Abstract
The JNK signaling cascade is critical for cellular responses to a variety of environmental and cellular stimuli. Although gene expression aspects of JNK signal transduction are well studied, there are minimal data on the physiological impact of JNK signaling. To bridge this gap, we investigated how JNK impacted physiology in HeLa cells. We observed that inhibition of JNK activity and JNK silencing with siRNA reduced the level of reactive oxygen species (ROS) generated during anisomycin-induced stress in HeLa cells. Silencing p38 had no significant impact on ROS generation under anisomycin stress. Moreover, JNK signaling mediated amplification of ROS production during stress. Mitochondrial superoxide production was shown to be the source of JNK-induced ROS amplification, as an NADPH oxidase inhibitor demonstrated little impact on JNK-mediated ROS generation. Using mitochondrial isolation from JNK null fibroblasts and targeting the mitochondrial scaffold of JNK, Sab, we demonstrated that mitochondrial JNK signaling was responsible for mitochondrial superoxide amplification. These results suggest that cellular stress altered mitochondria, causing JNK to translocate to the mitochondria and amplify up to 80% of the ROS generated largely by Complex I. This work demonstrates that a sequence of events exist for JNK mitochondrial signaling whereby ROS activates JNK, thereby affecting mitochondrial physiology, which can have effects on cell survival and death.
Keywords: JNK, MAP Kinases (MAPKs), Mitochondria, Reactive Oxygen Species (ROS), Signal Transduction
Introduction
The c-Jun N-terminal kinases (JNK) are serine/threonine protein kinases and members of the mitogen-activated protein kinase (MAPK) superfamily (1). There are three JNK isoforms in humans, JNK1, JNK2, and JNK3. JNK1 and JNK2 are ubiquitously expressed in human tissues, whereas JNK3 has specific locals in the brain, heart, and testes (1). JNK signaling is responsive to many environmental and cellular stimuli with prolonged JNK activity resulting in pro-apoptotic transcription and cell death (2). JNK can phosphorylate transcription factors, including c-Jun, and promote activator protein-1-mediated transcription (2).
JNK signaling contributes to apoptotic signaling through the phosphorylation of pro-apoptotic proteins (2). JNK can phosphorylate Bcl-2, triggering Bcl-2 migration from the mitochondrial membrane and dissipation of mitochondrial membrane potential (3, 4). The ability of JNK to utilize non-nuclear substrates for cell death induction indicates that JNK signaling could impact cell death beyond induction of pro-apoptotic transcription.
Recent research has identified a new subcellular location for JNK signaling. At the outer mitochondrial membrane, JNK can interact with and phosphorylate a scaffold, Sab, and this may be the means for anchoring JNK to the mitochondria for signaling events (5, 6). Mitochondrial translocation of JNK has been described in cellular and mammalian models for DNA damage (7, 8), liver injury (9), oxidative stress (10), anisomycin-induced stress (11), and cerebral ischemia (12). In human myeloid lymphoma cells treated with γ radiation to induce DNA damage, JNK translocates to the mitochondria of stressed cells and phosphorylates Bcl-xL contributing to apoptosis (7). During acetaminophen-induced liver injury, mitochondrial JNK signaling is responsible for decreased mitochondrial respiration (9). During oxidative stress in cardiomyocytes, JNK migrates to the mitochondria, and induces the release of cytochrome c (10). In anisomycin-treated primary cortical neurons, mitochondrial JNK was able to phosphorylate pyruvate dehydrogenase complex subunit Eα1 inhibiting pyruvate dehydrogenase activity resulting in decreased pyruvate flux into the mitochondria (11). Cerebral ischemia provides a more dynamic model of mitochondrial JNK signaling where under physiological conditions JNK1 is the primary mitochondrial isoform; however, JNK3 becomes the primary mitochondrial JNK isoform following middle cerebral artery occlusion (12). These data provide evidence that mitochondrial JNK signaling is a crucial event in cell death signaling.
JNKs are also responsive to mitochondrial signals. The most prominent mitochondrial inducers of JNK signaling are reactive oxygen species (ROS),2 which are by-products of cellular respiration and consequences of mitochondrial dysfunction (13, 14). The major sources of ROS generation are the mitochondrial respiratory complexes (15–17). Complex I produces superoxide radicals that are introduced into the matrix of the mitochondria, whereas Complex III-generated superoxide is released into the cytosol (17). As ROS levels increase in the mitochondria, damage occurs to the reactive cysteines in phosphatases and metabolic enzymes. Oxidation of respiratory complex constituents contribute to a loss in mitochondrial respiration (16). The culmination of oxidative stress in the mitochondria is dissipation of mitochondrial membrane potential and subsequent release cytochrome c (16).
JNK is activated by ROS through upstream kinases, primarily apoptosis signaling regulated kinase 1 (ASK1), a kinase responsive to mitochondrial ROS (18). Previous work in our laboratory demonstrated that JNK signaling contributes to the generation of ROS during stress. We found that INS-1 insulinoma rat cells treated with streptozotocin produced ROS, and use of a highly selective small molecule inhibitor of JNK (SR-3562 or Compound 9l) prevented ROS production (19). Congruent with our results, JNK-dependent ROS generation was observed in TNF-α stimulated murine embryonic fibroblasts (MEFs) lacking functional NF-κB signaling (20).
We propose that mitochondrial JNK (MitoJNK) signaling alters mitochondrial physiology promoting ROS generation during stress. In this work, we demonstrate in anisomycin-treated HeLa cells that JNK translocated to the mitochondria and amplified mitochondrial superoxide generation. MitoJNK altered mitochondrial activity through the inhibition of respiratory Complex I contributing to decreased respiratory function and increased ROS generation. MitoJNK signaling created a pro-oxidant producing environment in the mitochondria resulting in oxidative stress. The data presented herein will illustrate a role for MitoJNK signaling as an initiating event in cell death.
EXPERIMENTAL PROCEDURES
Materials
Anisomycin, apocynin, and N-acetylcysteine were purchased from MP Biomedical. Rotenone and antimycin A were purchased from Enzo Life Sciences International. Carboxin was purchased from Sigma. Control (nonsense), JNK, and p38 siRNAs were purchased from Cell Signaling Technology. Sab siRNAs were purchased from Novus Biologicals.
Cells and Cell Culture
HeLa cells (ATCC) and JNK1−/− and JNK2−/− murine embryonic fibroblasts (JNK null fibroblasts; kind gift from Dr. Roger Davis, Howard Hughes Medical Institute, University of Massachusetts) were grown under typical cell culture conditions (37 °C and 5% CO2) in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Only cells between 5 and 15 passages were used for experiments. Cells were treated with chemical agents once a confluence of 75–80% had been achieved to assure the experiments were conducted on actively growing cells.
Transfections and Gene Silencing
Knockdown of JNK, p38, and Sab expression was achieved by small interfering RNA-mediated gene silencing. HeLa cells transfected with siRNAs specific for JNK, p38, Sab, or control siRNAs were introduced into cells using the HiPerfect transfection reagent from Qiagen. Cells were transfected according to the manufacturer's suggestion. Briefly, HeLa cells were grown to ∼50% confluence, then the cells were transfected with 30 ng of siRNA in 100 μl of culture medium with 12 μl of HiPerfect reagent. Transfection complexes formed for 10 min at room temperature, then the mixture was added to cells. Knockdown was confirmed after 72 h using Western blot analysis of cell lysates.
