Abstract
Replicating amyloids, called prions, are responsible for transmissible neurodegenerative diseases in mammals and some heritable phenotypes in fungi. The transmission of prions between species is usually inhibited, being highly sensitive to small differences in amino acid sequence of the prion-forming proteins. To understand the molecular basis of this prion interspecies barrier, we studied the transmission of the [PSI+] prion state from Sup35 of Saccharomyces cerevisiae to hybrid Sup35 proteins with prion-forming domains from four other closely related Saccharomyces species. Whereas all the hybrid Sup35 proteins could adopt a prion form in S. cerevisiae, they could not readily acquire the prion form from the [PSI+] prion of S. cerevisiae. Expression of the hybrid Sup35 proteins in S. cerevisiae [PSI+] cells often resulted in frequent loss of the native [PSI+] prion. Furthermore, all hybrid Sup35 proteins showed different patterns of interaction with the native [PSI+] prion in terms of co-polymerization, acquisition of the prion state, and induced prion loss, all of which were also dependent on the [PSI+] variant. The observed loss of S. cerevisiae [PSI+] can be related to inhibition of prion polymerization of S. cerevisiae Sup35 and formation of a non-heritable form of amyloid. We have therefore identified two distinct molecular origins of prion transmission barriers between closely sequence-related prion proteins: first, the inability of heterologous proteins to co-aggregate with host prion polymers, and second, acquisition by these proteins of a non-heritable amyloid fold.
Keywords: Amyloid, Prions, Protein Folding, Translation Release Factors, Yeast, Saccharomyces, Sup35, Prion Interference, Prion Species Barrier
Introduction
Noncovalent polymerization of proteins coupled with their deep conformational rearrangement can result in the formation of amyloid fibers possessing a regular cross-β-sheet structure. Amyloid formation is associated with >30 different diseases in humans and other mammals, many of which are neurodegenerative in nature, e.g. Alzheimer, Parkinson, and Huntington diseases (1). Amyloid diseases are noninfectious, except for the prion diseases that are linked to the PrP protein and that include Creutzfeldt-Jakob disease, sheep scrapie, and other transmissible spongiform encephalopathies (2–5). Importantly, prion transmission between animal species is often impeded or blocked, even when the difference in sequence of the PrP proteins is small (6). For example, ovine prions cannot directly infect humans, but they can infect cattle, and bovine prions can then infect humans, although inefficiently (7, 8).
Prions have also been described in yeast; these prions are generally not detrimental and manifest themselves as genetic elements with unusual inheritance properties (9–15). Among yeast prions, the best studied is [PSI+], the prion determinant that gives rise to a nonsense suppressor phenotype as a consequence of the aggregation and partial inactivation of the translation termination factor Sup35 (eRF3) (16–18). The N-terminal domain of Sup35 is essential for the de novo appearance and maintenance of [PSI+] and is therefore referred to as the prion-forming domain. The function of the charged middle domain of Sup35 is unclear, whereas the C-terminal domain performs the essential translation termination activity of this protein (19, 20). In [PSI+] cells, Sup35 polymerizes via its prion domain (21). Similar to mammalian prions, [PSI+] can exist in multiple variants or strains, which can be distinguished by the strength of their nonsense suppressor phenotype and stability of inheritance. Strong [PSI+] variants show efficient nonsense suppression and high mitotic stability, whereas weak variants show inefficient nonsense suppression and low mitotic stability (22, 23). The dissimilarity in the properties of the [PSI+] variants reflects heritable differences in the structure of the underlying Sup35 prion polymers (24, 25).
The yeast [PSI+] prion has been used to demonstrate conservation of the prion properties of Sup35 proteins from different distantly related yeast species and the inability to transmit the infectious prion state between Sup35 proteins from these species. The observed prion species barrier appears to be linked to the inability of Sup35 molecules from different species to co-aggregate, a prerequisite for efficient transmission of the prion state (26–28). However, such a barrier for prion transmission can also exist between two sequence-related prion proteins that are able to co-aggregate or interact in polymerization. For example, when expressed in murine cell culture, hamster PrP blocked prion propagation of murine PrP, i.e. interacted with it but did not acquire the prion state (29). In yeast, polymerization of overproduced Sup35 can be seeded efficiently by an imperfect template, the prion form of Rnq1, but the resulting amyloid polymers of Sup35 are predominantly non-heritable due to their poor fragmentation by the molecular chaperone Hsp104 (30). The highly preferential formation of non-heritable polymers on a prion template made up of a different protein is a more general phenomenon and has been reported for other combinations of seeding and seeded proteins (31). This finding led us to propose that the formation of non-heritable polymers in place of heritable prion polymers may be the cause of transmission barriers between prion proteins able to interact (32).
