Abstract
Scorpion β-toxins bind to the extracellular regions of the voltage-sensing module of domain II and to the pore module of domain III in voltage-gated sodium channels and enhance channel activation by trapping and stabilizing the voltage sensor of domain II in its activated state. We investigated the interaction of a highly potent insect-selective scorpion depressant β-toxin, Lqh-dprIT3, from Leiurus quinquestriatus hebraeus with insect sodium channels from Blattella germanica (BgNav). Like other scorpion β-toxins, Lqh-dprIT3 shifts the voltage dependence of activation of BgNav channels expressed in Xenopus oocytes to more negative membrane potentials but only after strong depolarizing prepulses. Notably, among 10 BgNav splice variants tested for their sensitivity to the toxin, only BgNav1-1 was hypersensitive due to an L1285P substitution in IIIS1 resulting from a U-to-C RNA-editing event. Furthermore, charge reversal of a negatively charged residue (E1290K) at the extracellular end of IIIS1 and the two innermost positively charged residues (R4E and R5E) in IIIS4 also increased the channel sensitivity to Lqh-dprIT3. Besides enhancement of toxin sensitivity, the R4E substitution caused an additional 20-mV negative shift in the voltage dependence of activation of toxin-modified channels, inducing a unique toxin-modified state. Our findings provide the first direct evidence for the involvement of the domain III voltage-sensing module in the action of scorpion β-toxins. This hypersensitivity most likely reflects an increase in IIS4 trapping via allosteric mechanisms, suggesting coupling between the voltage sensors in neighboring domains during channel activation.
Keywords: Insect, Ion Channels, Neurotoxin, RNA Editing, Sodium Channels, Scorpion Toxin, Voltage-sensing Module
Introduction
Voltage-gated sodium (Nav) channels are essential for the initiation and propagation of action potentials in most excitable cells. They consist of a large pore-forming α-subunit that is associated with a variable number of smaller subunits in different excitable tissues (1). The α-subunit comprises four repeat homologous domains (I–IV), each having six membrane-spanning segments (S1–S6). The S1–S4 segments constitute the voltage-sensing module. S4 in each domain contains four to seven positively charged residues and moves outward in response to membrane depolarization, which initiates the channel activation process (1). The S5 and S6 segments and their connecting P loops compose the pore-forming module. Each reentrant P loop contains two short segments (SS1 and SS2) that span the membrane as a hairpin and form the lining of the pore.
Mammals produce functionally and pharmacologically diverse Nav α-subunits by selective expression of distinct sodium channel genes (at least nine in rat and human) in different tissues (2, 3). In insects such as Drosophila melanogaster and Blattella germanica, extensive alternative splicing and RNA editing of a single gene generate a broad array of variants that are diverse in gating and pharmacological properties (4–6).
Because of their pivotal role in excitability, Nav channels are targeted by a variety of toxins derived from plants and animals as part of their defense or preying strategies (1, 7–10). Among venomous animals, scorpions produce a rich repertoire of 61–76-residue-long peptide toxins that modify sodium channel gating upon binding to distinct extracellular receptor sites in the α-subunit (7, 8, 11–13). The toxins that affect Nav channels are divided into α- and β-classes according to their mode of action and binding properties (14, 15). α-Toxins inhibit channel fast inactivation in a voltage-dependent manner upon binding at receptor site 3, assigned mainly to domains I and IV. β-Toxins interact with receptor site 4, assigned to the voltage-sensing module of domain II and the pore module of domain III, and shift the voltage dependence of activation to more negative membrane potentials (11, 16–19). The sensitivity of mammalian Nav subtypes to scorpion β-toxins differs greatly. Rat brain (rNav1.2)2 and skeletal muscle (rNav1.4) channels are often more sensitive than the cardiac channel (rNav1.5) to β-toxins of South American scorpions such as Css4 from Centruroides suffusus suffusus, Ts1 from Tityus serrulatus, and TdVIII from Tityus discrepans (16–20). Moreover, Tz1 from Tityus zulianus is especially active at rNav1.4 compared with rNav1.2 and human Nav1.5 (18).
The prominent activity of Css4 on rNav1.2 and rNav1.4 compared with its very weak effect on rNav1.5 motivated Catterall and co-workers (16) to analyze each of the 16 extracellular loops of rNav1.5 in the background of rNav1.2. Their analysis revealed that IS5–SS1, IIS1–S2, IIS3–S4, and IIISS2–S6, particularly a G845N substitution in IIS3–S4, play a role in determining toxin preference, suggesting that scorpion β-toxins bind to the S3–S4 loop (16). In a similar fashion, differences in potency of the toxin Tz1 on rat muscle, cardiac, and neuronal Nav channels motivated Heinemann and co-workers (18) to swap rNav1.2a sequences in the background of rNav1.4. Their analysis revealed that three amino acid residues in the C-terminal pore loop (SS2–S6) of domain III determine the Tz1 preference for the skeletal muscle Nav channel (18).
To account for enhanced sodium channel activation by Css4, a voltage sensor-trapping model was proposed in which the S4 voltage sensor of domain II (IIS4) is trapped and stabilized in its outward activated position by the scorpion β-toxin (16, 21). Neutralization or reversal of gating charges in the voltage sensor of domain II enhances the action of scorpion β-toxins. According to the classical models of sodium channel gating, the voltage sensors of the sodium channel activate independently, and at least three of them have to be in an activated position for the channel to open (22, 23). For a scorpion β-toxin to shift the threshold of activation, more than one voltage sensor could be affected by the toxins. Bezanilla and co-workers (24) combined electrophysiological and spectroscopic measurements to determine the structural rearrangements induced by the scorpion β-toxin Ts1 on the rNav1.4 channel. Consistent with studies using Css4, Ts1 binding to the channel has been shown to be restricted to a single binding site in the voltage sensor of domain II, where it traps IIS4 in the activated state. Interestingly, Ts1 binding to S4 of domain II allosterically potentiates activation of the other three voltage sensors at more hyperpolarized potentials. However, it is unknown how voltage-sensing modules in other domains contribute to the voltage sensor trapping by β-toxins.
