Abstract
Iron-sulfur proteins play an essential role in a variety of biologic processes and exist in multiple cellular compartments. The biogenesis of these proteins has been the subject of extensive investigation, and particular focus has been placed on the pathways that assemble iron-sulfur clusters in the different cellular compartments. Iron-only hydrogenase-like protein 1 (IOP1; also known as nuclear prelamin A recognition factor like protein, or NARFL) is a human protein that is homologous to Nar1, a protein in Saccharomyces cerevisiae that, in turn, is an essential component of the cytosolic iron-sulfur protein assembly pathway in yeast. Previous siRNA-induced knockdown studies using mammalian cells point to a similar role for IOP1 in mammals. In the present studies, we pursued this further by knocking out Iop1 in Mus musculus. We find that Iop1 knock-out results in embryonic lethality before embryonic day 10.5. Acute, inducible global knock-out of Iop1 in adult mice results in lethality and significantly diminished activity of cytosolic aconitase, an iron-sulfur protein, in liver extracts. Inducible knock-out of Iop1 in mouse embryonic fibroblasts results in diminished activity of cytosolic but not mitochondrial aconitase and loss of cell viability. Therefore, just as with knock-out of Nar1 in yeast, we find that knock-out of Iop1/Narfl in mice results in lethality and defective cytosolic iron-sulfur cluster assembly. The findings demonstrate an essential role for IOP1 in this pathway.
Keywords: Gene Knock-out, Iron, Iron-Sulfur Protein, Mouse Genetics, Protein Assembly, Iop1
Introduction
Iron-sulfur cluster proteins are an important class of proteins that contain iron-sulfur clusters in a variety of configurations, including [2Fe-2S] and [4Fe-4S] (1, 2). The distinctive redox and biophysical properties of these clusters make them suitable for a range of functions. Consequently, iron-sulfur proteins have diverse roles and participate in a broad range of biologic processes, including Krebs cycle reactions, oxidative phosphorylation, translation, iron regulatory pathways, and purine metabolism. Notably, these proteins exist in many cellular locales, including the cytosol, mitochondria, and nucleus.
How iron-sulfur cluster proteins are assembled in different cellular compartments has been an area of active investigation (3, 4). Iron-sulfur cluster assembly for all compartments originates in the mitochondria, and the initial step is catalyzed by a cysteine desulfurase that removes elemental sulfur from cysteine. The incorporation of this sulfur into iron-sulfur clusters then occurs by mechanisms that depend, in part, on cell compartment-specific machineries. Such a pathway exists, for example, for cytosolic iron-sulfur proteins. Important insights into this pathway, the cytosolic iron-sulfur protein assembly (CIA)2 pathway, have been obtained from studies in Saccharomyces cerevisiae (3, 5). These studies have revealed a series of proteins that, in a stepwise fashion, assemble and deliver iron-sulfur clusters to target proteins. A current model is that the proteins Tah18 and Dre2 provide reducing equivalents for the assembly of iron-sulfur clusters on a scaffold that contains Cfd1 and Nbp35 (6–10). These clusters are then delivered to Nar1, which in conjunction with Cia1 then facilitates the insertion of the iron-sulfur clusters into appropriate apoproteins (11, 12). It should also be recognized that there is a role for other proteins, including proteins that have the following characteristics: they participate in iron-sulfur cluster biogenesis in the mitochondria, are predominantly mitochondrial in localization, but are also present at low levels in the cytosol (4, 13–17).
There is emerging evidence that this pathway is conserved in mammals (2, 3). Homologues for all of the aforementioned proteins have been identified in mammals, including humans. Functional data have been presented as well. We have employed siRNA to knockdown a human homologue of Nar1, IOP1/NARFL (18). These studies provided evidence for an important role for IOP1 in mammalian cytosolic iron-sulfur protein assembly. Other studies knocking down NBP35 in human cells in culture resulted in impaired cytosolic iron-sulfur cluster assembly (19). In both studies, mitochondrial iron-sulfur cluster assembly was unaffected. Functional studies implicating these human proteins have also been performed in yeast. Thus, human homologues of Dre2 (Ciapin1), Tah18 (Ndor1), and Cia1 (Ciao1) can functionally replace their yeast counterparts (8, 20).
It is notable in this regard that human IOP1 cannot functionally substitute for Nar1 in yeast (12). Moreover, humans possess an additional protein that is homologous to Nar1, which is called nuclear prelamin A recognition factor (NARF) and is distinct from IOP1/NARFL (21, 22). NARF was originally identified as a prelamin A-binding protein. These considerations leave open the possibility that IOP1/NARFL (hereafter referred to as IOP1) might be redundant in function or perhaps not even relevant to the mammalian cytosolic iron-sulfur assembly pathway.
In these studies, we have genetically deleted Iop1 in the mouse. We find that its deletion leads to lethality in the embryo, in the adult mouse, and in cells derived from these mice. We present evidence that Iop1 deletion leads to a defect in cytosolic iron-sulfur protein assembly. Collectively, these findings lend strong support to a critical role for IOP1 in mammalian cytosolic iron-sulfur cluster assembly.
