Abstract
Toll-like receptor 3 (TLR3), a member of the pathogen recognition receptors, is widely expressed in various cells and has been shown to activate immune signaling pathways by recognizing viral double-stranded RNA. Recently, it was reported that the activation of TLR3 induced apoptosis in some cells, but the detailed molecular mechanism is not fully understood. In this study, we found that in endothelial cells polyinosinic-polycytidylic acid (poly(I-C)) induced dose- and time-dependent cell apoptosis, which was elicited by TLR3 activation, as TLR3 neutralization and down-regulation repressed the apoptosis. Poly(I-C) induced the activation of both caspases 8 and 9, indicating that TLR3 triggered the signaling of both the extrinsic and intrinsic apoptotic pathways. Poly(I-C) up-regulated tumor necrosis factor-related apoptosis-inducing ligand and its receptors, death receptors 4/5, resulting in initiating the extrinsic pathway. Furthermore, poly(I-C) down-regulated anti-apoptotic protein, B cell lymphoma 2 (Bcl-2), and up-regulated Noxa, a key Bcl-2 homology 3-only antagonist of Bcl-2, leading to the priming of the intrinsic pathway. A p53-related protein, the transactivating p63 isoform α (TAp63α), was induced by TLR3 activation and contributed to the activation of both the intrinsic and extrinsic apoptotic pathways. Both the cells deficient in p63 gene expression by RNA interference and cells that overexpressed the N-terminally truncated p63 isoform α (ΔNp63α), a dominant-negative variant of TAp63α, by gene transfection, survived TLR3 activation. Taken together, TAp63α is a crucial regulator downstream of TLR3 to induce cell death via death receptors and mitochondria.
Keywords: Apoptosis, Caspase, Mitochondria, p63, Toll-like Receptors (TLR), Bcl-2, Death Receptor, Noxa, TRAIL
Introduction
Toll-like receptors (TLR)4 recognize different pathogen-associated molecular patterns, leading to the activation of innate immune response and the priming of subsequent adaptive immune response by controlling multiple functions of dendritic cells (1, 2). Expressed in conventional dendritic cells and in a variety of epithelial cells, including airway, uterine, corneal, vaginal, cervical, biliary, and intestinal epithelial cells (1), toll-like receptor 3 (TLR3) was the first member of the TLR family known as a pattern recognition receptor and has been demonstrated to detect a conserved viral molecular pattern, double-stranded RNA (dsRNA), now regarded as a universal viral pathogen-associated molecular pattern (3).
TLR3 has been reported to promote the cross-priming of the cytotoxic T cell response against viral infection (4). Immunization with virus-infected cells or cells containing synthetic dsRNA leads to a striking increase in cytotoxic T cell cross-priming against cell-associated antigens. TLR3 is expressed in the central nervous system and plays important roles in the control of herpes simplex virus 1, which spreads from the epithelium to the central nervous system via cranial nerves (5). However, several recent studies have reported conflicting conclusions about TLR3 in host immunity to viruses. TLR3-deficient mice have been shown to be more resistant to lethal infection and have a reduced incidence of liver disease associated with hepatotropic Punta Toro virus infection compared with wild-type mice (6). The recombinant strain Western Reserve vaccinia virus has been reported to be more deleterious to the wild-type mice than the TLR3-deficient mice (7). More importantly, TLR3 has been reported to mediate West Nile virus entry into the brain, causing lethal encephalitis by impairment of the blood-brain barrier (8).
p63 is a p53-related gene that was cloned in 1998 (9). The high level of sequence similarity between the p63 and p53 proteins, particularly in the DNA-binding domain, allows p63 to transactivate p53-responsive genes, leading to cycle cell arrest and apoptosis. The p63 gene expresses three alternatively spliced C-terminal isoforms (α, β, and γ) and can be transcribed from the promoter upstream of the first exon or from an alternative promoter located in intron 3. Thus, it produces at least six mRNA variants, which encode six different p63 protein isoforms (transactivating p63 (TAp63) α, β, and γ and N-terminally truncated p63 (ΔNp63) α, β, and γ) (9, 10). The TAp63 isoforms are able to bind to DNA through a p53 response element and activate the transcription of target genes to induce cell cycle arrest or apoptosis. However, the ΔNp63 isoforms, which lack the transactivation domain, can bind to DNA through a p53 response element and exert dominant-negative effects over p53, p73, and p63 activities either by competing for the DNA-binding sites or by direct protein interaction. Compared with p53, TAp63 isoforms differentially recognize responsive elements, resulting in target gene specificity (11). One important p63 isoform, TAp63α, induces apoptosis via the death receptor apoptotic pathway in which the expression of CD95, the TNF receptor, and the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptors is triggered and via the mitochondrial apoptotic pathway in which the expression of pro-apoptotic B cell lymphoma 2 (Bcl-2) family members, such as Bax, is up-regulated (12). p63 has been further implicated in many p53-independent pathways, including stem cell regeneration, epidermal morphogenesis, and limb development (13, 14). Therefore, p63 is not functionally redundant to p53.
Recently, TLR has been reported to elicit apoptosis in several cells. In melanoma cells, TLR3 induced the Toll/IL-1R domain-containing adapter inducing IFNβ (TRIF)-dependent activation of pro-apoptotic signaling, which was under the control of inhibitor of apoptosis proteins (15). In pancreatic β cells, dsRNA induced TLR3-dependent apoptosis by the activation of the interferon regulatory factor 3 (IRF3) pathway (16). TLR3 also triggers apoptosis of human prostate cancer cells through a protein kinase C (PKC) α-dependent mechanism (17). In human breast cancer cells, TLR3 induces apoptosis that involves the molecular adapter, TRIF, and type I IFN autocrine signaling (18). Moreover, TLR3-mediated cell death involves the activation of caspases and engages both extrinsic and intrinsic apoptotic pathways (19). IFNα has been reported to sensitize human umbilical vein endothelial cells to apoptosis induced by dsRNA (20). However, the detailed mechanism of TLR3-induced cell apoptosis is not fully understood.
In this study, we found that TRAIL death receptors 4/5 (DR4/5) and Noxa elicited the extrinsic and intrinsic apoptosis pathways, respectively, in TLR3-induced endothelial cell apoptosis, which was regulated by TAp63α, a p53-related protein. By analyzing the cells deficient in p63 gene expression and the cells overexpressing ΔNp63α, a dominant-negative variant of TAp63α, we have established that TAp63α, as a pivotal control point, functions downstream of TLR3 to elicit cell apoptosis.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
The immortalized human umbilical vein endothelial cells (HUVECs) expressing endothelial cell characteristic markers, endothelial nitric-oxide synthase, CD31, and Ve-cadherin (21) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were grown in DMEM containing 10% fetal calf serum (FCS) and antibiotics. Primary HUVECs were purchased from ScienCell Research Laboratories and maintained in endothelial culture medium (ScienCell, Carlsbad, CA) supplemented with 5% FCS, 1% endothelial cell growth supplement, and antibiotics. All cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C. Mouse anti-human phospho-nuclear factor-κB (NF-κB) p65 (Ser-536) (7F1) mAb, rabbit anti-human NF-κB p65 antibody, mouse anti-human caspase 8 and 9 mAbs, rabbit anti-human poly(ADP-ribose) polymerase (PARP), and Bcl-2 antibodies and human p53 siRNA were purchased from Cell Signaling Technology (Beverly, MA). Mouse anti-human TRAIL/TNFSF10, TNFα/TNFSF1A, and INFβ mAbs, recombinant human TNFα/TNFSF1A, INFβ, general caspase inhibitor Z-VAD-FMK, caspase 8 inhibitor Z-IETD-FMK, and caspase 9 inhibitor Z-LEHD-FMK were purchased from R&D Systems (Minneapolis, MN). Human TNFα ELISA kit, mouse anti-human β-actin mAb, and fluorescein isothiocyanate (FITC)-annexin V/propidium iodide (PI) apoptosis kits were purchased from Jingmei Co. (Shenzhen, China). Pro-apoptosis chemical staurosporine was purchased from Cayman Chemical (Ann Arbor, MI). Mouse anti-human TLR3 mAb (clone TLR3.7 with the function to neutralize TLR3) was purchased from HyCult Biotechnology (Uden, The Netherlands). Mouse anti-human Noxa mAb was purchased from Abcam (Cambridge, UK). Polyinosinic-polycytidylic acid (poly(I-C)), a synthetic analog of dsRNA was purchased from InvivoGen (San Diego). p53 inhibitor pifithrin-α and PARP inhibitors DPQ and PJ-34 were purchased from Alexis Biochemicals (San Diego). Human TLR3 shRNA plasmid was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Human TRAIL ELISA kit was purchased from Boster Biochemicals (Wuhan, China).
