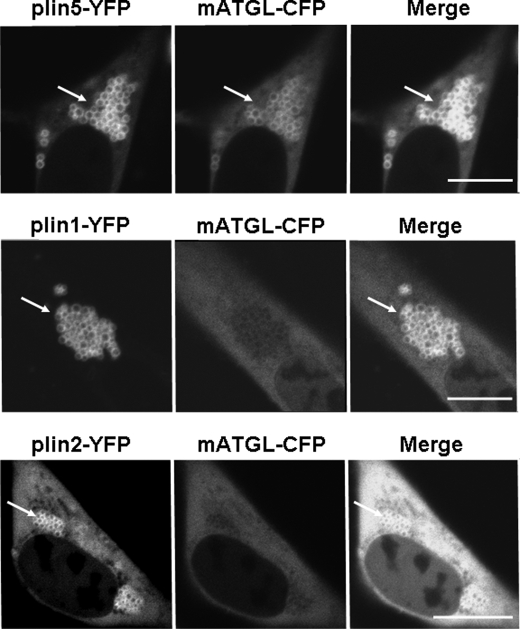
To explore protein/protein interactions between perilipin proteins and ATGL, we used CHO cell lines stably expressing fluorescent fusion proteins of perilipin 1, 2, and 5 and further co-transfected with an expression vector for an mATGL-CFP fusion protein. We opted to use catalytically inactive ATGL, mATGL (S47A) (32), to examine its interaction with perilipin proteins while eliminating the confounding factor of TAG hydrolysis. mATGL(S47A)-CFP was observed mostly in the cytoplasm when LDs were coated with perilipin 1-YFP, and cells were incubated under basal conditions (Fig. 1). Similarly, cells with perilipin 2-YFP-coated LDs showed a mostly diffuse mATGL(S47A)-CFP signal throughout the cytoplasm with minimal localization of mATGL to LD surfaces. Although mATGL did not extensively localize to perilipin 1-YFP- or perilipin 2-YFP-coated LDs, expression of wild-type ATGL-CFP resulted in a dramatic decrease in the number of LDs per cell, particularly in cells expressing perilipin 2-YFP (Fig. 2). These observations suggest that ATGL cycles rapidly on and off of LDs to catalyze lipolysis when LDs are coated with either perilipin 1 or 2. In contrast, mATGL(S47A)-CFP fluorescence was observed as thin rings on LD surfaces in cells expressing perilipin 5-YFP (Fig. 1). Similar data were obtained when wild-type ATGL-CFP was expressed in perilipin 5-YFP-expressing cells (Fig. 2); moreover, there was no apparent decrease in either the size or number of LDs. Among the three perilipins studied, only perilipin 5 promoted LD localization for ATGL, indicative of a possible direct interaction between these two proteins. Because recent published work suggests a complex interplay between perilipin 5 and CGI-58 (24), and also the possibility that CGI-58 interacts with ATGL (13), we next investigated all possible protein/protein interactions between perilipin 5, ATGL, and CGI-58.
FIGURE 1.
Localization of a catalytically inactive form of ATGL (mATGL(S47A)) to LDs is dependent on the perilipin protein composition. CHO-Flp-In cell lines were stably transfected with perilipin fusion proteins as follows: perilipin 5-YFP (plin5-YFP), perilipin 1-YFP (plin1-YFP), or perilipin 2-YFP (plin2-YFP) and co-transfected with mATGL-CFP. Cells were incubated overnight with 400 μm oleic acid before examination of live cells with confocal microscopy. Bar, 10 μm. White arrows indicate lipid droplets. Top, middle, and bottom rows show representative cells from four to six separate experiments.
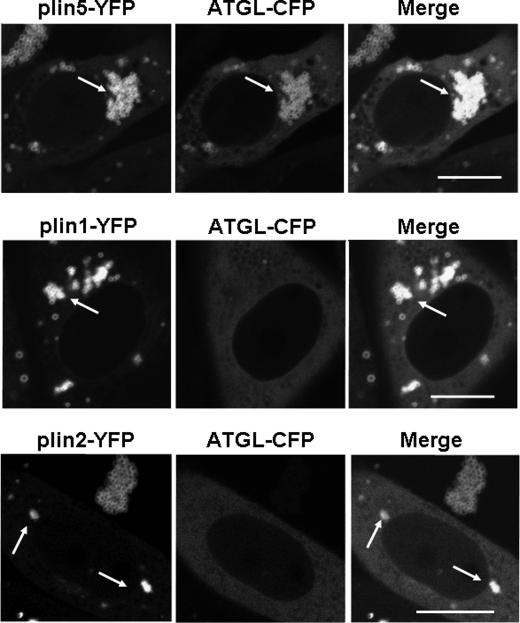
FIGURE 2.
Perilipin 1 and 2 (plin1-YFP and plin2-YFP) do not fully protect against TAG hydrolysis by wild-type ATGL, but perilipin 5 (plin5-YFP) does. Cells expressing YFP fusion proteins were incubated overnight with 400 μm oleic acid. The following day, live cells were examined with confocal microscopy. Bar, 10 μm. White arrows indicate lipid droplets. Top, middle, and bottom rows show representative cells from four to six separate experiments.