Cell Lysis and Western Blotting
To acquire protein for Western blot analysis after an experiment, cells were washed twice in PBS. Radioimmunoprecipitation assay buffer (RIPA; 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) supplemented with protease inhibitors (1 mm PMSF and Protease Inhibitor Mixture Set III (Calbiochem)) and phosphatase inhibitors (Phosphatase Inhibitor Mixture Set V (Calbiochem)) was added to the cells. Lysis buffer and cells were incubated at 4 °C for 5 min while rocking gently. Cells were scraped from the culture surface and transferred to a microcentrifuge tube. Following a 2-min incubation on ice, the cells were disrupted by brief sonication. The cell lysate was centrifuged at 14,000 × g for 15 min to remove cellular debris. The concentration of protein in the supernatant was quantified by BCA analysis using the Pierce kit protocol. For Western blot analysis, proteins were resolved by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were incubated with blocking buffer (1× TBST with 5% nonfat milk, 20 mm Tris-HCl, pH 6.7, 137 mm NaCl, 0.1% Tween 20, and 5% milk) for at minimum 1 h at room temperature or overnight at 4 °C. For phospho-Western blots, membranes were blocked with blocking buffer containing 5% bovine serum albumin (BSA) rather than nonfat milk. The membranes were incubated with primary antibodies specific for JNK (Cell Signaling Technology, number 9252), phospho-JNK (Thr183/Tyr185) (Cell Signaling Technology, number 9251), p38 (Cell Signaling Technology, number 9212), phospho-p38 (Cell Signaling Technology, number 9216), Sab (Novus Biologicals, H00009467-M01), or α-tubulin (Cell Signaling Technology, number 2144) at dilutions of 1:1000 in blocking buffer. Membranes were washed three times for 5 min in 1× TBST. Membranes were incubated with secondary antibodies in blocking buffer at 1:2000 for horseradish peroxidase (HRP)-conjugated antibodies (anti-rabbit IgG, HRP-linked (Cell Signaling Technology, number 7074 and anti-mouse IgG, HRP-linked, Cell Signaling Technology, number 7076) or 1:20,000 for fluorescently conjugated antibodies (anti-rabbit, DyLight 680 conjugate, number 5366, and anti-mouse, DyLight 800 conjugate, number 5151, Cell Signaling Technology). Western blots were developed using chemiluminescence or fluorescence detection using the Odyssey scanner (Li-Cor Biosciences).
Fluorescent Detection of ROS and Mitochondrial Superoxide
For the detection of whole cell ROS, we used dihydroethidium (DHE) staining of intracellular superoxide (21, 22). DMEM was exchanged with DMEM without phenol red 2 h prior to the start of the experiment. Cells were stained prior to anisomycin treatment with DHE or after anisomycin treatments depending on the experiment. For microscopy, cells were stained with 5 μm DHE for 10 min. The cells were then washed in DMEM without phenol red and with 2.5% FBS to reduce the fluorescent contributions of the dye and the serum. Cells were visualized in phenol red-free DMEM with 0.25% FBS. For fluorescence quantitation, cells (in 96-well plates) were stained and visualized as indicated above, except the DHE concentration was lowered to 50 nm to decrease background fluorescence in the plate. These data were confirmed using a second ROS-selective stain, 2′,7′-di-chlorofluorescein derivative (H2DCF-DA) (data not shown). For the detection of mitochondrial superoxide production, we utilized the mitochondrial sensitive dye MitoSOX Red (Invitrogen) (23). Staining was performed similar to DHE staining with the exception of concentration. For microscopic analysis, 10 μm MitoSOX Red was used, whereas 100 nm was used for fluorescent quantitation. To confirm mitochondrial localization of the dye, cells were counterstained with 5 μm MitoTracker Green (Invitrogen) for 10 min. Normalization of fluorescent data were done using counterstaining with Hoescht 33342 (1 μm) for 5 min. Fluorescent wavelength pairs for the individual dyes were 568/600 nm for DHE, 510/580 nm for MitoSOX Red, 490/516 nm for MitoTracker Green, and 350/461 nm Hoescht 33342 on a Spectromax M5e plate reader (Molecular Devices).
Preparation of Mitochondrial Fractions
Mitochondria were isolated as described by Palloti and Lenaz (24). Briefly, HeLa cells or JNK null fibroblasts (2 × 108 cells) were grown on 150-mm2 tissue culture plates as described above. The cells were washed twice at room temperature in PBS, and the cells were scraped gently from the culture surface. Cells were pelleted by centrifugation at 1000 × g for 15 min at room temperature. The pellet was resuspended in six times the pellet volume with ice-cold cell homogenization buffer (150 mm MgCl2, 10 mm KCl, 10 mm Tris-HCl, pH 6.7), and the suspension was placed on ice for 2 min. Using an ice-cold homogenizer, the cells were disrupted with up to 10 up-and-down strokes (disruption was confirmed by microscopy). To the disrupted cells, cell homogenization buffer with 0.25 m sucrose was added at one-third the volume of the suspension followed by gentle inversion to mix thoroughly. The nuclei were pelleted by centrifugation at 1,000 × g for 5 min at 4 °C. The supernatant was centrifuged at 5,000 × g for 10 min at 4 °C. The pellet was resuspended in ice-cold sucrose/Mg2+ buffer (150 mm MgCl2, 250 mm sucrose, 10 mm Tris-HCl, pH 6.7). The pellet was disrupted with an ice-cold Dounce homogenizer with a few strokes. The solution suspension was centrifuged at 5,000 × g for 10 min at 4 °C. Mitochondria found in the pellet were resuspended in the indicated buffers for the following experiments. The purity of the mitochondrial enrichments were determined using Western blot analysis for mitochondrial resident protein, cyclooxygenase IV (Cell Signaling Technology, number 5247), cytosolic protein, GAPDH (Cell Signaling Technology, number 3683), nuclear contamination with histone-H3 (Cell Signaling Technology, number 5192), and microsomal constituent, PDI (Cell Signaling Technology, number 2446). Mitochondrial fractions ascertained to be greater than 70% pure were used for protein and enzymatic analysis. To determine the functional capacity of the mitochondria, individual enrichments were examined with respect to citrate synthase activity. Citrate synthase catalyzes the conversion of oxaloacetate and acetyl-coenzyme A to citrate and reduces coenzyme-A. The sulfhydryl group on conenzyme-A is reacted with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), and the reaction mixture turns yellow; subsequently, this change can be measured spectrophotometrically by monitoring absorbance at a wavelength of 412 nm. Our reaction, modified from Ref. 25, contained 30 μg of mitochondria enrichment and was added to citrate synthase assay mixture (100 mm Tris-HCl, pH 8.0, 0.25 mm oxaloacetate, 50 μm acetyl-coenzyme A, 0.1 mm DTNB, and 0.1% Triton X-100. Absorbance was monitored at 412 nm for 5 min. Mitochondria were diluted to a concentration of 80 mg/ml, then frozen on dry ice/ethanol slurry and stored at −80 °C until use.
Detection of Mitochondrial Generated ROS in Mitochondrial Enrichments
For the detection of mitochondrial produced ROS in the presence of respiratory substrates and active JNK1α1 (Millipore, bisphosphorylated Thr183 and Tyr185), we employed an Amplex Red detection method using the Amplex Red Hydrogen Peroxide Detection kit protocol from Invitrogen. Briefly, 20 μg of isolated mitochondria were incubated with 30 μg of JNK1α1 for 15 min at 30 °C. The mitochondria/JNK mixture was then incubated with assay buffer and Amplex Red for 30 min at 30 °C. Hydrogen peroxide production by mitochondria was measured by fluorescence at 571 (excitation) and 585 nm (emission).