Reverse Transcription-PCR (RT-PCR)
Total RNA was extracted from 2 to 5 × 106 cells using TRIzol (Invitrogen), as described by the manufacturer. mRNA was reverse-transcribed with RevertAid (MBI Fermentas, Burlington Ontario, Canada) at 42 °C for 60 min, and the resulting cDNA was subjected to PCR (95 °C for 1 min followed by 25–35 cycles at 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 1 min and an extension for 10 min at 68 °C). PCR products were separated on 1.0% agarose gels and visualized with ethidium bromide. Forward and reverse primer pairs are listed (5′ to 3′) as follows: Bcl-2-F, CGTTTGGCAGTGCAATGGT, and Bcl-2-R, TTCTTGATTGAGCGAGCCTT; ΔNp63-F, TACCTGGAAAACAATGCCCA, and ΔNp63-R, ATGGCTGTTCCCCTCTACTC; DR4-F, CAATGGGAACATAGCCCTTTG, and DR4-R AAACACACCCTGTCCATGCA; DR5-F, TCTGATCACCCAACAAGCCCT, and DR5-R, ACAATCACCGACCTTGACCAT; GAPDH-F, AATCCCATCACCATCTTCCA, and GAPDH-R CCTGCTTCACCACCTTCTTG; IFNα-F, AGCTGCAAGTCAAGCTGCTCT, and IFNα-R, TTCTTCACAGCCAAGATGGA; IFNβ-F, TCTCCTCCAAATTGCTCTCCT, and IFNβ-R TACTCCTTGGCCTTCAGGTAA; IL-1β-F, TTGAAGCTGATGGCCCTAAAC, and IL-1β-R, CACCAAGCTTTTTTGCTGTG; IL-6-F, CCAGTACCCCCAGGAGAAGAT, and IL-6-R, TTGCCTTTTTCTGCAGGAAC; IL-8-F, TTGGCAGCCTTCCTGATTT, and IL-8-R, TCAAAAACTTCTCCACAACCC; MDM2-F, TTCGTGAGAATTGGCTTCC, and MDM2-R, GGCAGGGCTTATTCCTTTTCT; Noxa-F, CCAAACTCTTCTGCTCAGGAA, and Noxa-R, ATCACAGGTCATCTCCCTTCA; p21-F, TGGGGATGTCCGTCAGAA, and p21-R TTCCTCTTGGAGAAGATCAGC; p53-F, AGCTGTGGGTTGATTCCACAC, and p53-R TTTCTTCTTTGGCTGGGGA; p63α-F, ATGAACAAGCTGCCTTCTGTG, and p63α-R ACAGCATCAATAACACGCTCA; TAp63-F, GCCCATTGACTTGAACTTTG, and TAp63-R, GGGTCATCACCTTGATCTGGA; TLR3-F, AAAACCTTTGCCTTCTGCAC, and TLR3-R, GGAATCGTTACCAACCACATT; TNFα-F, TGACAAGCCTGTAGCCCATGTT, and TNFα-R, AGGGCAATGATCCCAAAGTAGA; and TRAIL-F, TTCACAGTGCTCCTGCAGTCT, and TRAIL-R, AGTTTATTTTGCGGCCCAGA.
Flow Cytometry
Human endothelial cells, grown to subconfluency, were harvested and washed with fluorescence-activated cell sorting (FACS) buffer (5 mmol/liter EDTA, 0.1% NaN3, and 1% FCS, in Dulbecco's phosphate-buffered saline (PBS)). After incubation with a monoclonal antibody against human TLR3 for 40 min on ice, the cells were stained with a FITC-labeled secondary antibody and examined for protein expression by flow cytometry (BD Biosciences).
Cell Death Assay
The death of cells was detected by FITC-annexin V/PI staining. In general, 1–2 × 106 cells were washed twice with PBS and then labeled with FITC-annexin V and PI in binding buffer, according to instructions provided by the manufacturer. Fluorescence signals of FITC and PI were detected on a FACScan (BD Biosciences). The log of FITC-annexin V fluorescence was displayed on the x axis, and the log of PI fluorescence was displayed on the y axis. For each analysis, 10,000 events were recorded.
Cytokine Measurements by ELISA
Production of human TNFα and TRAIL, found in the culture supernatants and cell lysates of human endothelial cells, in response to various concentrations of poly(I-C), was detected by ELISA kits, according to the manufacturer's standard protocols.
Immunoblot
Endothelial cells were lysed in 150 μl of 1× SDS sample buffer (62.5 mmol/liter Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, and 50 mmol/liter DTT), sonicated for 3 s, and then boiled for 5 min. The cell lysate was then centrifuged at 10,000 × g at 4 °C for 10 min. Total protein was electrophoresed on 4–12% gradient Tris-glycine precast gels (Invitrogen) and transferred onto Immobilon P membranes (Millipore). The membranes were blocked by incubation in 3% nonfat dry milk for 1 h at room temperature and then incubated with primary antibodies in PBS containing 0.01% Tween 20 overnight at 4 °C. After incubation with a horseradish peroxidase-conjugated secondary antibody, the protein bands were detected with SuperSignal chemiluminescent substrate-stable peroxide solution (Pierce) and BIOMAX-MR film (Eastman Kodak Co.). When necessary, the membranes were stripped with Restore Western blot stripping buffer (Pierce) and re-probed with antibodies against various cellular proteins.
siRNA Plasmid Construction and Transfection
For the construction of human p63 RNAi plasmid, a 21-oligonucleotide (GAGTGGAATGACTTCAACTTT) corresponding to nucleotides 2058–2078 in exon 14 of the human p63 mRNA (LOCUS NM_003722) was inserted into pRNAT-U6.1-Neo (GenScript, Piscataway, NJ) with BamHI and HindIII. The plasmid was transfected into endothelial cells with Lipofectamine 2000 (Invitrogen), according to the manufacturer's standard protocols.
