Abstract
HEL308 is a superfamily II DNA helicase, conserved from archaea through to humans. HEL308 family members were originally isolated by their similarity to the Drosophila melanogaster Mus308 protein, which contributes to the repair of replication-blocking lesions such as DNA interstrand cross-links. Biochemical studies have established that human HEL308 is an ATP-dependent enzyme that unwinds DNA with a 3′ to 5′ polarity, but little else is know about its mechanism. Here, we show that GFP-tagged HEL308 localizes to replication forks following camptothecin treatment. Moreover, HEL308 colocalizes with two factors involved in the repair of damaged forks by homologous recombination, Rad51 and FANCD2. Purified HEL308 requires a 3′ single-stranded DNA region to load and unwind duplex DNA structures. When incubated with substrates that model stalled replication forks, HEL308 preferentially unwinds the parental strands of a structure that models a fork with a nascent lagging strand, and the unwinding action of HEL308 is specifically stimulated by human replication protein A. Finally, we show that HEL308 appears to target and unwind from the junction between single-stranded to double-stranded DNA on model fork structures. Together, our results suggest that one role for HEL308 at sites of blocked replication might be to open up the parental strands to facilitate the loading of subsequent factors required for replication restart.
Keywords: DNA Enzymes, DNA Helicase, DNA Recombination, DNA Repair, DNA Replication
Introduction
DNA helicases are ubiquitous throughout evolution and represent several super-families of enzyme that contribute to critical DNA metabolic processes by unwinding the DNA double helix. Many DNA helicases act during DNA replication and repair and are vital for maintaining genomic stability (1). In both prokaryotes and eukaryotes, the widely studied RecQ helicase family plays a critical role in homologous recombination (HR)2 and the processing of stalled and collapsed replication forks (1, 2). The importance of these helicases is underlined by the fact that mutations in three human RecQ paralogs (Bloom's syndrome, Werner's syndrome, and RECQ4) result in devastating inherited human diseases associated with genomic instability (3–5). Numerous biochemical activities have been identified in the RecQ family that suggest multiple (non-exclusive) roles for these proteins at stalled and damaged forks and during repair. These include a contribution to the restart of replication at collapsed forks through early and late roles in HR, such as during end resection and D-loop disruption and in the migration and dissolution of recombination intermediates, respectively, as well as by catalyzing fork regression as part of template switch damage tolerance mechanisms (2, 4). Although most RecQ helicases can catalyze more than one of these reactions in vitro, it remains unclear precisely how these helicases process damaged replication forks in vivo. It should also be noted that several RecQ helicases are able to perform their helicase role in reverse, promoting strand reannealing in the absence of ATP (6–8). This might be critical for the eventual reversal of regressed forks following their extension during damage tolerance, or during Holliday junction (HJ) branch migration late in HR. In addition to the RecQ family, several other mammalian helicases have been implicated in the processing of damaged replication forks, including the products of the Fanconi anemia complementation group M and J genes (FANCM and FANCJ), whereas RTEL1 might promote the disassembly of inappropriate early recombination intermediates, similar to yeast Srs2, acting as an “antirecombinase” (9–12). Finally, in vitro replication fork regression activities have been reported for the yeast Rad5/human helicare-like transcription factor (HLTF) translocases (13–15).
The more recently identified HEL308 family of helicases have also been associated with maintenance of genome stability, from archaea through to mammals (16, 17). The discovery of a gene (mus308) encoding a single protein with novel N-terminal helicase and C-terminal polymerase domains in Drosophila (18, 19) spurred the search for further factors with similarity to the helicase, polymerase, or both portions of the fly protein. The identification of a putative homolog of mus308 in mammals, HEL308, was followed by the description of related proteins in archaea and Caenorhabditis elegans, and the HEL308 family members studied to date are all ATP-dependent 3′-5′ helicases (16, 17, 20, 21). Moreover, identification of PolQ, which harbors a protein containing similar helicase and polymerase domains to mus308, although the helicase is apparently inactive, and PolN, that harbors similarity to the polymerase portion of HEL308, also followed (22, 23). It has recently been suggested that human PolN and HEL308 might cooperate during the processing of damaged forks (24), whereas studies in C. elegans suggest that HEL308 might contribute to the Fanconi anemia pathway of replication-repair, acting in a pathway distinct from PolQ (20). Consistent with a key role in processing stalled or collapsed forks, Drosophila mus308 was originally identified in a screen for mutants with sensitivity to DNA interstrand cross-linking (ICL) agents (18). It is well established that ICL agents are a potent block to replication, and that one of the major ICL repair pathways in metazoans is triggered by replication fork collision with the ICL (25). In contrast to the Drosophila gene, loss of HEL308 in chicken DT-40 cells does not sensitize to ICLs (26), although knockdown of HEL308 in HeLa cells sensitizes to mitomycin C, suggesting a role for human HEL308 in DNA repair (24).
Despite its potential importance in replication and repair, limited studies of HEL308 have been forthcoming, and we know little of its likely cellular role. Here, we sought to characterize the biochemical activities of human HEL308 in detail, and to explore whether the protein acts in association with the DNA replication apparatus.