In this study, we analyzed the sensitivity of 10 B. germanica Nav (BgNav) variants (25) to a potent insect-selective Lqh-dprIT3 depressant toxin (26). The discovery of one variant that is hypersensitive to Lqh-dprIT3 led to the identification of the critical effect of Leu-1285 and Glu-1290 in IIIS1 on channel sensitivity to the toxin. We further show that substitutions of the two innermost positively charged residues (R4E and R5E) in IIIS4 also enhance the effect of Lqh-dprIT3. These findings demonstrate that the voltage-sensing module of domain III is critical for the action of the scorpion β-toxin.
EXPERIMENTAL PROCEDURES
Toxin Production and Functional Analysis
Of eight Lqh-dprIT3 variants produced by the scorpion Leiurus quinquestriatus hebraeus, the highly potent variant c, hereafter referred to as Lqh-dprIT3, was produced in recombinant form and analyzed as described previously (26).
Channel Mutagenesis
Site-directed mutagenesis of BgNav1-1 and BgNav1-1a was performed via PCR using oligonucleotide primers and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), and the modifications were verified by DNA sequencing.
Expression of BgNav Channels in Xenopus Oocytes
The procedures for oocyte preparation and cRNA injection are identical to those described previously (27). For robust expression of the BgNav channels, cRNA was co-injected into oocytes with D. melanogaster tipE cRNA (1:1 ratio), which enhances channel expression (28, 29).
Electrophysiological Recording and Analysis
The voltage dependence of activation and inactivation was measured using the two-electrode voltage clamp technique. Methods for two-electrode recording and data analysis were similar to those described previously (30). Sodium currents were measured with an OC725C oocyte clamp (Warner Instruments, Hamden, CT) and a Digidata 1322A interface (Axon Instruments Inc., Foster City, CA). Data were sampled at 50 kHz and filtered at 2 kHz. Leak currents were corrected by P/4 subtraction. pCLAMP 8.2 software (Axon Instruments Inc.) was used for data acquisition and analysis. The maximal peak sodium current was limited to <2.0 μA to achieve optimal voltage control.
The voltage dependence of sodium channel conductance (G) was calculated by measuring the peak current at test potentials ranging from −80 to +65 mV in 5-mV increments and divided by (V − Vrev), where V is the test potential and Vrev is the reversal potential for sodium ions. Peak conductance values were normalized to the maximal peak conductance (Gmax) and fit with a two-state Boltzmann equation of the form G/Gmax = (1 + exp(V − V1/2)/k)−1 or with the sum of two such expressions, where V is the potential of the voltage pulse, V1/2 is the voltage for half-maximal activation, and k is the slope factor.
The percentage of channel modification by Lqh-dprIT3 was determined by the percentage of channels with the voltage dependence of activation shifted to negative membrane potentials, which was derived from double Boltzmann fits of the conductance-voltage relationships. Data are presented as means ± S.D. Statistical significance was determined by one-way analysis of variance (p < 0.05).
RESULTS
Effects of Lqh-dprIT3 on Sodium Channel Splice Variant BgNav1-1
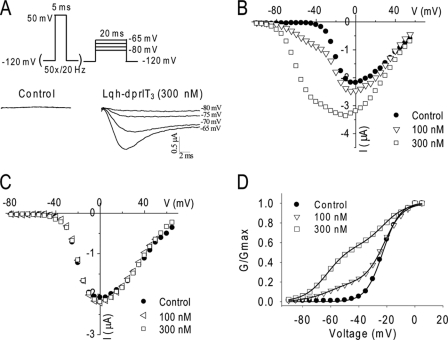
We first examined a well characterized cockroach sodium channel, BgNav1-1 (27), for the effect of Lqh-dprIT3. Previous studies showed that, for most of scorpion β-toxins, a strong depolarization prepulse is required for the toxins to induce a negative shift in the voltage dependence of activation (11, 16–19). For example, a 1-ms depolarizing prepulse to 50 mV followed by a depolarizing test pulse to −65 mV activates Css4-modified Nav1.2 channels but not unmodified channels (16). We initially used a similar protocol to examine the effect of Lqh-dprIT3 on BgNav1-1 channels and found that a brief depolarizing pulse was not sufficient to detect Lqh-dprIT3 action (data not shown). We then applied a 20-Hz train of 50 5-ms depolarizing prepulses to 50 mV as the conditional pulses, followed by a 20-ms depolarizing test pulse between −80 and −65 mV from a holding potential of −120 mV. As expected, in the absence of the toxin, no sodium currents were detected under any of the test pulses (Fig. 1A). However, in the presence of 300 nm Lqh-dprIT3, substantial sodium currents were detected at −75 to −65 mV (Fig. 1A), indicating that Lqh-dprIT3 modified the gating of BgNav1-1 channels.
FIGURE 1.
Effects of Lqh-dprIT3 on the BgNav1-1 sodium channel. A, sodium current traces in the absence (left) and presence (right) of Lqh-dprIT3 (300 nm). To measure the effect of Lqh-dprIT3, the following protocol was used: a 20-Hz train of 50 5-ms depolarizing prepulses to 50 mV as the conditional pulses, followed by a series of 20-ms depolarizing test pulses between −80 and −65 mV. The holding potential was −120 mV. B and C, current-voltage relations in the absence (●) and presence of 100 nm (△ and ◁) or 300 nm (□) Lqh-dprIT3 with (B) and without (C) the conditional depolarizing pulses. D, conductance-voltage relations in the absence (●) and the presence of 100 nm (△) or 300 nm (□) Lqh-dprIT3 with the conditional depolarizing pulses. The voltage dependence of activation was measured using the protocol described under “Experimental Procedures.”