Expression Plasmid Construction and Transfection
All enzymes used for the construction of recombinant plasmids were products of MBI Fermentas (Burlington Ontario, Canada). Total RNA was isolated from squamous carcinoma CNE1 cells using TRIzol reagent and reverse-transcribed. The 1767-bp cDNA of human ΔNp63α was produced by PCR using the primers sense (5′-GCTAACATGTTGTACCTGGAAAAC-3′) and antisense (5′-TCACTCCCCCTCCTCTTTGA-3′) and inserted into the pMD19-T vector (Takara, Japan). The white clones (after blue-white screening) were subject to PCR screening, and positive plasmids were expanded. The amplified cDNA was digested by restriction enzymes BamHI and HindIII and inserted into pcDNA3.1+. The ΔNp63α cDNA inserted into the constructs was sequenced and verified as identical to the published sequences. The transfection of cells with the plasmid was performed in a 6-well culture plate with 60–80% confluence of cells using Lipofectamine 2000, according to the manufacturer's standard protocols. Stable transfected cells were selected with 800 μg/ml G418 for 3 weeks.
Quantitative Real Time RT-PCR (qRT-PCR)
The qRT-PCR was performed according to the manufacturer's standard protocols. Briefly, total RNA from cells was isolated and reverse-transcribed as above. The cDNA was amplified using TaqMan Universal PCR master mix (Applied Biosystems) and an ABI Prism 7500 sequence detection system (Applied Biosystems). Amplification of the target genes was normalized using the amplification levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control. The efficiency of the PCR was tested by amplification of the target from serially diluted cDNA generated from the reverse transcription of a stock set of human RNA. Data analysis and calculations were performed using the 2−ΔΔCT comparative method, as described by the manufacturer. Gene expression is shown as the fold inductions of a gene measured in poly(I-C)-treated samples, relative to samples cultured with medium.
Statistical Analysis
All experiments were performed at least three times, and the representative results are shown. Results are expressed as the mean ± S.D. Differences between groups were examined for statistical significance using Student's t test, and p values equal to or less than 0.05 were considered statistically significant.
RESULTS
Poly(I-C) Induces Cell Apoptosis in Endothelial Cells
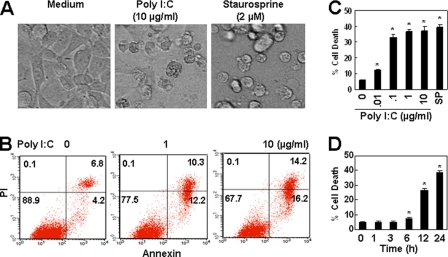
The activation of TLR3 induces apoptosis in various cells (15–20). We first detected the effect of poly(I-C), a synthetic dsRNA analog, on endothelial cell apoptosis. We found that the treatment of resting primary HUVECs with poly(I-C) for 24 h elicited a slight apoptotic effects (data not shown). However, when the cells, pretreated with 1 μg/ml poly(I-C) for 24 h, were re-stimulated with poly(I-C) for another 24 h, a significant fraction of cells underwent detachment (Fig. 1A). To quantitate the fraction of dead cells, we stained unfixed cells treated by poly(I-C) for 24 h with FITC-annexin V/PI and performed flow cytometry analysis. This assay detects all early apoptotic (annexin V-positive/PI-negative), late apoptotic (annexin V-positive/PI-positive), and necrotic (annexin V-negative/PI-positive) cells. Fig. 1B shows the annexin V/PI profiles of primary endothelial cells in response to the increasing concentrations of poly(I-C). Up to 30% of endothelial cells underwent cell apoptosis within 24 h by poly(I-C) treatment. In immortalized HUVECs, poly(I-C) induced dose- and time-dependent cell apoptosis (Fig. 1, C and D) without a poly(I-C) pretreatment, and up to 40% of the cells underwent cell apoptosis within 24 h.
FIGURE 1.
Poly(I-C) induces endothelial cell apoptosis. A, poly(I-C) induced the detachment of primary HUVECs. Cells, pretreated with 1 μg/ml poly(I-C) for 24 h, were re-stimulated with or without 10 μg/ml poly(I-C) for 24 h, and the cells in representative fields were photographed. Cells, treated with 2 μm staurosporine, were photographed as a positive control. B, poly(I-C) induced apoptosis in primary HUVECs. Cells, pretreated with 1 μg/ml poly(I-C) for 24 h, were re-stimulated with the indicated concentrations of poly(I-C) for 24 h. Unfixed cells were stained with FITC-annexin V/PI. Cell apoptosis was measured by flow cytometry analysis. C, poly(I-C) induced the dose-dependent cell apoptosis in immortalized HUVECs. Cells were treated with the indicated concentrations of poly(I-C) for 24 h. Cell apoptosis was measured as in B. Data are shown as percentages and represented as mean ± S.D. of triplicates. Cells treated with 1 μm staurosporine were used as a positive control. *, p < 0.05 compared with the control group. D, poly(I-C) (10 μg/ml) induced time-dependent cell apoptosis in immortalized HUVECs. *, p < 0.05 compared with the control group.
Poly(I-C) Induces TLR3-dependent Cell Apoptosis
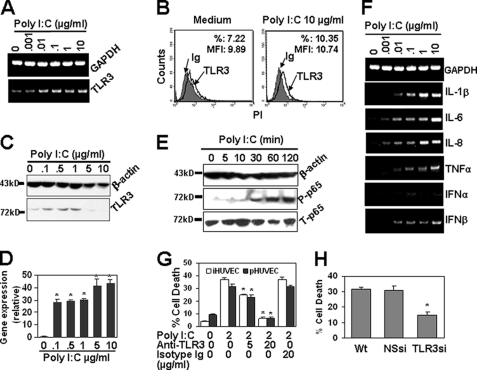
IFNα sensitizes freshly isolated HUVECs to double-stranded RNA-induced apoptosis (20), and we found poly(I-C) induced cell apoptosis in both primary and immortalized endothelial cells (Fig. 1). To determine the contribution of TLR3 in poly(I-C)-induced cell apoptosis, we first detected the TLR3 gene expression in immortalized HUVECs. As shown in Fig. 2, A and B, resting immortalized HUVECs expressed TLR3 gene transcript and protein. The expression of TLR3 was slightly up-regulated by treatment with poly(I-C) at both the gene and protein levels. In primary HUVECs, Western blot results show that the treatment with 0.1–1 μg/ml poly(I-C) up-regulated the expression of TLR3. However, TLR3 levels were low in 5–10 μg/ml poly(I-C)-treated cells (Fig. 2C), because most of the cells underwent apoptosis. Quantitative real time RT-PCR data show that poly(I-C) significantly up-regulated the gene expression of TLR3 in primary HUVECs (Fig. 2D). We then examined the potential of poly(I-C) to elicit TLR3-medicated signaling transduction. The results show that the treatment of immortalized HUVECs with 1 μg/ml poly(I-C) induced the phosphorylation of the NF-κB p65 subunit (Fig. 2E). Poly(I-C) treatment also up-regulated the transcripts coding for cytokines, including IL-1β, IL-6, IL-8, TNFα, and IFNβ, in immortalized HUVECs (Fig. 2F). When a TLR3-neutralizing antibody was administered to inhibit TLR3 activation in both primary and immortalized HUVECs, apoptosis induced by poly(I-C) was repressed significantly (Fig. 2G). Moreover, TLR3 shRNA transfection significantly inhibited poly(I-C)-induced cell apoptosis (Fig. 2H). Thus, we conclude that TLR3 contributed to the cell apoptosis induced by poly(I-C) in endothelial cells, although there are other pathogen-associated molecular pattern receptors, such as RIG-I and MDA5, which recognize dsRNA (22).
FIGURE 2.