EXPERIMENTAL PROCEDURES
Purification of Human HEL308
The pFastBac HTb vector with the cDNA encoding HEL308 (17) (kind gift of Federica Marini, Milan, Italy) was transformed into DH10Bac Escherichia coli. After transfection of the resulting recombinant bacmid into Sf21 insect cells, virus stock was generated. For protein production, 4.5 × 108 cells were infected with the His6-HEL308 baculovirus at a multiplicity of infection of 5. Two days post-infection, cells were collected by low speed centrifugation and washed twice with ice-cold PBS. The presence of HEL308 in the lysate was confirmed by electrophoresis through SDS-PAGE gels followed by blotting to nitrocellulose. The blots were hybridized with mouse monoclonal anti-HIS and mouse monoclonal anti-HEL308 antibodies (both from Santa Cruz Biotechnology). For fractionation, the cells were lysed in 20 ml of lysis buffer (20 mm sodium phosphate (pH 7.4), 500 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, 5 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, EDTA-free protease inhibitor mixture (Roche)) for 30 min on ice. The imidazole was added to the clarified lysate to a final concentration of 2 mm, and the lysate was subsequently loaded on a HiTrapHIS column (AKTA system). After washing the column with 10 volumes of lysis buffer containing 50 mm imidazole, the HEL308 protein was eluted by gradual increase of the salt concentration to 500 mm. The protein eluted at ∼150 mm imidazole. Next, the HEL308-containing fractions were pooled and separated on a gel filtration column (SUPERDEX200) calibrated with a gel filtration buffer (10 mm sodium phosphate (pH 7.4), 150 mm NaCl, 10% glycerol, 1 mm dithiothreitol). Finally, the protein was concentrated on the HiTrap heparin column calibrated with a gel filtration buffer. After washing the resin with 5 volumes of calibration buffer containing 200 mm NaCl, the protein was eluted by continuing increase in salt concentration to 1 m. The HEL308 eluted at ∼500 mm NaCl. Aliquots of eluted fractions were stored at −80 °C for further analysis. The purity of obtained eluates was verified by running the samples through SDS-PAGE gel followed by Coomassie staining. The protein concentration was determined by the Bradford method.
DNA Substrates and Helicase Assay
Oligonucleotides were 5′ end-labeled using T4 DNA kinase and [γ-32P]ATP. Free [γ-32P]ATP was removed from the labeled oligonucleotides with mini Quick Spin DNA columns (Roche). The set of DNA substrates was generated by annealing the appropriate combination of cold oligonucleotides to the labeled ones followed by substrate gel purification. (Substrates are shown schematically in supplemental Table S1, and their sequences are listed in supplemental Table S2.) The proper assembly of each of the generated DNA substrates was verified by restriction analysis (data not shown). Standard helicase reactions were performed at 37 °C in 20 μl containing 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 4 mm MgCl2, 4 mm ATP, 1 mm dithiothreitol, 100 mg/ml bovine serum albumin, ∼ 0.5 fmol DNA, and indicated concentrations of purified human HEL308, human replication protein A (RPA) (kind gift of Fumiko Esashi, Oxford, UK) or single-stranded DNA-binding protein (SSB) (Stratagene). Unless stated otherwise, reactions were carried out for 25 min and subsequently terminated by the addition of 6 μl of loading buffer (0.25% bromphenol blue, 0.25% xylene cyanol, 30% glycerol, 0.1% SDS, 0.17 m EDTA). DNA species were separated by electrophoresis through non-denaturing 10% polyacrylamide gels (Bio-Rad). The DNA unwinding rate was determined by scanning the dried gels with a Typhoon scanner and followed by quantification of the obtained signal using ImageQuant software.
GFP-HEL308 DNA Construct
The full-length human HEL308 cDNA was amplified with the following primers: FW BglII 5′-GTATTTTCAGAGATCTATGGATG and REV XmaI 5′-AAATGTGGTATCCCGGGTTAT (the BglII site is italicized and the XmaI site is underlined). To obtain expression of HEL308 with an N-terminal GFP tag, a GFP gene was introduced in front of HEL308 cDNA by cloning the PCR product into the pEGFP-C1 vector.
Cell Culture and Generation of the GFP-HEL308 U2OS Cells
U2OS cells were grown at 37 °C in DMEM containing 10% fetal calf serum. Cells expressing GFP-tagged HEL308 were generated by transforming 10 μg of the GFP-HEL308 DNA construct into ∼5 × 106 U2OS cells in 5 ml of standard medium with Lipofectamine 2000 (Invitrogen). Selection with 500 μg/ml of G418 was started 24 h after transfection. After 10 days, G418-resistant green clones were isolated, and the whole cell protein extracts were screened for the expression of GFP-HEL308 by immunoblot analysis with both rabbit polyclonal anti-GFP (Living Colors) and mouse monoclonal anti-HEL308 (Santa Cruz Biotechnology) antibodies.