EXPERIMENTAL PROCEDURES
Gene Targeting
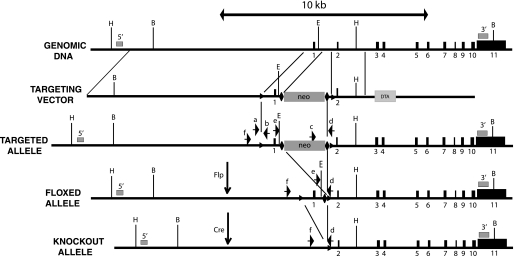
The construct targeting the mouse Iop1 gene was prepared by recombineering (23). In brief, a minitargeting vector was constructed in the vector pL451 (24). This minitargeting vector contained genomic DNA encompassing exon 1 of the mouse Iop1 gene with upstream and downstream loxP sites, a neomycin selection cassette flanked by FRT sites, and additional sequences downstream of exon 1. A retrieval plasmid was constructed in the vector pMC1-DTA (25). This retrieval plasmid contained sequences that flank 11.1 kb of genomic DNA sequence at the mouse Iop1 locus, as well as a diphtheria toxin A negative selection cassette. This retrieval plasmid was used to capture, by recombineering, 11.1 kb of mouse Iop1 genomic DNA containing exon 1 and exon 2 from C57BL/6 bacterial artificial chromosome clone RP23-377L13 (Invitrogen). The resulting product was then used, in the second recombineering step with the minitargeting vector, to generate the final targeting vector. This targeting vector contains an 8.0-kb 5′ arm, exon 1 with upstream and downstream loxP sites, a neomycin selection cassette flanked by FRT sites, and a 2.1-kb 3′ arm (see Fig. 1). Integrity of exons 1 and 2 was confirmed by DNA sequencing.
FIGURE 1.
Iop1 gene targeting strategy. Black boxes indicate exons. Numbers below boxes indicate exon number. Positions of 5′ and 3′ Southern probes are indicated by gray boxes and are denoted by 5′ and 3′, respectively. Positions of loxP and FRT sites are indicated by triangles and diamonds, respectively. Genotyping primers are denoted by arrows and are as follows: a = Iop LoxP5; b = Iop LoxP3; c = Iop Neo; d = Iop Com; e = Iop Frt; f = Iop Del. Restriction enzyme sites are as follows: B = BamHI; E = EcoRV; H = HpaI. neo, neomycin cassette; DTA, diphtheria toxin A-chain cassette.
EAP6 C57BL/6 ES cells were electroporated with the targeting vector and selected using G418 at the University of Pennsylvania School of Medicine Gene Targeting Facility. Screening was performed by Southern blotting. One correctly targeted clone was identified, and it was injected into BALB/c blastocysts to generate chimeras at the University of Pennsylvania School of Medicine Transgenic Core Facility. Chimeric male mice were then mated with albino C57BL/6J-Tyrc-2J female mice (The Jackson Laboratory), and germline transmission was assessed by a combination of coat color and PCR. Mice with germline transmission of the targeted allele were then mated with C57BL/6-Tg(ACTB-Flpe)2Arte mice (Taconic) to delete the neomycin cassette followed by further crossing with C57BL/6 mice to segregate the Flp allele, thereby creating mice (Iop1f/+) harboring an allele in which exon 1 has been flanked by loxP sites (“floxed”) mice. Iop1f/f mice were obtained by crossing Iop1f/+ mice.
Iop1+/− mice were generated by mating Iop1f/f mice with C57BL/6-Gt(ROSA)26Sortm16(Cre)Arte mice (Taconic) followed by further crossing to segregate the Cre allele. Iop1+/− mice were crossed. In some crosses, timed matings were performed. Iop1f/f; Rosa26-creERT2 mice were generated by crossing Iop1f/f mice with C57BL/6-Gt(ROSA)26Sortm9(cre/ESR1)Arte mice (Taconic) followed by a second cross with Iop1f/f mice. Iop1 knock-out was induced in Iop1f/f; Rosa26-creERT2 mice by administration of tamoxifen (MP Biomedicals) at a dose of 5 mg/day for up to 5 consecutive days by oral gavage.
All mice employed in this study were maintained in a C57BL/6 background. All animal procedures were approved by the Institutional Animal Care and Use Committees at the University of Pennsylvania in compliance with Animal Welfare Assurance.
Southern Blotting
Digoxigenin-labeled probes were generated by PCR using a PCR DIG probe synthesis kit (Roche Applied Science). For the 5′ probe (0.34 kb), the primers were 5′-CTC TCA CCA CAA GGA ATG-3′ and 5′-TTT TTA ATG GAG TCT ATC TGG-3′. For the 3′ probe (0.50 kb), the primers were 5′-AAG GCT CCG GAT ACA GAG-3′ and 5′-AAG AGT CTG GAA CCT CAG-3′. For both probes, bacterial artificial chromosome clone RP23-377L13 was employed as the template. Southern blotting was performed using DIG Easy Hyb, DIG wash and block buffer set, anti-digoxigenin-alkaline phosphatase conjugates, and CDP-Star substrate (all from Roche Applied Science).
PCR Genotyping
DNA was isolated from mouse tails (26). The following primers were employed for genotyping. For the 5′ loxP site (PCR product = 0.15 kb for wild type allele, 0.23 kb for floxed allele), the primers were: Iop LoxP5 primer = 5′-TTG AAA TGT AAA TAA AGA AAA TA-3′ and Iop LoxP3 primer = 5′-GCT GTT GAG ATG TTA AGA CTG GAG-3′. For the neomycin cassette (PCR product = 0.55 kb), the primers were: Iop Neo primer = 5′-CGC CTT CTT GAC GAG TTC TTC TG-3′ and Iop Com primer = 5′-GGC GGT CTG TCT GTA TCC CTC TC-3′. For the 3′ loxP site after neomycin cassette deletion (PCR product = 0.30 kb for wild type allele, 0.42 kb for floxed allele), the primers were: Iop Frt primer = 5′-CCT GGG ACC ATG GGA GTG TTT AG-3′ and Iop Com (see above). For the Iop1 knock-out allele (PCR product = 0.75 kb), the primers were: Iop Del primer = 5′-CTA ATA GAC TTG GGT TTG GTG GTG-3′ and Iop Com (see above).