Poly(I-C) induces TLR3-dependent apoptosis. A, poly(I-C) up-regulated gene expression of TLR3. Immortalized HUVECs, treated with the indicated concentrations of poly(I-C) for 24 h, were harvested, and the gene expression of TLR3 was measured by RT-PCR. GAPDH transcript was measured as a loading control. B, immortalized HUVECs expressed TLR3 protein. Cells, cultured in the presence or absence of 10 μg/ml poly(I-C) for 24 h, were harvested. The expression of TLR3 protein was measured by flow cytometry. Mouse isotype Ig was used as a control. C, poly(I-C) up-regulated the expression of TLR3 protein in primary HUVECs. Cells, treated with the indicated concentrations of poly(I-C) for 24 h, were harvested and lysed with sample buffer. Equal amounts of total proteins were electrophoresed and blotted for the detection of TLR3 protein expression. β-Actin protein was detected as a loading control. D, poly(I-C) significantly up-regulated the gene expression of TLR3 in primary HUVECs. Cells were treated as in Fig. 1B. The gene transcript of TLR3 was measured by qRT-PCR. GAPDH gene expression was detected as an endogenous control. *, p < 0.05 compared with the control group. E, poly(I-C) induced the phosphorylation of the NF-κB p65 subunit. Immortalized HUVECs, starved overnight with serum-free medium, were cultured in the presence of 2 μg/ml poly(I-C) at 37 °C and lysed at the indicated time points. Phospho-p65 (P-p65) and total p65 (T-p65) were detected by Western blot. F, RT-PCR results show that poly(I-C) up-regulated the gene expression of cytokines in immortalized HUVECs. G, TLR3 neutralization abrogated poly(I-C)-induced cell apoptosis. Immortalized HUVECs (iHUVEC) and 1 μg/ml poly(I-C) pretreated (37 °C for 24 h) primary HUVECs (pHUVEC) were cultured in the absence or presence of mouse anti-human TLR3 antibody at 37 °C for 1 h and then treated with or without 2 μg/ml poly(I-C) for 24 h. Cell apoptosis was tested as in Fig. 1B. Mouse isotype Ig was used as a control. Data are represented as mean ± S.D. of triplicates. *, p < 0.05 compared with the poly(I-C) treatment group. H, TLR3 down-regulation by RNA interference abrogated poly(I-C)-induced cell apoptosis in immortalized HUVECs. Wild-type (Wt), nonspecific shRNA (NSsi), and TLR3 shRNA (TLR3si)-transfected cells were treated with 2 μg/ml poly(I-C) for 24 h. Cell apoptosis was detected as in Fig. 1B. *, p < 0.05 compared with the control group.
Both the Intrinsic and Extrinsic Apoptosis Pathways Contribute to the Cell Apoptosis Induced by Poly(I-C)
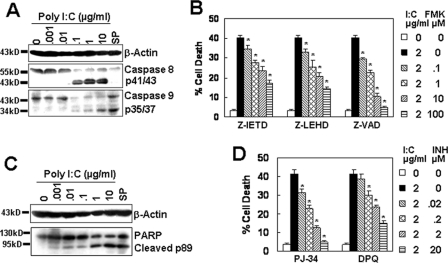
The following are the two main pathways known to contribute to the induction of apoptosis: a death receptor-initiated caspase 8-dependent pathway known as the extrinsic pathway; and a mitochondrion-initiated caspase 9-dependent pathway known as the intrinsic pathway (23). An understanding of the particular pathway involved in TLR3-induced cell death is important because it might provide insight into the potential factors that are responsible for initiating cell suicide and allow for the development of a more targeted therapy. To elucidate the detailed mechanism of TLR3-induced cell death in endothelial cells, we first examined the involvement of these two pathways by measuring the activation of caspases 8 and 9 and their common downstream molecule, PARP, which is a specific hallmark of apoptotic cell death. Western blot analysis shows that poly(I-C) treatment led to the cleavage of caspase 8, resulting in the release of cleaved fragments p41/43 (Fig. 3A), indicating the activation of the extrinsic pathway. Similarly, poly(I-C) treatment also led to the cleavage of caspase 9, resulting in the release of cleaved fragments p35/p37 (Fig. 3A), suggesting the activation of the intrinsic pathway. To prove further the involvement of caspases in TLR3-induced cell death, caspase 8-specific inhibitor Z-IEHD-FMK, caspase 9-specific inhibitor Z-LEHD-FMK, and pan-caspase inhibitor Z-VAD-FMK, which have been reported to improve cell survival in sepsis by preventing lymphocyte apoptosis (24), were administered to the culture to test the effect of caspase inhibition on poly(I-C)-induced cell death. As expected, the inhibition of caspase 8 or 9 led to the dose-dependent decrease of cell apoptosis, but the down-regulation of cell apoptosis was partial, and the levels of cell apoptosis were not down-regulated to that as low as the background levels (Fig. 3B). On the contrary, pan-caspase inhibitor Z-VAD-FMK completely repressed cell death to the background level (Fig. 3B). When we analyzed the cellular signaling downstream of the caspases, we found that poly(I-C) treatment led to the cleavage of PARP, resulting in the release of the cleaved fragment p89 (Fig. 3C). Additionally, PARP inhibitors dose-dependently repressed poly(I-C)-induced cell death (Fig. 3D). These results indicate that both the extrinsic and intrinsic pathways contributed to poly(I-C)-induced cell apoptosis.
FIGURE 3.
Poly(I-C) activates both the intrinsic and extrinsic pathways. A, poly(I-C) activated both caspases 8 and 9. Immortalized HUVECs, treated with the indicated concentrations of poly(I-C) for 24 h, were lysed with sample buffer. The cleaved fragments from caspases 8 and 9 were detected by Western blot. 2 μm staurosporine (SP) was used as a control. B, caspase inhibitors down-regulated the cell apoptosis induced by poly(I-C). Immortalized HUVECs were cultured in the absence or presence of caspase 8 inhibitor (Z-IETD-FMK), caspase 9 inhibitor (Z-LEHD-FMK), or pan-caspase inhibitor (Z-VAD-FMK) at 37 °C for 1 h, followed by the treatment with or without 2 μg/ml poly(I-C) for 24 h. Cell apoptosis was tested as in Fig. 1B. Data are represented as mean ± S.D. of triplicates. *, p < 0.05 compared with the poly(I-C) treatment group. C, Western blot results show that poly(I-C) activated the caspase downstream molecule PARP. D, PARP inhibitors down-regulated the cell apoptosis induced by poly(I-C). *, p < 0.05 compared with the poly(I-C) treatment group.
TNFα Does Not Contribute to the Initiation of the Extrinsic Pathway
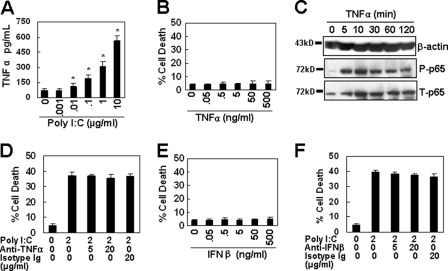
The extrinsic pathway was initiated by extracellular death ligands, such as TNFα, TRAIL, and FasL, binding to their cell surface death receptors, TNFRs, DR4/5, and Fas, respectively. As TNFα positively regulates brain-blood barrier permeability to promote virus entry into the central nervous system (8), and poly(I-C) up-regulated the expression of TNFα gene transcript (Fig. 2F), we paid special attention to the effect of TNFα on the cell apoptosis induced by TLR3 activation. ELISA analysis shows that the treatment of cells with increasing concentrations of poly(I-C) dose-dependently up-regulated TNFα secretion in the culture supernatant of immortalized HUVECs (Fig. 4A). However, when the cells were treated with increasing concentrations of TNFα, no cell apoptosis was detected in immortalized HUVECs (Fig. 4B), although the treatment of cells with TNFα induced the phosphorylation of NF-κB (Fig. 4C), which is one of the most important signaling molecules downstream of the TNF receptors (25). Furthermore, TNFα neutralization by monoclonal antibodies did not decrease the cell apoptosis induced by poly(I-C) in immortalized HUVECs (Fig. 4D). These results indicate that TNFα did not contribute to TLR3-induced cell death.