The toxin effect was also evident in analyses of current-voltage relation and conductance curves exhibiting a negative shift in the voltage dependence of channel activation after conditional prepulses in the presence of toxin (Fig. 1, B and D). In addition, a significant increase in peak current was observed at 300 nm but not at a lower concentration (100 nm) (Fig. 1B). Neither of these effects was observed without the prepulses (Fig. 1C). The voltage dependence of channel inactivation was not affected by the toxin (data not shown). At both 100 and 300 nm, the current-voltage and conductance-voltage relations determined from Lqh-dprIT3 effects were biphasic (Fig. 1D). Fitting the conductance curves with the sum of two Boltzmann relations revealed that the voltage dependence of activation of 40% (with 100 nm Lqh-dprIT3) and 80% (with 300 nm Lqh-dprIT3) of BgNav1-1 channels shifted 40 mV to a more hyperpolarizing membrane potential. The extent of the toxin-induced hyperpolarizing shift in activation was not concentration-dependent. Thus, like other scorpion β-toxins, Lqh-dprIT3 causes sodium channels to activate at subthreshold membrane potentials.
Examination of the Sensitivity of BgNav1 Variants to Lqh-dprIT3
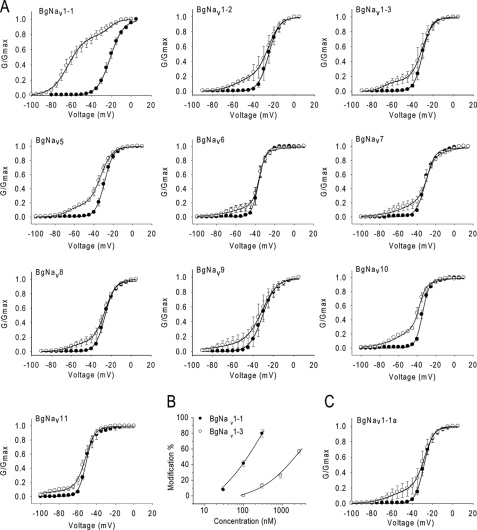
To identify specific regions of the channel that are involved in the enhanced response to Lqh-dprIT3, we examined different BgNav splice variants. We determined the voltage dependence of channel activation of nine additional BgNav splice variants in the absence or presence of Lqh-dprIT3 using the same protocol described in Fig. 1A. Because BgNav1-1 is a type 1 splice variant, we selected two additional splice variants of this type, BgNav1-2 and BgNav1-3, and seven variants each representing a different splice type. Besides differences in usage of alternative exons, these variants also contain four to eight unique amino acid changes likely due to RNA editing (25). Strikingly, Lqh-dprIT3 (300 nm) modified only 14–27% of the channels for all of the other splice variants compared with 80% for BgNav1-1 (Fig. 2A and Table 1), indicating that these variants are less sensitive to Lqh-dprIT3 compared with BgNav1-1. Dose-response analysis revealed that BgNav1-1 is 17-fold more sensitive to Lqh-dprIT3 compared with BgNav1-3 (Fig. 2B). The toxin-induced hyperpolarizing shift in the voltage dependence of channel activation ranged from −23 to −42 mV (Table 1).
FIGURE 2.
Effect of Lqh-dprIT3 on 10 BgNav variants. A, conductance-voltage relations in the absence (●) and presence (○) of 300 nm Lqh-dprIT3 for eight BgNav variants and the recombinant BgNav1-1a channel, originating from BgNav1-1. The conductance-voltage relation was measured using the protocol described under “Experimental Procedures.” B, dose-response curve of BgNav1-3 in comparison with BgNav1-1. C, conductance-voltage relation in the absence (●) and presence ○ of 300 nm Lqh-dprIT3 for the recombinant BgNav1-1a channel. Four amino acid changes in BgNav1-1 were reversed by site-directed mutagenesis generating BgNav1-1a (25).
TABLE 1.
Voltage dependence of activation of BgNav splices variants before and after application of 300 nm Lqh-dprIT3
Each value represents the mean ± S.D. for at least five oocytes.
| Toxin-free |
Lqh-dprIT3 (300 nm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| V0.5 | k | V0.5 | k1 | A1 | V0.5 | k2 | A2 | |
| % | % | |||||||
| BgNav1-1 | −23.2 ± 1.0 | 6.3 ± 0.3 | −21.0 ± 0.4 | 5.3 ± 0.3 | 20 ± 5a | −62.8 ± 2.6 | 7.9 ± 0.5 | 80 ± 5a |
| BgNav1-1a | −28.4 ± 2.1 | 4.1 ± 0.6 | −28.1 ± 1.5 | 5.0 ± 0.3 | 82 ± 5 | −60.0 ± 3.3 | 10.4 ± 1.6 | 18 ± 5 |
| BgNav1-2 | −25.2 ± 2.8 | 5.1 ± 1.1 | −25.5 ± 2.1 | 6.1 ± 1.4 | 81 ± 3 | −58.1 ± 1.2 | 8.6 ± 0.8 | 19 ± 3 |
| BgNav1-3 | −30.4 ± 2.1 | 4.3 ± 0.6 | −30.6 ± 1.5 | 5.5 ± 0.4 | 86 ± 4 | −68.7 ± 0.8 | 5.4 ± 0.3 | 14 ± 4 |
| BgNav5 | −35.9 ± 2.6 | 3.7 ± 0.5 | −35.7 ± 0.7 | 4.0 ± 0.4 | 77 ± 1 | −59.3 ± 3.0 | 11.5 ± 0.9 | 23 ± 1 |
| BgNav6 | −27.9 ± 2.4 | 4.1 ± 0.9 | −29.2 ± 2.6 | 5.2 ± 0.9 | 79 ± 5 | −59.0 ± 5.8 | 10.9 ± 2.3 | 21 ± 3 |
| BgNav7 | −31.7 ± 1.9 | 4.8 ± 0.4 | −32.5 ± 2.3 | 3.1 ± 0.6 | 79 ± 6 | −56.7 ± 2.8 | 11.8 ± 2.1 | 21 ± 5 |
| BgNav8 | −26.5 ± 1.0 | 4.8 ± 0.2 | −26.2 ± 0.9 | 5.5 ± 0.2 | 86 ± 3 | −60.0 ± 0.5 | 8.2 ± 0.2 | 14 ± 3 |
| BgNav9 | −30.7 ± 3.1 | 6.4 ± 1.2 | −31.0 ± 2.9 | 7.5 ± 1.4 | 82 ± 5 | −60.0 ± 2.6 | 10.1 ± 1.5 | 18 ± 5 |
| BgNav10 | −34.1 ± 2.4 | 3.5 ± 0.5 | −35.8 ± 0.7 | 3.9 ± 0.7 | 75 ± 6 | −58.6 ± 2.0 | 14.3 ± 2.6 | 25 ± 6 |
| BgNav11 | −49.3 ± 2.0 | −3.2 ± 0.5 | −43.2 ± 2.7 | 5.6 ± 0.7 | 73 ± 3 | −73.7 ± 3.0 | 2.9 ± 1.1 | 27 ± 3 |
a Significant differences from other splice variants (p < 0.05).