Localization of CGI-58 in Cells Expressing Perilipins 1, 2, or 5 and Determination of Protein/Protein Interactions between CGI-58 and ATGL
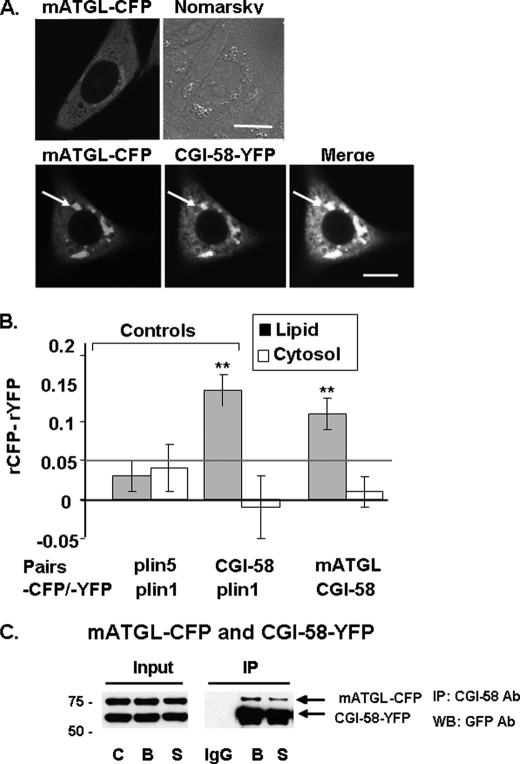
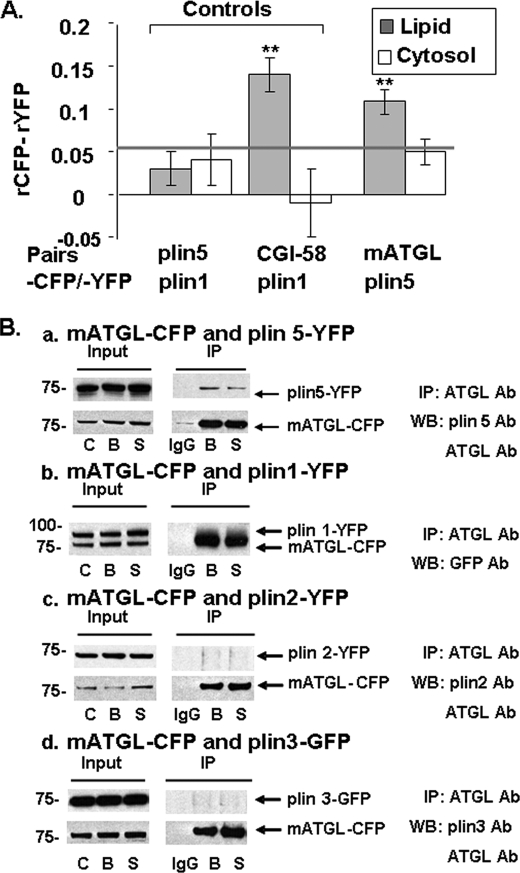
We observed that CGI-58-CFP is more efficiently recruited to LDs coated with perilipin 1-YFP or perilipin 5-YFP than those coated with perilipin 2-YFP (data not shown), confirming previous reports (21, 24). We then asked if CGI-58 and mATGL directly interact in live cells (Fig. 3). CHO-K1 cells expressing mATGL(S47A)-CFP alone or co-expressing mATGL(S47A)-CFP and CGI-58-YFP were incubated overnight with 400 μm oleic acid to increase LD formation and levels of endogenous perilipin 2 on LDs (35). Co-expression of CGI-58-YFP with mATGL(S47A)-CFP increased recruitment of mATGL to LDs, when compared with results obtained in cells expressing only mATGL(S47A)-CFP (Fig. 3A). These observations suggest that CGI-58 recruits ATGL to the surfaces of LDs via a protein/protein interaction. Similar data were obtained in CHO-K1 cells overexpressing perilipin 1 (data not shown). We used AFRET measurements to evaluate CGI-58 interactions with mATGL in situ (Fig. 3B), following the same strategy that we have reported previously (25). Because AFRET occurs when fluorescent probes are no more than 8–10 nm apart, it detects proteins in close proximity that likely interact. For negative controls, perilipin 5-CFP and perilipin 1-YFP were expressed together in cells and failed to show interactions by AFRET, as expected. For positive controls, we expressed CGI-58-CFP and perilipin 1-YFP and observed strong interactions by AFRET, as reported previously (25). Evidence for protein/protein interactions, as demonstrated by differences in anisotropy, was statistically significant for CGI-58/mATGL only at the LD surface and not in the cytoplasm (Fig. 3B). This interaction was confirmed by co-immunoprecipitation (co-IP) and, moreover, was demonstrated for both basal and PKA-activated conditions (Fig. 3C). Because of complexity in the interactions between perilipin 5, CGI-58, and ATGL, we next investigated if ATGL localizes to the LD surface by intermediary interaction with CGI-58 or by direct interaction with perilipin 5.