FIGURE 4.
TNFα and IFNβ do not trigger cell apoptosis. A, poly(I-C) up-regulated the expression of TNFα in immortalized HUVECs. Cells were treated with the indicated concentrations of poly(I-C) for 24 h. The supernatant was harvested to detect the TNFα secretion by ELISA. Data are represented as mean ± S.D. of triplicates. *, p < 0.05 compared with the medium control. B, TNFα (37 °C for 24 h) did not induce cell apoptosis in immortalized HUVECs. C, TNFα (50 ng/ml) induced NF-κB signaling in immortalized HUVECs. D, TNFα neutralization did not inhibit poly(I-C)-induced cell apoptosis in immortalized HUVECs. E, IFNβ (37 °C for 24 h) did not induce cell apoptosis in immortalized HUVECs. F, IFNβ neutralization did not inhibit poly(I-C)-induced cell apoptosis in immortalized HUVECs.
IFNβ Is Not Involved in Poly(I-C)-induced Cell Apoptosis
We found that poly(I-C) up-regulated the gene expression of interferon type I IFNβ (Fig. 2F). To determine whether IFNβ is involved in poly(I-C)-induced cell apoptosis, immortalized HUVECs were treated with recombinant IFNβ, and cell apoptosis was detected by FITC-annexin V/PI staining. The results show that IFNβ treatment did not induce cell apoptosis (Fig. 4E). Moreover, when immortalized HUVECs, pretreated with IFNβ-neutralizing antibody, were re-stimulated with poly(I-C), the cell apoptosis induced by poly(I-C) was not decreased (Fig. 4F), suggesting that IFNβ was not involved in poly(I-C)-induced cell death.
Extrinsic Pathway Is Triggered by TRAIL Reacting with DR4/5
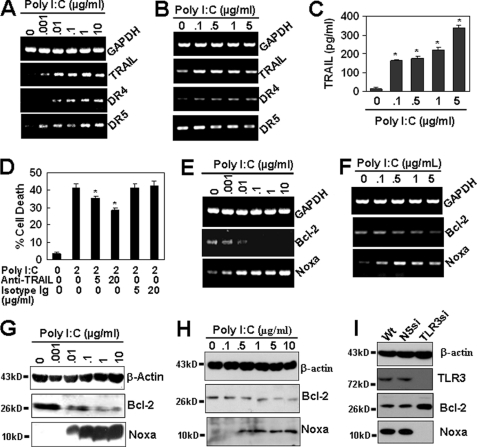
In the TNF family, TRAIL is another initiator that triggers cell death by reacting with its receptors, DR4 and DR5, via the extrinsic pathway (26). The recognition of multiple mycobacterial components by TLR2 and TLR4 induces the release of TRAIL from neutrophils (27), suggesting that the activation of TLRs is a source of TRAIL. In our study, we found that resting primary and immortalized HUVECs expressed lower levels of TRAIL, DR4, and DR5 (Fig. 5, A and B). The treatment of cells with poly(I-C) up-regulated the gene transcripts of TRAIL and its receptors (Fig. 5, A and B) in a dose-dependent manner, and ELISA results show that the up-regulation of TRAIL was significant in primary HUVECs (Fig. 5C), suggesting that TRAIL and DR4/5 counterparts may be involved in poly(I-C) induced cell death. This is supported by the observation that the TRAIL neutralization by monoclonal antibodies down-regulated the cell apoptosis induced by poly(I-C) from 41.5 to 28.3% (Fig. 5D). These results indicate that TRAIL reacting with DR4/5 triggered the extrinsic pathway.
FIGURE 5.
TRAIL-DR4/5 and Noxa trigger the extrinsic and intrinsic pathways, respectively. A and B, RT-PCR results show that poly(I-C) (37 °C for 24 h) up-regulated the gene expression of TRAIL, DR4, and DR5 in immortalized (A) and 1 μg/ml poly(I-C) pretreated primary (B) HUVECs. C, poly(I-C) up-regulated the protein expression of TRAIL in primary HUVECs. Cells, pretreated with 1 μg/ml poly(I-C), were re-treated with the indicated concentrations of poly(I-C) for 24 h. TRAIL in the cell lysates was assayed by ELISA. *, p < 0.05 compared with the control. D, TRAIL neutralization repressed the cell apoptosis induced by poly(I-C) in immortalized HUVECs. *, p < 0.05 compared with the poly(I-C) treatment group. E and F, RT-PCR results show the effect of poly(I-C) (37 °C for 24 h) on the gene expression of Bcl-2 and Noxa in immortalized (E) and primary (F) HUVECs. G and H, Western blot results show the effect of poly(I-C) on the protein expression of Bcl-2 and Noxa in immortalized (G) and primary (H) HUVECs. I, inhibition of TLR3 repressed the poly(I-C)-induced down-regulation of Bcl-2 and up-regulation of Noxa in immortalized HUVECs. Cells were transiently transfected with human TLR3 shRNA plasmid and then treated with 2 μg/ml poly(I-C) for 24 h. The protein expression of TLR3, Bcl-2 and Noxa was detected by Western blot.
Poly(I-C) Down-regulates the Anti-apoptotic Molecule, Bcl-2, and Up-regulates the Pro-apoptotic Molecule, Noxa
The Bcl-2 family of intracellular proteins is the central group of regulating factors of the intrinsic apoptosis pathway, and its opposing fractions of anti- and pro-apoptotic members arbitrate the life-or-death decision (28). To confirm the initiator for the activation of the intrinsic pathway, we first assessed the status of the pro-survival subfamily after the treatment with poly(I-C). As expected, in both primary and immortalized HUVECs, TLR3 activation by poly(I-C) dose-dependently down-regulated both the gene (Fig. 5, E and F) and the protein expression (Fig. 5, G and H) of Bcl-2, which is one of the most potential molecules to inhibit cell death in response to many cytotoxic insults (28). Other members in the pro-survival subfamily, such as Bcl-xl, were not regulated (data not shown). When we examined the pro-apoptotic subfamily, we found that poly(I-C) had little effect on the expression of Bax subfamily members (data not shown); however, a significant up-regulation of Bcl-2 homology 3 (BH3)-only subfamily member Noxa, which is an important mediator for p53 induced cell death (29), was found at the gene level in immortalized (Fig. 5E) and primary (Fig. 5F) cells and at the protein level in immortalized (Fig. 5G) and primary (Fig. 5H) cells. In addition, TLR3 inhibition by RNA interference repressed the down-regulation of Bcl-2 and the up-regulation of Noxa (Fig. 5I) in immortalized HUVECs. Taken together, these results suggest that the poly(I-C)-induced down-regulation of anti-apoptotic Bcl-2 and up-regulation of pro-apoptotic Noxa is related to TLR3 activation and may contribute to the trigger of the intrinsic pathway.