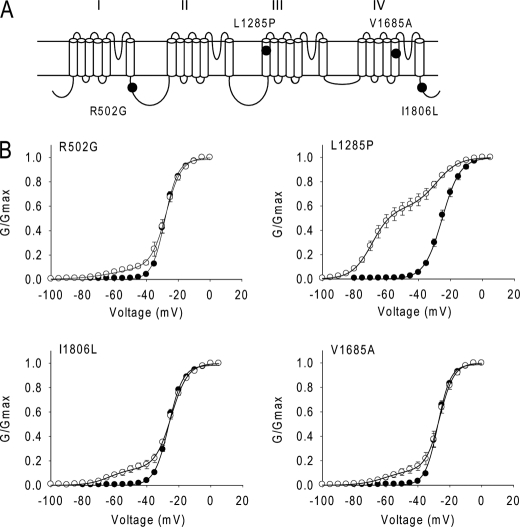
Because the type 1 splice variants BgNav1-2 and BgNav1-3 were less sensitive to Lqh-dprIT3 compared with BgNav1-1, we reasoned that the alternative exons common to these three variants were not responsible for the hypersensitivity to Lqh-dprIT3. Compared with BgNav1-2 and BgNav1-3, four amino acid residues are different in BgNav1-1: R502G, L1285P, V1685A, and I1806L (Fig. 3). Of these four differences, L1285P and V1685A have been shown to result from U-to-C RNA-editing events (25). We examined the effect of Lqh-dprIT3 on a recombinant channel, BgNav1-1a, in which these four unique residues were replaced with those of BgNav1-2 and BgNav1-3 (25). We found that the sensitivity of BgNav1-1a to Lqh-dprIT3 decreased prominently, with only 18% of the channels modified by 300 nm toxin (Fig. 2 and Table 2). This result suggested that one or more of these four amino acids were responsible for the hypersensitivity of BgNav1-1 to Lqh-dprIT3.
FIGURE 3.
L1285P is responsible for the hypersensitivity of BgNav1-1 to Lqh-dprIT3. A, the four amino acid changes in BgNav1-1 are indicated in the topology of the BgNav sodium channel protein. B, conductance curves in the absence (●) and presence (○) of 300 nm Lqh-dprIT3 for four BgNav1-1a channel mutants. Each of the four mutations was introduced into the BgNav1-1a channel. The conductance-voltage relation was measured using the protocol described under “Experimental Procedures.”
TABLE 2.
Voltage dependence of activation of BgNav1-1a and mutants before and after the application of 300 nm Lqh-dprIT3
Each value represents the mean ± S.D. for at least five oocytes.
| Toxin-free |
Lqh-dprIT3 (300 nm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| V0.5 | k | V0.5 | k1 | A1 | V0.5 | k2 | A2 | |
| % | % | |||||||
| BgNav1-1a | −28.4 ± 2.1 | 4.1 ± 0.6 | −28.1 ± 1.5 | 5.0 ± 0.3 | 82 ± 5 | −60.0 ± 3.3 | 10.4 ± 1.6 | 18 ± 5 |
| R502G | −28.3 ± 0.6 | 4.0 ± 0.2 | −27.5 ± 1.3 | 5.1 ± 0.1 | 86 ± 2 | −60.0 ± 3.5 | 11.6 ± 1.5 | 14 ± 2 |
| L1285P | −25.0 ± 1.0 | 5.4 ± 0.2 | −25.1 ± 1.6 | 5.5 ± 0.5 | 29 ± 4a | −64.3 ± 1.5 | 8.6 ± 0.9 | 71 ± 4a |
| V1685A | −27.5 ± 0.7 | 3.9 ± 0.2 | −26.0 ± 1.5 | 4.7 ± 0.3 | 86 ± 4 | −61.3 ± 2.0 | 7.4 ± 1.0 | 14 ± 4 |
| I1806L | −26.2 ± 0.5 | 4.4 ± 0.3 | −24.6 ± 1.2 | 5.1 ± 0.2 | 87 ± 3 | −63.5 ± 2.0 | 6.6 ± 0.8 | 13 ± 3 |
a Significant differences from BgNav1-1a (p < 0.05).