FIGURE 3.
CGI-58 interacts with mATGL (S47A) at the lipid droplet surface as determined by in situ AFRET and co-IP. Images were collected by confocal microscopy of live cells expressing pairs of fluorescent fusion proteins (A), and calculations for AFRET were performed for pairs of proteins indicated along the x axis (B). The gray line indicates the threshold of significance. Data are means ± S.E. from six to nine experiments. **, p < 0.01. C, CHO-K1 cells were co-transfected with mATGL(S47A)-CFP and CGI-58-YFP, and the cells were incubated with 400 μm oleic acid overnight. The following day, cells were incubated with either 5 μm triacsin C (B, basal conditions) or for 30 min with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C (S, stimulated conditions). Cell lysates were incubated with rabbit anti-CGI-58 IgG (B and S) or rabbit preimmune control IgG (IgG). Immunoprecipitates were analyzed by Western blot (WB) for ATGL and CGI-58 using a commercial GFP antibody (Ab) that cross-reacts with YFP. One of three similar experiments is shown.
CGI-58 Is Not Needed for ATGL Recruitment to the LD Surface When Perilipin 5 Is Present
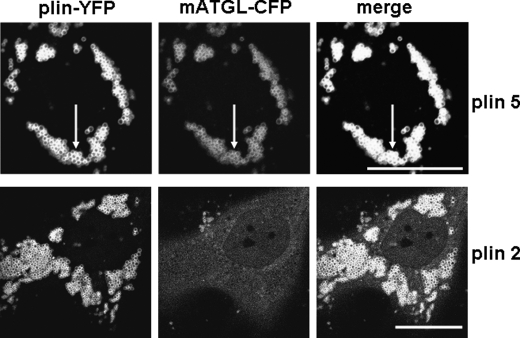
To rule out the possibility that ATGL is recruited to perilipin 5-coated LDs by intermediary binding activity of CGI-58, we studied protein localization in human skin fibroblasts from a patient with Chanarin-Dorfman syndrome, which lack functional CGI-58 (Fig. 4) (30). mATGL(S47A)-CFP was recruited to perilipin 5-YFP-coated LDs, indicating that CGI-58 is not required for the localization of ATGL to LDs. These experiments led to exploration of the possibility of a direct interaction between ATGL and perilipin 5.
FIGURE 4.
ATGL binds to perilipin 5 even in the absence of CGI-58. Human skin fibroblasts from a patient with Chanarin-Dorfman syndrome were co-transfected with mATGL(S47A)-CFP and either perilipin 5-YFP (plin5-YFP) or perilipin 2-YFP (plin2-YFP). Cells were incubated overnight with 400 μm oleic acid. The following day, live cells were examined with confocal microscopy as in Fig. 1. Bar, 10 μm. Representative cells are shown from two separate experiments. White arrows indicate lipid droplets.
Perilipin 5 and ATGL Interact, as Determined by AFRET and Co-IP
We collected AFRET data in CHO-K1 cells expressing perilipin 5-YFP and mATGL(S47A)-CFP. Interaction was detected between perilipin 5-YFP and mATGL(S47A)-CFP on LDs (Fig. 5A); no interaction was detected between these proteins in the cytoplasm. Comparable data were obtained from AFRET measurements using perilipin 5-YFP and wild-type ATGL-CFP constructs (data not shown). We confirmed these AFRET results by co-IP; co-IP of perilipin 5 with mATGL correlated well with the detection of protein/protein interactions by AFRET (Fig. 5B, panel a). Other pairs of proteins composed of mATGL(S47A) paired with each of perilipin 1-YFP, perilipin 2-YFP, and perilipin 3-YFP were investigated; mATGL-CFP did not co-IP with any of these other perilipins (Fig. 5B, panels b–d). Therefore, ATGL interacts with perilipin 5, but not perilipins 1–3. Interestingly, we did not find an interaction between perilipin 5 and GOS2, a protein recently found to interact with and inhibit ATGL (data not shown).
FIGURE 5.
ATGL binds uniquely to perilipin 5, as determined by in situ AFRET and co-IP. A, images were collected by confocal microscopy of live cells expressing pairs of fluorescent fusion proteins shown along the x axis, as in Fig. 2, and calculations for AFRET were performed (23). The gray line indicates the threshold of significance. Data are means ± S.E. from 6 to 12 experiments. *, p < 0.01. B, CHO-Flp-In cells constitutively expressing perilipin 5-YFP (panel a), perilipin 1-YFP (panel b), perilipin 2-YFP (panel c), or perilipin 3-YFP (panel d) were transfected with mATGL-CFP. Cells were incubated with 400 μm oleic acid overnight. The following day, cells were incubated with either 5 μm triacsin C (B, basal conditions) or for 30 min with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C (S, stimulated conditions). Cell lysates were incubated with rabbit anti-ATGL IgG (B and S) or rabbit preimmune control IgG (IgG). Immunoprecipitates were analyzed by Western blot (WB) using a commercial GFP antibody that cross-reacts with YFP or the appropriate perilipin antibody (Ab). One of three similar experiments is shown.