Respiratory Complex Enzyme Assays
For Complex I assays, we enriched for Complex I by isolating mitochondrial membranes similar to Ref. 25. Mitochondria were washed in hypotonic buffer (25 mm potassium phosphate, 5 mm MgCl2, pH 7.2). The mitochondria were sonicated with five 5-s pulses at 30% maximum power using a microtip sonicator. The membranous fraction was pelleted by centrifugation at 11,000 × g for 10 min at 4 °C. Complex I activity was measured using an assay to detect NADH:ubiquinone oxidoreductase. The Complex I-enriched fraction (30 μg) was added to a reaction mixture containing 25 mm potassium phosphate, 5 mm MgCl2, pH 7.2, with 2 mm KCN, 2.5 mg/ml of BSA, 0.15 mm NADH, 70 mm decylubiquinone, and 1 mm antimycin A. The rate of NADH oxidation was observed by monitoring the loss in absorbance at 340 nm. Rotenone (1 mm) was added to the reaction following 5 min to determine the amount of NADH oxidation dependent on rotenone-sensitive Complex I activity. Only Complex I enrichments with greater than 75% rotenone sensitive activity were used in our analysis. Complex III activity was monitored by using a ubiquinol2:cytochrome c reductase reaction (25). Mitochondria (50 μg) were added to a reaction mixture containing 25 mm potassium phosphate, 5 mm MgCl2, pH 7.2, with 2 mm KCN, 2.5 mg/ml of BSA, 1 mm rotenone, 500 nm carboxin, 0.6 mm n-dodecyl-β-d-maltoside, 15 μm oxidized cytochrome c, and 35 μm ubiquinol2. Reduction of cytochrome c was monitored by absorbance at 550 nm for 5 min. Complex III activity was confirmed by inhibition with 1 mm antimycin A. Ubiquinol2 was synthesized according to Ref. 26.
Oxygen Consumption
Oxygen consumption and respiratory states were monitored using the Seahorse 96F Flux Analyzer (Seahorse Biosciences). Briefly, HeLa cells were plated at a density of 5 × 105 cells per well, and placed in Seahorse assay media in the presence of glucose (25 mm) and glutamine (4 mm), glutamate/malate/ADP (8 mm), or pyruvate/malate/ADP (8 mm). Oxygen consumption was monitored for four 5-min intervals to obtain an accurate measure. Data were normalized to viable cell number following the assay. Viable cells were determined by trypan blue exclusion staining using the Bio-Rad TC10 Automated Cell Counter. Cells were removed from the plate, then 10 μl of the cells were added to 10 μl of 0.4% trypan blue solution (Bio-Rad). Viability was measured immediately following metabolic assay.
Determination of ATP Concentration
Intracellular ATP concentration was determined using the ATP Determination Kit protocol (Invitrogen), which is a luciferase-based method of ATP quantitation. Cells were washed twice in PBS, and lysed by addition of boiling H2O. Cellular debris was pelleted by centrifugation at 14,000 × g for 15 min at 4 °C. ATP quantitation was carried out immediately following lysis.
Replicates and Statistics
For cell-based studies, at least four biological replicates between passages 5 and 15 were used for HeLa cells. For mitochondrial harvests, a minimum of three biological replicates were used. Biochemical assays, fluorometric detection of superoxide, and other cellular measures were done with a minimum of five experimental replicates. To determine statistical significance a Student's paired t test was employed. Statistical significance is indicated by an asterisk in figures when the p value is greater than 0.05.
RESULTS
Anisomycin Activates JNK Signaling and Stimulates JNK-dependent ROS Production
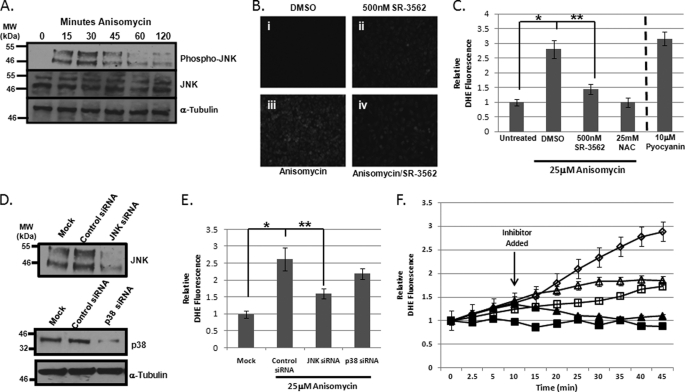
To evaluate the role of JNK in ROS evolution, we chose anisomycin-stressed HeLa cells as an established JNK responsive (27, 28). Using 25 μm anisomycin, we reduced HeLa cell viability by 50% within 4 h of anisomycin treatment. Western blot analysis utilizing an antibody selective for active JNK (JNK bisphosphorylated on threonine 183 (Thr183) and tyrosine 185 (Try185)) showed appreciable anisomycin-induced JNK activation between 15 and 30 min. JNK remained active through 60 min with a significant decrease between 30 and 60 min (Fig. 1A). Western blot analysis revealed there was no change in total JNK concentration during this time (Fig. 1A), and HeLa cell lysates were loaded equally as demonstrated in the α-tubulin Western blot (Fig. 1A). Because JNK activation occurred between 5 and 45 min, we chose to monitor ROS generation during the first 45 min of anisomycin treatment, as previous work from our laboratory had demonstrated that JNK-selective small molecule inhibitors, such as SR-3562 (or compound 9l) could inhibit ROS generation in a dose-dependent manner with an IC50 of ∼1 nm in INS-1 cells treated with streptozotocin (19). Following 35 min of DMSO treatment or anisomycin stress, HeLa cells were stained with DHE to detect whole cell superoxide generation. Microscopy images of DHE fluorescence revealed that treatment with 25 μm anisomycin increased intracellular ROS (Fig. 1B, iii) compared with DMSO-treated cells (Fig. 1B, i). Pre-treatment of cells with 500 nm SR-3562 (19) prior to DMSO treatment resulted in a slight increase in DHE fluorescence (Fig. 1B, ii), whereas pretreatment with 500 nm SR-3562 prevented ROS accumulation in anisomycin-stressed HeLa cells (Fig. 1B, iv). A concentration of 500 nm SR-3562 was used given that this was 100-fold greater than the concentration observed for complete inhibition of JNK-dependent ROS generation in anisomycin-stressed HeLa cells (supplemental Fig. S1). Quantitative analysis of the staining revealed that superoxide levels were increased in anisomycin-treated cells over 2.5 times that of untreated cells (Fig. 1C). The use of 500 nm SR-3562 prevented 75–80% of the superoxide production (**, p < 0.05) similar to the antioxidant, N-acetylcysteine (NAC, 25 mm) (Fig. 1C). Pyocyanin (10 μm) was used as a positive control for ROS evolution (Fig. 1C). To further implicate JNK signaling in ROS production, we silenced JNK expression using JNK-specific siRNAs; consequently, we were able to knockdown JNK expression by over 85% according to Western blot analysis (Fig. 1D), and scrambled siRNA had no impact on JNK expression (Fig. 1D). Similar to SR-3562, silencing JNK expression reduced superoxide production by 70% compared with cells transfected with scrambled siRNAs (Fig. 1E) (**, p < 0.05). As a control, we also silenced p38, which has been documented to have similar signaling effects as JNK signaling during cell death (29–31). In Fig. 1D, p38 knockdown was observed to be greater than 80% by Western blotting. Moreover, silencing p38 contributed minimally (∼5%) to ROS generation (Fig. 1E). These data indicate a role for JNK in ROS generation; however, we wanted to know if the kinetics of ROS generation changed as JNK became activated. To address this question, we pre-stained cells with DHE prior to the addition of 25 μm anisomycin, and monitored the increase in cellular fluorescence over 45 min of anisomycin stress. Treatment with 25 μm anisomycin increased ROS production over the 45 min (Fig. 1F, open diamonds), whereas inhibition with 25 mm NAC prevented ROS production (Fig. 1F, black squares). Pretreatment with 500 nm SR-3562 prior to anisomycin addition resulted in significantly less ROS production than anisomycin alone (Fig. 1F, open squares). Addition of 25 mm NAC 10 min following anisomycin resulted in inhibition of ROS generation (Fig. 1F, black triangles); meanwhile, inhibition of JNK by 500 nm SR-3562 prevented further generation of ROS after 10 min (Fig. 1F, open triangles) preventing the secondary generation of ROS seen in cells treated with only anisomycin (Fig. 1F, open diamonds).
