Abstract
Carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) is expressed in a variety of cell types and is implicated in carcinogenesis. Alternative splicing of CEACAM1 pre-mRNA generates two cytoplasmic domain splice variants characterized by the inclusion (L-isoform) or exclusion (S-isoform) of exon 7. Here we show that the alternative splicing of CEACAM1 pre-mRNA is regulated by novel cis elements residing in exon 7. We report the presence of three exon regulatory elements that lead to the inclusion or exclusion of exon 7 CEACAM1 mRNA in ZR75 breast cancer cells. Heterologous splicing reporter assays demonstrated that the maintenance of authentic alternative splicing mechanisms were independent of the CEACAM1 intron sequence context. We show that forced expression of these exon regulatory elements could alter CEACAM1 splicing in HEK-293 cells. Using RNA affinity chromatography, three members of the heterogeneous nuclear ribonucleoprotein family (hnRNP L, hnRNP A1, and hnRNP M) were identified. RNA immunoprecipitation of hnRNP L and hnRNP A1 revealed a binding motif located central and 3′ to exon 7, respectively. Depletion of hnRNP A1 or L by RNAi in HEK-293 cells promoted exon 7 inclusion, whereas overexpression led to exclusion of the variable exon. By contrast, overexpression of hnRNP M showed exon 7 inclusion and production of CEACAM1-L mRNA. Finally, stress-induced cytoplasmic accumulation of hnRNP A1 in MDA-MB-468 cells dynamically alters the CEACAM1-S:CEACAM1:L ratio in favor of the l-isoform. Thus, we have elucidated the molecular factors that control the mechanism of splice-site recognition in the alternative splicing regulation of CEACAM1.
Keywords: Cell Adhesion, RNA-binding Protein, RNA-Protein Interaction, RNA Splicing, RNA Interference (RNAi), CEACAM1, hnRNP
Introduction
Maintenance of cell polarity and tissue architecture is vital to the normal growth and differentiation program of epithelial cells (1). Disruption of cell polarity and tissue architecture, which is associated with uncontrolled cell proliferation and differentiation, is a feature of neoplastic transformation (2). A complex network of cell-cell and cell-extracellular matrix interactions mediated in part by adhesion molecules play an important role in epithelial tissue shape and function. Not surprisingly, many cancers have altered expression of various cell adhesion molecules. One of them is CEACAM1,4 whose aberrant expression has been linked to a variety of carcinomas (3, 4).
CEACAM1 is a glycosylated transmembrane protein and is a member of the CEA immunoglobulin superfamily (5). In humans, CEACAM1 pre-mRNA undergoes alternative splicing, giving rise to 11 splice variants (6). All splice variants include a highly conserved N-terminal ectodomain that defines the CEA gene family and is responsible for the homotypic adhesion function of these gene products (5, 7). CEACAM1 is expressed as a type I transmembrane protein in human tissues with 1, 3, or 4 extracellular Ig-like domains and either short or long cytoplasmic domains that engage distinct signal transduction pathways based on their different amino acid sequences (5). The two cytoplasmic domain splice variants, CEACAM1-L and CEACAM1-S, are identical in the first five amino acids but then differ starting at the splice junction (8). CEACAM1-S is produced by the usage of a proximal stop codon and leads to a cytoplasmic tail that is 12–14 amino acids long. CEACAM1-S binds to actin, tropomysin, calmodulin, and annexin II and contains serine and threonine residues that can be phosphorylated (9–11). Remarkably, the role of the short cytoplasmic tail mediates lumen formation via an apoptotic and cytoskeletal reorganization mechanism when mammary epithelial cells are grown in a three-dimensional model of mammary morphogenesis (11). In contrast, CEACAM1-L has two ITIM (immunoreceptor tyrosine-based inhibitory motif) regions that can be phosphorylated by a Src family kinase (12) followed by recruitment of the phosphatases SHP-1 or SHP-2 (13–15). The association with SHP-1 has been shown to inhibit lymphocyte activation events (13–15), suggesting that CEACAM1-L plays an important and primarily inhibitory mediator role in immune responses. Whether the ratio of splice isoforms contributes to the cellular phenotype is currently unclear. To that end we recently showed that an alteration of CEACAM1 splicing could be a common feature of breast cancer where an optimal ratio between the short and the long cytoplasmic domain splice variants might be required for normal tissue function (16).
Alternative splicing is an evolutionary strategy among metazoans for generating functionally diverse isoforms from a single gene through the selective joining of different exons (17). The decision of the splicing machinery to include or exclude a particular exon in the mature mRNA depends largely on the recognition and selection of the flanking splice-sites. Studies aimed at understanding the alternative splicing of a number of genes indicate that the auxiliary cis-acting elements, known as exon splicing enhancers (ESE) and exon splicing silencers (ESS), play an important role in the recognition of exons surrounding the regulated splice sites (18, 19). The function of auxiliary cis-acting elements provides the binding platform for splicing regulators, which in cooperation with other factors promote or inhibit the use of a nearby splice-site (20, 21). Regulation of alternative splicing is, therefore, a combinatorial phenomenon in which cooperative assembly of the activation and/or repression complexes near the regulated exons determine whether a particular exon will be included/excluded in the mRNA (22, 23). Importantly, deregulated splice variant expression has been identified as the cause of a number of genetic disorders, and certain forms of cancer have been linked to unbalanced isoform expression from genes involved in cell cycle regulation or apoptosis (24, 25).
The most common splicing regulators are mRNA-binding proteins that are members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family (26) and serine/arginine-rich (SR) protein families (27). hnRNPs rapidly associate with nascent transcripts and have been implicated in the repression of certain alternative splicing events (17). Three abundant hnRNPs, hnRNP L, hnRNP A1, and hnRNP M, are implicated in many alternative splicing events in human and several other eukaryotes and play a role in regulated splicing events by inhibiting the use of splice sites from the spliceosome (28, 29). Most of these hnRNPs can also form homophilic and heterophilic interactions with other hnRNPs (30, 31). Working antagonistically to the hnRNPs are the SR family of splicing factors, which promote inclusion of a particular exon by binding to ESEs to recruit other components of the splicing machinery to the 5′ and 3′ splice sites (32).
Previously, we proposed that regulatory cis-acting elements in exon 7 play an important role in the alternative splicing of CEACAM1 (16). We showed that replacement of these regulatory elements by human β-globin exon sequences resulted in exon 7-skipped mRNA as the predominant product. The present study addresses the mechanism that gives rise to CEACAM1-L and CEACAM1-S isoforms. Here we have identified three regions of exon 7 that can lead to activation or inhibition of exon 7 splicing. We show that the trans-acting auxiliary splicing factors hnRNP L, hnRNP A1, and hnRNP M bind exon 7 RNA specifically to control splice-site recognition. Finally, we show that deregulation of CEACAM1 splicing in breast cancer cells can be caused by stress-induced cytoplasmic accumulation of hnRNP A1.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfection
ZR75 cells were cultured in RPMI 1640 medium, and MDA-MB-468 cells were cultured in minimum Eagle's medium (MEM) supplemented with 1% sodium pyruvate, 0.15% sodium bicarbonate, 1 × non-essential amino acids, and 1 × penicillin-streptomycin-amphotericin B. HEK-293 cells were cultured in Dulbecco's modified MEM. All media were supplemented with 10% heat-inactivated FBS and grown at 37 °C in humidified air containing 5% CO2.Twenty-four hours before transfection, cells were seeded in 6-well plates (35 mm) at a density of 5 × 105 cells/well for ZR75 cells and 8 × 105 cells/well for HEK-293 cells. Cells were transfected with plasmid DNA (2–3 μg) and Lipofectamine 2000 (Invitrogen) or electroporation using Amaxa Nucleofector Kit V (Lonza) according to the manufacturer's instructions.
RNA Isolation and RT-PCR
Total RNA was isolated 24–48 h after mini-gene transfections and 72 h for shRNA transfections using RNeasy isolation kit (Qiagen) and treated with DNase I (Qiagen) as recommended by the manufacturer. Reverse transcription was carried out using 1 μg of total RNA using Oligo(dT)12–18 primer (Invitrogen) and Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's instructions. RT reactions were diluted 5-fold for PCR reactions. PCR amplification of wild-type exon 7, exon 7 with mutations, and constitutively expressed housekeeping gene glyceraldehyde phosphate dehydrogenase (GAPDH) transcripts were performed using either vector-specific or gene-specific primers (provided upon request). PCR was performed in a total volume of 20 μl containing 1 μl of first-strand cDNA solution, 0.1 mm dNTPs, 0.4 μl of Phire Hot Start DNA polymerase (Finnzymes), 4 μl of 5× PCR buffer, and 0.2 μm primers. The reactions were initiated by heating the samples to 98 °C for 30 s followed by 30 cycles of 98 °C for 5 s, 60 °C for 5 s, and 72 °C for 10 s followed by a final extension at 72 °C for 1 min. The products were analyzed on 2.25% agarose gels and visualized by ethidium bromide or SYBR safe DNA gel stain (Invitrogen). The relative amounts of the PCR products were analyzed by scanning the gels and determining the intensities of stained bands using ImageJ software Version 1.36b (National Institutes of Health).