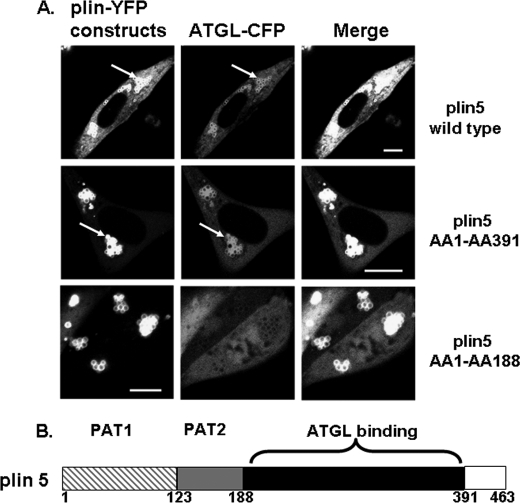
Identification of the ATGL-binding Site on Perilipin 5
To identify the sequence of perilipin 5, which binds ATGL, mATGL(S47A)-CFP was co-expressed with either one of two carboxyl-terminally truncated fluorescent perilipin 5 fusion proteins, including amino acids 1–391 or 1–188. As shown in Fig. 6, although wild-type and both forms of carboxyl-terminally truncated perilipin 5 targeted to LDs, mATGL(S47A)-CFP failed to localize to LDs coated with the shorter truncation mutation (amino acids 1–188). Therefore, in this cell model, ATGL requires perilipin 5 for lipid droplet localization and binds to a sequence between amino acids 188 and 391. A truncated protein containing amino acids 188–391 failed to target to LDs, indicating that essential LD-targeting motifs are contained within the first 188 amino acids of perilipin 5; thus, we were unable to test this truncated protein for ATGL binding (data not shown).
FIGURE 6.
A perilipin 5 domain between amino acids 188 and 391 is the site of ATGL binding. A, plasmids for fusion constructs of wild-type perilipin 5, perilipin 5 (amino acids 1–391), and perilipin 5 (amino acids 1–188) with YFP were co-transfected with ATGL(S47A)-CFP. Bar, 10 μm. White arrows indicate lipid droplets. Top, middle, and bottom rows show representative cells from four to six separate experiments. B, schematic view for perilipin 5 domains; PAT-1 (hatched gray) and PAT-2 (gray) are domains shared by all perilipin proteins (except plin4 lacks the PAT1 domain), the ATGL-binding site specific to perilipin 5 resides within the black region.
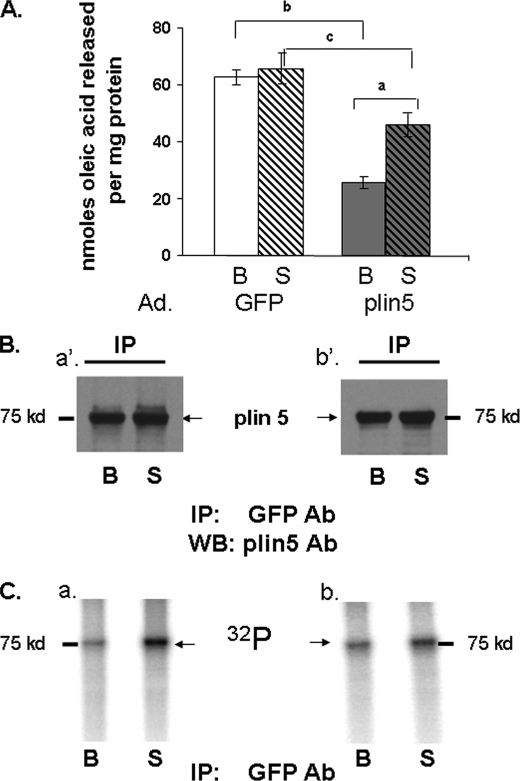
Effect of Perilipin 5 on Lipolysis
To evaluate the role of perilipin 5 in regulating lipolysis, we used AML12 mouse liver cells, which express endogenous ATGL but not perilipin 5 (16). These cells were transduced with adenovirus driving expression of either GFP or perilipin 5; lipolysis was studied under both basal and PKA-activated conditions. Cells were incubated overnight with radiolabeled oleic acid and then incubated in medium lacking supplemental fatty acids, with or without forskolin, to activate adenylyl cyclase for 2 h. Cells expressing perilipin 5 released lower amounts of fatty acids in basal conditions than control cells expressing GFP (Fig. 7A). After activation of PKA, lipolysis in perilipin 5-expressing cells increased by 2-fold over basal release of fatty acids; however, the total amount of fatty acids released from cells expressing perilipin 5 remained significantly lower than that of control cells expressing GFP. These data show that lipolysis in this model system is increased by activation of PKA.