FIGURE 1.
Anisomycin-induced stress initiates JNK-dependent ROS generation. A, JNK is activated by anisomycin in HeLa cells. JNK activation over the course of 2 h of 25 μm anisomycin stress was determined by Western blotting for bisphosphorylated JNK on Thr183 and Tyr185 (top). JNK abundance was monitored by Western blotting for total JNK (middle). α-Tubulin served as a loading control bottom. B, fluorescence microscopy of cellular ROS as detected by DHE. DMSO-treated cells (i) were compared with cells treated with 500 nm SR-3562 for 30 min (ii), cells stressed with 25 μm anisomycin for 45 min (iii), and cells pretreated for 30 min with 500 nm SR-3562 then stressed with 25 μm anisomycin (iv). C, fluorometric quantitation of cells stained with DHE to detect cellular ROS. Cells were stained with DHE following treatment with DMSO, 500 nm SR-3562 (30 min), 25 μm anisomycin (45 min), and 500 nm SR-3562 (30 min prior to anisomycin) and 25 μm anisomycin (45 min). 25 mm NAC, an antioxidant, was used as a negative control for ROS generation, and 10 μm pyocyanin, an inducer of ROS generation, was used as a positive control. D, Western blot analysis of JNK and p38 siRNA knockdowns of gene expression. Relative percent knockdown was estimated using densitometry. E, quantitation of cellular ROS in anisomycin-treated HeLa cells with JNK or p38 knockdown. After a 72-h siRNA transfection, cells were treated with 25 μm anisomycin for 45 min. ROS was detected using DHE staining and fluorescence was measured. F, kinetic profile of ROS generation in anisomycin-stressed HeLa cells. Cells were prestained with DHE prior to treatment with 25 mm NAC (black squares), 25 μm anisomycin (open diamonds), or 500 nm SR-3562 (open squares). Fluorescence was monitored for 45 min following the addition of the treatment. In two experiments, 25 mm NAC (black triangles) and 500 nm SR-3562 (open triangles) were added 10 min after 25 μm anisomycin.
JNK-dependent ROS Generation Is Fueled by Mitochondrial Superoxide
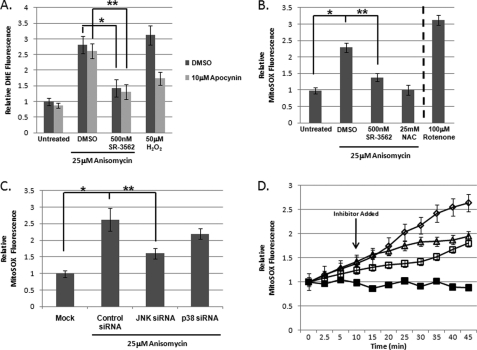
To determine whether JNK-mediated ROS production was generated in the mitochondria or in the cytosol, we chose to use apocynin to inhibit NADPH oxidases (32); these enzymes are responsible for most of the non-mitochondrial ROS generation. Prior to 25 μm anisomycin addition, cells were treated with 10 μm apocynin. Superoxide generation was monitored by DHE fluorescence following 45 min of anisomycin stress. There was only a small (statistically insignificant) difference between DMSO-treated and apocynin-treated cells following anisomycin-induced stress (Fig. 2A). Addition of 500 nm SR-3562 decreased DMSO-treated and apocynin-treated DHE fluorescence by ∼2-fold. These data suggested that JNK signaling was directing mitochondrial superoxide generation. As a control to ensure apocynin was inhibiting NADPH oxidases, we monitored the impact of apocynin on ROS generation during H2O2 stress. Apocynin treatment decreased ROS generation by 50% during H2O2-induced oxidative stress (Fig. 2A).
FIGURE 2.
JNK signaling amplifies mitochondrial superoxide production. A, JNK-dependent ROS is not generated by NADPH oxidases. Prior to anisomycin stress, cells were treated with either DMSO or 10 μm apocynin, and DHE fluorescence was measured 45 min following the addition of anisomycin. 50 μm Hydrogen peroxide (H2O2) was used as a positive control for NADPH oxidase-generated superoxide production. B, MitoSOX staining indicated that the JNK-mediated ROS was mitochondrial superoxide. MitoSOX fluorescence was used to determine mitochondrial superoxide production following 45 min of anisomycin-induced stress. Fluorescence was monitored in DMSO controls, cells were treated with 500 nm SR-3562, 25 μm anisomycin, and cells were pre-treated with 500 nm SR-3562 for 30 min prior to anisomycin addition. 25 mm NAC was included as a negative control for superoxide generation, whereas 100 μm rotenone served as a positive control for mitochondrial generated superoxide. C, JNK signaling, not p38, was required for mitochondrial superoxide generation during anisomycin stress. Following 72 h of gene silencing, mitochondrial superoxide levels were observed using MitoSOX Red. Mock transfected cells and cells containing JNK, p38, or control siRNAs were treated with anisomycin for 45 min prior to measurement. D, kinetic superoxide profiles demonstrate JNK signaling amplifies mitochondrial ROS generation. Mitochondrial superoxide production (MitoSOX fluorescence) was monitored over 45 min of anisomycin stress. Cells were pre-treated with 25 mm NAC (black squares) or 500 nm SR-3562 (open squares) 30 min prior to stress or 10 min (500 nm SR-3562, open triangles) following 25 μm anisomycin addition (25 μm anisomycin, open diamonds).
To probe the mitochondria for JNK-mediated superoxide production, we used the mitochondria selective, superoxide sensitive fluorophore, MitoSOX Red. Using the same approach as before, cells were pretreated with 500 nm SR-3562, 25 mm NAC, or DMSO, and then 25 μm anisomycin was added to the cultures for 45 min. Compared with anisomycin-treated cells, cells pre-treated with SR-3562 had a 70% decrease in MitoSox fluorescence (**, p < 0.05) indicative of decreased superoxide generation (Fig. 2B). NAC was also able to completely inhibit mitochondrial superoxide generation (Fig. 2B). As a positive control for detection of mitochondrial induced superoxide, rotenone-treated (100 μm, 4 h) cells were used to detect mitochondrial superoxide production (Fig. 2B).