Plasmids
All DNA constructs were generated by standard cloning procedures and confirmed by sequencing. Plasmids used in the analysis of cis-acting elements in the CEACAM1 primary transcript were made using the mini-gene splicing cassette containing genomic CEACAM1 exons 6–8 (CAM 6-7-8) (16). Linker-mutated RNA reporter constructs were prepared by replacing the exon 7 sequence indicated in Fig. 1A with the sequence AAGCATGCAA using overlap extension PCR (33). The first step PCR products were used as the template for the overlapping PCR with primers ET7 and EBGH-R. The amplified fragments were digested with NheI and HindIII, gel-purified, and ligated to plasmid CAM 6-7-8 linearized with appropriate restriction enzymes. All sequences were evaluated to ensure that premature termination codons did not arise during cloning. To construct heterologous substrates, BamHI- and EcoRI-containing oligonucleotides were inserted into plasmid RG6, generously provided by Dr. Tom Cooper (34). shRNAs for hnRNP A1 and enhanced green fluorescent protein were cloned into pBluescript vector with a human H1 promoter generously provided by Dr. Douglas Black. The GFP sequence targeted by the shRNA was ACGACTTCTTCAAGTCCGC. The T7-hnRNP A1 expression plasmid was generously provided by Dr. Javier Casares. The shRNAs targeting hnRNP L and luciferase and FLAG-hnRNP L were generously provided by Dr. Xiao-Feng Qin, Dr. Jiuyong Xie, and Dr. Kristin Lynch, respectively. Plasmid encoding hnRNP M4 and the vector control, pINX-N-FF-B, were generously provided by Dr. Russ Carstens (29).
FIGURE 1.
Identification of cis-acting regulatory elements in exon 7. A, shown is a schematic of scanning mutagenesis across exon 7 in CAM 6-7-8 reporter and derivative mini-genes. The splicing of CAM 6-7-8 yields CEACAM1 long and short cytoplasmic domain splice variants, CEACAM1-S and CEACAM1-L. The black rectangle represents exon 6, the gray rectangle represents exon 7, and the white rectangle represents exon 8 of CEACAM1. The black arrow represents the CMV promoter. The 10-nt block mutations are indicated: E7-1, bases 1–10; E7-2, bases 6–15; E7-3, bases 11–20; E7-4, bases 16–25; E7-5, bases 21–30; E7-6, bases 26–35; E7–7, bases 31–40; E7-8, bases 36–45; E7-9, bases 41–50. B and C, RT-PCR analysis of RNAs derived from ZR75 cells transiently transfected with indicated mini-genes or no plasmid mock control is shown. ERE-1, ERE-2, and ERE-3 refer to ERE 1, 2, and 3, respectively. M refers to a 100-bp DNA ladder (New England Biolabs). L and S refer to CEACAM1-L and CEACAM1-S. The data shown are representative of three independent experiments. The mean ± S.E. is shown for percent exon 7 inclusion calculated as the % CEACAM1-L/(CEACAM1-L + CEACAM1-S) mRNA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 versus CAM 6-7-8 control.
In Vitro Transcription
32P-Labeled in vitro transcription reactions were carried out as previously described (35). Labeled RNAs were transcribed from a T7-containing template where in all cases exon 7 size was maintained at 53 nt. Nonspecific negative control RNA (NS) was derived from the first 53 nt of pUC18 bacterial plasmid 5′-TCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGG-3′.For RNAs used in Fig. 7B, sequences were 5′-GGCAAGCGACCAGCGTGATCGGTGAAAACCTCTGTGTGTACGTCGAGGGCCT-3′ (E1:N), 5′-TCGCGCGTTTCGGTGATTCACAGAGCACAAACGACGGTGAAAACCTCTGT-3′ (N:E2:N), and 5′-TCGCGCGTTTCGGTGATGACGGTGAAAACCTCTCCTCAGTCTCCAACCACA-3′ (N:E3), where italicized nucleotides indicate exon 7 sequence. Large scale unlabeled transcription reactions for RNA affinity purification were carried out using AmpliScribe according to the manufacturer's instructions (Epicenter).
FIGURE 7.
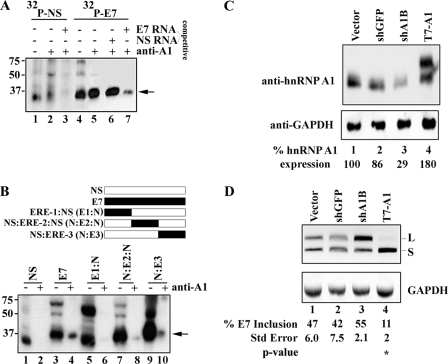
Binding and gene dosage analyses of hnRNP A1 in exon 7. A, unlabeled competitor RNA titrates hnRNP A1 association with exon 7 from nuclear extracts. Nuclear extract was incubated under splicing conditions with 0.28 pmol of uniformly labeled 32P-labeled NS RNA, 32P-labeled exon 7 RNA, and unlabeled competitor as indicated, treated with UV light, digested with RNase A, and then either directly resolved on a 12.5% SDS-PAGE gel (lanes 1 and 4) or immunoprecipitated with anti-hnRNP A1 (lanes 2, 3, 5, 6, and 7). B, hybrid NS and exon 7 RNA probes were used to narrow the hnRNP A1 binding site to the 3′ terminal end of exon 7. The schematic above the figure shows the NS RNA (white box), E7 RNA (black box), and the 20-nt ERE-1, ERE-2, and ERE-3 RNAs (black box) fused to NS RNA to make a 53-nt RNA molecule. C, depletion and overexpression of hnRNP A1 results in the increased production of CEACAM1-L and CEACAM1-S, respectively. Immunoblot analyses of HEK-293 cells using anti-hnRNP A1 is shown. D, RT-PCR analysis of mini-genes from C (sh-GFP is directed to GFP, sh-A1B is directed to hnRNP A1, and T7-A1 is over-expression) is shown. The mean ± S.E. for at least n = 3 is shown. Arrows denote cross-linking and/or immunoprecipitated hnRNP A1. **, p < 0.01 versus vector control.
RNA Mobility Shift Assay
Nuclear extracts were prepared from HeLa S3 cells obtained from National Cell Center essentially as described (36). Separation of splicing-related complexes were assembled in HeLa nuclear extracts, 12.5-μl total volume reactions, containing exon 7 RNA (10,000 cpm/μl) incubated in the absence or presence of unlabeled NS competitor RNA or unlabeled exon 7 RNA under conditions that support in vitro splicing as described previously (37). Reactions were incubated at 30 °C for 15 min. After the incubation, 2.5 μl of 4 μg/μl heparin and 2.5 μl of 5 × loading dye containing 1 × Tris borate EDTA (89 mm Tris-HCl, 89 mm boric acid, 2.5 mm EDTA), 20% glycerol, 0.25% bromphenol blue, and 0.25% xylene cyanol were added, and 3-μl aliquots of each reaction mixture were loaded onto a 2% horizontal low-melting agarose gel followed by the separation of RNA-containing complexes at 70 V for 4–6 h in Tris-glycine running buffer at room temperature (38). Gels were fixed in 10% acetic acid and 10% methanol for 30 min and then were dried under vacuum at 80 °C. The binding assay using recombinant His-hnRNP A1 was performed in 10-μl volumes containing gel-shift buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Nonidet P-40, 0.5 mm dithiothreitol, 1 mm MgCl2). Escherichia coli t-RNA was always added to the mixture before the addition of the protein (final concentration 0.01–0.09 μg/μl). The reaction mixtures were incubated at 30 °C for 15 min before the addition of 2 μl of dye and loading on a non-denaturing 5% acrylamide gel (29:1 acrylamide/bisacrylamide, 50 mm Tris-Cl, pH 8.8, 50 mm glycine, and 5% glycerol) run in Tris-glycine buffer (50 mm Tris, pH 8.8, and 50 mm glycine).
RNA Affinity Assay
RNAs were covalently linked to adipic acid dihydrazide-agarose beads as indicated previously (39, 40). The beads containing immobilized RNA were incubated with HeLa nuclear extract under splicing in a 25× reaction and incubated for 30 min at 30 °C. The bound beads were washed 4 times with 1 ml of buffer without ATP, MgCl2, or tRNA. The proteins were eluted in 75 μl of loading dye (80 mm Tris-Cl, pH 6.8, 0.1 m dithiothreitol, 2% SDS, 10% glycerol, and 0.2% bromphenol blue) by heating for 5 min at 65 °C. Eluted proteins were then separated in 12.5% SDS-polyacrylamide gels under denaturing conditions.