Immunofluorescence and Confocal Microscopy Imaging
Cells were trypsinized and seeded at ∼50% confluency on glass coverslips. Twenty-four hours later, cells were washed with PBS and subsequently incubated in medium with 10 μm camptothecin (CPT) (Sigma) and fixed at room temperature with 2% paraformaldehyde in PBS for 15 min at indicated times. The HEL308 protein was detected by direct fluorescence. For the immunofluorescence, the cells were permeabilized with 0.1% Triton in PBS and blocked with 0.15% BSA and 0.5% glycine in PBS. Mouse monoclonal antibodies were used to identify RPA (anti-RPA, 1:1000, Calbiochem). For the visualization of immunoconjugated proteins, Alexa Fluor 555-conjugated goat anti-mouse immunoglobin (1:1000) was used. FANCD2 and Rad51 were detected by rabbit polyclonal antibodies (Abcam and kind gift of Stephen West, Cancer Research UK, respectively, both: 1:1000) and with goat anti-rabbit Alexa Fluor 555-conjugated immunoglobulin (1:1000) (Invitrogen). All antibodies were diluted in blocking buffer and incubated with cells at 37 °C under humid conditions. Finally, the slides were mounted with DAPI Vectashield mounting medium. Nuclear staining patterns were visualized with a Zeiss LSM META510 confocal laser-scanning microscope. To detect GFP or Alexa Fluor 488 fluorescence, the images were recorded with a 488-nm Ar laser. The 543 nm HeNe laser was used to detect Cy3 or 555 Alexa Fluor. DAPI-stained chromatin was identified by Diode 405 laser.
HEL308 Colocalization with Active Replication Sites
The sites of active replication were labeled by-15 min cell incubation in medium containing 50 μm iododeoxyuridine (IdU) before the CPT treatment. After fixing cells, HEL308 was detected by immunofluorescence using rabbit polyclonal anti-GFP (Living Colors) (1:500) antibodies, washed with 0.1% Triton in PBS, blocked, and incubated with goat anti-rabbit Alexa Fluor 488-conjugated (1:1000) (Invitrogen) antibodies. Following washing with 0.1% Triton in PBS, cells were incubated in 8% paraformaldehyde for 20 min and in 4% paraformaldehyde for 15 min at room temperature. DNA was denatured for 15 min in 4 m HCl, followed by five PBS washes and blocking. The incorporated IdU was detected with mouse monoclonal anti-bromodeoxyuridine antibodies (BD Biosciences) (1:200) and with the goat anti-mouse Cy3-conjugated antibodies (Sigma) (1:1000) according to the standard immunofluorescence procedure.
RESULTS
HEL308 Relocalizes to Sites of Stalled Replication along with Rad51 and FANCD2 following Camptothecin Treatment
The requirement for human HEL308 for normal resistance to replication-blocking lesions such as those induced by mitomycin C and CPT suggests a role for HEL308 at damaged forks (24). Therefore, we investigated the ability of HEL308 to form foci upon CPT treatment. To this end, a construct encoding N-terminally GFP-tagged HEL308 was stably integrated into U2OS cells. Colonies obtained after selection were screened for the expression of the GFP-HEL308 fusion protein with anti-GFP and anti-HEL308 antibodies. A protein of ∼170 kDa, corresponding to GFP-HEL308, was detected in stably transfected clones by both antibodies. Importantly, no free GFP was detected in these cells (supplemental Fig. S1).
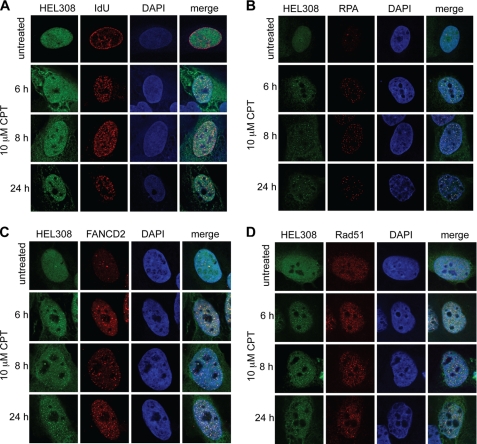
First, the colocalization of HEL308 with sites of ongoing replication was assessed prior to and following DNA damage. We determined the colocalization of GFP-HEL308 with sites of IdU incorporation prior to and following treatment with CPT, which induces replication fork stalling and collapse (Fig. 1A). A striking colocalization of GFP-HEL308 with IdU patches was observed following CPT treatment. The diffuse signal of GFP-HEL308 changed to focus-like structures, and more than 90% of the GFP-HEL308 foci colocalized with IdU incorporation patches. This persisted for at least 8 h following CPT treatment, with the number of foci in decline by 24 h. Moreover, GFP-HEL308 also colocalized with RPA foci following CPT treatment (Fig. 1B), again in about 90% of cases, where the kinetics of foci formation and resolution were similar to those observed with IdU. Together, this data indicates that HEL308 is recruited to sites of stalled replication following DNA damage.
FIGURE 1.