Mouse Embryonic Fibroblast (MEF) Generation
Murine fibroblasts were obtained from day 13.5 Iop1f/f; Rosa26-creERT2 embryos by the primary explant technique (27) and then immortalized by transfection with pSG5-T, which contains the coding sequence for the SV40 large T antigen (28). MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Iop1 knock-out was induced with 250 nm 4-hydroxytamoxifen (Sigma) for 24 h.
Real Time PCR
RNA isolation and real time PCR were performed as described previously using a SYBR Green master mix (Applied Biosystems) (18). The following primers were employed to measure Iop1 and ferritin heavy chain (Fth) messages in mouse RNA samples: Iop1, 5′-GGG CTC CCT GGT CAA AGA C-3′ and 5′-TGG TAG ATC TTG TCA GGA GTC AGA AG-3′; Fth, 5′-AAG ATG GGT GCC CCT GAA G-3′ and 5′-CCA GGG TGT GCT TGT CAA AGA-3′. Dissociation curve analysis revealed a single peak. The sequences of other real time PCR primers has been described (29). Relative quantification was performed employing the ΔΔCt method and β-actin as the endogenous control.
Antibodies and Western Blotting
Antibodies to p38 (sc-728), JNK (sc-571), ERK (sc-93), and β-tubulin (sc-9104) were obtained from Santa Cruz Biotechnology. Antibodies against caspase 3 (catalogue number 9665), phospho-p38 (Thr-180/Tyr-182) (catalogue number 4631), phospho-JNK/SAPK (Thr-183/Tyr-185) (catalogue number 9251), and phospho-ERK1/2 (Thr-202/Tyr-204) (catalogue number 4370) were obtained from Cell Signaling Technologies. Antibodies against microtubule-associated light chain 3 (LC3; catalogue number AM1800) were obtained from Abgent. Antibodies against iron regulatory protein 2 (IRP2) were obtained from Novus Biologicals (NB100-1797). Antibodies to IOP1-(417–476) and cytochrome c have been described (18, 22). Western blotting was performed essentially as described (22). Typically, 30 μg of cell lysate were subjected to SDS-PAGE, and the proteins were transferred to polyvinylidene difluoride membranes. The membranes were blocked with 10% nonfat milk in phosphate-buffered saline containing 0.05% Tween 20 and probed with primary antibodies followed by horseradish peroxidase-coupled secondary antibodies. Detection was performed using SuperSignal substrate (Pierce).
Extract Preparation, Enzyme Assays, and EMSA
Liver extracts from Iop1f/f or Iop1f/f; Rosa26-creERT2 mice were prepared as follows. Three days after the initial tamoxifen dose, mice were sacrificed by CO2 inhalation. Livers were isolated, rinsed in PBS, and then minced and homogenized in ice-cold 100 mm Hepes, pH 7.4, 250 mm sucrose, 0.01% digitonin, and mammalian protease inhibitor mixture (Sigma). After 10 min on ice, the suspension was centrifuged at 300 × g for 5 min at 4 °C to remove tissue debris. The supernatant was then centrifuged at 10,000 × g for 10 min at 4 °C. The resulting supernatant was designated as the cytosolic fraction. The cell pellets were further washed three times in the same buffer employed for homogenization with a 1-min centrifugation at 3,000 × g after each wash. The pellet was then lysed in 100 mm Hepes, pH 7.4, 0.5% Triton X-100, and mammalian protease inhibitor mixture. After 10 min on ice, the lysate was centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant from this centrifugation was designated as the mitochondrial fraction. For MEFs, cytosolic and mitochondrial extracts were prepared as described previously (16, 18). Protein concentrations of extracts were measured using a DC protein assay kit (Bio-Rad).
Aconitase, lactate dehydrogenase, citrate synthase, and xanthine oxidase activities were measured as described (16, 18). An MTT assay was performed using a kit from ATCC (catalogue number 30-1010K). EMSA was performed as described (18).
Fluorescence Microscopy
Cells were exposed to 500 nm MitoTracker Red CMXRos (Invitrogen) for 30 min. Images were obtained using an Olympus BX60 fluorescence microscope.
Statistical Analysis
Student's t test was used for statistical analysis. Significant differences were considered when p < 0.05. Data are presented as the mean ± S.D.
RESULTS
Mus musculus possesses orthologues of the two members of the human IOP family, which are IOP1/Narfl and IOP2/Narf, therefore making this species appropriate for the study of IOP1. To pursue this, our strategy was to knock out the Iop1 gene in the mouse, as outlined in Fig. 1. The murine Iop1 gene in chromosome 17 contains 11 exons. Exon 1 contains coding sequence for amino acids 1–22 of Iop1, which includes the initiator methionine codon. We chose to target this exon and associated upstream promoter sequences for deletion by flanking it with loxP sites. It might be noted that upon deletion of exon 1, the next available exonic ATG is present in exon 2, and this ATG is out of frame with respect to the Iop1 coding sequence. We therefore conclude that this strategy should yield a null allele. The targeting vector contains 5′ and 3′ homology arms, as well as neomycin positive selection and diphtheria toxin negative selection cassettes.