Consistent with this hypothesis, Chen et al. (33) observed that Sup35 proteins from Saccharomyces paradoxus and Saccharomyces bayanus co-aggregated very efficiently with the prion form of Saccharomyces cerevisiae Sup35 without acquiring the prion state. This implied that these heterologous Sup35 proteins efficiently adopted a non-heritable amyloid structure. However, a further study of these proteins by Chen et al. (34) showed higher [PSI+] transmission and lower Sup35 co-aggregation than originally reported. Hence, the possibility of co-aggregation without prion transmission cannot be reliably concluded from these studies.
To check our hypothesis of the formation of non-heritable amyloid folds, we analyzed the co-polymerization of four closely related Sup35 proteins with the [PSI+] form of S. cerevisiae Sup35 and established whether the [PSI+] prion could be efficiently transmitted to the heterologous proteins. In several cases, we observed efficient co-polymerization without concomitant prion transmission, confirming our hypothesis. In addition, our study shows that heterologous Sup35 proteins can have a dominant-negative effect on [PSI+] prion propagation. These new data allow us to make the first detailed reconstruction of the molecular events responsible for [PSI+] elimination when two closely related prion proteins are expressed in the same cell, and they point to the formation of a non-heritable fold by heterologous Sup35 as being critical for prion transmission barriers.
EXPERIMENTAL PROCEDURES
Media, Strains, Plasmids, and Genetic Methods
Yeast cells were grown at 30 °C on either complete (yeast extract/peptone/dextrose (YPD)) or synthetic complete medium containing 2% glucose. For assaying the [PSI+] suppressor phenotype, selective media with a decreased concentration (0.07 mg/ml) of adenine sulfate or modified YPD (YPDred: 0.5% yeast extract, 2% peptone, and 4% glucose) were used because these media promote accumulation of red pigment in the ade2 mutants. To eliminate the [PSI+] determinant, the cells were grown from single cells to colonies on medium containing 3 mm guanidine hydrochloride. DNA transformation of the yeast cells was performed using the lithium acetate method (35).
Centromeric and multicopy plasmids carrying the hybrid SUP35 genes were constructed on the basis of the pRS315-SUP35-SE plasmid. To create this plasmid, the S. cerevisiae genomic XhoI-XbaI fragment encompassing the SUP35 gene was inserted into the same sites of the pRS315 polylinker. Then, the region encoding the prion domain (amino acid residues 1–120) was replaced with a short fragment containing sites for SmaI, BglII, SacI (Ecl136II), and NarI. The region encoding residues 1–120 was amplified from the genomic DNA of the yeast S. cerevisiae, Saccharomyces mikatae, Saccharomyces kudriavzevii, S. paradoxus, and S. bayanus and inserted into the BglII and Ecl136II sites. In the constructs obtained, the coding regions were joined seamlessly, but Met-124 was replaced with alanine to exclude translation initiation at this residue. The 5′-noncoding region was slightly altered: the region of nucleotides −11 to −3 was replaced with sequence GATCCCCGGGAGATCT. The 3-HA versions of these SUP35 constructs encoded proteins with the 3-HA tag (amino acid sequence GLINIFYPYDVPDYAGYPYDVPDYAGSYPYDVPDYAAQIP) inserted after amino acid residue 251.
The S. cerevisiae strain 22V-H63-ΔS35 (MATa ade2-1 SUQ5 kar1 lys1 his3 ura3 leu2 cyhR ΔSUP35) and its [PSI+] derivatives were used (36). This strain carries the chromosomal deletion of SUP35 and the pRS316-SUP35 URA3 plasmid with S. cerevisiae SUP35. To check the ability of the hybrid proteins to acquire a prion state, we replaced the pRS316-SUP35 plasmid with the pRS315 LEU2 plasmids bearing the hybrid SUP35 genes. The [psi−] colonies of strain 22V-H63-ΔS35 are distinguished by their red color and adenine requirement because the weak serine-inserting tRNA suppressor SUQ5 (SUP16) cannot suppress the ade2-1 ochre mutation in the absence of the [PSI+] determinant. This allowed selection of [PSI+] in the transformants of this strain by the appearance of the colonies with white or pink color, depending on the [PSI+] variant. To quantify the mitotic stability of [PSI+] based on the hybrid Sup35 proteins, three colonies of each [PSI+] isolate were suspended in water and plated onto YPDred medium. Plates were incubated for 3 days, and the percentage of red colonies was determined.
[PSI+] Transmission and Curing
The efficiency of [PSI+] transmission to the hybrid Sup35 proteins was determined as follows. 48 transformants for each combination of SUP35 type and [PSI+] variant were grown on synthetic complete medium lacking leucine but containing uracil to allow the cells to lose spontaneously the original URA3 SUP35-cer plasmid pRS316-SUP35. The efficiency of transmission was then counted among the Ura− clones as the proportion of the Ade+ clones. To ensure that the Ade− clones did not contain any phenotypically silent “ultra-weak” hybrid [PSI+], the presence of Sup35 polymers was checked by semidenaturing detergent-agarose gel electrophoresis in about one-third of the Ade− clones, and the polymers were never found.