We also silenced JNK or p38, and then monitored mitochondrial superoxide generation. Compared with cells treated with 25 μm anisomycin and transfected with scrambled siRNAs, we found that silencing JNK inhibited mitochondrial superoxide production by 66% (p < 0.05), and silencing p38 had little impact (<5% decrease) on mitochondrial superoxide generation (Fig. 2C). To confirm that JNK induction of mitochondrial superoxide generation occurred in the same temporal domain as JNK activation, we monitored the kinetics of mitochondrial superoxide production during anisomycin-induced stress. Cells were prestained with MitoSOX prior to anisomycin-induced stress. As observed earlier, anisomycin-induced stress resulted in increased mitochondrial superoxide production during the 45 min of stress (Fig. 2D, open diamonds); this production was prevented by pretreatment with 25 mm NAC (Fig. 2D, black squares). Pretreatment with 500 nm SR-3562 also prevented generation of superoxide comparable with cells treated with only anisomycin, but more than cells treated with NAC (Fig. 2D, open squares). The addition of 500 nm SR-3562 10 min following anisomycin introduction prevented a second phase of superoxide generation at the mitochondria (Fig. 2D, open triangles). These data indicate that JNK signaling may be altering mitochondria to promote superoxide generation.
Mitochondrial JNK Signaling Directly Induces ROS Production in Response to Stress
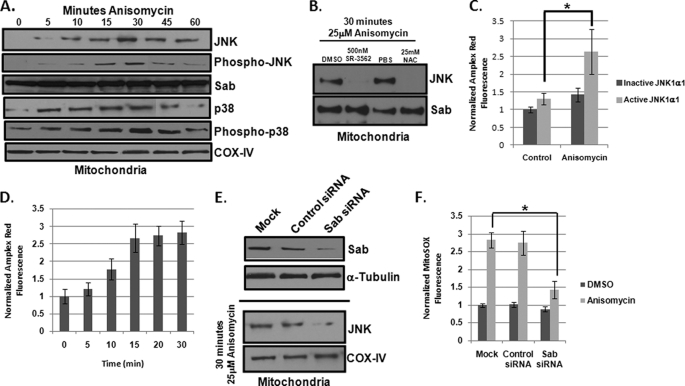
Given that JNK can translocate to the mitochondria (9, 11), we examined if mitochondrial JNK was present during the same time superoxide production was enhanced during anisomycin-induced stress. For this, we isolated mitochondria from HeLa cells at 0, 5, 10, 15, 30, 45, and 60 min post-anisomycin addition. Western blot analysis indicated that JNK was translocated to the mitochondria by 5–10 min post-stress, and MitoJNK was maximal after 30 min (Fig. 3A); moreover, mitochondrial localized JNK was active as indicated by the phospho-JNK (Thr183/Tyr185) Western blot (Fig. 3A). Interestingly, only one JNK species was found on the mitochondria (Fig. 3A). We also examined whether p38 was present at the mitochondria. In Fig. 3A, p38 has similar mitochondrial translocation kinetics to JNK. As expected, this p38 species was bisphosphorylated on Thr180 and Tyr182, indicative of active p38 (Fig. 3A). The relative abundance of the putative mitochondrial scaffold for JNK, Sab, remained constant during the observed 60 min (Fig. 3A). Equivalent mitochondrial loading was assured by Western analysis of relative cyclooxygenase IV (Fig. 3A). In addition, mitochondrial preparation purity was obtained through Western blot analysis; nuclear contamination (histone-H3) was less than 5%, cytosolic contamination (GAPDH) was less than 5%, and microsomal/lysosomal (PDI) contamination was less than 10% on average (data not shown).
FIGURE 3.
Mitochondrial JNK signaling specifically enhances superoxide production. A, anisomycin stress-induced JNK translocation to the mitochondria of HeLa cells. Mitochondria were isolated from HeLa cells at up to 60 min following anisomycin treatment. Mitochondria enrichments were analyzed by Western blotting for the presence of JNK, pJNK, and p38. Cyclooxygenase IV (COX IV) served as a loading control for mitochondrial preaparations. Sab was used as an additional mitochondrial control. B, 500 nm SR-3562 and 25 mm NAC prevented JNK translocation to the mitochondria. Following 30 min of 25 μm anisomycin treatment, mitochondria were isolated from cells that were pretreated with DMSO, 500 nm SR-3562, PBS, and 25 mm NAC. Western blot analysis was performed for the presence of JNK and Sab was used as a mitochondrial loading control. C, addition of active human JNK1α1 to mitochondria isolated from JNK null fibroblasts induced ROS production. Mitochondria from DMSO or anisomycin-treated JNK null fibroblasts were incubated with inactive or active recombinant human JNK1α1. ROS generation was monitored after 30 min of incubation at 30 °C by Amplex Red hydrogen peroxide detection assay. D, anisomycin stress altered mitochondria to promote JNK-mediated ROS production. Mitochondrial enrichments were obtained from JNK null fibroblasts treated with 25 μm anisomycin for up to 30 min. Mitochondria from each time point were then incubated with active JNK1α1 and ROS generation was detected by Amplex Red assay. E, siRNA-mediated knockdown of Sab expression. HeLa cells were transfected with siRNAs specific for Sab or a scrambled control siRNA. Following 72 h of siRNA treatment, protein abundance was monitored by Western blotting for Sab. α-Tubulin served as a loading control. Following mock, control siRNA, or Sab siRNA silencing, cells were treated with 25 μm anisomycin for 30 min, and then mitochondria were harvested. Western blots were performed for JNK. F, mitochondrial superoxide levels were decreased in anisomycin-stressed HeLa cells with reduced levels of Sab. HeLa cells were transfected with control or Sab-specific siRNAs for 72 h. Cells were then treated with 25 μm anisomycin for 45 min and mitochondrial superoxide was monitored using MitoSOX Red.
Given that SR-3562 and NAC were able to inhibit JNK-induced mitochondrial superoxide production, we examined if these two molecules interfered with JNK translocation to the mitochondria. Cells were preincubated with 500 nm SR-3562 or 25 mm NAC prior to 30 min of 25 μm anisomycin, and then mitochondria were isolated from cells. Pre-treatment with either SR-3562 or NAC completely blocked JNK translocation to the mitochondria (Fig. 3B). The absence of either molecule resulted in successful translocation of JNK to the mitochondria (Fig. 3B). Sab was used as a mitochondrial loading control (Fig. 3B).
To determine whether the interaction of JNK with the mitochondria was sufficient for superoxide production, we isolated mitochondria from JNK-null (JNK1−/−, JNK2−/−) MEFs. Although wild type (WT) MEFs were sensitive to anisomycin stress, JNK null MEFs demonstrated no change in cell viability in the presence of 25 μm anisomycin after 4 h (supplemental Fig. S2A). Moreover, JNK null MEFs did not generate ROS to the levels of WT MEFs during anisomycin stress (supplemental Fig. S2B). They exhibited an ∼15% increase in ROS in the presence of anisomycin (supplemental Fig. S2B). A similar trend was observed with mitochondrial superoxide production (supplemental Fig. S2C). Isolated JNK null mitochondria were incubated with either inactive or active recombinant JNK1α1, as previous studies have demonstrated that JNK1α1 could impact mitochondrial physiology (9, 10). We next added the mitochondrial substrate glutamate/ADP and monitored ROS generation by Amplex Red detection of H2O2. Incubation with inactive JNK1α1 did not increase ROS production, and incubation of activated JNK1α1 produced a slight increase in ROS (∼10%) by unstressed mitochondria (Fig. 3C). Taking into consideration that anisomycin-induced cells did not experience a JNK-mediated increase in superoxide generation until after 10 min of stress, we stressed the JNK null MEFs with anisomycin for 15 min, harvest the mitochondria, and monitored ROS generation in the presence and absence of active JNK1α1. We found that active JNK1α1 was able to increase superoxide production by ∼2-fold (*, p < 0.05) on stressed mitochondria, whereas inactive JNK1α1 was unable to increase ROS production with stressed mitochondria (Fig. 3C). Complementation of JNK null MEFs with JNK1α1 (supplemental Fig. S3A) also partially restored the ability of JNK null MEFs to induce ROS generation in response to anisomycin stress (supplemental Fig. S3B). To examine if stress-induced mitochondrial changes were required for mitochondrial JNK signaling, we isolated mitochondria from JNK null MEFs exposed to 25 μm anisomycin for 0, 5, 10, 15, 20, and 30 min and treated the mitochondria with active JNK1α1. We found that it required 15 min for the mitochondria to become “primed” for mitochondrial JNK signaling (Fig. 3D).