FIGURE 7.
Perilipin 5 effect on lipolysis and identification of perilipin 5 as a phosphorylated protein. A, AML12 cells were transduced with an adenoviral (Ad) construct for the expression of GFP or perilipin 5 (plin5) constructs for 48 h prior to lipolysis measurements. Cells were loaded overnight with [3H]oleic acid and 400 μm unlabeled oleic acid. Supplemental fatty acids were removed, and 5 μm triacsin C with (S) or without (B) 20 μm forskolin were added to the medium for 2 h. Release of fatty acids is shown for both basal (B) and stimulated (S) conditions. Data represent means ± S.E. (n = 16) (a, p < 0.01, for PKA stimulated value compared with basal value; b, p < 0.01; and c, p < 0.05 for cells expressing GFP compared with cells expressing perilipin 5. B and C, AML12 cells were transduced with adenoviral perilipin 5-YFP. Cells were incubated with 400 μm oleic acid overnight. 48 h after transduction, cells were loaded with [32P]orthophosphate for 2 h and then incubated for 30 min with (S) or without (B) 20 μm forskolin and 0.5 mm IBMX. Cell lysates were incubated with a commercial GFP antibody (Ab), and immunoprecipitates were analyzed by Western blot (WB) for perilipin 5. Two separate experiments were performed and are shown as indicated by panels a′ and b′ for Western blots (B), and panels a and b for autoradiograms (C).
Perilipin 5 Is Phosphorylated by PKA
Because ATGL is not a substrate for PKA (7), we hypothesized that either perilipin 5 or CGI-58 is phosphorylated by PKA. Indeed, it was recently demonstrated that release of CGI-58 from perilipin 1 is regulated by PKA (12, 21) and that perilipin 5 recruits CGI-58 to LD (24). To assess whether either perilipin 5 or CGI-58 is phosphorylated by PKA, AML12 cells transduced with adenovirus for perilipin 5-YFP and differentiated 3T3-L1 adipocytes expressing endogenous CGI-58 were each incubated with [32P]orthophosphate, in the presence or absence of forskolin and IBMX. In lysates of AML12 cells expressing perilipin 5-YFP, antibodies raised against either GFP or perilipin 5 immunoprecipitated a protein of ∼75 kDa (Fig. 7B), consistent with the size of perilipin 5-YFP. Incorporation of 32P into this band was noted (Fig. 7C) in basal conditions; the signal was increased by 2-fold following incubation of cells with forskolin and IBMX. In lysates of differentiated 3T3-L1 adipocytes, an anti-CGI-58 antibody immunoprecipitated a protein with an apparent molecular mass slightly greater than 37 kDa, consistent with the molecular mass of CGI-58. However, we failed to detect 32P in the same region on the autoradiogram (data not shown). These data indicate that perilipin 5 is a constitutively phosphorylated protein and a downstream target of PKA.
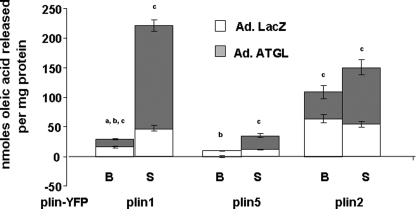
Perilipin 1 and 5 Modulate Lipolysis Catalyzed by ATGL
To evaluate the role of perilipins in regulating the activity of ATGL, studies were performed in CHO cell lines stably expressing fluorescent fusion proteins of perilipin 1, 2, and 5 under basal and PKA-activated conditions. The cells were transduced with either an adenovirus driving the expression of ATGL-CFP or a control adenovirus driving expression of β-galactosidase (Fig. 8) (25); the control cells express endogenous lipases, including very low levels of ATGL (36, 37). The cells were incubated overnight with radiolabeled oleic acid and then incubated in medium lacking supplemental fatty acids, with or without forskolin, for 2 h. CHO cells expressing β-galactosidase and endogenous lipases exhibited differences in basal and stimulated lipolysis corresponding to the type of perilipin protein coating the LDs and confirmed our earlier report (25). Among the three perilipins studied, perilipin 5 was as effective as perilipin 1 in inhibiting lipolysis under basal conditions, although perilipin 2 was the least effective; moreover, only perilipin 1 cells increased lipolysis in response to the activation of PKA. Overexpression of ATGL significantly increased basal lipolysis in cells expressing perilipin 2 or 1 but not perilipin 5. Following activation of PKA, lipolysis in perilipin 1 cells increased by more than 7-fold. Although the amount of fatty acids released from perilipin 5 cells remained significantly lower than levels of fatty acids released from either perilipin 1 or 2 cells, activation of PKA leads to ∼3-fold increased lipolysis in perilipin 5 cells. In contrast, activation of PKA had no effect on lipolysis in perilipin 2 cells, whether or not ATGL was overexpressed. It is also important to note that increased lipolysis was observed in cells overexpressing ATGL without parallel overexpression of CGI-58, suggesting that endogenous CGI-58 was sufficient to serve as a co-activator of ATGL.