The frequency of [PSI+] loss caused by the presence of hybrid Sup35 proteins was defined as the proportion of [psi−] cells in colonies arising from single [PSI+] cells and determined as follows. For each combination of hybrid Sup35 protein and [PSI+] variant, three transformants were taken, each suspended in liquid synthetic complete medium and streaked to single cells on synthetic complete medium plates containing leucine (i.e. non-selective for plasmids with hybrid SUP35) but lacking uracil and low in adenine. After 4 days of growth, white or sectored colonies were scored as [PSI+] and red colonies as [psi−]. All red colonies were unable to grow in the absence of adenine.
Preparation of Yeast Cell Lysates
The yeast cultures were grown in liquid medium to A600 = 1. The cells were harvested, washed in water, and lysed by vortexing with glass beads in buffer containing 30 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 10 mm dithiothreitol. To prevent proteolytic degradation, 25 mm EDTA, 10 mm phenylmethylsulfonyl fluoride, and Complete protease inhibitor mixture (Roche Applied Science) were added. The cell debris was removed by centrifugation at 10,000 × g for 5 min.
Centrifugation
To separate Sup35 polymer and monomer fractions, 100 μl of yeast cell lysate was centrifuged through 100 μl of a 10% sucrose cushion at 100,000 × g (48,000 rpm in a TLA-100.1 rotor, Beckman Optima TL ultracentrifuge) for 15 min at 4 °C.
Electrophoresis
Sup35 amyloid polymers were analyzed by electrophoresis on horizontal 1.8% agarose gels with 25 mm Tris, 250 mm glycine, and 0.1% SDS (semidenaturing detergent-agarose gel electrophoresis) (37). To analyze Sup35 polymers and monomers in a single gel, the standard SDS-PAGE system (38) was modified as described (39). Yeast cell lysates were mixed with sample buffer, incubated for 1 min at room temperature, and loaded onto the gel, and electrophoresis was run for 30 min. The Sup35 monomers were separated, whereas polymers stopped at the start of the stacking gel. To dissolve and analyze the polymers, sample buffer was loaded into the wells, and the gel was sealed and boiled for 5 min. The separation was continued after boiling. After electrophoresis, the proteins were transferred to a Hybond ECL nitrocellulose membrane and decorated with antibody to the Sup35 N-terminal and middle domains. The bound antibodies were detected using the GE Healthcare ECL system.
RESULTS
Design and Functional Activity of Hybrid Sup35 Proteins
To establish the mechanisms that prevent transmission of inherited prion folds between sequence-related prion proteins, we used Sup35 from yeast of the Saccharomyces sensu stricto group, namely S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus (40, 41). Previous studies have shown that the Sup35 N-terminal domain plays a key role in the “species barrier” (26, 33). In this study, to exclude any possible influence of the middle and C-terminal domains of Sup35 on the interactions between the heterologous Sup35 molecules, we used hybrid SUP35 genes. These genes were created from S. cerevisiae SUP35 by replacing the sequence encoding the N-terminal domain (residues 1–120) with the corresponding regions from heterologous SUP35 genes (Fig. 1). The encoded hybrid proteins were designated as Sup35-par, Sup35-mik, Sup35-kud, and Sup35-bay, respectively, and all constructs were verified by DNA sequencing. The Sup35-kud sequence has not been previously reported, and in contrast to the S. bayanus Sup35 sequence used by Chen et al. (33), the Sup35-bay sequence used here did not differ from S. cerevisiae Sup35 (Sup35-cer) at position 17 (serine). The amino acid similarities of the N-terminal regions of these proteins to the Sup35-cer N-terminal region are 94, 89, 86, and 78%, respectively. Importantly, the prion domains of Sup35-par and Sup35-mik align with Sup35-cer with no gaps, whereas Sup35-kud has one gap and Sup35-bay lacks one oligopeptide repeat and has two single-residue insertions (Fig. 1A).
FIGURE 1.
Sup35 proteins used. A, sequence alignment of Sup35 prion domains from S. cerevisiae (cer), S. mikatae (mik), S. kudriavzevii (kud), S. paradoxus (par), and S. bayanus (bay). Only differing amino acid residues are shown. Gaps are shown by highlighted dashes. B, scheme of the hybrid Sup35 proteins. The Sup35 N-terminal domain (N) was replaced with analogous domains from the indicated species. In one set of constructs, the 3-HA tag was placed after residue 251. M, middle domain; C, C-terminal domain.
To differentiate between the different Sup35 proteins, we created an additional set of Sup35 constructs by inserting the 3-HA tag after amino acid residue 251, i.e. just before the C-terminal domain (Sup35-3-HA); addition of the 3-HA tag reduced the electrophoretic mobility of the Sup35 protein. All hybrid Sup35 proteins, including those with the 3-HA tag, supported viability of the otherwise Sup35-deficient 22V-H63-ΔS35 [psi−] strain. Cells producing Sup35 without a 3-HA tag showed the expected non-suppressed phenotype, although the red colony color was less pronounced than in the [psi−] control (Fig. 2, left panel). This phenotypic effect may be related to a minor change in the 5′-untranslated region, which led to a small decrease in Sup35 levels (see Fig. 4, cer lane, and supplemental Fig. S1). Cells producing Sup35 with the 3-HA tag showed an Ade+ phenotype and white colony color, suggesting that the 3-HA tag reduced activity of the Sup35-3-HA proteins in translation termination.