p53 Does Not Contribute to Poly(I-C)-induced Cell Apoptosis
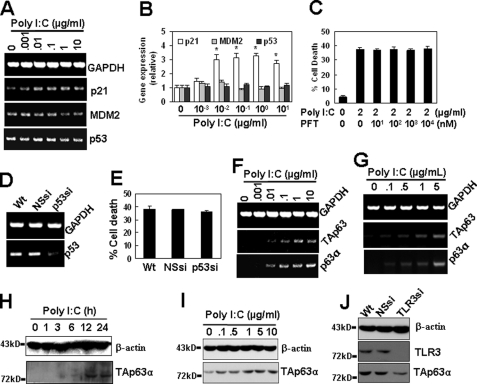
The tumor suppressor gene, p53, has been identified as a transcription regulator of Bcl-2 and Noxa (29, 30). Therefore, we wished to determine whether poly(I-C)-mediated cell death is regulated by p53. As p21 and murine double minute 2 (MDM2) are two important p53 target genes (31, 32), we first detected the expression of these genes induced by TLR3 activation in immortalized HUVECs. The RT-PCR results show that poly(I-C) treatment up-regulated the gene expression of p21 but not the expression of MDM2 (Fig. 6, A and B); furthermore, poly(I-C) treatment did not regulate p53 gene expression (Fig. 6, A and B). Then, we chose a p53-specific inhibitor pifithrin-α, which has been reported to protect mice from the side effects of cancer therapy by inactivating p53 (33), to test whether p53 is involved in poly(I-C)-induced cell death in immortalized HUVECs. Unexpectedly, p53 inhibition by pifithrin-α did not repress the cell death induced by poly(I-C) (Fig. 6C). Moreover, when p53 was down-regulated by RNA interference (Fig. 6D), the cell apoptosis induced by poly(I-C) was not down-regulated in immortalized HUVECs (Fig. 6E). These results indicate that p53 did not contribute to the cell death induced by poly(I-C).
FIGURE 6.
Roles of p53 and p63 in poly(I-C)-induced cell apoptosis. A, RT-PCR results show that poly(I-C) up-regulated p21 but neither MDM2 nor p53 gene expression. B, quantitative expression of p21, MDM2, and p53 in A by qRT-PCR. *, p < 0.05 compared with the control group. C, effect of p53 inhibitor pifithrin-α (PFT) on poly(I-C)-induced cell apoptosis in immortalized HUVECs. D, p53 expression was down-regulated by RNA interference in immortalized HUVECs. Cells were transiently transfected with p53 siRNA. p53 gene expression was detected 48 h after the transfection. E, effect of p53 down-regulation on poly(I-C)-induced cell apoptosis in immortalized HUVECs. Cells were transiently transfected with p53 siRNA and cultured for 24 h and then were treated with poly(I-C) for 24 h for the detection of cell apoptosis. F and G, poly(I-C) up-regulated the gene expression of TAp63α. Immortalized (F) and 1 μg/ml poly(I-C) pretreated primary (G) HUVECs were treated with the indicated concentrations of poly(I-C) for 24 h. The gene expression of p63 was measured by RT-PCR using a primer pair in the N terminus to detect the transactivation domain and a primer pair in the C terminus to detect p63α, -β, and -γ splices. H and I, Western blot results show that poly(I-C) up-regulated the protein expression of TAp63α in immortalized (H) and primary (I) HUVECs. J, Western blot results show that the TLR3 down-regulation by TLR3 shRNA transient transfection inhibited the expression of TAp63α in primary HUVECs.
TAp63α Is Involved in Poly(I-C)-induced Cell Apoptosis
TAp63α, a p53-related protein, has been reported to induce apoptosis via death receptors and mitochondria (12). Thus, we postulated that it may contribute to poly(I-C)-induced cell death. To confirm this hypothesis, we analyzed the expression and the activation of p63 in resting and poly(I-C)-treated cells. The results show that the resting cells expressed low levels of TAp63α transcript and protein but that poly(I-C) treatment led to the up-regulation of TAp63α gene expression in both immortalized (Fig. 6F) and primary (Fig. 6G) HUVECs and that the protein expression in both immortalized (Fig. 6H) and primary (Fig. 6I) cells was up-regulated in a dose-dependent manner. These up-regulations were TLR3-dependent, because TLR3 down-regulation by RNA interference inhibited the observed up-regulations (Fig. 6J). These results indicate that p63 may be involved in the regulation of TLR3-induced cell death.
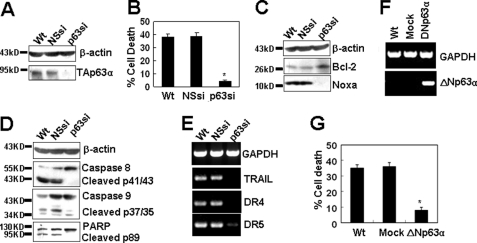
The biological significance of our findings depends on whether the loss of p63 is sufficient to suppress the cell apoptosis induced by TLR3 activation. To test this point, p63 expression was inhibited by RNA interference in immortalized HUVECs (Fig. 7A). Poly(I-C) up-regulated the expression of p63 in wild-type (Wt) and nonspecific shRNA (NSsi)-transfected cells but did not up-regulate p63 expression in p63 shRNA (p63si)-transfected cells (Fig. 7A), suggesting that p63 expression was efficiently inhibited by RNA interference. When the cells were treated with poly(I-C), the inhibition of p63 with specific shRNA virtually abrogated the poly(I-C)-induced cell apoptosis compared with that in wild-type cells, whereas cell apoptosis was not regulated in nonspecific shRNA-transfected cells (Fig. 7B). As expected, the inhibition of p63 by shRNA abrogated poly(I-C)-induced down-regulation of Bcl-2 and poly(I-C)-induced up-regulation of Noxa (Fig. 7C), resulting in the inactivation of caspase 9 (Fig. 7D). To our surprise, p63 inhibition also abrogated the activation of caspase 8 (Fig. 7D), indicating that TAp63α also regulated the extrinsic pathway, which was further demonstrated by the observation that p63 shRNA inhibited the expression of TRAIL and DR4/5 (Fig. 7E). Western blot results also show that p63 RNA interference resulted in the inhibition of the activation of the molecule, PARP (Fig. 7D). Furthermore, the overexpression of ΔNp63α (Fig. 7F), which is a p63 dominant-negative variant, by gene transfection, inhibited the cell apoptosis induced by poly(I-C) (Fig. 7G). Taken together, these results indicated that TAp63α is a critical regulator for the cell apoptosis induced by TLR3 activation.
FIGURE 7.
Modulation of p63 expression represses poly(I-C)-induced cell apoptosis. A, p63 was down-regulated by RNA interference in immortalized HUVECs. Wild-type (Wt), nonspecific shRNA (NSsi), and p63 shRNA (p63si) stably transfected cells were treated with 2 μg/ml poly(I-C) for 24 h. TAp63α protein was measured by Western blot. B, p63 down-regulation repressed poly(I-C)-induced cell apoptosis in immortalized HUVECs. *, p < 0.05 compared with the Wt group. C, Western blot results show that p63 RNA interference inhibited the down-regulation of Bcl-2 and the up-regulation of Noxa in immortalized HUVECs. D, Western blot results show that p63 RNA interference inhibited the activation of caspases 8 and 9 and PARP in immortalized HUVECs. E, RT-PCR results show that p63 RNA interference inhibited the expression of TRAIL, DR4, and DR5 in immortalized HUVECs. F, ΔNp63α was up-regulated by gene transfection in immortalized HUVECs. Cells were transiently transfected with pCDNA3+ (Mock) or pCDNA3+/ΔNp63α and harvested at 48 h post-transfection. The gene expression of ΔNp63α was detected by RT-PCR. G, ΔNp63α overexpression inhibited poly(I-C)-induced cell apoptosis in immortalized HUVECs. Cells were transfected as in F and cultured for 24 h, treated with 2 μg/ml poly(I-C) for another 24 h, and then assayed for cell apoptosis. *, p < 0.05 compared with control groups.