Mass Spectrometry
Excised gel bands were cut into small (∼1 mm) pieces and destained with 200 mm ammonium bicarbonate in 50% UV grade acetonitrile (Burdick and Jackson, Muskegon, MI). The destained gel pieces were reduced with dithiothreitol and alkylated with iodoacetamide. The proteins were then digested by overnight treatment at 30 °C with sequencing grade modified trypsin (Promega) in 200 mm ammonium bicarbonate. The digestion supernatant was collected, and the gel pieces were further extracted with 2 washes of 1% double-distilled, PPB/Teflon grade acetic acid (Sigma-Aldrich) in 60% acetonitrile. The combined supernatant and extract were combined and reduced to a volume of ∼20 μl in a vacuum centrifuge. The digested peptides were analyzed by LC/MS using an Agilent 6420 Q-ToF mass spectrometer with an Agilent 1200 nano-flow HPLC (Agilent Technologies, Santa Clara, CA) at the City of Hope Mass Spectrometry core facility. Separation was carried out using the Chip Cube source and a high capacity protein identification chip. Samples were loaded onto a 160-nl trapping column at 4 μl/min in 99% buffer A (0.1% formic acid in water). The trapping column was then switched online with a 75-μm × 150-mm analytical column and eluted directly into the mass spectrometer. The analytical gradient ran from 8% buffer B (0.1% formic acid in 90% acetonitrile/10% water) to 30% buffer B over 45 min. Mass spectometry data were collected in a data-dependent fashion. From each full mass scan, up to six precursor ions were selected for fragmentation. After being selected, each ion was placed on an exclusion list for 30 s. Both full mass and fragment ion spectra were collected at a rate of 4 Hz. Data were converted to mzData using Agilent's supplied tool. Converted data files were searched against the ENSEMBL human data base and the cRap data base using X!Tandem. The preliminary search assumed tryptic specificity, quantitative carbamidomethylation of cysteine, and possible oxidation of methionine. Refinement searches considered non-tryptic cleavage, deamidation of asparagine and glutamine, oxidation of tryptophan, and double oxidation of methionine and tryptophan. All searches used a mass tolerance of 50 ppm for both precursors and fragments.
Western Blot Analysis
Cells were incubated in radioimmune precipitation assay buffer (10 mm sodium phosphate, pH 7.2, 150 mm NaCl, 1% Nonidet P40, 1% sodium deoxycholate, 0.1% SDS, 2 mm EDTA, 1 mm DTT, 1 mm PMSF, 100 units/ml Benzonase) and protease inhibitors on ice for 20 min, and the lysate was cleared by centrifugation and kept at −80 °C (41). Proteins (5 μl aliquot from RNA affinity assay or 10 μg cell lysate) were separated by SDS-gel electrophoresis, transferred to nitrocellulose membranes, and probed with antibodies for hnRNP A1, hnRNP L, hnRNP H, or hnRNP K (each at 1 μg/ml) followed by infrared-labeled secondary antibody (LI-COR Biosciences, Lincoln, NE). GAPDH was detected with 1:5000 diluted mouse anti-GAPDH primary antibody (Abcam, Cambridge, UK). Signals were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences).
UV Cross-linking and Immunoprecipitation Assays
For UV-cross-linking experiments, radiolabeled RNAs (100,000 cpm/μl) were incubated in equimolar HeLa nuclear extract or S100 cytoplasmic extract under conditions that support in vitro splicing (in 12.5 μl total volume, 25% extract). Protein concentrations were measured by the Bradford Assay. Reaction mixtures were incubated for 15 min at 30 °C, placed on Parafilm wrapped around a heat sink metal block on ice, and irradiated at 254 nm at a distance of ∼0.5 cm (Spectroline, model ENF-240C, 115 V, 60 Hz, 0.20 Amps) for 30 min. Samples were then treated with RNase A (4 mg/ml final concentration) at 37 °C for 20 min. Proteins were boiled for 5 min in cracking buffer (80 mm Tris-Cl, pH 6.8, 0.1 m dithiothreitol, 2% sodium dodecyl sulfate, 10% glycerol, and 0.2% bromphenol blue), and separated on 12.5% SDS-polyacrylamide gels. Binding of proteins to the RNA was visualized using Kodak XAR-5 film after 24 h.
Immunoprecipitation was performed as described with slight modifications (42). Briefly, radiolabeled RNAs (100,000 cpm/μl) subjected to cross-linking and RNase A digestion were mixed with 175 μl of NET2–150 buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Nonidet P-40, 0.5 mm dithiothreitol, 1 mm MgCl2). 1 μl of specified antibody was added, and the mixture was rotated at 4 °C for 24 h. Anti-hnRNP A1 (4B10) was provided by Dr. Gideon Dreyfuss, and anti-hnRNP L, anti-hnRNP H, and anti-hnRNP K were purchased from Santa Cruz Biotechnology. The next day, 30 μl of activated protein G-Sepharose CL-4B (Amersham Biosciences) in NET2–150 buffer was added to the reaction samples and rotated at room temperature for 2 h. Samples were stringently washed 3 times in NET2–500 buffer (with 500 mm NaCl) and 2 times in NET2–150 buffer. Proteins were boiled for 5 min in cracking buffer and separated on 12.5% SDS-polyacrylamide gels. Binding of proteins to the RNA was visualized using Kodak XAR-5 film after 24 h.
Immunofluorescence
Twenty-four hours before treatment, cells were seeded in an 8-well chamber slide at a density of 4 × 104 cells/well for MDA-MB-468. Then cells were treated with 600 mm sorbitol or left in growth medium for 2 h the following day. Cells were fixed with 4% p-formaldehyde in PBS for 15–30 min at room temperature followed by incubation for 10 min in 0.2% Triton X-100. For detection of nuclear material, DAPI staining was used at a 1:2000 dilution. Endogenous hnRNP A1 and hnRNP U (Santa Cruz Biotechnology, 3G6) were visualized with a 1:1000 dilution using an Olympus IX81 Inverted microscope with Q Imaging Retiga 2000R cooled CCD camera with Image Pro Plus Version 6.3 imaging software and 40× magnification.
RESULTS
Identification and Characterization of Exon Regulatory Element (ERE) Controlling Alternative Splicing of Exon 7
We recently showed that breast cancer specimens displayed expression of both short and long cytoplasmic domain splice variants of CEACAM1 as compared with normal breast tissues that predominately express the S-isoform, suggesting that alteration of CEACAM1 splicing could be a common feature of breast cancer. We also demonstrated that regulatory cis-acting elements in exon 7 play a role in the alternative splicing of CEACAM1-L and CEACAM1-S (16). To further study the regulatory factors involved in breast cancer and to narrow the regions of exon 7 that are important for controlling alternative splicing, we performed scanning mutagenesis across the exon following the method described recently by Salati and co-workers (40). In this method a 10-nt block random sequence (see “Experimental Procedures”) was sequentially introduced spanning exon 7 in the context of our pre-mRNA reporter CAM 6-7-8 (Fig. 1A). To prevent disruption of the 3′ splice-site signal that could potentially activate an unknown cryptic splice site, the last three nucleotides at the 3′-end were not disrupted. Because mRNAs harboring premature termination (nonsense) codons can be subjected to selective degradation, we confirmed that the 10-nt block random sequence does not contain a premature stop codon. Moreover, this sequence does not contain any of the previously reported ESEs or ESSs as verified by two independent Web-based programs (43, 44).
To determine the effects of nucleotide substitutions on splicing regulation, 9 mini-gene reporter constructs (E7-1 to E7-9) were transiently expressed in ZR75 breast cancer cells, and after 24 h, total RNA was isolated followed by RT-PCR with plasmid-specific primers. The wild-type reporter CAM 6-7-8 produced both CEACAM1-S and long CEACAM1-L splice variants (Fig. 1B). In contrast, the splicing of E7-1-, E7-2-, and E7-3-driven transcripts produced predominantly the CEACAM1-S isoform, suggesting that mutations in the first 20-nt disrupted a splicing responsive element that regulates exon 7 alternative splicing (Fig. 1B, compare lane 3 with lanes 4–6). A similar analysis with splicing reporters E7-4, E7-5, and E7-6 that carry sequence mutations from nt 16 to 35 produced predominantly the CEACAM1-L isoform (Fig. 1, B, lane 3 compared with lanes 7–9, and C), indicative of the presence of a second splicing regulatory element. The mutations of the sequence from nt 36 to 50 (E7-7, E7-8, and E7-9) resulted in both short and long splice isoforms, albeit with different S:L ratios compared with the CAM 6-7-8 control. We note that although mutations of nt 31–40 and 41–50 favored exon 7 inclusion, mutations of nt 36–45 shifted splicing toward exon 7 excluded mRNA (Fig. 1B, compare lane 3 with lanes 10–12). These results indicate that the spatial organization of nt 31–50 contains a third splicing regulatory element with putative overlapping signals that can repress and/or promote exon 7 splicing. We observed no significant differences in phenotypes across other cell lines tested, i.e. HeLa and HEK-293 cells (data not shown). Together these results indicate that mechanistic control of alternative splicing events for CEACAM1 are regulated by three EREs that can be grouped into ERE-1, ERE-2, and ERE-3 for exon 7 RNA (Fig. 1B).
Exon 7 Contains ESE and ESS Elements
Previous studies have demonstrated that splicing regulatory elements control alternative splicing by virtue of short sequence motifs of 4–6 bases (e.g. ESE and ESS) (45, 46). To further characterize putative ESEs and ESSs located in exon 7, we constructed additional mini-gene reporters that contained minimal sequence mutations (Fig. 2A). Because substitution of nt 6 through nt 15 in E7-2 showed the greatest abrogation of splicing regulation, we reasoned that these nucleotides might confer ESE activity to ERE-1. Although not particularly purine-rich, a correlative feature seen in other ESEs, we noticed that the preferred binding site involving the CAGC sequence found in wild-type exon 7 in this region related to several ESE sites (e.g. CAGC, GAGC, CAGCC, GCAGC, ACAGC, AGCAG, CAGCA, CCAGC, and CCCAG; see Table 2 for a complete list in Ref. 47) associated with SRp55, SRp40, SC35, and SF2/ASF when spatially arranged around a tandem 3′ splice site (Fig. 2A) (47). Although this genetic organization is notably absent surrounding exon 7, the fact that the region of overlap between E7-2 and E7-3 in Fig. 1A showed such a strong shift to CEACAM1-S strongly suggested nt 11–14 was an important target for ESE activity.