FIGURE 2.
Phenotypes of the cells producing hybrid Sup35. Left panel, [psi−] cells producing only the indicated Sup35 proteins. Right panel, cells producing Sup35-cer in the prion variant indicated at the top and hybrid Sup35 indicated on the side. The presence of the 3-HA tag in hybrid Sup35 is indicated at the bottom.
FIGURE 4.
Electrophoretic analysis of co-polymerization of hybrid Sup35 proteins. Yeast cells with the indicated [PSI+] variants were transformed with centromeric plasmids producing hybrid Sup35-3-HA proteins. Cell lysates were loaded onto gels without boiling and run for half a distance. The whole gels were then boiled, and the electrophoretic separation was continued. The gels were blotted, and the blots were stained with antibody to the Sup35-cer N-terminal and middle domains. Sx-3HA, hybrid Sup35-3-HA proteins; Sc, Sup35-cer lacking the tag. For ease of comparison, the values for [PSI+] loss and transmission are given below the gels. The W1 panel also shows lysates of the cells having lost the plasmid encoding Sup35-cer (asterisk).
Sup35 from Different Saccharomyces Species Can Form Prions in S. cerevisiae Cells
The concept of the prion transmission barrier implies that heterologous proteins are able to acquire a prion state. To establish this for the hybrid Sup35 proteins, we attempted to obtain their prion state through overproduction of these proteins. The 22V-H63-ΔS35 [psi−] [PIN+] strain, carrying a centromeric LEU2-based plasmid encoding one or other of the hybrid Sup35 proteins, was transformed with a multicopy URA3-based plasmid encoding the same hybrid Sup35, and Ade+ Leu+ clones were selected. In most cases, the Ade+ phenotype was lost following growth on the guanidine hydrochloride-containing medium, which is indicative of the [PSI+] prion being responsible for the phenotype. This was confirmed by the presence of SDS-resistant polymers of hybrid Sup35 proteins in the guanidine hydrochloride-curable Ade+ clones (supplemental Fig. S2). The newly generated hybrid [PSI+] isolates varied in suppressor efficiency and the degree of mitotic stability. The [PSI+] strains based on Sup35-mik and Sup35-bay were mainly of the weak type (supplemental Table SI), the latter in agreement with previous results (33).
Expression of Hybrid Sup35 Proteins Affects the [PSI+] Phenotype and Propagation
To study the interaction of hybrid Sup35 proteins with the [PSI+] prion based on Sup35-cer, we used four independent [PSI+] variants of strain 22V-H63-ΔS35: strong (S), weak-1 (W1), weak-2 (W2), and weak-3 (W3). This strain harbors the SUP35 chromosomal deletion and a centromeric URA3-based plasmid encoding Sup35-cer (36). Cells with these [PSI+] variants were transformed with centromeric LEU2-based plasmids encoding one or other of the hybrid Sup35 proteins or Sup35-cer as a control. The phenotypes of resulting transformants were assessed on medium lacking Leu and Ura to ensure coexpression of the hybrid and wild-type Sup35 proteins in the same cell.
In contrast to wild-type Sup35 controls, all transformants producing hybrid Sup35 without the 3-HA tag showed no [PSI+] nonsense suppression phenotype, i.e. manifested antisuppression (Fig. 2). This indicates that a significant proportion of Sup35 in these cells remained soluble and functional, leading to efficient translation termination. The antisuppressor effect of the hybrid Sup35-3-HA proteins was much less pronounced, which suggests that the soluble Sup35 was represented mainly by hybrid Sup35. In several cases, expression of the hybrid Sup35 proteins caused the [PSI+] loss with frequencies being dependent on both the [PSI+] variant and hybrid Sup35. For example, weak [PSI+] variants in the presence of Sup35-bay or Sup35-mik produced [psi−] cells with up to 24% frequency (the proportion of [psi−] cells in a colony), whereas expression of Sup35-kud caused no detectable [PSI+] loss (Fig. 3 and supplemental Table SII). The presence of the 3-HA tag did not significantly alter the measured [PSI+] loss for any hybrid Sup35 (supplemental Table SII), which is consistent with the 3-HA tag not affecting Sup35 polymerization. The ability of the hybrid Sup35 proteins to interfere with [PSI+] propagation indicates a physical interaction between the hybrid Sup35 molecules and the prion form of Sup35-cer.
FIGURE 3.

Frequencies of [PSI+] transmission and loss. Upper, [PSI+] transmission to the indicated Sup35 alleles. Lower, [PSI+] loss in the presence of the indicated Sup35 alleles. The [PSI+] variants used are shown. All hybrid Sup35 proteins lacked the 3-HA tag, but the [PSI+] loss was similar in the presence of hybrid Sup35 with 3-HA (supplemental Table SII). The data in the original digital form are presented in supplemental Tables SII and SIII.