FIGURE 8.
Control of lipolysis by perilipins in the presence and absence of ectopic ATGL. CHO-Flip-In cells stably expressing perilipin 1-YFP (plin1-YFP), perilipin 2-YFP (plin 2-YFP), or perilipin 5-YFP (plin5-YFP) were transduced with an adenoviral (Ad) ATGL-CFP construct for 48 h prior to lipolysis measurements. Control cells were infected with an adenoviral construct for β-galactosidase (LacZ). Data are represented in stacked columns; the full length of the column represents the amount of fatty acid released in cells overexpressing ATGL (gray and white bars combined); length of white bars represent the amount of fatty acids released in cells with LacZ (endogenous lipase); length of gray bars represent the amount of free fatty acids released due to the presence of exogenous ATGL. Lipolysis was measured as described in Fig. 6. Data represent means ± S.E. (n = 12) (a, p < 0.01, for PKA-stimulated value compared with basal value in cells expressing LacZ; b, p < 0.01 PKA stimulated value compared with basal value in ATGL-overexpressing cells; c, p < 0.01 for lipolysis value in cells expressing LacZ compared with lipolysis value in cells overexpressing ATGL.
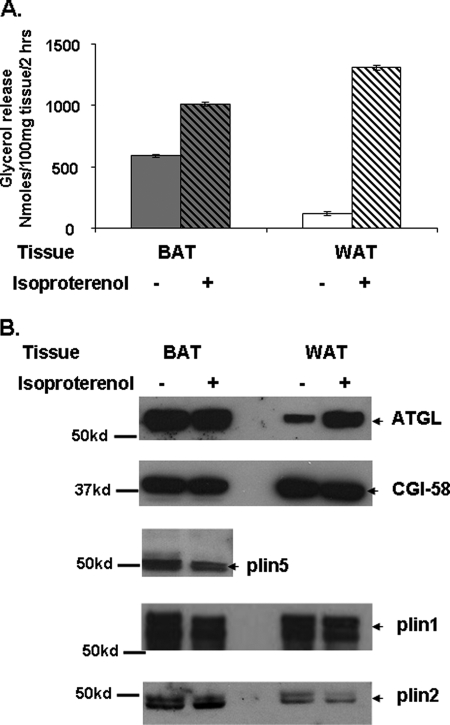
Lipolysis was also assessed in pieces of BAT from overnight fasted mice. BAT contains adipocytes with multilocular lipid droplets, which are coated with both perilipin 1 and 5. A 2-fold increase in glycerol release was observed in BAT tissue samples upon activation of PKA relative to basal glycerol release (Fig. 9). In contrast, white adipose tissue samples (WAT) containing adipocytes with unilocular LDs and expressing mostly perilipin 1 showed an 11-fold increase in glycerol release following activation of PKA relative to basal glycerol release. Importantly, ATGL content of the LD fraction from BAT was unchanged following PKA activation (Fig. 9). In contrast, ATGL was actively recruited to the LD fraction from WAT in parallel with activation of lipolysis. These data suggest that both ATGL recruitment to lipid droplets and lipolysis are significantly affected by the perilipin 5 content of lipid droplets in BAT. Interestingly, we did not observe changes in CGI-58 content in the LD fraction of either BAT or WAT following PKA activation (Fig. 9).
FIGURE 9.
Increased ATGL is recruited to LDs in WAT but not BAT following stimulation of tissues with a β-adrenergic receptor agonist. A, 100 mg of WAT and BAT from overnight-fasted mice were incubated for 2 h with adenosine deaminase (1 unit/ml) and phenylisopropyladenosine (100 nm) (basal conditions) or with adenosine deaminase (1 unit/ml) and isoproterenol (10 μm) (stimulated conditions) prior to measurement of glycerol content of media samples. B, fat cakes containing lipid droplets were isolated by ultracentrifugation from 100 mg of BAT and WAT lysates after incubations as described in A. Equal amounts of total fat cake protein were loaded as follows: for ATGL and CGI-58, 45 μg of total protein (same blot was re-used to identify ATGL and CGI-58); for plin5, 20 μg of total protein; and for plin 1, 5 μg of total protein were loaded. Experiments were performed twice.
DISCUSSION
A major finding of this study is that perilipins at the surfaces of LDs regulate ATGL binding and TAG hydrolase activity by different mechanisms. Because ATGL is ubiquitously expressed, these findings suggest that tissue-specific regulation of lipolysis is a function of the differing perilipin composition of LDs in various tissues.