DISCUSSION
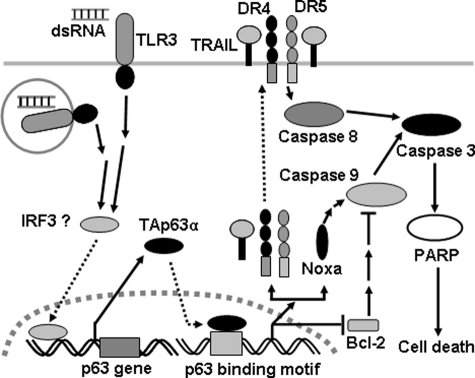
The results presented in this study are compatible with the model outlined in Fig. 8. TLR3, expressed on the cell surface or in the endosomal membrane, is activated by endogenous dsRNA or its exogenous analogs, such as poly(I-C), leading to the activation of a signal pathway to trigger the expression of TAp63α, which translocates to the nucleus and binds to DNA through p53- or p63-responsive elements to up-regulate the expression of the pro-apoptotic Bcl-2 family member, Noxa, and down-regulate anti-apoptotic Bcl-2. By engaging anti-apoptotic Bcl-2 homologs (34), Noxa elicits the activation of downstream caspase 9 and initiates the intrinsic cell apoptosis pathway. The down-regulation of Bcl-2 induced by TAp63α also contributes to the cell apoptosis initiated by this BH3-only molecule. Additionally, TAp63α also up-regulates the expression of TRAIL and its two receptors, DR4 and DR5. The interaction of TRAIL and DR4/5 accumulates and activates caspase 8, leading to the initiation of the extrinsic pathway. The activation of caspase 8 and caspase 9 elicits the activation of the downstream factors, caspase 3 and PARP, and finally induces cell apoptosis. Taken together, TAp63α, as a critical regulator downstream of TLR3, functions to induce cell apoptosis via both death receptors and mitochondria. These results suggest that TAp63α is a potential target for the prevention and control of the injury induced by TLR3 activation.
FIGURE 8.
Model of the pathway by which activated TLR3 induces TAp63α-dependent cell apoptosis. When activated by endogenous dsRNA or exogenous poly(I-C), TLR3 induces the up-regulation of TAp63α, which translocates into the nucleus and binds to the p53- or p63-responsive elements to initiate the transcription of TRAIL, DR4, DR5, and Noxa and repress the transcription of Bcl-2. The interaction of TRAIL with its receptors DR4/5 activates caspase 8 and initiates the extrinsic apoptotic pathway, whereas the up-regulation of Noxa and the down-regulation of Bcl-2 activate caspase 9 and initiate the intrinsic apoptotic pathway. Both caspase 8 and caspase 9 elicit the activation of their common downstream molecules, such as caspase 3 and PARP, and ultimately induce cell death. Taken together, the activation of TLR3 by poly(I-C) induces cell death via death receptors and mitochondria by up-regulating TAp63α.
Immune homeostasis is essential for the normal function of the immune system, and its breakdown leads to fatal inflammatory diseases. The evolutionarily conserved TLR family plays a key role in the detection and removal of invading pathogens to maintain homeostasis. The activation of TLRs by their respective ligands triggers the well characterized myeloid differentiation factor 88 (MyD88)-dependent and/or TRIF-dependent signaling cascades, leading to the activation of downstream signaling molecules, the production of pro-inflammatory cytokines (1, 35), and the occasional induction of cell death (36), which is another way to protect the host against the spread of microbes. TLR2 induces apoptosis in macrophages via the molecular adapter, MyD88, signaling pathway and the extrinsic Fas-associated death domain-caspase 8 pathway (37, 38). The apoptosis induced by TLR4 in macrophages is via the TRIF pathway but not MyD88 pathways (39). It has been reported that synthetic dsRNA induces the apoptosis of human breast cancer cells in a TLR3-dependent manner, which involves the activation of caspases in the extrinsic apoptotic pathway (18). Similar to our observations in which TLR3 activates both caspases 8 and 9, TLR3 has also been reported to induce the activation of the intrinsic and extrinsic apoptotic pathways in melanoma (19) and endothelial cells (20).
p63 protein is widely expressed in human and murine tissues, particularly in the basal cells of many epithelial tissues, such as the epidermis, cervix, urothelium, and prostate (9). The p63 transcription isotype, TAp63α, is expressed in the heart, testis, kidney/adrenal, thymus, brain, and cerebellum (9). In our study, we found that resting endothelial cells expressed lower levels of TAp63α, but the treatment with poly(I-C) up-regulated the expression of TAp63α in a dose-dependant manner. As TAp63α is a mediator of apoptosis via both death receptors and mitochondria (12), we presumed that TAp63α may be a mediator of TLR3-induced cell death. This presumption was proven by the observation that the inhibition of TAp63α expression by RNA interference repressed the cell death of endothelial cells. Therefore, we conclude that TAp63α is a linker between TLR3 and the caspase-dependent cell death.
Noxa is a BH3-only antagonist of Bcl-2 and functions in mediating p53-dependent apoptosis (40). It has also been reported that Noxa plays an important role in the induction of apoptosis by single-stranded RNA viruses and dsRNA in a p53-independent manner (41). In this study, we reported that TLR3 activation up-regulated the expression of both the gene transcript and protein of Noxa, which was also p53-independent but TAp63α-dependent, because the down-regulation of TAp63α by RNA interference repressed the up-regulation of Noxa. Furthermore, TLR3 activation down-regulated the expression of the anti-apoptosis molecule, Bcl-2. Taken together, TAp63α is a mediator of the activation of the intrinsic pathway induced by TLR3.
Viruses and agonists of TLR7 and TLR9 have been reported to induce the expression of TRAIL in plasmacytoid dendritic cells, endowing the plasmacytoid dendritic cells with the ability to kill virus-infected cells and the TRAIL-sensitive tumor cells (42). The detailed signaling pathway to up-regulate TRAIL expression by viral infection and TLR activation is unclear. In our study, we found that TLR3 activation up-regulated the expression of TRAIL and its receptors DR4/5. However, we did not know whether TLR3-induced up-regulation of TRAIL was dependent on TAp63α, even though TAp63α has been reported to induce the expression of Fas and the TRAIL receptors DR4/5 (12). As the inhibition of TAp63α decreased cell death to the background level, we tested the effect of TAp63α RNA interference on the expression of TRAIL and its receptors, and we found that the down-regulation of TAp63α repressed the up-regulation of TRAIL and its receptors DR4/5. These results suggested that TAp63α is also a mediator of the activation of the extrinsic pathway.
The signaling pathway of the pro-inflammation and antiviral infection induced by TLR3 activation has been clearly elucidated. The recognition of dsRNA by TLR3 in the endosomal membrane or on the cell surface recruits TRIF to the receptor, which induces pro-inflammatory cytokines and type I IFNs (1). In our studies, we found that the recognition of poly(I-C) by TLR3 up-regulated the p53 family member, TAp63α, and then initiated both the intrinsic and extrinsic pathways in a caspase-dependent manner that finally resulted in cell death. These results indicate that TLR3 may induce cell death and an antiviral immune response via distinct signaling pathways, as TAp63α inhibition did not regulate the expression of cytokines, including IL-1β, TNFα, IL-6, IL-8, and IFNβ in endothelial cells (data not shown). Thus, we conclude that TAp63α is an important regulator for TLR3-induced cell apoptosis and may be an available target for the prevention and control of virus-induced cell and tissue damage.