Hybrid Sup35 Proteins Can Co-polymerize with Sup35-cer
To establish whether the hybrid Sup35 proteins are able to form polymers in the presence of the prion form of Sup35-cer, we used a novel electrophoretic technique that allows quantitative analysis of the prion polymer and monomer fractions of a cell lysate in one gel (39). This method avoids the problem encountered by the more common centrifugation-based assay, namely that soluble Sup35 can appear in the high molecular weight fraction through its association with ribosomes or other translation factors (18). In these experiments, we used hybrid Sup35-3-HA proteins, which allowed us to distinguish them from wild-type Sup35-cer by reduced electrophoretic mobility.
The hybrid Sup35 proteins showed varying degrees of polymerization depending on which hybrid Sup35 and [PSI+] variant were used. Efficient polymerization was observed for Sup35-par in all [PSI+] variants, except for W3, and for Sup35-mik in the W1 and W2 [PSI+] variants (Fig. 4 and supplemental Fig. S3). Centrifugation analysis of the same strains demonstrated a similar extent of Sup35 polymerization (supplemental Fig. S4).
In several cases, coexpression of the hybrid Sup35 proteins resulted in a significant increase in soluble Sup35-cer levels (Fig. 4). This can be explained in part by the presence of [psi−] cells in the population. However, in some cases, the proportion of soluble Sup35-cer appeared to be higher than the observed proportion of [psi−] cells. For example, coexpression of Sup35-mik, Sup35-bay, or Sup35-par along with Sup35-cer in the W3 [PSI+] strain resulted in appearance of ∼10% of [psi−] cells. The levels of soluble Sup35-cer in the W3 [PSI+] cells expressing Sup35-mik or Sup35-bay were ∼10–15%, and therefore, they could be accounted for by the presence of [psi−] cells. However, the soluble fraction of Sup35-cer in the cells expressing Sup35-par was higher by ∼20%, and this value can be attributed to the inhibition of Sup35-cer polymerization by the Sup35-par protein. Similar increased levels of soluble Sup35-cer were caused by Sup35-mik in the W1 and W2 [PSI+] variants and by Sup35-bay in the W1 [PSI+] variant (Fig. 4).
In contrast to other Sup35 proteins, the Sup35-kud and Sup35-bay proteins showed low levels of polymerization, if any at all. However, although coexpression of Sup35-kud caused no loss of the [PSI+] prion, coexpression of Sup35-bay eliminated weak [PSI+] with 13–24% efficiency (Fig. 3), suggesting that Sup35-bay interacted with the prion form of Sup35-cer, whereas Sup35-kud did not.
It should be noted that the co-polymerization of hybrid Sup35 proteins with Sup35-cer was equivalent to the levels of their polymerized form only when [PSI+] transmission was absent or negligible. When [PSI+] was transmitted with a significant frequency, in a certain proportion of cells, hybrid Sup35 acquired a prion fold and formed inheritable polymers independently of Sup35-cer. The proportion of such cells equaled the transmission efficiency and was close to zero in most cases, excluding expression of Sup35-par in S, W1, and W2 [PSI+] cells (supplemental Table SIII). The Sup35-par co-polymerization in S and W1 [PSI+] cells could be estimated as close to 100% and in W2 [PSI+] cells as 40% (supplemental “Methods”).
Intermolecular Transmission of the [PSI+] Prion State
To establish the frequency of the [PSI+] transmission from Sup35-cer to the hybrid Sup35 proteins, it is required to induce loss of the URA3 plasmid encoding Sup35-cer and then score the [PSI+] phenotype. Usually, the loss of URA3 plasmids is achieved by growing cells on 5-fluoroacetic acid-containing medium. However, we have observed that 5-fluoroacetic acid caused significant loss of weak [PSI+] (supplemental Table SIV), which can result in underestimation of the transmission frequency of these [PSI+] variants. This forced us to rely on the spontaneous loss of the URA3 SUP35-cer plasmid.
No [PSI+] transmission to Sup35-mik, Sup35-bay, and Sup35-kud was detected, whereas the control transmission to Sup35-cer occurred with 100% frequency. Sup35-par acquired the S [PSI+] variants with 68% frequency and the weak [PSI+] variants with 5–13% frequency (Fig. 3 and supplemental Table SIII).
DISCUSSION
To define the molecular basis of interspecies prion transmission barriers, we employed the yeast [PSI+] prion model. Hybrid Sup35 proteins were created using S. cerevisiae Sup35, but the N-terminal prion-forming domain was replaced with the corresponding sequence from four closely related Saccharomyces species, S. mikatae, S. kudriavzevii, S. paradoxus, and S. bayanus. By coexpressing these Sup35 molecules in an S. cerevisiae strain bearing one of four [PSI+] prion variants, we have established the ability of hybrid Sup35 molecules to co-polymerize with Sup35-cer and the efficiency with which different [PSI+] variants can be transmitted to the hybrid Sup35 proteins. In addition, we have found that coexpression of hybrid Sup35 proteins can destabilize native [PSI+].