HEL308 colocalizes with sites of DNA replication as well as RAD51 and FANCD2 following replication fork damage. Confocal images of fixed GFP-HEL308-expressing U2OS cells treated with 10 μm CPT for the stated times. A, shown are cells pulse-labeled with IdU before CPT treatment. HEL308 protein is depicted in green (HEL308 column). The pattern in red represents sites of replication (IdU). DNA staining is depicted in blue (DAPI). The column on the right displays the merged images, in which yellow/purple demonstrates colocalization of HEL308 and IdU. B, cells (not subject to IdU pulse) as in A, except the pattern in red depicts subcellular localization of RPA, FANCD2 (C), or RAD51 (D).
We next determined whether the sites of HEL308 recruitment are associated with DNA repair and replication restart. When the colocalization of HEL308 with FANCD2 was assessed, we discovered that there is a significant overlap between the localization of HEL308 and FANCD2 following CPT treatment, with ∼80% of foci colocalizing (Fig. 1C). As FANCD2 has been associated with the promotion of fork repair by HR (27–29), we also studied colocalization of HEL308 with Rad51, and this followed a similar pattern to FANCD2, where over 80% of HEL308 foci colocalized with RAD51 8 h following CPT (Fig. 1D). Together, this data suggests that HEL308 is involved in the processing of stalled forks that require recombination-mediated processes for their restart.
Purification of the Human HEL308 Protein
For HEL308 purification, we used a DNA construct encoding human HEL308 cDNA with an N-terminal hexahistidine tag (see “Experimental Procedures”). The cDNA was placed under the transcriptional control of the polyhedrin promoter in recombinant baculovirus, which was subsequently used to infect Sf21 cells. Highly purified protein was obtained by sequential His-tag affinity, gel filtration, and heparin affinity chromatography (supplemental Fig. S2, A and B). The presence of HEL308 protein in the eluates was confirmed by immunoblots using anti-HIS and anti-HEL308 antibodies (not shown). The elution profile of the gel filtration column suggests that HEL308 also forms a multimer and possibly a trimer (supplemental Fig. 2C). A HEL308 active site mutant (HEL308K365M) was also purified by the same procedure, and no helicase activity was observed on any of the critical DNA substrates used throughout this work using the mutant protein (supplemental Fig. S3).
HEL308 Unwinds DNA Structures with 3′ Single-stranded DNA (ssDNA) Regions
It has previously been shown in vitro that human HEL308 is an ATPase-dependent 3′ to 5′ helicase, using partial DNA duplexes as substrates (17). We confirmed the 3′-5′ directionality of HEL308 (Fig. 2A) and also determined that a minimal length of between 7–15 nt of 3′ ssDNA tail is required for HEL308 to unwind a replication fork-like structure (supplemental Fig. S4).
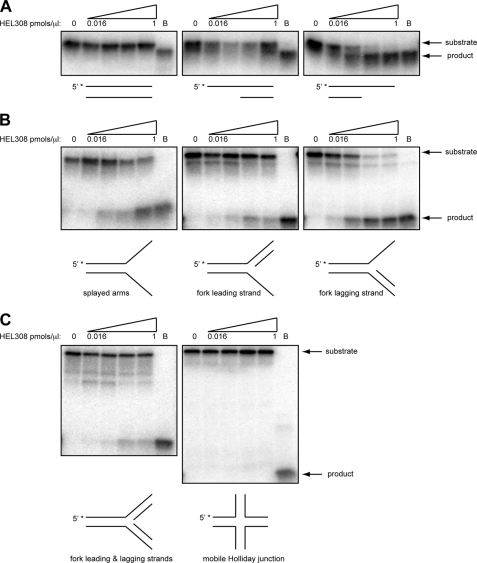
FIGURE 2.
HEL308 requires a region of 3′-ssDNA to unwind substrates. A, the double-stranded blunt, 5′ overhang and 3′ overhang substrates listed in supplemental Table 1 were 5′ end-labeled on the upper strand (5′ with asterisk), mock-treated (0 pmol/μl lane), or treated with increasing concentrations from 0.016 to 1 pmol/μl of HEL308 (0.016, 0.06, 0.25, and 1.00 pmol/μl). In the lanes B, substrates were boiled to denature them prior to loading on the gel. The position of the substrate and fully unwound/denatured products are marked. B, the model fork substrates listed in supplemental Table 1 with no additional model nascent strand (left panel), or containing either a leading (center panel) or lagging nascent strand (right panel) were treated as in A. C, a model fork substrate with both nascent leading and lagging strands and a model mobile HJ structure were incubated with HEL308, as for the substrates in A.