Uniquely, perilipin 5 is the only perilipin to directly bind ATGL; the ATGL-binding site was identified between amino acids 188 and 391 of the perilipin 5 sequence. Shortly after we submitted our manuscript, another research group identified a comparable sequence of perilipin 5 as a binding site for ATGL (38). Perilipin 5 is most highly expressed in oxidative tissues, such as smooth and skeletal muscle, BAT, and to a lesser extent liver, and therefore, it is likely a primary regulator of lipolysis in these tissues, as we have demonstrated for BAT. Our data show that although perilipin 5 recruits ATGL to LDs, it reduces the TAG hydrolase activity of ATGL unless PKA is activated. Our data also show that perilipin 5 is a substrate for PKA, suggesting that phosphorylation of perilipin 5 enables lipolysis. Elucidation of the mechanism by which lipolysis is increased requires further study. We and others have shown that perilipin 5 binds both ATGL and CGI-58, a co-activator of ATGL; however, a recent parallel study (38) suggests that a single molecule of perilipin 5 cannot simultaneously bind both ATGL and CGI-58. Thus, it is possible that phosphorylation of perilipin 5 promotes release of CGI-58 from the perilipin scaffold and is required to facilitate the interaction of CGI-58 with ATGL and subsequent activation of lipolysis. This proposed mechanism is similar to that elucidated for perilipin 1; perilipin 1 binds CGI-58 under basal conditions, but activation of PKA and the ensuing phosphorylation of perilipin 1 releases CGI-58 (12, 21, 22) to promote the interaction of CGI-58 with ATGL. Alternatively, phosphorylation of perilipin 5 may be required to optimally position ATGL and CGI-58 bound to neighboring molecules of perilipin 5 for activation of lipolysis.
Neither perilipin 1 nor 2 interacts directly with ATGL; moreover, overexpression of perilipin 1 or 2 in cells reduces the localization of ATGL to LDs. Although expression of perilipin 1 is limited to adipocytes in WAT and BAT and steroidogenic cells, perilipin 2 is ubiquitously expressed. Our data provide confirmation of a previous report that overexpression of perilipin 2 reduces LD association of ATGL and lipolysis, as demonstrated in human embryonic kidney cells (15). In another study, binding of endogenous ATGL to perilipin 1-coated LDs increased concomitantly with major increases in lipolysis when PKA was activated in cultured human adipocytes (14). We have confirmed the observation that activation of PKA increases ATGL recruitment to LDs in adipocytes of murine WAT and showed that activation of PKA does not promote a binding interaction of ATGL with phosphorylated perilipin 1. We hypothesize that perilipin 1 and 2 and ATGL compete for binding sites on the surfaces of LDs. Thus, hydrolysis of TAG stored in perilipin 1- or 2-coated LDs is controlled by a balance of three parameters as follows: the amount of ATGL present, the amount of CGI-58 available to co-activate ATGL, and the relative coverage of the surfaces of LDs by perilipin 1 or 2. Moreover, in adipocytes, PKA-mediated phosphorylation of perilipin 1 may promote TAG hydrolysis by two possible mechanisms as follows: 1) conformational changes in phosphorylated perilipin 1 may reveal binding sites on the surfaces of LDs for ATGL, and 2) release of CGI-58 from the perilipin scaffold increases the interaction of the lipase co-activator with ATGL. Finally, phosphorylated perilipin 1 recruits hormone-sensitive lipase to LDs through a direct binding interaction (25) to promote diacylglycerol hydrolysis, driving lipolysis forward.
The mechanism by which perilipin 1 and 2 reduce the binding of ATGL to LDs could be overcome by the overexpression of CGI-58. Thus, our experiments confirm a critical role of CGI-58 to recruit ATGL to LDs coated with perilipins 1 or 2 (21). We demonstrated a direct interaction between CGI-58 and ATGL, confirming previous reports (12, 36). In contrast, CGI-58 is not required for the binding interaction between ATGL and perilipin 5, because these interactions were observed in fibroblasts from humans lacking functional CGI-58.
In summary, our studies suggest that perilipin 5 plays a negative regulatory role in LD hydrolysis by binding and inhibiting ATGL activity at the LD surface under basal conditions. Even when PKA is activated, the release of fatty acid is only modestly increased. Based on our studies, we propose that perilipin 5 plays a critical role in oxidative tissues to protect mitochondria from a rapid influx of fatty acid during lipolysis. Consistent with this mechanism, smooth, cardiac, and skeletal muscle increase their LD and perilipin 5 content concomitant with fatty acid uptake during fasting (17–19). Further perilipin 5 gain and loss of function studies in mice are warranted to test this hypothesis.
Supplementary Material
Acknowledgment
We thank Dr. Martin Woodle for careful review and helpful editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1RO1 DK 075017 (to C. S.) and R01 DK54797 (to D. B.) and by Intramural Research Programs of NIDDK. This work was also supported by Career Development Award 1-05-CD-17 from the American Diabetes Association (to C. S.), the Geriatric Research, Education and Clinical Center, Baltimore Veterans Affairs Health Care Center, and Clinical Nutrition Research Unit of Maryland Grant DK072488.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TAG
- triacylglyceride
- ADFP
- adipose differentiation-related protein
- AFRET
- anisotropy forster resonance energy transfer
- LD
- lipid droplet
- ATGL
- adipocyte triglyceride lipase
- CFP
- cyan fluorescent protein
- BAT
- brown adipose tissue
- WAT
- white adipose tissue
- IP
- immunoprecipitation
- IBMX
- isobutylmethylxanthine.