FIGURE 2.
Identification of an ESE and ESS encoded within ERE-1 and ERE-2 of exon 7. A, nucleotide comparison of target ESE and ESS sequences is shown. E7-2 ′ and E7-5′ contain the wild-type sequence except where indicated with substitutions. RNAs were of equal length in relation to exon 7 but for simplicity only those nucleotides under study are depicted. B and C, RT-PCR analysis and quantitation of RNAs derived from ZR75 cells were as described in the Fig. 1 caption. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 versus CAM 6-7-8 control.
To test whether the CAGC motif contributed to the ESE activity in ERE-1, a splicing reporter E7-2′ was constructed and transiently expressed in ZR75 cells (Fig. 2, B and C). We observed that compared with the splicing of E7-2 mRNA, E7-2′ showed an ∼25-fold increase in CEACAM1-L mRNA (Fig. 2, B and C, compare lanes 4 and 5). The fact that exon 7 inclusion was higher in CAM 6-7-8 versus the E7-2 ′ reporter (66 versus 25%; Fig. 2, B and C, lanes 3 versus 5) suggested that the sequences flanking the CAGC motif also contribute to ESE activity.
Similarly, as E7-5 showed the greatest abrogation of CEACAM1-S in ERE-2, we next considered whether this exon region encoded an ESS. Examination of this region identified a cluster of CA-rich elements (nt-22–25 and nt-29–32) that could be a major cis-acting RNA signal. Recent studies show intronic polymorphic CA-repeat regions in the human endothelial nitric-oxide synthase gene bind specifically to hnRNP L (48). To test the hypothesis that E7-5 abolished an ESS, we chose to study the first CA motif in exon 7 that overlapped with E7-4 and E7-5. We constructed a derivative of E7-5, called E7-5′ (Fig. 2A), and this mutant was transiently expressed in ZR75 cells (Fig. 2, B and C). Although we observed production of CEACAM1-L mRNA for the E7-5′ construct, we unexpectedly observed the additional expression of CEACAM1-S mRNA (Fig. 2B, compare lanes 6 and 7). We interpret this finding as indicating the ESS is comprised of at least these nucleotides but include other nucleotides that play a role in controlling the production of CEACAM1-S mRNA and are part of a larger ESS. In sum, we conclude the existence of an ESE in ERE-1 and ESS in ERE-2, and both are determinants for regulation of exon 7 splicing.
Exon 7 EREs Can Function in a Heterologous Splicing Substrate in Vivo
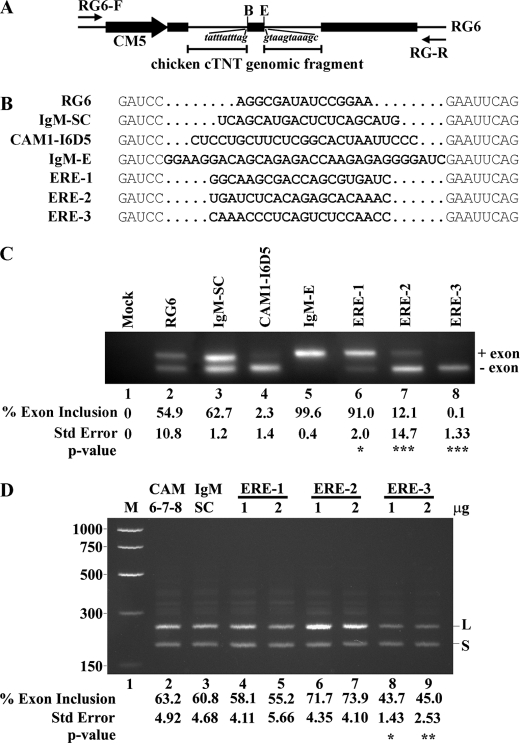
We next investigated whether exon 7 could direct CEACAM1 alternative splicing independent of flanking intron 6 and intron 7 RNA. We constructed mini-gene splicing reporters that contained ERE-1, ERE-2, or ERE-3 sequences expressed in the middle exon flanked by introns from chicken cTNT (cardiac troponin T (34)) and tested whether we could recapitulate CEACAM1 splicing in breast cancer cells (Fig. 3). For comparison, enhancer and scrambled sequences from the IgM substrate (49) and an intronic splicing silencer identified in CEACAM1 intron 65 denoted CAM1-I6D5 were included (Fig. 3B). The wild-type and scrambled reporters (RG6 and IgM-SC) produced both short and long splice variants when expressed in ZR75 cells (Fig. 3C, lanes 2 and 3). In contrast, the splicing of ERE-1 strongly influenced variable exon 7 inclusion, similar to the control enhancer reporter IgM-E (Fig. 3C, lanes 5 and 6). When wild-type ERE-2 or ERE-3 was tested, we observed strong splicing repression similar to the control CAM1-I6D5 (Fig. 3C, lanes 7 and 8). To further strengthen our conclusion that these 20-nt fragments (e.g. ERE-1, ERE-2, and ERE-3) can adopt functional representative structures in vivo, we forced their expression in HEK-293 cells and looked again at changes in CAM 6-7-8 mini-gene splicing (Fig. 3D). We hypothesized that endogenous splicing factors, titrated by these ERE-containing constructs, would be unavailable to regulate exon 7 and thereby cause similar splicing phenotypes to that observed previously (Fig. 1). In the case of ERE-1, we observed a dose-dependent shift from CEACAM1-L to CEACAM1-S mRNA (Fig. 3D, lanes 4 and 5, compared with lanes 2 and 3). By contrast, either concentration of ERE-2 plasmid could enhance activation of exon 7 inclusion to produce CEACAM1-L mRNA (Fig. 3D, lanes 6 and 7 compared with lanes 2 and 3). Quite surprisingly, overexpression of ERE-3 does not lead to CEACAM1-L mRNA as expected with a marked shift in exon 7 inclusion from 63.2 to 45.0% toward CEACAM1-S (Fig. 3D, lane 9 compared with lane 2). Apparently, the pool of available splicing factors persists in fine balance, and the sequestration by ERE-3 over-expression frees this restriction, allowing unrestrained access of an unidentified splicing factor to interact with ERE-2 which leads to an exon 7 exclusion phenotype. We conclude that maintenance of authentic alternative splicing mechanisms are independent of the CEACAM1 intron sequence context and are fine-tuned to respond to changes in the cellular micro-environment.
FIGURE 3.
Exon 7 EREs can function in a heterologous context. A, shown is a diagram of middle exon replacement strategy in RG6 mini-gene. Black rectangles represent exons of RG6, black lines represent chicken cardiac troponin T (cTNT) introns, and the black arrow represents the CMV promoter and restriction sites unique to the middle exon (B is BamHI, and E is EcoRI). Flanking intronic sequences to the variable exon are shown in italics. The locations of vector-specific primers flanking the first and last exons are indicated with small black arrows. B, shown is a nucleotide comparison between constructs to show purine content and similarity in variable exon length. Bold nucleotides represent unique sequence inserted into middle exon of RG6. IgM-Sc and IgM-E are scrambled control (IgM-SC) and enhancer sequences from IgM substrate, respectively. CAM1-I6D5 contains an intronic splicing silencer from intron 6 of CEACAM1. C, RT-PCR after transfection in ZR75 cells with the indicated mini-genes is shown. D, titration by ERE-containing constructs alters CEACAM1 alternative splicing. RT-PCR after transfection in HEK-293 cells of CAM 6-7-8 mini-gene influenced by overexpression of indicated constructs is shown. M is Benchtop PCR marker (Promega). The mean ± S.E. for at least n = 3 is shown for percent exon 7 inclusion. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus RG6 or CAM 6-7-8 control.
A Specific Protein Complex Assembles on Exon 7
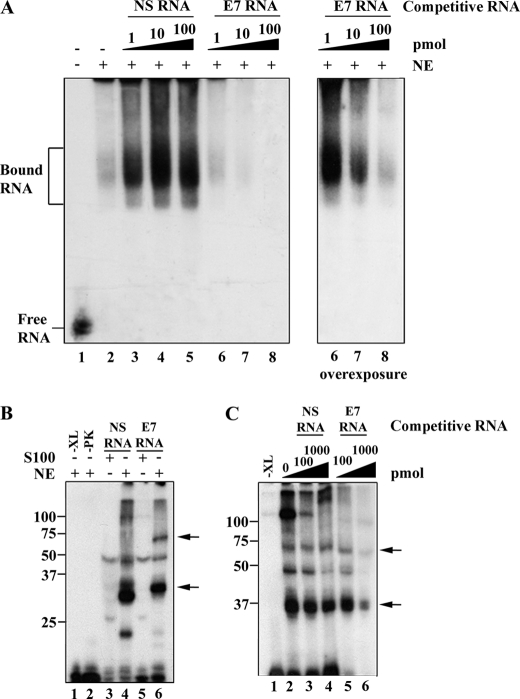
To identify proteins that associate with exon 7, we used an electrophoretic mobility shift assay designed to capture spliceosome-containing complexes (38) and UV-induced cross-linking assays using nuclear extracts prepared from HeLa cells, a cell line that faithfully recapitulates CEACAM1 splicing when transfected with mini-gene CAM 6-7-8 (data not shown). We generated RNA probes containing the exon 7 sequence as well as a control nonspecific (NS) RNA sequence (see “Experimental Procedures”). As shown by our native RNA mobility shift assay, slow migrating complexes are visible upon incubation of 32P-labeled exon 7 RNA with nuclear extract (Fig. 4A, lane 2). The specificity of the gel-shift complex was tested by the addition of unlabeled NS and exon 7 RNA. We found that assembly of an exon 7-dependent complex is nearly completely abolished by the addition of unlabeled exon 7 RNA, as low as 1-fold, but is not inhibited by the addition of excess NS RNA (Fig. 4A, lanes 3–5 and 6–8). To rule out the possibility that the splicing complexes were degraded and to prove that these complexes were out-competed for when treated with specific competitor, we show the splicing complexes can be inhibited in a dosage-dependent manner (Fig. 4A, right panel). We note the addition of our NS RNA titrated endogenous RNA nucleases, which had the effect of stabilizing the splicing complex (Fig. 4A, lanes 3–5).