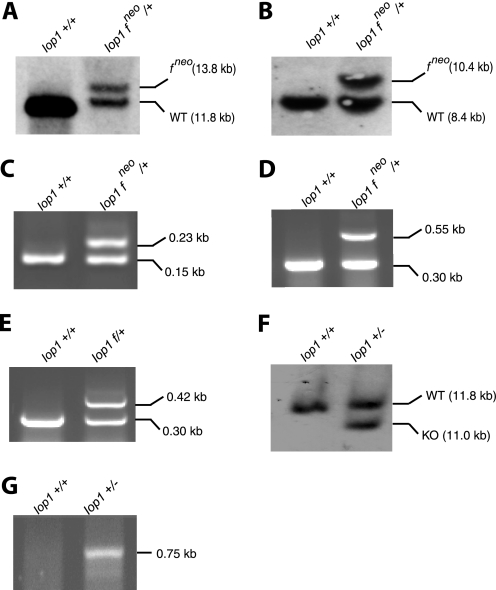
C57BL/6 ES cells were electroporated with the targeting construct and then selected using G418. Of 300 clones screened by Southern blotting, one clone was identified with correct 5′ recombination and 3′ recombination events. Southern blotting with a 5′ probe reveals the expected 13.8-kb band upon hybridization with HpaI-digested DNA (Fig. 2A), whereas Southern blotting with a 3′ probe reveals the expected 10.4-kb band upon hybridization with BamHI/EcoRV-digested DNA (Fig. 2B).
FIGURE 2.
Southern blotting and PCR confirmation of Iop1 gene targeting. A and B, Southern blots of ES cell DNA digested with HpaI (A) or BamHI/EcoRV (B) and hybridized with 5′ (A) or 3′ (B) Southern probes, respectively. The positions of the targeted allele (which contains the neomycin cassette) are indicated by fneo. C–E and G, PCR was performed on tail DNA to assess the presence of the 5′ loxP site (Iop LoxP5 and Iop LoxP3 primers) (C) and the neomycin cassette (Iop Neo and Iop Com primers; Iop Frt primer was also included to allow detection of the wild type allele at 0.30 kb) (D), the 3′ loxP site following deletion of the neomycin cassette (Iop Frt and Iop Com primers) (E), and the Iop1 knock-out allele (Iop Del and Iop Com primers) (G). The sizes of the PCR products are as indicated. Iop1 genotypes of the DNA samples are indicated above the gels. Iop1 f denotes the presence of the floxed allele in the absence of the neomycin cassette. F, Southern blot of tail DNA digested with HpaI and hybridized with the 5′ Southern probe.
This ES cell clone was injected into BALB/c blastocysts, resulting in eight male chimeras. The male chimeras were mated to C57BL/6 females, and two of the male chimeras yielded litters with germline transmission, as assessed by black coat color and PCR genotyping. PCR analysis of tail DNA revealed the integrity of the 5′ loxP site as well as the presence of the neomycin cassette (Fig. 2, C and D, respectively). The neomycin cassette, which is flanked by FRT sites, was removed by breeding correctly targeted mice with mice expressing Flp recombinase. Its removal was confirmed by PCR (Fig. 2E). We then obtained Iop1f/f mice, which were fertile and phenotypically normal, consistent with the absence of adverse effects from the loxP sites.
We generated Iop1+/− mice by breeding the Iop1f/f mice with mice expressing Cre recombinase. Southern blotting of HpaI-digested DNA with the 5′ probe reveals an 11.8-kb band corresponding to the wild type allele and an additional 11.0-kb band indicative of deletion of exon 1 and the associated upstream promoter (Fig. 2F). PCR analysis using primers that flank the original 5′ and 3′ loxP sites is also consistent with the expected deletion (Fig. 2G). These mice were fertile and without any gross abnormality.
We crossed Iop1+/− heterozygous mice to determine whether we could obtain Iop1−/− mice. Of 55 live births, none were Iop1−/− (Table 1). In addition, Iop1+/− mice were born in numbers lower than the expected Mendelian frequency, suggesting that Iop1 haploinsufficiency has a deleterious effect during embryogenesis. We examined embryos at embryonic day 10.5 (n = 42) and likewise failed to observe any Iop1−/− embryos. Interestingly, the ratio of Iop1+/− heterozygotes to wild type mice at embryonic day 10.5 is higher than that observed at birth, further supporting a deleterious effect of Iop1 haploinsufficiency during embryogenesis. From these studies, we conclude that knock-out of Iop1 results in embryonic lethality in the mouse at a day earlier than embryonic day 10.5.
TABLE 1.
Genotypes of embryos and pups produced from Iop1+/− intercrosses
Values are given as a percentage of total analyzed at a given stage. In parentheses, the data show the number observed with a given genotype/total number analyzed at a given stage. E10.5, embryonic day 10.5.
| Genotype | % expected | Stage of development |
|
|---|---|---|---|
| E10.5 (4 litters) | Live birth (10 litters) | ||
| Iop1+/+ | 25 | 44 (19/42) | 64 (35/55) |
| Iop1+/− | 50 | 55 (23/42) | 36 (20/55) |
| Iop1−/− | 25 | 0 | 0 |
It is formally possible that Iop1 serves a essential functional role only during embryogenesis but not later in life. To address this, as well as circumvent the embryonic lethality of Iop1 knock-out, we generated a temporally inducible conditional knock-out of Iop1. We crossed the Iop1f/f mice with mice bearing a tamoxifen-inducible Cre recombinase (creERT2). The latter mouse line expresses the modified Cre from the Rosa26 locus, which allows a broad tissue expression (30). We then employed this Iop1f/f; Rosa26-creERT2 mouse line to globally delete Iop1 in the adult mouse. Toward this end, we treated adult mice with tamoxifen to induce Iop1 deletion. We observed 100% lethality after 5 days following tamoxifen treatment (n = 4). Iop1f/f controls, in contrast, were completely viable and phenotypically normal following the same treatment (n = 4).