L1285P in IIIS1 Is Responsible for the Hypersensitivity of BgNav1-1 to Lqh-dprIT3
To determine which of the four amino acid substitutions in BgNav1-1 is responsible for the hypersensitivity to Lqh-dprIT3, we substituted each of the four amino acids in the background of BgNav1-1a with those found in BgNav1-1 (i.e. R502G, L1285P, V1685A, and I1806L) and determined the effect of Lqh-dprIT3. Whereas none of the substitutions had an effect on the voltage dependence of channel activation in the absence of toxin (Table 2), 300 nm Lqh-dprIT3 caused a shift in all four channel mutants (Fig. 3). The fraction of toxin-modified channels was 71% for the L1285P mutant compared with BgNav1-1. In contrast, the fraction of toxin-modified channels for the other three mutants was 13–14%, similar to that for BgNav1-1a (Fig. 3 and Table 2). These results indicated that the L1285P substitution was responsible for the hypersensitivity of BgNav1-1 to Lqh-dprIT3.
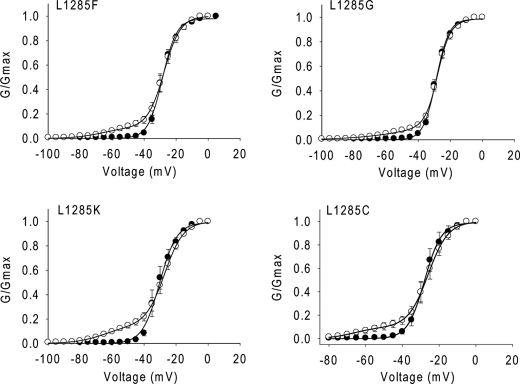
To further determine the impact of the side chain of the residue at position 1258 on Lqh-dprIT3 action, we substituted Leu-1285 with an aromatic residue (Phe), two neutral residues (Gly and Cys), and a positively charged Lys residue in the background of BgNav1-1a. In contrast to L1285P, none of these substitutions increased the channel sensitivity to Lqh-dprIT3 (Fig. 4 and Table 3). In addition, none of these substitutions significantly altered the voltage dependence of channel activation. However, the L1285G substitution caused a 13-mV positive shift in the voltage dependence of steady-state inactivation (data not shown).
FIGURE 4.
Substitutions of Leu-1285 with other residues does not alter the hypersensitivity of BgNav1-1 to Lqh-dprIT3. Shown are conductance curves in the absence (●) and presence (○) of 300 nm Lqh-dprIT3 for four BgNav1-1a channel mutants with different side chain substitutions at position 1285. The conductance-voltage relation was measured using the protocol described under “Experimental Procedures.”
TABLE 3.
Voltage dependence of activation of BgNav1-1a and L1285 substitutions before and after the application of 300 nm Lqh-dprIT3
Each value represents the mean ± S.D. for at least five oocytes.
| Toxin-free |
Lqh-dprIT3 (300 nm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| V0.5 | k | V0.5 | k1 | A1 | V0.5 | k2 | A2 | |
| % | % | |||||||
| BgNav1-1a | −28.4 ± 2.1 | 4.1 ± 0.6 | −28.1 ± 1.5 | 5.0 ± 0.3 | 82 ± 5 | −60.0 ± 3.3 | 10.4 ± 1.6 | 18 ± 5 |
| L1285P | −25.0 ± 1.0 | 5.4 ± 0.2 | −25.1 ± 1.6 | 5.5 ± 0.5 | 29 ± 4a | −64.3 ± 1.5 | 8.6 ± 0.9 | 71 ± 4a |
| L1285F | −28.6 ± 1.1 | 4.2 ± 0.2 | −25.0 ± 1.0 | 4.8 ± 0.2 | 87 ± 4 | −61.6 ± 3.1 | 8.3 ± 0.2 | 13 ± 4 |
| L1285G | −28.7 ± 0.8 | 3.9 ± 0.3 | −27.8 ± 1.2 | 4.0 ± 0.6 | 85 ± 3 | −50.6 ± 6 | 10.1 ± 2.0 | 15 ± 3 |
| L1285K | −30.5 ± 2.1 | 4.6 ± 0.4 | −27.5 ± 1.5 | 5.9 ± 0.6 | 86 ± 2 | −67.2 ± 1.9 | 8.5 ± 1.2 | 14 ± 2 |
| L1285C | −28.7 ± 0.3 | 4.1 ± 0.3 | −25.9 ± 1.6 | 4.6 ± 0.2 | 85 ± 2 | −59.6 ± 3.1 | 11.0 ± 1.6 | 15 ± 2 |
a Significant differences from BgNav1-1a (p < 0.05).
Charge Reversal or Neutralization of a Negatively Charged Residue, Glu-1290, Increases the Channel Sensitivity to Lqh-dprIT3
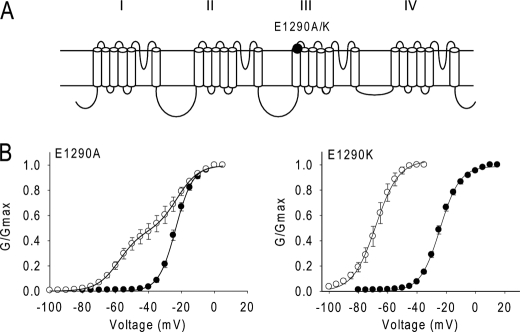
Because of the prominent effect of the L1285P substitution in IIIS1 on Lqh-dprIT3 action, we analyzed the effect of alanine substitution of two nearby, negatively charged residues (Glu-1290 and Asp-1291) at the extracellular end of IIIS1. Glu-1290 is highly conserved in voltage-gated ion channels, including potassium, calcium and sodium channels. The E1290A substitution did not alter the voltage dependence of activation in the absence of toxin. However, the E1290A channel was also more sensitive to Lqh-dprIT3 compared with BgNav1-1a, similar to the effects of the L1285P substitution. 52% of the E1290A channels were modified by 300 nm Lqh-dprIT3 versus 18% of the BgNav1-1a channels (Fig. 5 and Table 4). Substitution of Asp-1291 with alanine did not alter the channel sensitivity to Lqh-dprIT3 (data not shown). Because charge neutralization at Glu-1290 enhanced the effect of Lqh-dprIT3, we further characterized the role of this residue by its substitution with the positively charged amino acid lysine. The E1290K mutation did not alter the voltage dependence of activation in the absence of toxin, but Lqh-dprIT3 (300 nm) caused a complete shift of the conductance curve channel by −40 mV (Fig. 5 and Table 4).