FIGURE 4.
A spliceosome regulatory complex specifically assembles on exon 7. A, unlabeled competitor RNA can titrate an exon 7-associated nuclear complex. Left panel, 32P-labeled exon 7 RNA (0.28 pmol) was incubated in the presence (+) (lanes 2–8) or absence (−) (lane 1) of HeLa nuclear extract (NE) under splicing conditions and resolved on a native gel. Unlabeled competitor RNA was also added to the reactions as indicated. Right panel, an overexposed membrane (lanes 6–8) of the left panel is shown. B, UV light-induced cross-linking to exon 7 reveals nuclear and cytoplasmic proteins. S100 extract (containing cytoplasmic proteins) cross-linking reactions were loaded in lanes 3 and 5. The HeLa extract (containing nuclear proteins)-cross-linking reactions were loaded in lanes 1, 2, 4, and 6. Mock-treated reactions that did not include UV cross-linking but did include proteinase K (PK) were loaded in lanes 1 and 2, respectively. C, unlabeled competitor RNA titrates cross-linked proteins associated with exon 7. HeLa nuclear extract was incubated with no unlabeled RNA competitor (lane 2), NS competitor (lanes 3 and 4) or exon 7 competitor RNA (lanes 5 and 6) as indicated. A mock-treated reaction that did not include UV cross-linking was loaded as a control (lane 1). The protein size markers (kDa) are indicated to the left, and arrows denote sites of cross-linking bands of interest.
To more directly investigate the role of exon 7-associated proteins, we used UV photocross-linking to covalently attach bound proteins to the labeled RNA. Two proteins cross-linked to exon 7 RNA in HeLa extract, the most prominent migrating close to the 34- and 75-kDa protein standards (Fig. 4B, lane 6) as compared with the control NS RNA (Fig. 4B, lane 4). We note that many auxiliary splicing factors bind weakly to RNA in the molecular weight range of 34-kDa, and this could account for the cross-linked protein observed in the NS RNA sample, which has a similar migration pattern to the exon 7 cross-link. Importantly, the S100 extracts (the cytoplasmic fraction of the HeLa splicing extract preparation) did not show the 34- and 75-kDa proteins cross-linked to exon 7. We can conclude these proteins are nuclear in origin (Fig. 4B, compare lanes 5 and 6).
To rule out the possibility that we had identified only nonspecific protein interactions associating with exon 7 RNA, we next addressed binding specificity by using competition experiments with either excess unlabeled NS RNA or exon 7 RNA (Fig. 4C). Our data show that the cross-linked proteins we identified were specifically competed for upon introduction of excess unlabeled exon 7 RNA, whereas control NS RNA had no significant influence on the association of these protein factors (Fig. 4C, lanes 6 compared with 4). This assay also revealed the presence of a cross-linked doublet, indicating more than one protein migrated in the range we estimate as 34-kDa. Taken together, we demonstrate the presence of a group of unknown proteins that bind specifically to exon 7.
Identification of hnRNP L, hnRNP A1, and hnRNP M Association with Exon 7 RNA
To identify the protein components of the exon 7-specific complex, we performed RNA affinity isolation as described recently (40). We observed protein bands migrating in the proximity of the UV cross-links in the exon 7 RNA sample, notably in the same molar ratio, as compared with the control sample (Fig. 5A). The regions of the gel close to the 75- and 34-kDa protein were excised and analyzed by LC-MS/MS. Multiple peptides corresponding to hnRNP L (17), hnRNP A1 (3), and hnRNP M (7) were identified in the screen. These splicing factors mediate alternative splicing through high affinity repeat elements. In the case of exon 7, cytidine-adenosine elements in ERE-2 suggested a putative RNA-protein interaction with hnRNP L (75 kDa). By contrast, the high affinity SELEX-derived UAGRG(A/U) hnRNP A1 (34 kDa) binding site was notably absent in exon 7. In the case of hnRNP M (75 kDa), our interest to investigate this further derived from its putative role as a surface expression receptor for soluble CEACAM5/CEA (50).
FIGURE 5.

RNA affinity purification of hnRNPs associated with exon 7. A, RNAs produced by transcription were used to link to adipic acid beads followed by incubation with 250 μg of HeLa nuclear extract under splicing conditions followed by SDS-PAGE electrophoresis and mass spectrometry or analyses by silver staining as shown. B, a Western blot of samples used for SDS-PAGE gel from panel A with specified antibodies is shown.
The binding of hnRNP L and hnRNP A1 to exon 7 regulatory elements was verified using specific antibodies in Western blot analyses (Fig. 5B, lanes 2 and 3). By contrast, hnRNP M could not be verified by this assay, indicating a potential protein-protein and not protein-RNA interaction. To control for non-specificity, the blots were probed for hnRNP K and hnRNP H activity (Fig. 5B, lanes 2 and 3). We could not detect any significant difference in levels for these two proteins. Our native purification conditions could not distinguish between proteins associated through a splicing regulatory complex or through direct RNA interaction. To address how these splicing factors associate with exon 7, we next performed RNA immunoprecipitations of these hnRNP splicing factors.
An hnRNP L ESS in Exon 7 RNA
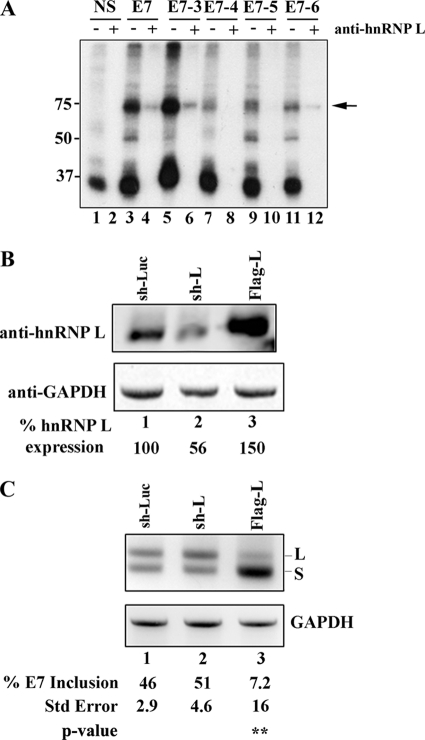
Our linker-mutagenesis screen suggested the presence of an ESS that regulates the ERE-2 region of exon 7. We also observed a 75-kDa cross-linked protein that by LC-MS/MS and Western blot analyses was likely hnRNP L. We were, therefore, interested in determining the exon 7 binding motif using a modified RNA immunoprecipitation assay using anti-hnRNP L antibodies. We addressed the identity of the 75-kDa cross-link observed in Fig. 4 by testing the ability of exon 7 to precipitate hnRNP L (Fig. 6A, lane 4 versus lane 2). Having established that a binding site for hnRNP L existed along exon 7, we asked whether sequences comprising ERE-2 could pull down hnRNP L. To distinguish among the possible binding sites, we made RNA transcripts derived from templates used in our linker mutagenesis screen (e.g. E7-3 through E7-6) and compared the binding of these constructs to our control wild-type and NS RNA. Our data demonstrate the inability of E7-4 and E7-5 RNA to interact with hnRNP L, suggesting we had identified the core sequences in the hnRNP L binding motif, nt 22–30 (Fig. 6A, lanes 8 and 10, compared with lane 4). To test whether hnRNP L mediates exon 7 skipping in vivo, we performed RNAi and overexpression analysis studies (Fig. 6, B and C). Plasmid-encoded shRNAs to control luciferase (sh-Luc) or hnRNP L (sh-L) or overexpression hnRNP L plasmid (FLAG-L) were transiently co-expressed with mini-gene CAM 6-7-8 in HEK-293 cells, and total protein was isolated and tested for the success of down-regulation and overexpression of hnRNP L. Our results show that we could down-regulate the expression of hnRNP L to 56% of wild type (Fig. 6B, lane 2, compared with lane 1). When total RNA was isolated followed by RT-PCR, we observed the production of CEACAM1-L mRNA under limiting hnRNP L expression, whereas strong production of CEACAM1-S mRNA under high levels of this protein (51% exon 7 inclusion and 7.2% exon 7 inclusion, respectively; Fig. 6C, lanes 2 and 3). Thus, we were able to modulate the switch in isoform production simply by controlling the levels of a trans-acting factor regulating exon 7.
FIGURE 6.