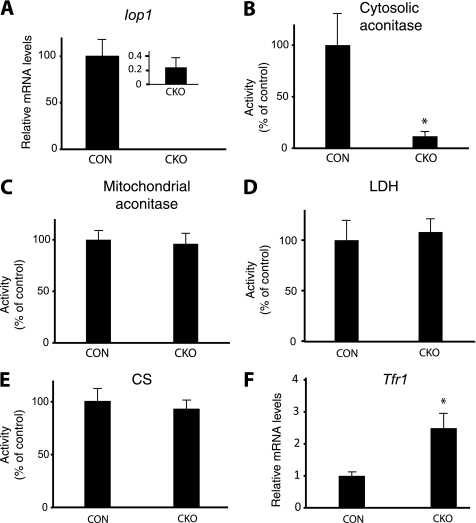
We examined mice at an earlier time point. We treated Iop1f/f and Iop1f/f; Rosa26-creERT2 mice with tamoxifen. Three days after the initial tamoxifen dose, we isolated liver RNA and confirmed, by real time PCR, loss of the Iop1 message (Fig. 3A). We prepared liver extracts and measured the activities of a number of enzymes. We find that Iop1 knock-out results in substantial loss of activity of the iron-sulfur cluster protein cytosolic aconitase (Fig. 3B). Specificity for cytosolic iron-sulfur proteins is supported by the observation that mitochondrial aconitase activity is preserved (Fig. 3C). Further support for specificity is provided by the observation that the activities of two non iron-sulfur enzymes, lactate dehydrogenase (a cytosolic enzyme) and citrate synthase (a mitochondrial enzyme), are not significantly changed (Fig. 3, D and E).
FIGURE 3.
Acute deletion of Iop1 in adult mice. Control (CON; Iop1f/f) or conditional knock-out (CKO; Iop1f/f; Rosa26-creERT2) mice at 6–7 weeks of age were treated with tamoxifen for three consecutive days. After the last tamoxifen dose, livers were harvested, and both RNA and protein extracts were prepared. A and F, levels of Iop1 (A) or Tfr1 (F) message were quantitated by real time PCR (n = 3). B–E, activities of cytosolic aconitase (B), mitochondrial aconitase (C), lactate dehydrogenase (LDH) (D), or citrate synthase (CS) (E) were measured (n = 3). Data are presented as mean ± S.D. * indicates p < 0.01.
Upon loss of its active site iron-sulfur cluster, cytosolic aconitase is converted to IRP1. IRP1 binds to iron response elements (IREs) in the mRNAs encoding proteins involved in iron homeostasis (31, 32). Multiple IREs reside in the 3′-untranslated region (UTR) of the transferrin receptor 1 (Tfr1) mRNA. Binding of IRP1 to this IRE serves to stabilize the Tfr1 mRNA. Measurements of Tfr1 message in the liver reveal an increase upon Iop1 knock-out, consistent with conversion of cytosolic aconitase to Irp1 (Fig. 3F).
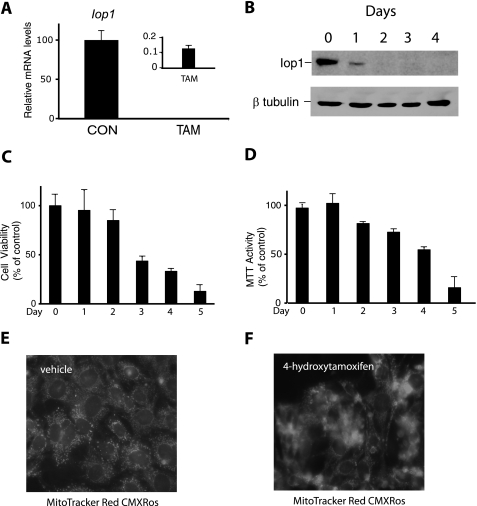
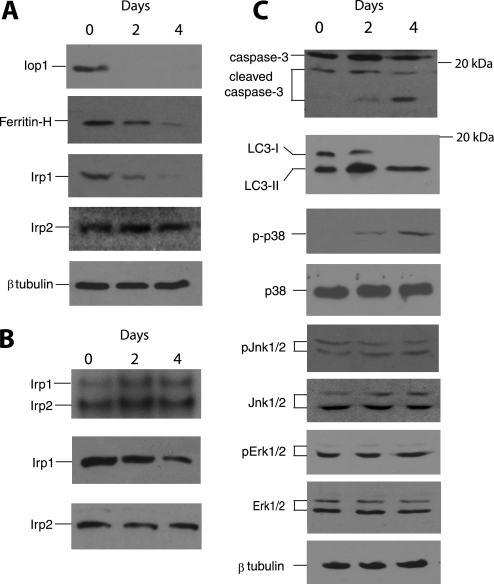
We isolated and then immortalized MEFs from Iop1f/f; Rosa26-creERT2 embryos. When these cells were then treated with 4-hydroxytamoxifen, we observed a dramatic reduction in Iop1 message (Fig. 4A). Examination of extracts by Western blotting using anti-IOP1 antibodies reveals a substantial loss of the protein after 1 day and almost no detectable Iop1 by 2 days (Fig. 4B). These results are consistent with the deletion producing a null allele.
FIGURE 4.