FIGURE 5.
Neutralization and charge reversal of Glu-1290 enhance the channel sensitivity to Lqh-dprIT3. A, sodium channel topology indicating the substitutions of Glu-1290. B, conductance curves in the absence (●) and presence (○) of 300 nm Lqh-dprIT3 are shown. The conductance-voltage relation was measured using the protocol described under “Experimental Procedures.”
TABLE 4.
Voltage dependence of activation of BgNav1-1a and mutants before and after the application of 300 nm Lqh-dprIT3
Each value represents the mean ± S.D. for at least five oocytes.
| Toxin-free |
Lqh-dprIT3 (300 nm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| V0.5 | k | V0.5 | k1 | A1 | V0.5 | k2 | A2 | |
| % | % | |||||||
| BgNav1-1a | −28.4 ± 2.1 | 4.1 ± 0.6 | −28.1 ± 1.5 | 5.0 ± 0.3 | 82 ± 5 | −60.0 ± 3.3 | 10.4 ± 1.6 | 118 ± 5 |
| E1290A | −24.5 ± 1.8 | 4.9 ± 0.3 | −24.0 ± 3.5 | 6.9 ± 0.2 | 48 ± 12a | −60.0 ± 2.4 | 6.1 ± 0.5 | 52 ± 12a |
| E1290K | −24.2 ± 1.2 | 6.6 ± 0.4 | −68.1 ± 2.8 | 7.5 ± 0.4 | ||||
a Significant differences from BgNav1-1a (p < 0.05).
Effects of Reversal of the Gating Charges in Domain III on Channel Activation and Toxin Action
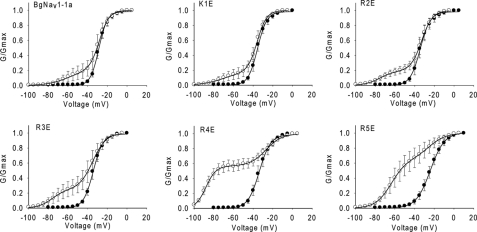
To further investigate the role of the voltage-sensing module of domain III in channel sensitivity to Lqh-dprIT3, we evaluated the effects on toxin action of charge reversal of the five positively charged residues in IIIS4. We individually substituted these residues with glutamic acid, generating K1E, R2E, R3E, R4E, and R5E. In the absence of toxin, the K1E, R2E, R3E, and R4E substitutions caused 5–7-mV negative shifts in the voltage dependence of activation, whereas the R5E substitution caused a 4-mV positive shift (Table 5). We did not observe a significant change in the slope of the activation curve for the K1E, R2E, R3E, and R4E channel mutants, although the R5E channel mutant demonstrated a slightly shallower slope compared with the parental channel (Fig. 6 and Table 5).
TABLE 5.
Voltage dependence of activation of BgNav1-1a and mutants before and after the application of 300 nm Lqh-dprIT3
Each value represents the mean + S.D. for at least five oocytes.
| Toxin-free |
Lqh-dprIT3 (300 nm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| V0.5 | k | V0.5 | k1 | A1 | V0.5 | k2 | A2 | |
| % | % | |||||||
| BgNav1-1a | −28.4 ± 2.1 | 4.1 ± 0.6 | −28.1 ± 1.5 | 5.0 ± 0.3 | 82 ± 5 | −60.0 ± 3.3 | 10.4 ± 1.6 | 18 ± 5 |
| K1E | −35.5 ± 2.5 | 4.7 ± 0.4 | −35.8 ± 3.9 | 4.8 ± 0.3 | 82 ± 4 | −70.0 ± 6.1 | 10.7 ± 2.1 | 18 ± 4 |
| R2E | −34.3 ± 2.8 | 5.1 ± 0.3 | −32.9 ± 2.1 | 5.5 ± 0.4 | 81 ± 4 | −75.5 ± 4.5 | 7.4 ± 0.8 | 19 ± 4 |
| R3E | −34.2 ± 2.5 | 5.0 ± 0.2 | −34.7 ± 2.7 | 6.8 ± 0.3 | 75 ± 5 | −76.1 ± 8.7 | 7.0 ± 0.6 | 25 ± 5 |
| R4E | −33.7 ± 3.8 | 5.7 ± 0.5 | −26.9 ± 3.6 | 8.1 ± 0.6 | 43 ± 8a | −87.9 ± 3.6 | 5.1 ± 0.5 | 57 ± 8a |
| R5E | −24.0 ± 1.9 | 7.2 ± 0.4 | −25.0 ± 3.9 | 8.6 ± 0.8 | 35 ± 7a | −63.6 ± 5.0 | 7.9 ± 0.6 | 65 ± 7a |
a Significant differences from BgNav1-1a (p < 0.05).
FIGURE 6.
Effects of charge reversal of the five positively charged residues in IIIS4 on the action of Lqh-dprIT3. Shown are conductance curves in the absence (●) and presence (○) of 300 nm Lqh-dprIT3. The conductance-voltage relation was measured using the protocol described under “Experimental Procedures.”
The R4E and R5E channel mutants were more sensitive to Lqh-dprIT3 compared with BgNav1-1a and the K1E, R2E, and R3E channel mutants. At 300 nm, the toxin modified >50% of the R4E and R5E channels but only 18–25% of the other channels. For the R5E channels, Lqh-dprIT3 (300 nm) shifted the voltage dependence of activation of modified R5E channels by 40 mV in the hyperpolarizing direction, similar to the extent of the shift for the BgNav1-1a, K1E, R2E, and R3E channels (Fig. 6 and Table 5). However, Lqh-dprIT3 caused a −60-mV shift in voltage dependence of activation for the modified R4E channels, which started to activate at membrane potentials as negative as −100 mV.