The work was supported in part by the Opening Project of State Key Laboratory for Molecular Virology and Genetic Engineering Grant 2008-K-0002, China.
- TLR
- toll-like receptor
- BH3
- Bcl-2 homology 3
- ΔNp63α
- N-terminally truncated p63 isoform α
- DR4/5
- death receptor 4/5
- dsRNA
- double-stranded RNA
- NF-κB
- nuclear factor κB
- PARP
- poly (ADP-ribose) polymerase
- PI
- propidium iodide
- poly(I-C)
- polyinosinic-polycytidylic acid
- TAp63α
- transactivating p63 isoform α
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- TRIF
- Toll/IL-1R domain-containing adapter inducing IFNβ
- HUVEC
- human umbilical vein endothelial cell
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- qRT
- quantitative PCR
- F
- forward
- R
- reverse.
REFERENCES
- 1. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 2. Iwasaki A., Medzhitov R. (2004) Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 3. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 4. Schulz O., Diebold S. S., Chen M., Näslund T. I., Nolte M. A., Alexopoulou L., Azuma Y. T., Flavell R. A., Liljeström P., Reis e Sousa C. (2005) Nature 433, 887–892 [DOI] [PubMed] [Google Scholar]
- 5. Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C. L., Casrouge A., Zhang X. X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J. L. (2007) Science 317, 1522–1527 [DOI] [PubMed] [Google Scholar]
- 6. Gowen B. B., Hoopes J. D., Wong M. H., Jung K. H., Isakson K. C., Alexopoulou L., Flavell R. A., Sidwell R. W. (2006) J. Immunol. 177, 6301–6307 [DOI] [PubMed] [Google Scholar]
- 7. Hutchens M., Luker K. E., Sottile P., Sonstein J., Lukacs N. W., Núñez G., Curtis J. L., Luker G. D. (2008) J. Immunol. 180, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang T., Town T., Alexopoulou L., Anderson J. F., Fikrig E., Flavell R. A. (2004) Nat. Med. 10, 1366–1373 [DOI] [PubMed] [Google Scholar]
- 9. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 10. Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006) Cell Death Differ. 13, 962–972 [DOI] [PubMed] [Google Scholar]
- 11. Osada M., Park H. L., Nagakawa Y., Yamashita K., Fomenkov A., Kim M. S., Wu G., Nomoto S., Trink B., Sidransky D. (2005) Mol. Cell. Biol. 25, 6077–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Lena A. M., Candi E., Terrinoni A., Catani M. V., Oren M., Melino G., Krammer P. H., Stremmel W., Müller M. (2005) EMBO J. 24, 2458–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 14. Senoo M., Pinto F., Crum C. P., McKeon F. (2007) Cell 129, 523–536 [DOI] [PubMed] [Google Scholar]
- 15. Weber A., Kirejczyk Z., Besch R., Potthoff S., Leverkus M., Häcker G. (2010) Cell Death Differ. 17, 942–951 [DOI] [PubMed] [Google Scholar]
- 16. Dogusan Z., García M., Flamez D., Alexopoulou L., Goldman M., Gysemans C., Mathieu C., Libert C., Eizirik D. L., Rasschaert J. (2008) Diabetes 57, 1236–1245 [DOI] [PubMed] [Google Scholar]
- 17. Paone A., Starace D., Galli R., Padula F., De Cesaris P., Filippini A., Ziparo E., Riccioli A. (2008) Carcinogenesis 29, 1334–1342 [DOI] [PubMed] [Google Scholar]
- 18. Salaun B., Coste I., Rissoan M. C., Lebecque S. J., Renno T. (2006) J. Immunol. 176, 4894–4901 [DOI] [PubMed] [Google Scholar]
- 19. Salaun B., Lebecque S., Matikainen S., Rimoldi D., Romero P. (2007) Clin. Cancer Res. 13, 4565–4574 [DOI] [PubMed] [Google Scholar]
- 20. Kaiser W. J., Kaufman J. L., Offermann M. K. (2004) J. Immunol. 172, 1699–1710 [DOI] [PubMed] [Google Scholar]
- 21. Gifford S. M., Grummer M. A., Pierre S. A., Austin J. L., Zheng J., Bird I. M. (2004) J. Endocrinol. 182, 485–499 [DOI] [PubMed] [Google Scholar]
- 22. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 23. Hotchkiss R. S., Nicholson D. W. (2006) Nat. Rev. Immunol. 6, 813–822 [DOI] [PubMed] [Google Scholar]
- 24. Hotchkiss R. S., Tinsley K. W., Swanson P. E., Chang K. C., Cobb J. P., Buchman T. G., Korsmeyer S. J., Karl I. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14541–14546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malinin N. L., Boldin M. P., Kovalenko A. V., Wallach D. (1997) Nature 385, 540–544 [DOI] [PubMed] [Google Scholar]
- 26. Ashkenazi A. (2002) Nat. Rev. Cancer. 2, 420–430 [DOI] [PubMed] [Google Scholar]
- 27. Kemp T. J., Ludwig A. T., Earel J. K., Moore J. M., Vanoosten R. L., Moses B., Leidal K., Nauseef W. M., Griffith T. S. (2005) Blood 106, 3474–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 29. Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 30. Wu Y., Mehew J. W., Heckman C. A., Arcinas M., Boxer L. M. (2001) Oncogene. 20, 240–251 [DOI] [PubMed] [Google Scholar]
- 31. Zhao B. X., Chen H. Z., Lei N. Z., Li G. D., Zhao W. X., Zhan Y. Y., Liu B., Lin S. C., Wu Q. (2006) EMBO J. 25, 5703–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 33. Komarov P. G., Komarova E. A., Kondratov R. V., Christov-Tselkov K., Coon J. S., Chernov M. V., Gudkov A. V. (1999) Science 285, 1733–1737 [DOI] [PubMed] [Google Scholar]
- 34. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 35. Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 36. Rasschaert J., Ladrière L., Urbain M., Dogusan Z., Katabua B., Sato S., Akira S., Gysemans C., Mathieu C., Eizirik D. L. (2005) J. Biol. Chem. 280, 33984–33991 [DOI] [PubMed] [Google Scholar]
- 37. Aliprantis A. O., Yang R. B., Weiss D. S., Godowski P., Zychlinsky A. (2000) EMBO J. 19, 3325–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. López M., Sly L. M., Luu Y., Young D., Cooper H., Reiner N. E. (2003) J. Immunol. 170, 2409–2416 [DOI] [PubMed] [Google Scholar]
- 39. Ruckdeschel K., Pfaffinger G., Haase R., Sing A., Weighardt H., Häcker G., Holzmann B., Heesemann J. (2004) J. Immunol. 173, 3320–3328 [DOI] [PubMed] [Google Scholar]
- 40. Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000) Science 288, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 41. Sun Y., Leaman D. W. (2005) J. Biol. Chem. 280, 15561–15568 [DOI] [PubMed] [Google Scholar]
- 42. Chaperot L., Blum A., Manches O., Lui G., Angel J., Molens J. P., Plumas J. (2006) J. Immunol. 176, 248–255 [DOI] [PubMed] [Google Scholar]