Acute deletion of Iop1 in MEFs. Iop1f/f; Rosa26-creERT2 MEFs were treated with either vehicle (ethanol as control (CON)) or 4-hydroxytamoxifen (TAM). A, RNA was isolated 48 h after treatment, and Iop1 mRNA levels were assessed by real time PCR (n = 3). B, whole cell extracts were prepared from tamoxifen-treated Iop1f/f; Rosa26-creERT2 MEFs at the indicated times and then examined by Western blotting using the indicated antibodies. C and D, cell viability was assessed by either trypan blue exclusion (C) or MTT assay (D) (n = 3 for each). Data are presented as mean ± S.D. E and F, Iop1f/f; Rosa26-creERT2 MEFs were treated with vehicle (E) or 4-hydroxytamoxifen (F). Five days later, MitoTracker Red CMXRos (500 nm) was added for 30 min before examination by fluorescence microscopy. Images are at 400× magnification.
We measured the viability of these MEFs following tamoxifen treatment. By either trypan blue exclusion or MTT assay, we observed reduced viability occurring as early as 3 days after tamoxifen treatment, with almost complete loss of viability at 5 days (Fig. 4, C and D). The decrease in viability was delayed with respect to loss of Iop1 protein. Examination of cells using the fluorescent dye MitoTracker Red CMXRos at 5 days after tamoxifen treatment confirmed substantial loss of viability, with only scattered cells maintaining mitochondrial integrity (compare Fig. 4F with Fig. 4E).
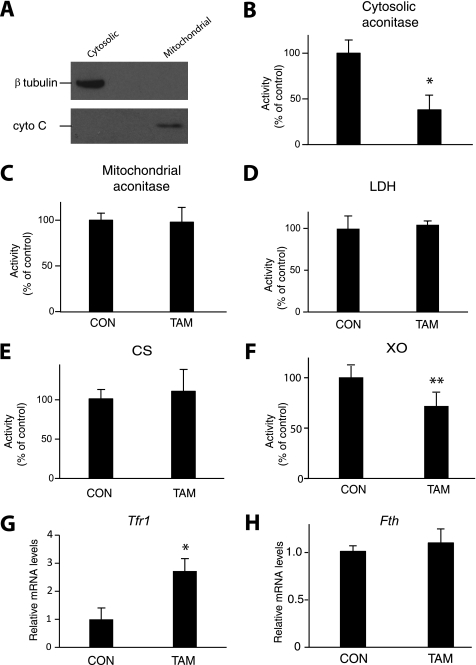
The delay between loss of Iop1 protein and loss of cell viability provided a window in which to pursue molecular studies. We harvested cells at 3 days after tamoxifen treatment, a time at which Iop1 protein expression has been lost, but importantly, before cell viability is almost completely comprised. We prepared cytosolic and mitochondrial fractions, which show the expected presence of tubulin and cytochrome c, respectively (Fig. 5A). We then measured enzyme activities. As shown in Fig. 5B, we find that Iop1 knock-out results in substantial loss of activity of cytosolic aconitase (38% of control, p < 0.01). We did not find any difference in activity in mitochondrial aconitase, nor did we observe changes in cytosolic lactate dehydrogenase or mitochondrial citrate synthase (Fig. 5, C–E). We examined xanthine oxidase, a cytosolic iron-sulfur cluster enzyme distinct from cytosolic aconitase, and find that its activity was also decreased (Fig. 5F).
FIGURE 5.
Assay of enzymes and Tfr1 message in extracts following Iop1 deletion in MEFs. Iop1f/f; Rosa26-creERT2 MEFs were treated with either vehicle (ethanol as control (CON)) or 4-hydroxytamoxifen (TAM). A, 3 days later, cytosolic and mitochondrial extracts (10 μg) were analyzed by Western blotting using anti-β-tubulin or anti-cytochrome c antibodies. B–F, activities of cytosolic aconitase (B), mitochondrial aconitase (C), lactate dehydrogenase (LDH) (D), citrate synthase (CS) (E), and xanthine oxidase (XO) (F) were measured (n = 3). * indicates p < 0.01, and ** indicates p < 0.05. G and H, levels of Tfr1 (G) and Fth (H) message were quantitated by real time PCR (n = 3). * indicates p < 0.01. Data are presented as mean ± S.D.
Measurements of Tfr1 mRNA reveal an increase upon Iop1 deletion, consistent with increased Irp1 activity due to loss of iron-sulfur cluster (Fig. 5G). In contrast, no significant change in ferritin heavy chain mRNA levels was seen following Iop1 deletion (Fig. 5H). Irp1 binds to an IRE in the 5′-UTR of the ferritin heavy chain mRNA, thereby serving to inhibit translation (31, 32). We find that Iop1 knock-out leads to decreased ferritin heavy chain protein (Fig. 6A, second panel from top). We as well as others have observed that the apo form of IRP1 is less stable than the iron-replete form (18, 33, 34). Consistent with this, Irp1 protein levels also decline upon Iop1 knock-out (Fig. 6A, third panel from top) whereas Irp2 protein levels remain the same (Fig. 6A, fourth panel from top).
FIGURE 6.
Activation of selective intracellular pathways following Iop1 deletion in MEFs. A, Iop1f/f; Rosa26-creERT2 MEFs were treated with 4-hydroxytamoxifen for the indicated times. Cytosolic extracts were examined by Western blotting using the indicated antibodies. B, Iop1f/f; Rosa26-creERT2 MEFs were treated with 4-hydroxytamoxifen for the indicated times. Cytosolic extracts (20 μg) were incubated with a 32P-labeled RNA probe containing the ferritin heavy chain 5′ IRE and subjected to 5% PAGE (top panel). Irp1- and Irp2-induced gel mobility shifts are as indicated. Western blotting of extracts for Irp1 and Irp2 are shown in the bottom two panels. C, Iop1f/f; Rosa26-creERT2 MEFs were treated with 4-hydroxytamoxifen for the indicated times. Whole cell extracts (top two panels) or cytosolic extracts (all other panels) were examined by Western blotting using the indicated antibodies. The phosphorylated forms of p38, Jnk1/2, and Erk1/2 are denoted by p-p38, pJnk1/2, and pErk1/2, respectively. Positions of select molecular mass markers (in kDa) are indicated to the right of some panels.