In the majority of cases, hybrid Sup35 proteins showed different patterns of physical and genetic interaction with the [PSI+] form of Sup35-cer, and this behavior also depended on the [PSI+] variant being examined. The lack of, or very weak, interaction as assayed by all of the tests used was observed only for Sup35-kud in all [PSI+] variants and for Sup35-bay and Sup35-mik in the S [PSI+] variants.
Hybrid Sup35 Proteins Acquire a Non-heritable Fold from Sup35-cer Prion Seeds
Two proteins were able to co-polymerize efficiently with the prion form of Sup35-cer: Sup35-mik in the W1 and W2 [PSI+] variants and Sup35-par in the S, W1, and W2 [PSI+] variants. However, no transmission of [PSI+] to Sup35-mik was detected in these cases. This allowed us to characterize the fold(s) acquired by Sup35-mik as non-heritable because they could not propagate in the absence of original prion based on Sup35-cer. Such behavior is also typical of the non-heritable amyloids of Sup35, which cannot propagate in the absence of the [PIN+] prion (30).
The S, W1, and W2 [PSI+] variants were transmitted to Sup35-par with a frequency of 10–70% (Fig. 3 and supplemental Table SIII). Thus, in these cases, in 30–90% of cells, Sup35-par did not acquire the prion state and efficiently co-polymerized with Sup35-cer in a non-heritable fold. Despite the significant [PSI+] transmission values observed in these experiments, the probability (p) with which a Sup35-par molecule acquires the prion fold upon joining to a Sup35-cer prion polymer is extremely low. In these experiments, Sup35-par binds to Sup35-cer prion polymers very many times (n) in every cell lineage. Because [PSI+] is heritable, if the prion fold is acquired by Sup35-par in just one such binding event in a cell, [PSI+] should be present in all progeny of this cell. Therefore, p should be very low; otherwise, the final efficiency of the [PSI+] transmission (P) should approach 100%. p may be estimated as p = P/n, when P is significantly <100%. We estimate n as being of the order of tens of thousands or larger, and so p is <10−4. More precise expression for p is p = 1 − (1 − P)1/n (supplemental “Methods”).
In five combinations of the Sup35 type/[PSI+] variant, we observed a significant [PSI+] loss (10–24%) even though co-polymerization of hybrid Sup35 proteins with Sup35-cer was barely detectable. This suggests that hybrid Sup35 molecules in these cases joined the Sup35-cer polymer but as a single molecule or a small number of molecules. It is likely that hybrid Sup35 acquired an amyloid fold, but certainly this was not a prion fold.
Previously, we have shown that prions preferentially seed non-prion amyloids when the seeding and seeded proteins are different (30, 31). This work shows that such a preference exists even for closely related Sup35 proteins. These observations suggest a certain common feature of prion folds, which makes their formation unfavorable. This feature could relate to another characteristic property of prions: in contrast to normal protein folds and non-heritable amyloid folds (30), prion folds are recognized by chaperones, such as Hsp104 and/or other chaperones, which results in the fragmentation of prion polymers. The chaperone recognition is likely to be related to the exposure of hydrophobic amino acid residues on the surface of amyloid (42), which is unfavorable in terms of energy because hydrophobic residues normally tend to be buried inside of a molecule to minimize their contact with water.
Molecular Model for the Interaction of Heterologous Sup35 with the Prion Form of Sup35-cer
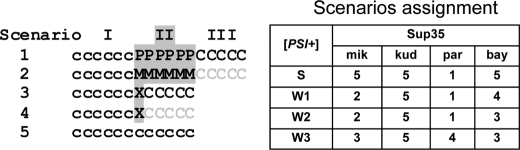
All of the heterologous Sup35 proteins studied, except for Sup35-kud in all [PSI+] variants and Sup35-bay in S [PSI+] variants (Fig. 5, Scenario 5), interacted with the prion forms of Sup35-cer as judged by their co-polymerization and induced [PSI+] elimination. Elimination of native [PSI+] induced by heterologous Sup35 is a novel effect described in this work, and uncovering its mechanisms requires a detailed analysis of how Sup35-cer polymerizes in the presence of closely related Sup35 molecules. [PSI+] loss can result from binding of heterologous Sup35 to the Sup35-cer prion polymer end and inhibition of further Sup35-cer polymerization. This should be manifested in the increased levels of soluble Sup35-cer, and such an effect was most pronounced for Sup35-mik and Sup35-bay in W1 and W2 [PSI+] and for Sup35-par in W3 [PSI+] (Fig. 5, Scenarios 2 and 4). However, in other cases, the levels of soluble Sup35-cer were not substantially increased. Therefore, in these cases, joining of a heterologous Sup35 molecule to the end of a Sup35-cer prion polymer did not preclude binding of Sup35-cer to the terminal heterologous Sup35 and its further polymerization. However, in such a heterotypic seeding, one should expect highly preferential formation of a non-prion fold by Sup35-cer (Fig. 5, Scenarios 1 and 3).