To elucidate if mitochondrial JNK signaling was responsible for increased superoxide generation in a cellular model, we targeted Sab. Using siRNA specific for Sab, we knocked down Sab protein levels by greater than 70% in HeLa cells compared with mock transfected and cells transfected with scrambled siRNAs (Fig. 3E). To determine whether Sab knockdown was sufficient to prevent JNK translocation to the mitochondria, Sab was silenced, and then mitochondria were harvested from cells treated with DMSO or 25 μm anisomycin for 30 min. Western blot analysis demonstrated that mock and control siRNA-transfected cells had increased JNK on the mitochondria compared with cells that had decreased Sab (Fig. 3E). These results suggest that Sab knockdown can be used to evaluate MitoJNK signaling in cells. We next monitored mitochondrial superoxide generation in the presence and absence of Sab during anisomycin-induced stress. Cells with reduced Sab protein levels had decreased ROS production (>60%) compared with mock transfected cells or cells transfected with scrambled siRNAs (*, p < 0.05) (Fig. 3F). These results indicated that mitochondrial JNK signaling was sufficient to induce increased superoxide production in stressed cells. Additionally, silencing Sab enhanced cell viability in the presence of 25 μm anisomycin compared with what was observed with JNK knockdown (supplemental Fig. S4).
Mitochondrial JNK Signaling Inhibits Mitochondrial Respiration through Inhibition of Respiratory Complexes
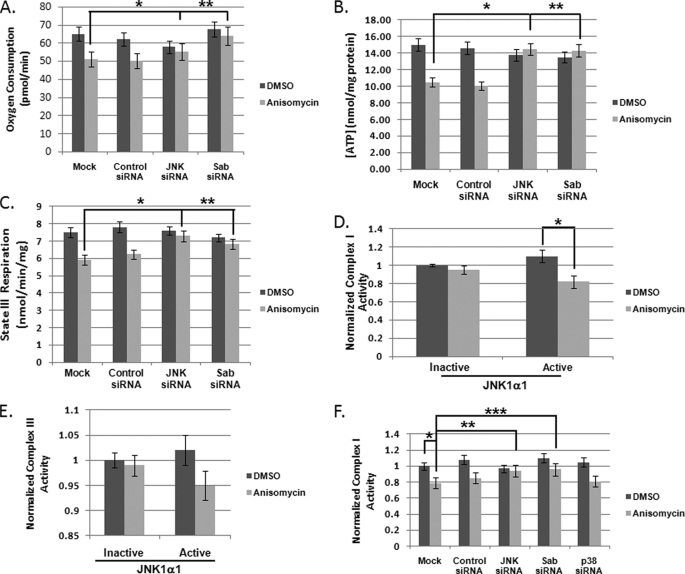
To understand the mechanism by which MitoJNK induced mitochondrial superoxide production, we evaluated the impact of anisomycin stress on mitochondrial metabolism. The major consequences of anisomycin-induced cell death were a decrease in oxygen consumption (Fig. 4A) coupled to a decrease in cellular ATP concentration (Fig. 4B). Silencing JNK expression or Sab expression prevented these events (Fig. 4, A and B). Conversely, silencing p38 had little impact on the decrease in cellular respiration and ATP concentration (data not shown). We also monitored the impact of mitochondrial JNK signaling on state III respiration to determine whether this was the potential source of decreased oxygen consumption and ATP production. State III respiration was examined in the presence and absence of JNK and Sab in anisomycin-treated HeLa cells. Anisomycin treatment led to an ∼20% decrease in state III respiration (Fig. 4C) in the mock or control siRNA-treated cells (*, p < 0.05), which was almost completely alleviated by the reduction in JNK or Sab expression (Fig. 4C), indicating that state III respiration was a contributing factor to decreased mitochondrial metabolism. Finally, to address how MitoJNK may be altering mitochondrial physiology to generate ROS, we analyzed the enzymatic activities of respiratory Complexes I and III of stressed mitochondria from JNK null MEFs in the presence and absence of active JNK1α1, given that JNK null MEFs did not have decreased oxygen consumption in the presence of 25 μm ansiomycin (supplemental Fig. S2D). Fig. 4D presents the normalized Complex I activity after anisomycin treatment (gray bars) compared with DMSO (black bars) in the presence of inactive or active JNK1α1. Anisomycin treatment had no effect on Complex I activity when inactive JNK1α1 was present. However, in the presence of active JNK1α1 Complex I was inhibited nearly 20% (*, p < 0.05) when glutamate/ADP were substrates. Complex III is inhibited ∼5% by the introduction of mitochondrial JNK (Fig. 4E). To determine whether Complex I inhibition was the MitoJNK-dependent cellular mechanism for superoxide generation, we evaluated mitochondrial respiratory complex function with reduced Sab on the mitochondria. Compared with untreated cells, anisomycin-treated cells demonstrated Complex I inhibition of greater than 20% (*, p < 0.05) (Fig. 4F); moreover, decreasing JNK expression prevented Complex I inhibition (**, p < 0.05) (Fig. 4F); however, silencing p38 had no impact on Complex I function (Fig. 4F). Silencing Sab also prevented Complex I inhibition (***, p < 0.05) (Fig. 4F). These data implicate mitochondrial JNK signaling in Complex I inhibition as the potential mechanism for increasing mitochondrial superoxide production during anisomycin stress.
FIGURE 4.
Mitochondrial JNK activity altered mitochondrial physiology promoting superoxide production. A, mitochondrial JNK decreased cellular oxygen consumption. HeLa cells were plated in assay media and treated with 25 μm anisomycin 30 min prior to oxygen consumption measurement. Seventy-two h prior to the experiment siRNAs were used to silence JNK, Sab, or p38. Oxygen consumption was normalized to cell viability immediately following the assay. B, mitochondrial JNK signaling induced decreased ATP production. ATP was extracted from cells following 2 h of anisomycin-induced stress. ATP was quantitated using a luciferase-based assay from Invitrogen. C, mitochondrial JNK signaling decreased State III respiration. Sab and JNK knockdown were achieved by a 72-h incubation with siRNAs specific for each gene. State III respiration was measured following 45 min of anisomycin stress using the Seahorse XF-96 Analyzer with media supplemented with mitochondrial substrates (glutamate/malate/ADP (8 mm) or pyruvate/malate/ADP (8 mm)). D, mitochondrial JNK signaling induced inhibition of respiratory Complex I. Mitochondria were isolated from DMSO and 25 μm anisomycin-treated JNK null fibroblasts. Mitochondria were then incubated with inactive and active JNK1α1. Complex I activity was detected spectrophotometrically. E, mitochondrial JNK is responsible for a decrease in Complex III activity. Mitochondria were isolated from DMSO and 25 μm anisomycin-treated JNK null fibroblasts. Mitochondria were incubated with inactive and active JNK1α1. Complex III activity was monitored by spectrophotometric methods. F, knockdown of JNK or Sab prevents Complex I inhibition in anisomycin-treated HeLa cells. Following gene knockdowns, mitochondria were isolated after 45 min of anisomycin stress. Complex I enzyme activity was then measured.