We performed electrophoretic mobility shift assays with cytoplasmic extracts prepared from these cells using a 32P-labeled RNA probe containing the ferritin heavy chain IRE from the 5′-UTR. Consistent with previous observations employing extracts from murine cells (35–37), we observed two bands with this assay; the more slowly migrating band corresponds to Irp1-bound probe, and the more rapidly migrating one corresponds to Irp2-bound probe (Fig. 6B, top panel). After Iop1 knock-out (for example, at 2 days following 4-hydroxytamoxifen treatment), we find an increase in the Irp1 gel shift (compare the top band in the first two lanes of the top panel) despite the observation that Irp1 protein levels decrease (Fig. 6B, middle panel). We also note a slight increase in the Irp2 gel shift (Fig. 6B, top panel), suggesting a minor contribution from this Irp isoform. Taken together, the data support the acquisition of functional Irp1 activity following Iop1 knock-out.
To further explore loss of viability upon Iop1 knock-out in MEFs, we examined a number of stress-induced pathways. We find that Iop1 loss results in induction of apoptosis, as evidenced by conversion of full-length caspase 3 to cleaved caspase 3 (Fig. 6C, top panel). Caspase 3 is a key effector caspase in apoptosis, and its activation occurs by cleavage (38). Interestingly, we also find evidence of induction of autophagy, as seen by the conversion of microtubule-associated light chain 3 I (LC3 I) to LC3 II (Fig. 6C, second panel from top). This conversion occurs during the formation of autophagosomes (39, 40). Protein kinases can also be induced by stress (41). Indeed, we find evidence of activation of p38, a major stress-induced kinase (Fig. 6C, third panel from top). Selectivity for this particular member of the MAPK family of protein kinases is provided by the observation that Jnk and Erk were not activated under the same conditions (Fig. 6B, fifth and seventh panels from top, respectively). A link between p38 activation and autophagy has been described by others (42). These observations suggest that activation of selective intracellular signaling pathways occurs upon Iop1 loss.
DISCUSSION
A number of studies in recent years have pointed to a role for IOP1 in the mammalian CIA pathway (16, 18). We report here the results of knocking out Iop1 in the mouse. Based on the results, we make two important conclusions. First, we conclude that Iop1 is a protein essential for life in mammals. This is a significant observation because there exists a homologous protein, Narf/Iop2 (21, 22). Narf shares a number of notable residues with Iop1: first, a set of cysteine residues that, in bacterial hydrogenases, corresponds to an H-cluster that chelates a distinctive active site iron-sulfur cluster; and second, a set of cysteine residues in the N-terminal half of the protein that is present in a ferredoxin-like domain. In contrast to Iop1, which is predominantly cytoplasmic, Narf is a nuclear protein (21). Aside from its capacity to specifically bind to prenylated prelamin A, essentially nothing is known regarding the function of Narf. Importantly, the present studies indicate that the activities of Iop1 and Narf are not redundant and that loss of Iop1 cannot be compensated for by Narf.
The second conclusion from this study is that Iop1 plays an essential role in cytosolic iron-sulfur cluster biogenesis. The inducible genetic ablation approach that is employed in both the in vivo studies and the in vitro studies examining MEFs support an essential role for Iop1 in this pathway. They differ in approach from previous knockdown studies employing siRNA, which inherently have the possibility of off-target effects. The selectivity of Iop1 for the CIA pathway is reinforced by the absence of effects of Iop1 deletion on mitochondrial aconitase activity or on that of non iron-sulfur enzymes, such as lactate dehydrogenase or citrate synthase.
The present study contributes to an expanding literature on the role of specific proteins in the mammalian cytosolic iron-sulfur cluster assembly pathway (1, 2). This, in turn, will form the basis for understanding this pathway in the context of human disease. To our knowledge, mutations in the CIA pathway have yet to be linked to human diseases. There is, however, clear evidence already for a link between iron-sulfur cluster biogenesis and human disease (2, 43). Specific human diseases arise from mutations in key proteins involved in iron-sulfur cluster biogenesis, and these diseases target a multitude of organ systems, including hematopoietic, neural, skeletal muscular, and cardiac tissues. The present studies would strongly suggest that complete loss of function mutations in human IOP1 would be incompatible with life. However, the possibility of hypomorphic mutations in IOP1 being linked to human disease remains an intriguing possibility.
Acknowledgments
We thank Dr. Tobias Raabe of the University of Pennsylvania Gene Targeting facility for performing the ES cell electroporation, Dr. Jean Richa of the University of Pennsylvania Transgenic core facility for performing the blastocyst injections, Adrian Flores for assistance in generation of the Iop1 targeting vector, Dr. Neal Copeland (NCI, National Institutes of Health) for recombineering reagents, and Dr. James Alwine (University of Pennsylvania) for the gift of the pSG5-T plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM71459 (to F. S. L.).
This article was selected as a Paper of the Week.
- CIA
- cytosolic iron-sulfur protein assembly
- IOP1
- iron-only hydrogenase-like protein 1
- IRP
- iron regulatory protein
- IRE
- iron response element
- MEF
- mouse embryonic fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NARF
- nuclear prelamin A recognition factor
- NARFL
- nuclear prelamin A recognition factor-like protein
- FRT
- FLP recognition target
- FLP
- Flippase recombinase.