REFERENCES
- 1. DeFronzo R. A. (2010) Diabetologia 53, 1270–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martinez-Botas J., Anderson J. B., Tessier D., Lapillonne A., Chang B. H., Quast M. J., Gorenstein D., Chen K. H., Chan L. (2000) Nat. Genet. 26, 474–479 [DOI] [PubMed] [Google Scholar]
- 3. Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., Londos C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., Yamada N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S. P., Laurin N., Himms-Hagen J., Rudnicki M. A., Levy E., Robert M. F., Pan L., Oligny L., Mitchell G. A. (2001) Obes. Res. 9, 119–128 [DOI] [PubMed] [Google Scholar]
- 6. Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 7. Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. (2002) J. Biol. Chem. 277, 4806–4815 [DOI] [PubMed] [Google Scholar]
- 8. Radner F. P., Streith I. E., Schoiswohl G., Schweiger M., Kumari M., Eichmann T. O., Rechberger G., Koefeler H. C., Eder S., Schauer S., Theussl H. C., Preiss-Landl K., Lass A., Zimmermann R., Hoefler G., Zechner R., Haemmerle G. (2010) J. Biol. Chem. 285, 7300–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 10. Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. (2004) J. Biol. Chem. 279, 47066–47075 [DOI] [PubMed] [Google Scholar]
- 11. Miyoshi H., Perfield J. W., 2nd, Obin M. S., Greenberg A. S. (2008) J. Cell. Biochem. 105, 1430–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. (2009) J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Cell Metab. 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 14. Bezaire V., Mairal A., Ribet C., Lefort C., Girousse A., Jocken J., Laurencikiene J., Anesia R., Rodriguez A. M., Ryden M., Stenson B. M., Dani C., Ailhaud G., Arner P., Langin D. J. (2009) J. Biol. Chem. 284, 18282–18291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. (2007) J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 16. Bell M., Wang H., Chen H., McLenithan J. C., Gong D. W., Yang R. Z., Yu D., Fried S. K., Quon M. J., Londos C., Sztalryd C. (2008) Diabetes 57, 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. (2007) Biochim. Biophys. Acta 1771, 210–227 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. (2006) J. Biol. Chem. 281, 14232–14240 [DOI] [PubMed] [Google Scholar]
- 19. Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., Finck B. N., Bickel P. E. (2006) Diabetes 55, 3418–3428 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi T., Omatsu N., Matsushita S., Osumi T. (2004) J. Biol. Chem. 279, 30490–30497 [DOI] [PubMed] [Google Scholar]
- 21. Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., Brasaemle D. L. (2004) J. Biol. Chem. 279, 42062–42071 [DOI] [PubMed] [Google Scholar]
- 22. Yamaguchi T., Omatsu N., Omukae A., Osumi T. (2006) Mol. Cell. Biochem. 284, 167–173 [DOI] [PubMed] [Google Scholar]
- 23. Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. (2007) J. Biol. Chem. 282, 5726–5735 [DOI] [PubMed] [Google Scholar]
- 24. Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. (2009) J. Biol. Chem. 284, 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D., Gong D., Londos C., Yamaguchi T., Holm C., Rizzo M. A., Brasaemle D., Sztalryd C. (2009) J. Biol. Chem. 284, 32116–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X., Lu X., Lombès M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. (2010) Cell Metab. 11, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bickel P. E., Tansey J. T., Welte M. A. (2009) Biochim. Biophys. Acta 1791, 419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. (1991) J. Biol. Chem. 266, 11341–11346 [PubMed] [Google Scholar]
- 29. Tansey J. T., Huml A. M., Vogt R., Davis K. E., Jones J. M., Fraser K. A., Brasaemle D. L., Kimmel A. R., Londos C. (2003) J. Biol. Chem. 278, 8401–8406 [DOI] [PubMed] [Google Scholar]
- 30. Williams M. L., Coleman R. A., Placezk D., Grunfeld C. (1991) Biochim. Biophys. Acta 1096, 162–169 [DOI] [PubMed] [Google Scholar]
- 31. Green H., Kehinde O. (1975) Cell 5, 19–27 [DOI] [PubMed] [Google Scholar]
- 32. Smirnova E., Goldberg E. B., Makarova K. S., Lin L., Brown W. J., Jackson C. L. (2006) EMBO 7, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wolins N. E., Rubin B., Brasaemle D. L. (2001) J. Biol. Chem. 276, 5101–5108 [DOI] [PubMed] [Google Scholar]
- 34. Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 35. Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. (2005) J. Biol. Chem. 280, 42841–42847 [DOI] [PubMed] [Google Scholar]
- 36. Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004) J. Biol. Chem. 279, 3787–3892 [DOI] [PubMed] [Google Scholar]
- 37. Magra A. L., Mertz P. S., Torday J. S., Londos C. (2006) J. Lipid Res. 47, 2367–2373 [DOI] [PubMed] [Google Scholar]
- 38. Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z., Zhou L. (2011) J. Biol. Chem. 286, 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.