HEL308 Unwinds Forks with a Nascent Lagging Strand in Preference to Splayed Arms or Partial Duplex
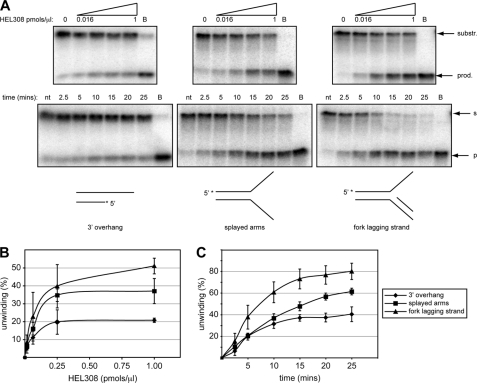
The ability to displace nascent strands from replication fork-like substrates and to bind HJ-like structures has been shown for the HEL308-like protein StoHjm from Sulfolobus tokodaii (30). Additionally, structure-specific annealing and the ability to regress model replication forks has been shown for this protein (31). Related biochemical activities, notably unwinding of the lagging strand on model fork substrates, have been reported for Hel308a from Methanothermobacter thermautotrophicus and Hjm from Pyrococcus furiosus (31–33). These findings prompted us to investigate whether HEL308 could process a variety of model structures that arise during replication and HR repair (Fig. 2, B and C). HEL308 was unable to process either model four-way HJ or chicken foot-like structures but could efficiently unwind a splayed arms structure (Fig. 2B and data not shown). Indeed, the splayed arms structure was unwound more efficiently than the partial duplex with a 3′ overhang (Fig. 3) suggesting that HEL308 prefers branched structures akin to those found at replication forks. To investigate this further, we analyzed if HEL308 could process model fork structures that contain either a nascent leading and/or lagging strand (Figs. 2B and 3). Interestingly, the presence of a lagging strand stimulated the ability of HEL308 to unwind the splayed arm (Figs. 2B and 3). This effect was specific to the nascent lagging strand because the presence of a nascent leading strand had little effect on the ability of HEL308 to unwind the splayed arms (Fig. 2B), whereas the presence of both leading and lagging nascent strands inhibited HEL308 activity (C). The ability of HEL308 to preferentially unwind forks with a nascent lagging strand suggests that HEL308 might act at damaged replication forks in which DNA replication on the leading strand template has been stalled, resulting in polymerase uncoupling and the continued DNA synthesis on the lagging strand template.
FIGURE 3.
HEL308 preferentially unwinds model fork substrates with a ssDNA junction. A, top row, the 3′ overhang partial duplex, splayed arms, and model fork with nascent lagging strand substrates listed in supplemental Table 1 were 5′ end-labeled on the upper strand (5′ with asterisk), mock-treated (0 pmol/μl lane), or treated with increasing concentrations from 0.016 to 1 pmol/μl of HEL308 (0.016, 0.06, 0.25, and 1.00 pmol/μl). Bottom row, substrates were treated with a fixed concentration (0.5 pmol/μl) of HEL308 for the stated times. Lanes marked nt contain control substrate not treated with enzyme, and those marked B include boiled fully denatured substrate. The DNA substrates analyzed are schematically depicted at the bottom of the corresponding gels. The position of substrate (s or substr.) and generated products (p or prod.) are marked next to the gels. B and C, quantification of the data obtained in A expressed in terms of unwinding as a function of HEL308 concentration in B and as a function of incubation time using the fixed concentration of HEL308 in C. The experiments were repeated in triplicate, and the error bars show mean ± S.D.
RPA Stimulates HEL308 and Does Not Prevent Strand Annealing
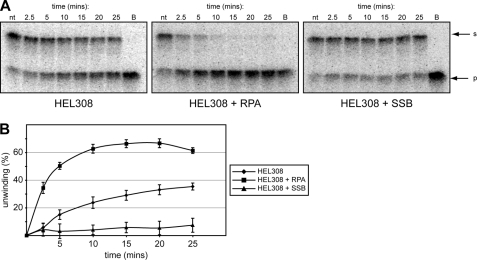
It has been shown previously that the basic DNA unwinding activity of human HEL308 is stimulated by RPA protein (17). In reactions containing the splayed arm DNA substrate, 0.5 pmols/μl HEL308 and 1.2 ng RPA, RPA stimulated HEL308 helicase activity with stimulation peaking at between 15 and 20 min (Fig. 4, A and B). At this time, RPA increased the displacement of the labeled strand by 2.5- to 3.0-fold. One possible explanation for the RPA stimulatory effect on HEL308 is that RPA can inhibit reannealing of the separated strands by binding to unwound ssDNA produced by HEL308. To exclude this possibility, we performed the reaction in the presence of the bacterial SSB protein (1 ng). In contrast to RPA, no stimulatory effect of E. coli SSB was detected at any time during the reaction (Fig. 4). Additionally, no HEL308 ATP-independent annealing activity was detected using a splayed arm substrate (data not shown). This suggests that RPA directly stimulates the helicase activity of HEL308 rather than inhibiting strand reannealing.
FIGURE 4.
RPA stimulates unwinding of model fork structures by HEL308. A, the splayed arms substrate listed in supplemental Table 1 was 5′ end-labeled mock-treated (0 pmol/μl lane) or treated with a fixed concentration (0.5 pmol/μl) of HEL308 for the stated times. Left panel reactions contained HEL308 only, center panel reactions contained 1.2 ng human RPA, and right panel reactions contained 1.0 ng of E. coli SSB protein. The nt controls in the center and right panels are without HEL308 but contain RPA and SSB, respectively. Lanes marked B show boiled substrate. The positions of substrate (s) and generated products (p) are marked next to the gels. B, quantification of the data obtained in A. For each experimental condition, the percentage of substrate unwinding as a function of time is shown. Each experiment was performed in triplicate. The error bars represent mean ± S.D.