Binding and gene dosage analyses of hnRNP L in exon 7. A, 32P-labeled RNA was prepared using the DNA templates described in Fig. 1. Nuclear extract was incubated under splicing conditions with 0.28 pmol of uniformly labeled RNA as indicated, treated with UV light, digested with RNase A, and then either directly resolved on a 12.5% SDS-PAGE gel (odd lanes) or immunoprecipitated with anti-hnRNP L (even lanes). B, depletion and overexpression of hnRNP L results in the increased production of CEACAM1-L and CEACAM1-S, respectively. Immunoblot analyses of HEK-293 cells using anti-hnRNP L is shown. C, RT-PCR analysis of mini-genes from B (sh-Luc is directed to luciferase, sh-L is directed to hnRNP L, and FLAG-L is overexpression) is shown. The mean ± S.E. for at least n = 3 is shown. The arrow denotes cross-linking and/or immunoprecipitated hnRNP L. **, p < 0.01 versus sh-Luc control.
An hnRNP A1 ESS in Exon 7 RNA
To test whether hnRNP A1 interacts specifically with exon 7 or through an auxiliary splicing factor, we immunoprecipitated the cross-linked extracts with anti-hnRNP A1 in the presence of competitor RNA (Fig. 7A). We reasoned that a competition analysis would show conclusively whether hnRNP A1 could bind to exon 7 in light of the absence of a canonical binding site. To start, we considered whether exon 7 can act as a ligand for hnRNP A1. This was evidenced by a significant pulldown as compared with our NS RNA control (Fig. 7A, compare lane 5 to 2). Next we showed that the introduction of excess unlabeled exon 7 could significantly decrease the amount of hnRNP A1 pulldown (Fig. 7A, lane 7 compared with lane 3). This, however, was not the case when unlabeled NS RNA was used as a competitor for exon 7-hnRNP A1 binding (Fig. 7A, lane 6). This control was important to establish that cross-links observed in the NS sample do not associate with exon 7 (Fig. 4B, lane 4). Taken together we conclude that hnRNP A1 associates with exon 7 RNA.
Having established that our control RNA did not bind hnRNP A1, we prepared hybrid RNA probes that contained a partial sequence from exon 7 and a partial sequence from the NS RNA (Fig. 7B, upper panel, and see “Experimental Procedures”). Using this approach we were able to narrow the binding site of hnRNP A1 to within the last 18 nt of exon 7 as demonstrated for the NS:ERE-3 probe where the signal for hnRNP A1 is stronger than for the other constructs tested (Fig. 7B, compare lanes 6, 8, and 10 compared with lane 4). To also test whether hnRNP A1 mediates exon 7 skipping in vivo, we performed RNAi and overexpression analysis studies (Fig. 7, C and D). Plasmid encoded shRNAs to control green fluorescent protein (sh-GFP) or hnRNP A1 (shA1B) or overexpression hnRNP A1 plasmid (T7-A1) were transiently co-expressed with mini-gene CAM 6-7-8 in HEK-293 cells, and total protein was isolated and tested for the degree of down-regulation and overexpression of hnRNP A1. Our results show that we could down-regulate the expression of hnRNP A1 to 29% of wild type (Fig. 7C, lane 3 compared with lane 2). RT-PCR of these samples showed a modest increase of CEACAM1-L mRNA from 42% in control samples to 55% when hnRNP A1 expression was limiting (Fig. 7D, lane 2 compared with lane 3). In contrast, a high increase of CEACAM1-S mRNA was observed when hnRNP A1 was overexpressed (88% exon 7 exclusion; Fig. 7D, lane 4).
hnRNP A1 Requires No Other Cellular Factors to Bind Exon 7 RNA
Because the binding site for hnRNP A1 was non-canonical, we were curious whether hnRNP A1 can interact directly with exon 7 in the absence of other cellular factors using an RNA-protein gel mobility shift assay with recombinant hnRNP A1 protein and naked RNA (Fig. 8A). As our control, we chose the regulatory splice elements in CD44 variant exon (v.5) containing a canonical hnRNP A1 binding site (51). RNA-protein mixtures were fractionated on a native polyacrylamide gel. The RNA mobility shift data shows that hnRNP A1 binds to CD44 v.5 RNA with an apparent Kd of ∼0.9 μm, whereas when we tested exon 7 RNA we observed an apparent Kd of ∼1.6 μm (Fig. 8B). At the highest concentration tested (26 pmol), the amount of hnRNP A1 complexed is less than the CD44 v.5 substrate, suggesting the binding site in exon 7 is not as efficient at binding hnRNP A1 (Fig. 8A, compare lane 10 to lane 5).
FIGURE 8.
HnRNP A1-exon 7 electromobility shift assay. A, 20 fmol of CD44 variant exon v.5 RNA or 20 fmol exon 7 RNA incubated in the presence of varying amounts of His-hnRNP A1 protein (0.113–0.9 μg) and 1.0 μg of yeast tRNA to quench nonspecific interactions is shown. Reactions were electrophoresed on a 5% acrylamide gel and set to an autoradiograph. B, the apparent KD determination where the RNA-protein complexes (protein concentration for which 50% of the input is shifted to an RNP complex) were calculated as a function of increasing hnRNP A1 concentrations. The Kd values were calculated using Prism Software Version 4 (GraphPad Software). The mean ± S.E. for n = 3 was too small to be visualized with p value < 0.05 versus control.
hnRNP M Activates Alternative Splicing of CEACAM1-L mRNA
Our screen for splicing factors that associate with exon 7 RNA also revealed a putative association with hnRNP M (collectively isoforms M1-M4), a protein with unclear dual functions as a pre-mRNA-binding protein (29), as well as a surface receptor in colon cancer cells and tissue (52). For example, in CEA-stimulated THP-1 macrophages, activated endothelial cells increase cell-cell adhesion of colorectal cancer cells through the binding of surface expression of hnRNP M (also called CEA-R) (50). Our laboratory's long interest in CEA (53) led us to investigate whether we could establish a direct relationship between hnRNP M and CEACAM1, which contains the PELPK sequence previously characterized (Fig. 9) (54). Transient transfection of plasmid-encoded hnRNP M compared with hnRNP A1 (now serving as our positive control) or vector alone in HEK-293 cells demonstrated a strong shift to CEACAM1-L mRNA (Fig. 9, A and B, lane 5 compared with lanes 3 and 4). Thus, hnRNP M has the opposite effect of hnRNP L and A1 on exon 7. This evidence shows that exon 7 splicing involves spliceosome-containing hnRNP M, defining one mechanism that leads to the production of CEACAM1-L mRNA.
FIGURE 9.

HnRNP M activates alternative splicing of CEACAM1-L mRNA. A and B, shown is transient co-transfection of CAM 6-7-8 (lane 3) and plasmid-encoded hnRNP M (lane 6) compared with hnRNP A1 (lane 5) or vector (lane 4) alone in HEK-293 cells. RT-PCR analysis and quantitation of RNAs were exactly as described in caption to Fig. 1. *, p < 0.05; **, p < 0.01 versus CAM 6-7-8.
Deregulation of CEACAM1 Splicing Caused by Stress-induced Cytoplasmic Accumulation of hnRNP A1
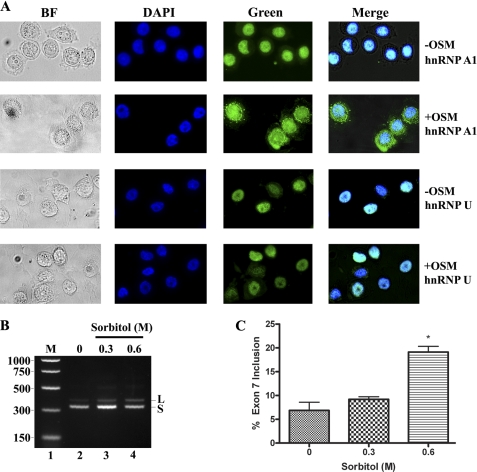
Mammalian cells have evolved mechanisms to respond to cellular stressors, i.e. heat shock, osmotic shock, or oxidative stress by accumulating hnRNP A1 to cytoplasmic granules (55, 56). These are cytoplasmic domains that contain translationally arrested mRNAs that in the case of phosphorylated hnRNP A1 can be activated by the MKK3/6-p38 signal transduction pathway (56). We hypothesized that if modulation of alternative splicing regulation in vivo correlates with functional hnRNP A1, then disruption of its cellular localization by stress should reveal deregulation of CEACAM1 splicing from CEACAM1-S mRNA to CEACAM1-L mRNA (Fig. 10). First, we show that upon exposure of MDA-MB-468 breast cancer cells to a high osmolarity medium, hnRNP A1 relocalizes to stress granules in the cytoplasm as compared with non-treated cells (Fig. 10A, compare panels −OSM versus +OSM, hnRNP A1). We included staining to non-shuttling protein hnRNP U as a control and detected no evidence of cytoplasmic accumulation of stress granules. We next isolated RNA from stress-induced cells and observed deregulation of CEACAM1 splicing as predicted (Fig. 10, B and C). The percent of exon 7 inclusion increases almost 3-fold (7.5–19.7%) in response to osmotic shock stress as compared with untreated cells (Fig. 10B, lanes 4 compared with 2). This provides additional evidence for the existence of a hnRNP A1 binding site in exon 7.
FIGURE 10.