DISCUSSION
JNK signaling has an established role in the initiation of pro-apoptotic signaling through the activation of transcription factors and phosphorylation of apoptosis-related proteins (2). Recently, mitochondrial translocation of JNK has been implicated in the initiation of apoptosis in several systems (7–12, 33). Our study extends those observations to demonstrate that mitochondrial JNK signaling can also have an impact on mitochondrial physiology leading to increased superoxide generation. We found that JNK increased cellular and mitochondrial superoxide production by up to 80%, and that 20% of generated ROS occurred prior to JNK-mediated superoxide production and in the absence of JNK (Fig. 1, C and E). This was corroborated in anisomycin stressed JNK null MEFs where we observed a 15–20% increase in ROS following anisomycin treatment (supplemental Fig. S2B). This non-JNK-mediated ROS may be the signal that activates JNK in the presence of anisomycin; similarly, mitochondrial oxidants, ROS and reactive nitrogen species, can activate JNK signaling (16, 34–36). Moreover, NAC prevents JNK activation under a variety of stresses (14), and we demonstrate in this work that NAC prevented ROS generation in response to anisomycin stress, including JNK-induced mitochondrial superoxide (Figs. 1 and 2). Therefore, mitochondrial JNK translocation may be a response to this early ROS generation. It has been demonstrated that ROS-induced JNK activation occurs through ASK-1, which is responsive to mitochondrial oxidants, namely ROS generated by mitochondrial respiratory complexes (2, 18, 35, 36). ASK-1 phosphorylates and activates JNK kinases MKK-4 and MKK-7 contributing to the bisphosphorylation of JNK (2, 18).
Given that anisomycin is a translational inhibitor and subsequently, a broad spectrum stress, it should be acknowledged that other kinases may contribute to superoxide production. To address these concerns, we utilized p38 siRNA to eliminate the possibility that p38 contributed significantly to ROS generation (Figs. 1 and 2). To complement this, we utilized a highly selective small molecule inhibitor of JNK that was shown not to inhibit related MAPKs, such as p38 and ERKs. This small molecule inhibitor, SR-3562 (19), was able to inhibit detectable ROS generation following JNK activation (Figs. 1 and 2). Similarly, the use of siRNAs, which specifically silence JNK had a similar result as SR-3562 (Figs. 1 and 2). Additionally, complementation of JNK null MEFs with JNK1α1 was sufficient to restore ROS generation during anisomycin stress (supplemental Fig. S3B). Taken together, it is likely in the time frame investigated in these studies that the JNK-dependent superoxide production was attributable to JNK signaling and not to the action of distinctly different protein kinase signaling pathways.
It has been proposed that JNK can induce ROS generation (19, 20, 37), and that this amplification of superoxide production is part of the cellular response to mitochondrial factors released early in cellular stress (37). This observation is born out in our experiment involving the incubation of active JNK with mitochondria from unstressed and stressed JNK null MEFs. Activated JNK was unable to initiate superoxide production until the cells had been stressed for more than 10 min (Fig. 3D). This would indicate that certain alterations are made to the mitochondria to promote MitoJNK signaling. This could be the result of recruitment of protein kinases to the mitochondria, as phosphorylation is a key regulator of mitochondrial physiology (38, 39). The changes required for MitoJNK signaling could also be a direct result of early changes in oxidative potential in the mitochondria, which may result in the modification of proteins in an oxidative related manner, such as by glutathionylation (40, 41) or by modification of oxidized lipids, namely 4-hydroxyneonal (42, 43). These modifications have been demonstrated to alter mitochondrial physiology during oxidative stress (40–43). Additionally, the precursory changes to the mitochondria may also include oxidative damage to mitochondria enzymes or DNA (16). The alteration could also be a result of caspase cleavage events (37). It is most likely that a combination of these events are required for mitochondrial JNK signaling, as these alterations individually do not explain the need for 10 min to initiate JNK mitochondrial ROS generation. Another intriguing speculation is that during the very early temporal events, JNK activation may be protective to the cell (28). Although our experiments were not designed to specifically address this point, it is worth noting. Identification of these mitochondrial alterations required for mitochondrial JNK signaling is a subject of current studies in our laboratory.
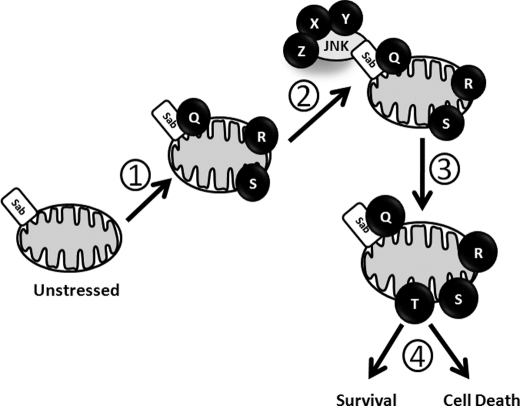
In our proposed model (Fig. 5), proteins Q, R, and S are localized to the mitochondria (Step 1) during the first 10 min of stress in the cell. After 10 min, activated JNK was translocated to the mitochondria (Step 2) and interacted with its scaffold, Sab (5, 6), and other unknown proteins (X, Y, Z). Given that SR-3562 inhibited JNK translocation to the mitochondria (Fig. 3B), it is interesting to hypothesize that JNK catalytic activity is required for translocation to the mitochondria. Moreover, because Sab is also a substrate of JNK (5, 6), one can speculate that JNK mitochondrial translocation is dependent upon the ability of JNK to phosphorylate Sab. JNK can then use established Bcl-2 proteins and BH3 proteins as substrates (2–4) or it can initiate new signaling cascades that result in the inhibition of respiratory Complex I. After 60 min activated JNK leaves the mitochondrial membrane (Step 3) after altering mitochondrial physiology (T), and cell death or survival functions (Step 4) are initiated.
FIGURE 5.
Model of JNK mitochondrial signaling. (1) early cellular stress alters mitochondria physiologically and architecturally, such as the recruitment of yet unidentified proteins (illustrated as Q, R, and S) and translocation of the mitochondrial JNK signaling complex. (2) stress-induced mitochondrial changes promote JNK mitochondrial signaling leading to a stress responsive mitochondrial phenotype (illustrated as T) (3). (4) pro-oxidant metabolism and oxidative stress could contribute to cell death or metabolic alterations and may be essential to cellular survival.
The inhibition of Complex I as a JNK-mediated event is an interesting observation, as tyrosine phosphorylation is thought to be the primary post-translational mechanism for controlling respiratory complex activity (44, 45). Complex I is a 45-subunit protein complex that resides in the inner mitochondrial membrane with projections into the mitochondria inner/outer membrane space and the mitochondrial matrix (45). JNK has been demonstrated, along with Sab, to localize on the outer mitochondrial membrane during stress (6, 9, 10) suggesting that MitoJNK likely has an “outside-in” signaling direction. It may be possible that JNK activates mitochondrial localized tyrosine kinases that use Complex I subunits as substrates to regulate activity (45). It could also be suggested that JNK modification of Bcl-2 and the subsequent loss of mitochondrial membrane potential (2) leads to the decrease in enzymatic activity, as well. The mechanism of JNK-mediated Complex I inhibition remains a point of active investigation.