Human HEL308 Does Not Directly Displace Nascent Lagging Strands from Model Replication Fork Structures
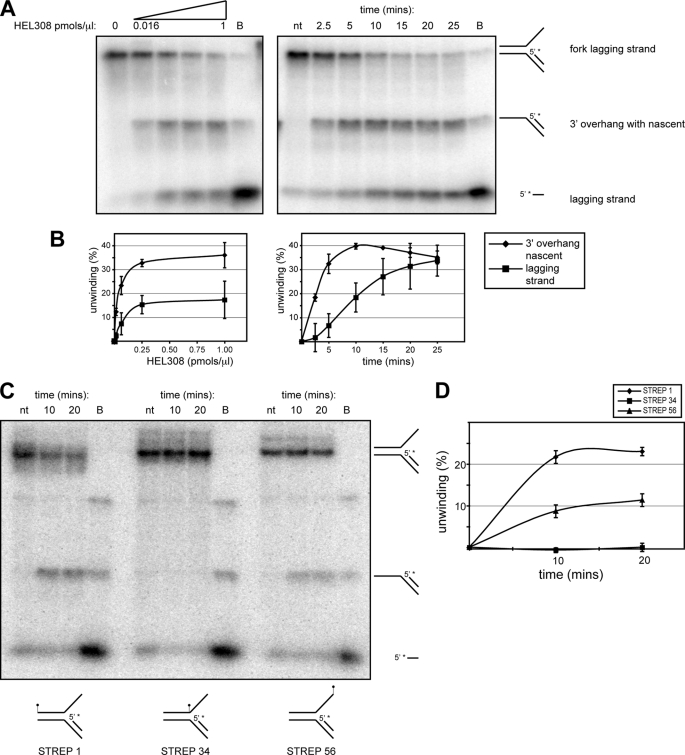
Several archaeal HEL308-like proteins show the ability to remove the nascent lagging strand from model fork structures (21, 33). To investigate if HEL308 also possessed this activity, we analyzed HEL308 activity on replication forks in which the nascent lagging strand was labeled. Using this substrate, we did indeed observe HEL308-mediated removal of the nascent lagging strand (Fig. 5, A and B). However, this product appeared to be the result of a secondary reaction in which the parental strands of the fork are first separated generating a partial duplex with a 3′ overhang, which is then subsequently unwound by HEL308 to release the labeled nascent lagging strand.
FIGURE 5.
HEL308 does not initially displace the nascent lagging strand from model fork structures. A, left panel, the model fork substrate with a 5′-labeled nascent lagging strand (5′ with asterisk) (supplemental Table 1) was mock-treated (0 pmol/μl lane) or treated with increasing concentrations from 0.016 to 1 pmol/μl of HEL308 (0.016, 0.06, 0.25, and 1.00 pmol/μl) Right panel, substrates were treated with a fixed concentration (0.5 pmol/μl) of HEL308 for the stated times. Lanes marked nt contain control not treated with enzyme, and those marked B include boiled substrate. The migration positions of the different products are shown to the right of the gels. B, quantification of the data obtained in A, where the ♦ represents quantification of the generation of the 3′-overhang intermediate (detected as a band halfway between the top and the bottom of the gels), whereas ■ represents generation of displaced nascent lagging strand (band at the bottom of the gels), as a function of HEL308 concentration (left graph) or as a function of time (right graph). In some cases, the error bars (mean ± S.D.) are obscured by a symbol. Each experiment was performed in triplicate. C, unwinding of model fork substrate with 5′-labeled nascent lagging strand, here containing streptavidin-biotin conjugates placed at the 5′ terminus of the upper strand (STREP 1), at the ss-to-dsDNA junction of the upper strand (STREP 34), or at the 3′ terminus of the upper strand (STREP 56). D, quantification of the signal from the gel shown in C, indicating formation of the 3′ overhang intermediate from the substrates with differently located streptavidin. Each experiment was performed in triplicate. Error bars represent mean ± S.D.
The above data indicate that HEL308 preferentially unwinds the parental strands of model replication forks and that this activity can be stimulated by the presence of RPA and a nascent lagging strand but not a nascent leading strand. To examine the mechanism by which the presence of a nascent lagging strand stimulates HEL308 activity, we modified the fork lagging strand substrate to contain a biotin-bound streptavidin conjugate at one of three positions. Blocking access to the end of 3′ ssDNA overhang slowed but did not abolish the reaction (STREP 56 substrate) (Fig. 5C). However, when the access to the ss-dsDNA junction was blocked by streptavidin, we observed total inhibition of the reaction (STREP 34 substrate) (Fig. 5C). Streptavidin present on the dsDNA end of the substrate had the least inhibitory effect on the reaction (STREP 1 substrate). These data indicate that an accessible ss-dsDNA junction is crucial for HEL308 loading onto DNA, and that the protein does not unwind the nascent lagging strand directly from the fork. Note that control experiments with DNA with biotin only (without streptavidin conjugation) did not produce a difference in unwinding, regardless of where the biotin was placed on the parental leading strand (not shown).