Cytoplasmic accumulation of hnRNP A1 in stress-induced breast cancer cells dynamically shifts the production of CEACAM1 from the S-isoform to the L-isoform. A, HnRNP A1 localizes to stress granules in the cytoplasm. MDA-MB-468 cells cultured on an 8-well chamber slide were left untreated (−OSM) or treated (+OSM) with 0.6 m sorbitol for 2 h at 37 °C. The cells were fixed and immunostained with anti-hnRNP A1 or anti-hnRNP U antibodies to detect the endogenous proteins. BF, bright field. B and C, RT-PCR analysis and quantitation of RNA derived from cells from panel A were exactly as described in caption to Fig. 1. M is Benchtop PCR marker. *, p < 0.05 versus 0 m control.
DISCUSSION
Because we previously identified cis-regulatory elements in exon 7 of CEACAM1 (16), it was likely that exon 7 was capable of recruiting various trans-regulatory proteins to direct splice-site recognition. Herein we identified three splicing regulatory domains in CEACAM1 exon 7 that associate with a splicing complex made up of hnRNP proteins that oppositely effect the alternative splicing of this exon.
The critical observation that led to this study was that sequential exon 7 mutations in a mini-gene transfected into ZR75 cells revealed numerous cis-sequence elements that produced a phenotypic reduction of CEACAM1 isoforms (Fig. 1). Notably, our dissection of exon 7 revealed a tripartite structure that is required for the production of CEACAM1-L and CEACAM1-S isoforms. The result for the ESE located in ERE-1 suggested the recruitment of a splicing factor required to promote exon inclusion. This was encouraging given that most splicing enhancers are located near the 3′ splice site and are not active if located within 100 nt of the exon-intron boundary (57). In ERE-1, we established its spatial proximity to within the first 15 nt of the 3′ splice-site of intron 6. We also demonstrated the presence of an ESS in ERE-2 and ERE-3, which was downstream of the ESE in ERE-1, demonstrating a molecular arrangement for exon 7 similar to observations for HIV-1 tat exon 2 and 3 (58, 59). There it was shown that juxtaposition of the ESE/ESS binding sites represses splicing by hnRNP A/B protein binding, inhibiting the bridging interactions between adjacent SR proteins, SC35, and other essential splicing factors such as U2AF65/U2AF35. We further showed that cis elements within exon 7 were authentic in that they could direct CEACAM1 alternative splicing independent of flanking introns 6 and 7 RNA using a reporter constructed of a middle exon flanked by introns from chicken cTNT (Fig. 3).
We next identified hnRNP L and hnRNP A1 as specific trans-acting factors that associate with exon 7 using RNA affinity purification and RNA immunoprecipitation pulldown assays. In contrast to the HIV-1 tat exon study, we found only hnRNP proteins bound to CEACAM1 exon 7. Although our proteomics analyses provided a clue that hnRNP M was in the spliceosome-containing complex (Fig. 4), we could not establish a physical interaction of hnRNP M with exon 7 using our RNA immunoprecipitation pulldown. We conclude that hnRNP M regulates CEACAM1 alternative splicing via heterophilic protein-protein interaction such as the association that exists for hnRNP L with hnRNP A2 (60).
The discovery that exon 7 contained an hnRNP A1 target sequence was surprising considering the obvious lack of similarity to the high affinity canonical sequences identified by the selection/amplification approach (UAGRG(A/U)) (61). This could be explained by the fact that typically in the selection of high affinity binding sites for hnRNP A1, non-physiological non-equilibrium techniques are used. In the case of CEACAM1, we used competition of exon 7 ligand to titrate hnRNP A1 binding (Fig. 7) to first show the authenticity of this interaction. However, to rule out possible contributions to hnRNP A1 from other protein-protein interactions in the splicing complex, we used purified hnRNP A1 and performed EMSA studies using naked exon 7 RNA (Fig. 8). Our results show that no other cellular factors are required for hnRNP A1 to bind exon 7 RNA.
How do hnRNP A1, hnRNP L, or hnRNP M mechanistically regulate CEACAM1 splicing? A likely scenario should involve hnRNP heterophilic protein-protein interactions similar to hnRNP L association with hnRNP A2 as they bind the 3′ UTR of Glut1 mRNA (60). Whether the association depends on tethering to the RNA or is free in complex during exon definition remains unclear. From our data (Fig. 3D), a most intriguing clue comes from the apparent regulation that hnRNP L and hnRNP A1 exert over each other. Our view is that splicing reverts to a normal CEACAM1-S phenotype when overexpression of ERE-3 RNA causes sequestration of hnRNP A1. In this event, hnRNP L is allowed unrestrained access to the middle portion of exon 7 (ERE-2). It is worth mentioning that the converse is not true, i.e. overexpression of ERE-2 does not lead to the CEACAM1-S isoform. This is suggestive that hnRNP A1 negatively regulates hnRNP L, and the latter plays the dominant role in the formation of CEACAM1-S in breast tissues. In other considerations, in vitro and in vivo studies show that hnRNP A1 has the potential to influence 5′ splice-site selection, albeit in pre-mRNAs that contain multiple 5′ splice sites (62) or 3′ splice site selection by a cooperative spreading model elaborated recently by Okunola and Krainer (63). Here it was shown that hnRNP A1 binds to RNA initially at the 3′-end of the ESS HIV-1 tat exon 3 RNA. This signals the spreading in a 3′ to the 5′ direction, antagonizing the binding of any SR protein-ESE interaction, inhibiting splicing at the alternative 3′ splice site (63). We demonstrate the existence of a strong ESE upstream of the hnRNP A1 binding site in exon 7, and therefore, it is tempting to speculate that production of CEACAM1-S proceeds in a similar manner. However, because no SR proteins were identified in our study, an alternate mechanism may operate. By contrast, the mechanisms by which hnRNP L and hnRNP M stimulate exon repression or activation have yet to be elucidated, but it is likely that RNA sequences are marked by these hnRNPs for removal or inclusion by the splicing machinery. It is worth noting that yeast two-hybrid and GST pulldown experiments can detect physical interactions between hnRNP L, hnRNP-I/PTB, and hnRNP E2 (64). It is possible that cooperative binding of hnRNP M to hnRNP L and hnRNP A1 might increase the overall stability of the CEACAM1 exon 7 regulatory complex and may be important for mediating exon 7 inclusion. Further mapping the precise sites of association of these hnRNPs will be required to better understand the potential contribution of these proteins to the overall activity of CEACAM1 splicing.
Because CEACAM1-S is the major splice form in human breast epithelial cells and we now show that hnRNP L and hnRNP A1 play a role in its production, it is interesting to ask if these hnRNPs have a general role in the maintenance of human epithelial cells. It is known that overall expression levels of hnRNP A1 is tightly regulated during development (65) and that increased expression of hnRNPs in general has also been reported in human lung carcinogenesis (66), colorectal cancer (67), and other cancers (68). It is unknown currently whether this deregulation is a consequence of, or a driving force for, tumorigenesis. We showed that differentially expressed CEACAM1 in various human tissues and alteration in the splicing pattern of CEACAM1 is linked to tumorigenesis (16). We also have shown that forced expression of CEACAM1-S in the mammary epithelial cell line MCF7, with minimal expression of the long splice variant, reverts the growth of these cells to a normal acinar morphology in three-dimensional culture (69). Thus, it is likely that a role for hnRNP L or hnRNP A1 exists in the expression of the short cytoplasmic domain splice variant for normal breast morphogenesis, and inappropriate expression of the L-isoform may disrupt this process. In this respect, the L-isoform is induced by IFN-γ,5 a cytokine associated with inflammation. Further studies will be needed to fully assess the potential value of this biomarker splicing candidate.
Acknowledgments
We thank Zhifang Zhang for statistical assistance, Gideon Dreyfuss for the initial aliquots of hnRNP A1 antibodies, Doug Black for His-tagged hnRNP A1, Roger Moore in the City of Hope Mass Spectrometry Core facility, Jocelyn Huey for exon 7 mutants, Celeste Cheung for hnRNP M assays, Mariko Lee at the City of Hope Microscopy Core facility, and Ren-Jang Lin, Ravinder Singh, and Kristin Lynch for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA84202.
K. J. Dery, S. Gaur, M. Gencheva, Y. Yen, J. E. Shively, and R. K. Gaur, unpublished data.
- CEACAM1
- carcinoembryonic antigen (CEA)-related cell adhesion molecule 1
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- hnRNP A1
- hnRNP L, and hnRNP M, alternative splicing, exon splicing silencers
- CEACAM1-L
- CEACAM1-S, exon 7
- ESE
- exon splicing enhancers
- ESS
- exon splicing silencers
- NS
- nonspecific
- nt
- nucleotides.