DISCUSSION
The differential sensitivities of BgNav sodium channel splice variants from B. germanica have been valuable in elucidating the channel regions involved in channel function and resistance to pyrethroid insecticides (4, 5). Here, we examined the sensitivity of 10 BgNav channel variants to the insect-selective scorpion depressant β-toxin Lqh-dprIT3. This screening unexpectedly revealed that substitutions at the voltage-sensing module of domain III influenced the action of the β-toxin. The binding site of the β-toxin Css4 has been shown to involve the voltage-sensing module of domain II and the pore module of domain III (16, 19, 31, 32). We found that a Leu-to-Pro substitution at position 1285 in IIIS1, resulting from an RNA-editing event, was responsible for the hypersensitivity of the sodium channel variant BgNav1-1 to Lqh-dprIT3. Further site-directed mutagenesis identified additional residues whose substitution affected the action of Lqh-dprIT3, including a negatively charged residue at the extracellular end of IIIS1 (Glu-1290) and the two innermost positively charged gating residues in IIIS4. Charge reversal of either Glu-1290 or each of the two gating charges in IIIS4 did not hinder but rather increased the channel sensitivity to the toxin.
A preliminary three-dimensional model of Css4 docking at rNav1.2a, constructed using as template the crystal structure of the bacterial voltage-gated potassium channel KvAP (21), suggested that the toxin binds at a crevice between S1–S2 and S3–S4 in domain II, thus controlling IIS4 movement during activation. When the two outermost positively charged residues in IIS4 of rNav1.2 were neutralized or charge-reversed, Css4 activity increased, most likely due to their increased mobility during activation, thus enhancing the trapping by the toxin (31).
The voltage-sensing module of domain III has not previously been implicated in the binding and/or action of scorpion β-toxins. The increased sensitivity to Lqh-dprIT3 by the L1285P and E1290A/K substitutions in IIIS1 might indicate that the Lqh-dprIT3 toxin binds at the crevice juxtaposed to S1 of domain III in the channel mutants. Alternatively, alterations in domain III may indirectly facilitate trapping of the voltage sensor in domain II by the toxin. The enhanced channel sensitivity to Lqh-dprIT3 when Leu-1285 was substituted specifically with proline (Table 2) seems more consistent with the latter explanation. Proline inserts a kink in the distal part of the S1 α-helix, which would disrupt the local arrangement of IIIS1, including Glu-1290. Because these alterations increased rather than decreased the effect of Lqh-dprIT3, we speculate that the unmodified IIIS1 in BgNav1 imposes some structural constraint that limits the full effect of the toxin in binding to the domain II voltage sensor and/or in the voltage sensor trapping, and this constraint is lifted by the above substitutions.
The most dramatic effect inspected was when the two innermost gating charges in domain III were substituted with negative charges. This result is quite different from the effects of substituting negative charges in the S4 voltage sensor of domain II. In domain II, neutralization of the two outermost charged residues markedly enhanced β-scorpion toxin activation, and neutralization of the three innermost positively charged residues had no effect (31). In contrast, we observed that reversal of the three outermost positively charged residues in IIIS4 had no effects on toxin activity, and reversal of the two innermost charges greatly enhanced the effect of the toxin. It is unlikely that the two innermost charges directly interact with the toxin because these residues are embedded in the membrane even when the channel is in the activated state. The effects on toxin sensitivity are also unlikely due to effects on the voltage dependence of activation because the effect of the R4E substitution was similar to those of the K1E, R2E, and R3E substitutions (a hyperpolarizing shift), and the R5E substitution had the opposite effect (a small depolarizing shift). For these reasons, we believe that effects of the R4E and R5E substitutions are most likely mediated by allosteric interactions with the S4 voltage sensor of domain II.
The percentage of modified channels was dependent on the concentration of toxin, whereas the magnitude of the toxin-induced shift in activation was concentration-independent. These results suggest that the channels can exist in only two states: unmodified and completely modified. The substitutions in IIIS1 (L1285P, E1290A, and E1290K) and the R5E substitution in IIIS4 increased the percentage of channels that were modified, suggesting that these alterations shifted the equilibrium for toxin modification. In contrast, the R4E substitution in IIIS4 increased both the percentage of modified channels and the magnitude of the shift in activation, indicating that this mutation also altered the modified state of the channel. These differential effects are more consistent with an allosteric modulation of channel gating than a direct interaction between the substituted residues and the toxin. Bezanilla and co-workers (33) proposed that the voltage sensors of the sodium channel are intrinsically tightly coupled. They have shown that binding of the scorpion β-toxin Ts1 to S4 of domain II allosterically activates the movement of all the other voltage sensors in the sodium channel, with the maximal effect on the neighboring voltage sensors of domains I and III rather than on the distant S4 of domain IV. Our results suggest that alterations in the voltage sensor of domain III affect this interaction and that mutations in domain III are likely to enhance the effect of the toxins on IIS4. The role of the voltage-sensing module of domain III in the action of Lqh-dprIT3 on insect sodium channels, as revealed in this study, may be conserved in mammalian sodium channel interaction with other scorpion β-toxins. Future research should test this possibility.
This work was supported, in whole or in part, by National Institutes of Health Grant GM057440 (to K. D.). This work was also supported by United States-Israel Binational Agricultural Research and Development Grant IS-4066-07 (to D. G., M. G., and K. D.), National Science Foundation Grant IBN 9808156 (to K. D.), and Israeli Science Foundation Grant 107/08 (to M. G. and D. G.).