FIGURE 5.
Molecular scenarios for Sup35 polymerization observed in this work. Scenarios are designated as indicated on the right. Polymers are represented as sequences of letters corresponding to Sup35 molecules and reflecting their origin: c and C denote cerevisiae, P denotes paradoxus, M denotes mikatae, and X denotes various hybrid Sup35 proteins. Lowercase letters designate the prion fold, and uppercase letters indicate the non-heritable fold. For simplicity, polymer growth is shown only on the right. The prion fold reproduction may occur only in area I after its fragmentation by Hsp104. Gray letters in area III reflect the lack or reduced probability of polymerization. In Scenario 5, hybrid Sup35 does not incorporate into a polymer.
Despite this, the [PSI+] propagation would be expected to continue if Hsp104 breaks the co-polymer within the original (i.e. seeding) prion stretch of Sup35-cer (denoted by lowercase c in Fig. 5), thus creating new polymer ends that can seed Sup35-cer prion formation. The [PSI+] prion can propagate in such a way only if the incorporation of hybrid Sup35 into Sup35-cer polymers occurs less frequently than the polymer fragmentation mediated by Hsp104. The fragmentation frequency may be roughly estimated as an inverse value of the average number of Sup35 molecules in a polymer. So, S [PSI+] variants with a polymer size of ∼20 Sup35 molecules (18) would be rapidly eliminated if the probability (h) of incorporation of hybrid Sup35 compared with Sup35-cer exceeds 1/20, or 5%.
Thus, the probability h is a key parameter defining the efficiency of prion elimination. In our experiments, h was always low because we never observed complete [PSI+] loss. Furthermore, low h implies that, in prion polymers made up of two different proteins, these proteins are not randomly mixed but rather are arranged in stretches of similar molecules. Consequently, the levels of co-polymerization of heterologous Sup35 are proportional to both the probability h to initiate such a stretch and the average number of these molecules in a stretch.
The expression of Sup35-bay and Sup35-mik induced loss of both the W1 and W2 [PSI+] variants with comparable frequency, although the amount of co-polymerized Sup35-mik was much higher compared with Sup35-bay. This leads us to suggest that these proteins joined onto Sup35-cer polymers with comparable frequency, but Sup35-mik formed relatively long homopolymeric stretches (Fig. 5, Scenario 2), whereas Sup35-bay joined as a single or a small number of molecules (Scenarios 3 and 4). The latter also applies to Sup35-par and Sup35-mik in W3 [PSI+] cells.
It may appear paradoxical that Sup35-par eliminated [PSI+] inefficiently despite its efficient co-polymerization with Sup35-cer in the S, W1, and W2 [PSI+] cells. This could be explained, at least in part, by the observation that the levels of soluble Sup35-par in the considered cells were significantly lower than those of any other hybrid Sup35 proteins we examined. The probability h should be proportional to the levels of soluble heterologous Sup35; thus, h and, correspondingly, the [PSI+] curing were low in these cases.
Implications for Elimination of Prions and Amyloids
In this work, we observed that even minor incorporation of heterologous Sup35 could cause a significant loss of native [PSI+]. It appears very likely that altered variants of the Sup35 prion-forming domain can be found that are able to join to the Sup35-cer polymer with higher frequencies and that would thus be able to rapidly eliminate the S. cerevisiae [PSI+] prion. Such alleles may be generated via mutagenesis of the native or heterologous Sup35 prion-forming domains. In a similar way, altered alleles of the mammalian prion and amyloid proteins may be found that would eliminate mammalian prions or block formation of amyloids. It is important to note that, although the induced prion curing can rely on two mechanisms, the conversion to a non-heritable amyloid fold and interference with polymerization via polymer end capping, the inhibition of amyloid formation can rely only on the latter. The inhibition of mammalian prion propagation by a heterologous protein has already been described in studies with murine neuroblastoma cells, where hamster PrP and several of its mutants profoundly interfered with prion propagation by mouse PrP (29).
Supplementary Material
Acknowledgments
We thank Dr. Gennady Naumov for the samples of S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus and Alexander K. Buell, Alexander Alexandrov, and Michael Agaphonov for productive discussions and help with the preparation of the manuscript.
Footnotes
The work was supported by the Wellcome Trust (to V. V. K. and M. F. T.) and the Russian Foundation for Fundamental Research (to M. D. T.-A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Figs. S1–S4, and Tables SI–SIV.