REFERENCES
- 1. Kuespert K., Pils S., Hauck C. R. (2006) Curr. Opin. Cell Biol. 18, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nittka S., Günther J., Ebisch C., Erbersdobler A., Neumaier M. (2004) Oncogene 23, 9306–9313 [DOI] [PubMed] [Google Scholar]
- 3. Hsieh J. T., Luo W., Song W., Wang Y., Kleinerman D. I., Van N. T., Lin S. H. (1995) Cancer Res. 55, 190–197 [PubMed] [Google Scholar]
- 4. Riethdorf L., Lisboa B. W., Henkel U., Naumann M., Wagener C., Löning T. (1997) J. Histochem. Cytochem. 45, 957–963 [DOI] [PubMed] [Google Scholar]
- 5. Gray-Owen S. D., Blumberg R. S. (2006) Nat. Rev. Immunol. 6, 433–446 [DOI] [PubMed] [Google Scholar]
- 6. Barnett T. R., Drake L., Pickle W., 2nd (1993) Mol. Cell. Biol. 13, 1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Obrink B. (1997) Curr. Opin. Cell Biol. 9, 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCuaig K., Turbide C., Beauchemin N. (1992) Cell Growth Differ. 3, 165–174 [PubMed] [Google Scholar]
- 9. Edlund M., Wikström K., Toomik R., Ek P., Obrink B. (1998) FEBS Lett. 425, 166–170 [DOI] [PubMed] [Google Scholar]
- 10. Schumann D., Chen C. J., Kaplan B., Shively J. E. (2001) J. Biol. Chem. 276, 47421–47433 [DOI] [PubMed] [Google Scholar]
- 11. Chen C. J., Kirshner J., Sherman M. A., Hu W., Nguyen T., Shively J. E. (2007) J. Biol. Chem. 282, 5749–5760 [DOI] [PubMed] [Google Scholar]
- 12. Brümmer J., Neumaier M., Göpfert C., Wagener C. (1995) Oncogene 11, 1649–1655 [PubMed] [Google Scholar]
- 13. Chen C. J., Shively J. E. (2004) J. Immunol. 172, 3544–3552 [DOI] [PubMed] [Google Scholar]
- 14. Nagaishi T., Pao L., Lin S. H., Iijima H., Kaser A., Qiao S. W., Chen Z., Glickman J., Najjar S. M., Nakajima A., Neel B. G., Blumberg R. S. (2006) Immunity 25, 769–781 [DOI] [PubMed] [Google Scholar]
- 15. Chen T., Zimmermann W., Parker J., Chen I., Maeda A., Bolland S. (2001) J. Leukoc. Biol. 70, 335–340 [PubMed] [Google Scholar]
- 16. Gaur S., Shively J. E., Yen Y., Gaur R. K. (2008) Mol. Cancer 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 18. Blencowe B. J. (2000) Trends Biochem. Sci. 25, 106–110 [DOI] [PubMed] [Google Scholar]
- 19. Pozzoli U., Sironi M. (2005) Cell. Mol. Life Sci. 62, 1579–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kan J. L., Green M. R. (1999) Genes Dev. 13, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh R., Valcárcel J. (2005) Nat. Struct. Mol. Biol. 12, 645–653 [DOI] [PubMed] [Google Scholar]
- 22. Hertel K. J. (2008) J. Biol. Chem. 283, 1211–1215 [DOI] [PubMed] [Google Scholar]
- 23. Wang Z., Burge C. B. (2008) RNA 14, 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faustino N. A., Cooper T. A. (2003) Genes Dev. 17, 419–437 [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Blanco M. A. (2006) Prog. Mol. Subcell. Biol. 44, 47–64 [DOI] [PubMed] [Google Scholar]
- 26. Han S. P., Tang Y. H., Smith R. (2010) Biochem. J. 430, 379–392 [DOI] [PubMed] [Google Scholar]
- 27. Lin S., Fu X. D. (2007) Adv. Exp. Med. Biol. 623, 107–122 [DOI] [PubMed] [Google Scholar]
- 28. Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J. F., Revil T., Chabot B. (2007) Adv. Exp. Med. Biol. 623, 123–147 [DOI] [PubMed] [Google Scholar]
- 29. Hovhannisyan R. H., Carstens R. P. (2007) J. Biol. Chem. 282, 36265–36274 [DOI] [PubMed] [Google Scholar]
- 30. Cartegni L., Maconi M., Morandi E., Cobianchi F., Riva S., Biamonti G. (1996) J. Mol. Biol. 259, 337–348 [DOI] [PubMed] [Google Scholar]
- 31. Cobianchi F., Karpel R. L., Williams K. R., Notario V., Wilson S. H. (1988) J. Biol. Chem. 263, 1063–1071 [PubMed] [Google Scholar]
- 32. Expert-Bezançon A., Sureau A., Durosay P., Salesse R., Groeneveld H., Lecaer J. P., Marie J. (2004) J. Biol. Chem. 279, 38249–38259 [DOI] [PubMed] [Google Scholar]
- 33. Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990) Biotechniques 8, 528–5352357375 [Google Scholar]
- 34. Orengo J. P., Bundman D., Cooper T. A. (2006) Nucleic Acids Res. 34, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dery K. J., Gusti V., Gaur S., Shively J. E., Yen Y., Gaur R. K. (2009) Methods Mol. Biol. 555, 127–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim D. S., Gusti V., Pillai S. G., Gaur R. K. (2005) RNA 11, 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konarska M. M., Sharp P. A. (1986) Cell 46, 845–855 [DOI] [PubMed] [Google Scholar]
- 39. Griffith B. N., Walsh C. M., Szeszel-Fedorowicz W., Timperman A. T., Salati L. M. (2006) Biochim. Biophys. Acta 1759, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szeszel-Fedorowicz W., Talukdar I., Griffith B. N., Walsh C. M., Salati L. M. (2006) J. Biol. Chem. 281, 34146–34158 [DOI] [PubMed] [Google Scholar]
- 41. Nayler O., Hartmann A. M., Stamm S. (2000) J. Cell Biol. 150, 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cloutier P., Toutant J., Shkreta L., Goekjian S., Revil T., Chabot B. (2008) J. Biol. Chem. 283, 21315–21324 [DOI] [PubMed] [Google Scholar]
- 43. Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X. H., Chasin L. A. (2004) Genes Dev. 18, 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suyama M., Harrington E. D., Vinokourova S., von Knebel Doeberitz M., Ohara O., Bork P. (2010) Nucleic Acids Res. 38, 7916–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miriami E., Margalit H., Sperling R. (2003) Nucleic Acids Res. 31, 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Akerman M., Mandel-Gutfreund Y. (2006) Nucleic Acids Res. 34, 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hui J., Hung L. H., Heiner M., Schreiner S., Neumüller N., Reither G., Haas S. A., Bindereif A. (2005) EMBO J. 24, 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watakabe A., Tanaka K., Shimura Y. (1993) Genes Dev. 7, 407–418 [DOI] [PubMed] [Google Scholar]
- 50. Aarons C. B., Bajenova O., Andrews C., Heydrick S., Bushell K. N., Reed K. L., Thomas P., Becker J. M., Stucchi A. F. (2007) Clin. Exp. Metastasis 24, 201–209 [DOI] [PubMed] [Google Scholar]
- 51. Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., König H. (2000) J. Biol. Chem. 275, 35353–35360 [DOI] [PubMed] [Google Scholar]
- 52. Laguinge L., Bajenova O., Bowden E., Sayyah J., Thomas P., Juhl H. (2005) Anticancer Res. 25, 23–31 [PubMed] [Google Scholar]
- 53. Shively J. E., Beatty J. D. (1985) Crit. Rev. Oncol. Hematol. 2, 355–399 [DOI] [PubMed] [Google Scholar]
- 54. Gangopadhyay A., Thomas P. (1996) Arch. Biochem. Biophys. 334, 151–157 [DOI] [PubMed] [Google Scholar]
- 55. Guil S., Long J. C., Cáceres J. F. (2006) Mol. Cell. Biol. 26, 5744–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Cáceres J. F. (2000) J. Cell Biol. 149, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Graveley B. R., Hertel K. J., Maniatis T. (1998) EMBO J. 17, 6747–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zahler A. M., Damgaard C. K., Kjems J., Caputi M. (2004) J. Biol. Chem. 279, 10077–10084 [DOI] [PubMed] [Google Scholar]
- 59. Zhu J., Mayeda A., Krainer A. R. (2001) Mol. Cell 8, 1351–1361 [DOI] [PubMed] [Google Scholar]
- 60. Hamilton B. J., Nichols R. C., Tsukamoto H., Boado R. J., Pardridge W. M., Rigby W. F. (1999) Biochem. Biophys. Res. Commun. 261, 646–651 [DOI] [PubMed] [Google Scholar]
- 61. Burd C. G., Dreyfuss G. (1994) EMBO J. 13, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mayeda A., Krainer A. R. (1992) Cell 68, 365–375 [DOI] [PubMed] [Google Scholar]
- 63. Okunola H. L., Krainer A. R. (2009) Mol. Cell. Biol. 29, 5620–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. (2000) J. Mol. Biol. 298, 395–405 [DOI] [PubMed] [Google Scholar]
- 65. Kamma H., Horiguchi H., Wan L., Matsui M., Fujiwara M., Fujimoto M., Yazawa T., Dreyfuss G. (1999) Exp. Cell Res. 246, 399–411 [DOI] [PubMed] [Google Scholar]
- 66. Carpenter B., MacKay C., Alnabulsi A., MacKay M., Telfer C., Melvin W. T., Murray G. I. (2006) Biochim. Biophys. Acta 1765, 85–100 [DOI] [PubMed] [Google Scholar]
- 67. Ma Y. L., Peng J. Y., Zhang P., Huang L., Liu W. J., Shen T. Y., Chen H. Q., Zhou Y. K., Zhang M., Chu Z. X., Qin H. L. (2009) J. Proteome Res. 8, 4525–4535 [DOI] [PubMed] [Google Scholar]
- 68. Patry C., Bouchard L., Labrecque P., Gendron D., Lemieux B., Toutant J., Lapointe E., Wellinger R., Chabot B. (2003) Cancer Res. 63, 7679–7688 [PubMed] [Google Scholar]
- 69. Kirshner J., Chen C. J., Liu P., Huang J., Shively J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]