REFERENCES
- 1. Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 2. Ye H., Rouault T. A. (2010) Biochemistry 49, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 4. Rouault T. A., Tong W. H. (2005) Nat. Rev. Mol. Cell Biol. 6, 345–351 [DOI] [PubMed] [Google Scholar]
- 5. Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. (2010) J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roy A., Solodovnikova N., Nicholson T., Antholine W., Walden W. E. (2003) EMBO J. 22, 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stehling O., Netz D. J., Niggemeyer B., Rösser R., Eisenstein R. S., Puccio H., Pierik A. J., Lill R. (2008) Mol. Cell Biol. 28, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Netz D. J., Stümpfig M., Doré C., Mühlenhoff U., Pierik A. J., Lill R. (2010) Nat. Chem. Biol. 6, 758–765 [DOI] [PubMed] [Google Scholar]
- 9. Netz D. J., Pierik A. J., Stümpfig M., Mühlenhoff U., Lill R. (2007) Nat. Chem. Biol. 3, 278–286 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. (2008) Mol. Cell Biol. 28, 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balk J., Aguilar Netz D. J., Tepper K., Pierik A. J., Lill R. (2005) Mol. Cell Biol. 25, 10833–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balk J., Pierik A. J., Netz D. J., Mühlenhoff U., Lill R. (2004) EMBO J. 23, 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tong W. H., Rouault T. (2000) EMBO J. 19, 5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tong W. H., Rouault T. A. (2006) Cell Metab. 3, 199–210 [DOI] [PubMed] [Google Scholar]
- 15. Tong W. H., Jameson G. N., Huynh B. H., Rouault T. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song D., Tu Z., Lee F. S. (2009) J. Biol. Chem. 284, 35297–35307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Land T., Rouault T. A. (1998) Mol. Cell 2, 807–815 [DOI] [PubMed] [Google Scholar]
- 18. Song D., Lee F. S. (2008) J. Biol. Chem. 283, 9231–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hausmann A., Aguilar Netz D. J., Balk J., Pierik A. J., Mühlenhoff U., Lill R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srinivasan V., Netz D. J., Webert H., Mascarenhas J., Pierik A. J., Michel H., Lill R. (2007) Structure 15, 1246–1257 [DOI] [PubMed] [Google Scholar]
- 21. Barton R. M., Worman H. J. (1999) J. Biol. Chem. 274, 30008–30018 [DOI] [PubMed] [Google Scholar]
- 22. Huang J., Song D., Flores A., Zhao Q., Mooney S. M., Shaw L. M., Lee F. S. (2007) Biochem. J. 401, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Copeland N. G., Jenkins N. A., Court D. L. (2001) Nat. Rev. Genet. 2, 769–779 [DOI] [PubMed] [Google Scholar]
- 24. Liu P., Jenkins N. A., Copeland N. G. (2003) Genome Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yagi T., Ikawa Y., Yoshida K., Shigetani Y., Takeda N., Mabuchi I., Yamamoto T., Aizawa S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9918–9922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laird P. W., Zijderveld A., Linders K., Rudnicki M. A., Jaenisch R., Berns A. (1991) Nucleic Acids Res. 19, 4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu J. (2005) in Current Protocols in Molecular Biology (Ausubel F. M. ed) pp. 28.1.1–28.1.8, John Wiley & Sons, Inc., New York [Google Scholar]
- 28. Yu Y., Alwine J. C. (2002) J. Virol. 76, 3731–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X., Sutherland S., Takeda K., Fong G. H., Lee F. S. (2010) Blood Cells Mol. Dis. 45, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., Schoor M., Jaenisch R., Rajewsky K., Kühn R., Schwenk F. (2003) Nucleic Acids Res. 31, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rouault T. A. (2006) Nat. Chem. Biol. 2, 406–414 [DOI] [PubMed] [Google Scholar]
- 32. Muckenthaler M. U., Galy B., Hentze M. W. (2008) Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- 33. Clarke S. L., Vasanthakumar A., Anderson S. A., Pondarré C., Koh C. M., Deck K. M., Pitula J. S., Epstein C. J., Fleming M. D., Eisenstein R. S. (2006) EMBO J. 25, 544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J., Fillebeen C., Chen G., Biederbick A., Lill R., Pantopoulos K. (2007) Mol. Cell Biol. 27, 2423–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. (2004) Science 306, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 36. Thomson A. M., Rogers J. T., Leedman P. J. (2000) J. Biol. Chem. 275, 31609–31615 [DOI] [PubMed] [Google Scholar]
- 37. Henderson B. R., Kühn L. C. (1995) J. Biol. Chem. 270, 20509–20515 [DOI] [PubMed] [Google Scholar]
- 38. Riedl S. J., Shi Y. (2004) Nat. Rev. Mol. Cell Biol. 5, 897–907 [DOI] [PubMed] [Google Scholar]
- 39. He C., Klionsky D. J. (2009) Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ravikumar B., Futter M., Jahreiss L., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Narayanan U., Renna M., Jimenez-Sanchez M., Sarkar S., Underwood B., Winslow A., Rubinsztein D. C. (2009) J. Cell Sci. 122, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raman M., Chen W., Cobb M. H. (2007) Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 42. McClung J. M., Judge A. R., Powers S. K., Yan Z. (2010) Am. J. Physiol. Cell Physiol. 298, C542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheftel A., Stehling O., Lill R. (2010) Trends Endocrinol. Metab. 21, 302–314 [DOI] [PubMed] [Google Scholar]