REFERENCES
- 1. Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 2. Horwich A. L., Weissman J. S. (1997) Cell 89, 499–510 [DOI] [PubMed] [Google Scholar]
- 3. Prusiner S. B., Scott M. R., DeArmond S. J., Cohen F. E. (1998) Cell 93, 337–348 [DOI] [PubMed] [Google Scholar]
- 4. Collinge J. (2001) Annu. Rev. Neurosci. 24, 519–550 [DOI] [PubMed] [Google Scholar]
- 5. Caughey B., Chesebro B. (2001) Adv. Virus Res. 56, 277–311 [DOI] [PubMed] [Google Scholar]
- 6. Scott M., Groth D., Foster D., Torchia M., Yang S. L., DeArmond S. J., Prusiner S. B. (1993) Cell 73, 979–988 [DOI] [PubMed] [Google Scholar]
- 7. Bruce M. E., Will R. G., Ironside J. W., McConnell I., Drummond D., Suttie A., McCardle L., Chree A., Hope J., Birkett C., Cousens S., Fraser H., Bostock C. J. (1997) Nature 389, 498–501 [DOI] [PubMed] [Google Scholar]
- 8. Hill A. F., Desbruslais M., Joiner S., Sidle K. C., Gowland I., Collinge J., Doey L. J., Lantos P. (1997) Nature 389, 448–450, 526 [DOI] [PubMed] [Google Scholar]
- 9. Wickner R. B. (1994) Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 10. Coustou V., Deleu C., Saupe S., Begueret J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sondheimer N., Lindquist S. (2000) Mol. Cell 5, 163–172 [DOI] [PubMed] [Google Scholar]
- 12. Du Z., Park K. W., Yu H., Fan Q., Li L. (2008) Nat. Genet. 40, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nemecek J., Nakayashiki T., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1892–1896 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patel B. K., Gavin-Smyth J., Liebman S. W. (2009) Nat. Cell Biol. 11, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1996) EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 17. Patino M. M., Liu J. J., Glover J. R., Lindquist S. (1996) Science 273, 622–626 [DOI] [PubMed] [Google Scholar]
- 18. Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. (2003) J. Biol. Chem. 278, 49636–49643 [DOI] [PubMed] [Google Scholar]
- 19. Ter-Avanesyan M. D., Kushnirov V. V., Dagkesamanskaya A. R., Didichenko S. A., Chernoff Y. O., Inge-Vechtomov S. G., Smirnov V. N. (1993) Mol. Microbiol. 7, 683–692 [DOI] [PubMed] [Google Scholar]
- 20. Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N. (1994) Genetics 137, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1997) Mol. Cell Biol. 17, 2798–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. (1996) Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kochneva-Pervukhova N. V., Chechenova M. B., Valouev I. A., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (2001) Yeast 18, 489–497 [DOI] [PubMed] [Google Scholar]
- 24. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 25. Krishnan R., Lindquist S. L. (2005) Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kushnirov V. V., Kochneva-Pervukhova N. V., Chechenova M. B., Frolova N. S., Ter-Avanesyan M. D. (2000) EMBO J. 19, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santoso A., Chien P., Osherovich L. Z., Weissman J. S. (2000) Cell 100, 277–288 [DOI] [PubMed] [Google Scholar]
- 28. Chien P., Weissman J. S. (2001) Nature 410, 223–227 [DOI] [PubMed] [Google Scholar]
- 29. Priola S. A., Caughey B., Race R. E., Chesebro B. (1994) J. Virol. 68, 4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salnikova A. B., Kryndushkin D. S., Smirnov V. N., Kushnirov V. V., Ter-Avanesyan M. D. (2005) J. Biol. Chem. 280, 8808–8812 [DOI] [PubMed] [Google Scholar]
- 31. Urakov V. N., Vishnevskaya A. B., Alexandrov I. M., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (2010) Prion 4, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kushnirov V. V., Vishnevskaya A. B., Alexandrov I. M., Ter-Avanesyan M. D. (2007) Prion 1, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen B., Newnam G. P., Chernoff Y. O. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2791–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen B., Bruce K. L., Newnam G. P., Gyoneva S., Romanyuk A. V., Chernoff Y. O. (2010) Mol. Microbiol. 76, 1483–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 36. Shkundina I. S., Kushnirov V. V., Tuite M. F., Ter-Avanesyan M. D. (2006) Genetics 172, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bagriantsev S. N., Kushnirov V. V., Liebman S. W. (2006) Methods Enzymol. 412, 33–48 [DOI] [PubMed] [Google Scholar]
- 38. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 39. Kushnirov V. V., Alexandrov I. M., Mitkevich O. V., Shkundina I. S., Ter-Avanesyan M. D. (2006) Methods 39, 50–55 [DOI] [PubMed] [Google Scholar]
- 40. Naumova E. S., Korshunova I. V., Jespersen L., Naumov G. I. (2003) FEMS Yeast Res. 3, 177–184 [DOI] [PubMed] [Google Scholar]
- 41. Naumova E. S., Bulat S. A., Mironenko N. V., Naumov G. I. (2003) Antonie Leeuwenhoek 83, 155–166 [DOI] [PubMed] [Google Scholar]
- 42. Alexandrov I. M., Vishnevskaya A. B., Ter-Avanesyan M. D., Kushnirov V. V. (2008) J. Biol. Chem. 283, 15185–15192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.