DISCUSSION
Through the expression of fluorescently tagged constructs in human cells, we observed that HEL308 protein localizes to sites of replication (coincident with IdU patches and RPA foci) following treatment with an agent (CPT) that arrests and collapses forks. In support of the notion that these represent sites of fork restart and induced HR, Rad51 also colocalized with GFP-HEL308 focus sites, as did FANCD2. Together, this places HEL308 at the sites of damaged replication forks. Consistently, deletion of HEL308 in human cells has previously been reported to induce sensitivity to replication-blocking lesions, namely the ICLs induced by mitomycin C and CPT (24).
To explore the potential mechanistic basis of HEL308 action, we performed a detailed biochemical analysis of the activity of HEL308 on model substrates that resemble those thought to be associated with stalled replication forks. Here, we observed a strong preference for unwinding substrates that model a fork bearing a nascent lagging strand, consistent with a general requirement for a 3′ ssDNA overhang for HEL308 unwinding. Furthermore, and consistent with a role at replication forks, human RPA but not bacterial SSB stimulated this activity, indicating a specific functional interaction between RPA and HEL308. Note, however, that to date we have not been able to detect any direct physical interaction between HEL308 and RPA using purified proteins, although such an interaction has recently been reported for archaeal HEL308 (34). We also sought to determine whether the unwinding on model substrates with a lagging nascent strand represented initial unwinding and release of the nascent lagging strand oligonucleotide or wholesale unwinding of the fork structure with subsequent release of the nascent lagging strand oligonucleotide. By placing biotin-streptavidin conjugates at several positions along the substrate, we discovered that we could inhibit unwinding by locating the conjugate at the ss-to-dsDNA junction. Coupled to the reaction kinetics, our experiments suggest that the enzyme unwinds from the junction point and does not initially displace the nascent lagging strand oligonucleotide.
Although a previous study had defined the polarity of the helicase activity of mammalian HEL308 (17), nothing was known of its preference for structures that arise at damaged replication forks. Previous studies using archaeal HEL308-like proteins (HEL308a from M. thermautotrophicus and Hjm from P. furiosus), however, also identified an ability to preferentially unwind a nascent lagging strand containing a model fork structure (21, 33), although in this case the authors were not able to distinguish whether this was the primary activity of the protein or whether the unwinding of the nascent lagging strand was a consequence of initial substrate unwinding at the ss-to-dsDNA junctions, as our studies suggest. Several other activities that were observed in the archaeal enzymes do not appear to be conserved in human HEL308, most notably the branch migration activity of Hjm from P. furiosus and the fork regression activity of Hjm from S. tokodaii (31, 33).
The ability of the HEL308 family to unwind model substrates with a nascent lagging strand, in a RPA-stimulated manner, supports a role for HEL308 at stalled replication forks. Interestingly, introduction of archaeal HEL308a into E. coli dnaE486 mutants produced a synthetic lethality phenotype, phenocopying the effects of RecQ expression (21). This suggests that HEL308a and RecQ might be recruited to and process similar structures produced at stalled forks. Indeed, HEL308a can process several structures that are substrates for E. coli RecQ, including splayed arm fork structures and mobile HJs (16, 21). Moreover, the HEL308 activities presented in the current study overlap in some aspects with those of mammalian RecQ helicases (1), such as the ability to unwind splayed-arm substrates, suggesting possible redundancy in some processes. However, there are major differences in the substrate preference for human HEL308 versus well characterized mammalian RecQ factors; for example, the ability of the latter to branch migrate HJs and related structures, as exemplified by Bloom's syndrome protein (1). Furthermore, we were unable to detect ATP-independent strand-reannealing activities with HEL308, as reported for several RecQ family members (6–8). A further bacterial helicase with a role on fork processing is UvrD (35), and indeed no eukaryotic functional homologs of this protein have been identified, raising the possibility that HEL308 fulfils a similar role to this protein in vivo. However, it should also be borne in mind that archaeal HEL308 is unable to complement E. coli uvrD mutants for UV sensitivity (16).
Taken together, our results favor a role for human HEL308 at stalled and collapsed replication forks. Based upon its biochemical activity, we propose that HEL308 is recruited to abnormal replication structures such as those that contain a nascent lagging strand but no nascent leading strand. Such structures need to be processed for replication to restart with a canonical replication fork (36). We found no evidence that HEL308 directly removes a nascent lagging strand at the fork, but rather showed that a nascent lagging strand stimulates HEL308 to unwind the parental strands. Such an activity may be important to allow access for additional factors such as members of the RecQ helicase family to promote either fork regression or HR, both processes that can facilitate replication restart.
Supplementary Material
Acknowledgments
We thank Federica Marini, Stephen C. West, and Fumiko Esashi for HEL308 constructs, anti-Rad51 antibodies, and RPA protein, respectively. We thank Blanka Sengerová for helpful comments on the manuscript.
This work was supported by Cancer Research UK grants (to P. J. M. and L. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
- HR
- homologous recombination
- HJ
- Holliday junction
- ICL
- interstrand cross-linking
- RPA
- replication protein A
- SSB
- single-strand binding
- CPT
- camptothecin
- IdU
- iododeoxyuridine
- FANC
- Fanconi anemia complementation
- ssDNA
- single-stranded DNA.