- rNav
- rat Nav
- BgNav
- B. germanica Nav.
REFERENCES
- 1. Catterall W. A. (2000) Neuron 26, 13–25 [DOI] [PubMed] [Google Scholar]
- 2. Goldin A. L. (1999) Ann. N.Y. Acad. Sci. 868, 38–50 [DOI] [PubMed] [Google Scholar]
- 3. Goldin A. L., Barchi R. L., Caldwell J. H., Hofmann F., Howe J. R., Hunter J. C., Kallen R. G., Mandel G., Meisler M. H., Netter Y. B., Noda M., Tamkun M. M., Waxman S. G., Wood J. N., Catterall W. A. (2000) Neuron 28, 365–368 [DOI] [PubMed] [Google Scholar]
- 4. Dong K. (2007) Invert. Neurosci. 7, 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong K. (2010) in Insect Pharmacology (Gilbert L. I., Gill S. S. eds) pp. 25–27, Elsevier, Oxford, United Kingdom [Google Scholar]
- 6. Soderlund D. M. (2010) Sodium Channels, Elsevier B. V., Oxford, United Kingdom [Google Scholar]
- 7. Catterall W. A. (1992) Physiol. Rev. 72, S15–S48 [DOI] [PubMed] [Google Scholar]
- 8. Gordon D. (1997) Invert. Neurosci. 3, 103–116 [DOI] [PubMed] [Google Scholar]
- 9. Nicholson G. M. (2007) Toxicon 49, 490–512 [DOI] [PubMed] [Google Scholar]
- 10. Terlau H., Olivera B. M. (2004) Physiol. Rev. 84, 41–68 [DOI] [PubMed] [Google Scholar]
- 11. Cestèle S., Catterall W. A. (2000) Biochimie 82, 883–892 [DOI] [PubMed] [Google Scholar]
- 12. Gordon D. (1997) in Toxins and Signal Transduction (Gutman Y., Lazarovici P. eds) pp. 119–149, Harwood, Amsterdam, The Netherlands [Google Scholar]
- 13. Martin-Eauclaire M. F., Couraud F. (1995) in Handbook Neurotoxicology (Chang L. W., Dyer R. S. eds) Marcel Dekker, New York [Google Scholar]
- 14. Gordon D., Karbat I., Ilan N., Cohen L., Kahn R., Gilles N., Dong K., Stühmer W., Tytgat J., Gurevitz M. (2007) Toxicon 49, 452–472 [DOI] [PubMed] [Google Scholar]
- 15. Gurevitz M., Karbat I., Cohen L., Ilan N., Kahn R., Turkov M., Stankiewicz M., Stühmer W., Dong K., Gordon D. (2007) Toxicon 49, 473–489 [DOI] [PubMed] [Google Scholar]
- 16. Cestèle S., Qu Y., Rogers J. C., Rochat H., Scheuer T., Catterall W. A. (1998) Neuron 21, 919–931 [DOI] [PubMed] [Google Scholar]
- 17. Cohen L., Ilan N., Gur M., Stühmer W., Gordon D., Gurevitz M. (2007) J. Biol. Chem. 282, 29424–29430 [DOI] [PubMed] [Google Scholar]
- 18. Leipold E., Hansel A., Borges A., Heinemann S. H. (2006) Mol. Pharmacol. 70, 340–347 [DOI] [PubMed] [Google Scholar]
- 19. Marcotte P., Chen L. Q., Kallen R. G., Chahine M. (1997) Circ. Res. 80, 363–369 [DOI] [PubMed] [Google Scholar]
- 20. Tsushima R. G., Borges A., Backx P. H. (1999) Pflugers Arch. 437, 661–668 [DOI] [PubMed] [Google Scholar]
- 21. Cestèle S., Yarov-Yarovoy V., Qu Y., Sampieri F., Scheuer T., Catterall W. A. (2006) J. Biol. Chem. 281, 21332–21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armstrong C. M., Bezanilla F. (1977) J. Gen. Physiol. 70, 567–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hodgkin A. L., Huxley A. F. (1952) Cold Spring Harb. Symp. Quant. Biol. 17, 43–52 [DOI] [PubMed] [Google Scholar]
- 24. Campos F. V., Chanda B., Beirão P. S., Bezanilla F. (2007) J. Gen. Physiol. 130, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song W., Liu Z., Tan J., Nomura Y., Dong K. (2004) J. Biol. Chem. 279, 32554–32561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strugatsky D., Zilberberg N., Stankiewicz M., Ilan N., Turkov M., Cohen L., Pelhate M., Gilles N., Gordon D., Gurevitz M. (2005) Biochemistry 44, 9179–9187 [DOI] [PubMed] [Google Scholar]
- 27. Tan J., Liu Z., Nomura Y., Goldin A. L., Dong K. (2002) J. Neurosci. 22, 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng G., Deák P., Chopra M., Hall L. M. (1995) Cell 82, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 29. Warmke J. W., Reenan R. A., Wang P., Qian S., Arena J. P., Wang J., Wunderler D., Liu K., Kaczorowski G. J., Van der Ploeg L. H., Ganetzky B., Cohen C. J. (1997) J. Gen Physiol. 110, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan J., Liu Z., Wang R., Huang Z. Y., Chen A. C., Gurevitz M., Dong K. (2005) Mol. Pharmacol. 67, 513–522 [DOI] [PubMed] [Google Scholar]
- 31. Cestèle S., Scheuer T., Mantegazza M., Rochat H., Catterall W. A. (2001) J. Gen. Physiol. 118, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cohen L., Karbat I., Gilles N., Ilan N., Benveniste M., Gordon D., Gurevitz M. (2005) J. Biol. Chem. 280, 5045–5053 [DOI] [PubMed] [Google Scholar]
- 33. Chanda B., Asamoah O. K., Bezanilla F. (2004) J. Gen. Physiol. 123, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]