REFERENCES
- 1. Wu L., Hickson I. D. (2006) Annu. Rev. Genet. 40, 279–306 [DOI] [PubMed] [Google Scholar]
- 2. Bernstein K. A., Gangloff S., Rothstein R. (2010) Annu. Rev. Genet. 44, 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rossi M. L., Ghosh A. K., Bohr V. A. (2010) DNA Repair 9, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chu W. K., Hickson I. D. (2009) Nat. Rev. Cancer 9, 644–654 [DOI] [PubMed] [Google Scholar]
- 5. Liu Y. (2010) DNA Repair 9, 325–330 [DOI] [PubMed] [Google Scholar]
- 6. Machwe A., Xiao L., Groden J., Matson S. W., Orren D. K. (2005) J. Biol. Chem. 280, 23397–23407 [DOI] [PubMed] [Google Scholar]
- 7. Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macris M. A., Krejci L., Bussen W., Shimamoto A., Sung P. (2006) DNA Repair 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 9. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., Hiom K. (2008) J. Biol. Chem. 283, 36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A. (2008) Mol. Cell 29, 141–148 [DOI] [PubMed] [Google Scholar]
- 11. Barber L. J., Youds J. L., Ward J. D., McIlwraith M. J., O'Neil N. J., Petalcorin M. I., Martin J. S., Collis S. J., Cantor S. B., Auclair M., Tissenbaum H., West S. C., Rose A. M., Boulton S. J. (2008) Cell 135, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacKay C., Toth R., Rouse J. (2009) Biochem. Biophys. Res. Commun. 390, 187–191 [DOI] [PubMed] [Google Scholar]
- 14. Blastyák A., Hajdú I., Unk I., Haracska L. (2010) Mol. Cell. Biol. 30, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., Haracska L. (2007) Mol. Cell 28, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodman I. L., Bolt E. L. (2009) Biochem. Soc. Trans. 37, 74–78 [DOI] [PubMed] [Google Scholar]
- 17. Marini F., Wood R. D. (2002) J. Biol. Chem. 277, 8716–8723 [DOI] [PubMed] [Google Scholar]
- 18. Boyd J. B., Sakaguchi K., Harris P. V. (1990) Genetics 125, 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris P. V., Mazina O. M., Leonhardt E. A., Case R. B., Boyd J. B., Burtis K. C. (1996) Mol. Cell. Biol. 16, 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muzzini D. M., Plevani P., Boulton S. J., Cassata G., Marini F. (2008) DNA Repair 7, 941–950 [DOI] [PubMed] [Google Scholar]
- 21. Guy C. P., Bolt E. L. (2005) Nucleic Acids Res. 33, 3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marini F., Kim N., Schuffert A., Wood R. D. (2003) J. Biol. Chem. 278, 32014–32019 [DOI] [PubMed] [Google Scholar]
- 23. Seki M., Marini F., Wood R. D. (2003) Nucleic Acids Res. 31, 6117–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moldovan G. L., Madhavan M. V., Mirchandani K. D., McCaffrey R. M., Vinciguerra P., D'Andrea A. D. (2010) Mol. Cell. Biol. 30, 1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Räschle M., Knipscheer P., Knipsheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., Walter J. C. (2008) Cell 134, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshimura M., Kohzaki M., Nakamura J., Asagoshi K., Sonoda E., Hou E., Prasad R., Wilson S. H., Tano K., Yasui A., Lan L., Seki M., Wood R. D., Arakawa H., Buerstedde J. M., Hochegger H., Okada T., Hiraoka M., Takeda S. (2006) Mol. Cell 24, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) Blood 100, 2414–2420 [DOI] [PubMed] [Google Scholar]
- 28. Hussain S., Wilson J. B., Medhurst A. L., Hejna J., Witt E., Ananth S., Davies A., Masson J. Y., Moses R., West S. C., de Winter J. P., Ashworth A., Jones N. J., Mathew C. G. (2004) Hum. Mol. Genet. 13, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 29. Wang X., Andreassen P. R., D'Andrea A. D. (2004) Mol. Cell. Biology 24, 5850–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu S., Li Z., Wang Z., Ma X., Sheng D., Ni J., Shen Y. (2008) Biochem. Biophys. Res. Commun. 376, 369–374 [DOI] [PubMed] [Google Scholar]
- 31. Li Z., Lu S., Hou G., Ma X., Sheng D., Ni J., Shen Y. (2008) J. Bacteriology 190, 3006–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujikane R., Komori K., Shinagawa H., Ishino Y. (2005) J. Biol. Chem. 280, 12351–12358 [DOI] [PubMed] [Google Scholar]
- 33. Fujikane R., Shinagawa H., Ishino Y. (2006) Genes Cells 11, 99–110 [DOI] [PubMed] [Google Scholar]
- 34. Woodman I. L., Brammer K., Bolt E. L. (2011) DNA Repair 10, 306–313 [DOI] [PubMed] [Google Scholar]
- 35. Cadman C. J., Matson S. W., McGlynn P. (2006) J. Mol. Biol. 362, 18–25 [DOI] [PubMed] [Google Scholar]
- 36. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.