The inhibition of respiratory Complex I is potentially the mechanism driving JNK-mediated superoxide production (16, 46), as inhibitors of mitochondrial respiratory Complex I, namely rotenone, have been used to generate superoxide demonstrating that Complex I inhibition is a suitable mechanism to induce oxidative stress (15, 16, 46). Although JNK has been demonstrated to also inhibit the pyruvate dehydrogenase complex, it is unlikely that decreased pyruvate flux was the mechanism for reduced Complex I activity, because glutamate/ADP was used as substrates in the assay (Fig. 4) (11). Additionally, HeLa cells display metabolic flexibility. Glutamine, not glucose, is a major mitochondrial metabolite with respect to anaplerosis of the TCA cycle and ATP production, whereas glucose serves a more biosynthetic purpose (47, 48). This manifests in the form of aerobic glycolysis with a majority of glucose-derived pyruvate being diverted to lactate rather than into the mitochondria via the pyruvate dehydrogenase complex (47, 48). Therefore, it is likely that JNK-mediated Complex I inhibition occurs by a mitochondrial means, either signaling or physiological.
The increase of ROS generation by MitoJNK signaling can be a crucial contributing factor to the initiation of cell death. The increased production of ROS can deplete mitochondrial antioxidants contributing to oxidative damage in the mitochondria (16, 46). Oxidative stress in the mitochondria can cause oxidation of cardiolipin contributing to the loss of mitochondrial membrane potential and the release of cytochrome c (16). Moreover, JNK-mediated superoxide production may also lead to the inhibition of phosphatases, such as MAPK phosphatase 1 (MKP1) (13). The reactive cysteine of MAPK phosphatase 1 is sensitive to oxidative stress (13), and oxidation of this cysteine prevents phosphatase activity leading to enhanced half-lives of JNK (and other MAPK) phosphorylation events on the mitochondria. Because MAPK phosphatase 1 activity has been shown to protect cells from death (49), it is interesting to speculate that oxidative inhibition of this phosphatase may be crucial to JNK-mediated cell death.
In this work, we have found that upon stress activation JNK was translocated to the mitochondria resulting in the amplification of mitochondrial superoxide production. The JNK-mediated ROS production may be achieved through MitoJNK signaling-induced inhibition of respiratory Complex I. These findings represent a new niche for JNK signaling. JNK regulation of mitochondrial physiology could be crucial to establishing disease pathology during neurodegeneration, heart disease, and other degenerative human diseases.
Supplementary Material
Acknowledgments
We thank Drs. Lisa Cherry, J. D. Laughlin, and Mariana Figuera-Losada for helpful conversations and comments regarding the manuscript. We acknowledge Dr. Roger Davis (HHMI, University of Massachusetts) for the kind gift of JNK knock-out MEFs used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grant NS057153 (to P. V. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Methods.
- ROS
- reactive oxygen species
- MitoJNK
- mitochondrial JNK
- MEF
- murine embryonic fibroblast
- DMSO
- dimethyl sulfoxide
- DHE
- dihydroethidium
- NAC
- N-acetylcysteine.
REFERENCES
- 1. Weston C. R., Davis R. J. (2007) Curr. Opin. Cell Biol. 19, 142–149 [DOI] [PubMed] [Google Scholar]
- 2. Dhanasekaran D. N., Reddy E. P. (2008) Oncogene 27, 6245–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J. J., Lee J. H., Ko Y. G., Hong S. I., Lee J. S. (2010) Oncogene 29, 561–575 [DOI] [PubMed] [Google Scholar]
- 4. Lei K., Nimnual A., Zong W. X., Kennedy N. J., Flavell R. A., Thompson C. B., Bar-Sagi D., Davis R. J. (2002) Mol. Cell. Biol. 22, 4929–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiltshire C., Gillespie D. A., May G. H. (2004) Biochem. Soc. Trans. 32, 1075–1077 [DOI] [PubMed] [Google Scholar]
- 6. Wiltshire C., Matsushita M., Tsukada S., Gillespie D. A., May G. H. (2002) Biochem. J. 367, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim M. J., Lee K. H., Lee S. J. (2008) FEBS J. 275, 2096–2108 [DOI] [PubMed] [Google Scholar]
- 8. Kharbanda S., Saxena S., Yoshida K., Pandey P., Kaneki M., Wang Q., Cheng K., Chen Y. N., Campbell A., Sudha T., Yuan Z. M., Narula J., Weichselbaum R., Nalin C., Kufe D. (2000) J. Biol. Chem. 275, 322–327 [DOI] [PubMed] [Google Scholar]
- 9. Hanawa N., Shinohara M., Saberi B., Gaarde W. A., Han D., Kaplowitz N. (2008) J. Biol. Chem. 283, 13565–13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aoki H., Kang P. M., Hampe J., Yoshimura K., Noma T., Matsuzaki M., Izumo S. (2002) J. Biol. Chem. 277, 10244–10250 [DOI] [PubMed] [Google Scholar]
- 11. Zhou Q., Lam P. Y., Han D., Cadenas E. (2008) J. Neurochem. 104, 325–335 [DOI] [PubMed] [Google Scholar]
- 12. Zhao Y., Herdegen T. (2009) Mol. Cell Neurosci. 41, 186–195 [DOI] [PubMed] [Google Scholar]
- 13. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 14. Shen H. M., Liu Z. G. (2006) Free Radic. Biol. Med. 40, 928–939 [DOI] [PubMed] [Google Scholar]
- 15. Castello P. R., Drechsel D. A., Patel M. (2007) J. Biol. Chem. 282, 14186–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamanaka R. B., Chandel N. S. (2010) Trends Biochem. Sci. 35, 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murphy M. P. (2009) Biochem. J. 417, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K., Takeda K., Minowa O., Miyazono K., Noda T., Ichijo H. (2001) EMBO Rep. 2, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamenecka T., Jiang R., Song X., Duckett D., Chen W., Ling Y. Y., Habel J., Laughlin J. D., Chambers J., Figuera-Losada M., Cameron M. D., Lin L., Ruiz C. H., LoGrasso P. V. (2010) J. Med. Chem. 53, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ventura J. J., Cogswell P., Flavell R. A., Baldwin A. S., Jr., Davis R. J. (2004) Genes Dev. 18, 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forkink M., Smeitink J. A., Brock R., Willems P. H., Koopman W. J. (2010) Biochim. Biophys. Acta 1797, 1034–1044 [DOI] [PubMed] [Google Scholar]
- 22. Hawkins B. J., Madesh M., Kirkpatrick C. J., Fisher A. B. (2007) Mol. Biol. Cell 18, 2002–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mukhopadhyay P., Rajesh M., Haskó G., Hawkins B. J., Madesh M., Pacher P. (2007) Nat. Protoc. 2, 2295–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pallotti F., Lenaz G. (2007) Methods Cell Biol. 80, 3–44 [DOI] [PubMed] [Google Scholar]
- 25. Kirby D. M., Thorburn D. R., Turnbull D. M., Taylor R. W. (2007) Methods Cell Biol. 80, 93–119 [DOI] [PubMed] [Google Scholar]
- 26. Trumpower B. L., Edwards C. A. (1979) J. Biol. Chem. 254, 8697–8706 [PubMed] [Google Scholar]
- 27. Bagowski C. P., Besser J., Frey C. R., Ferrell J. E., Jr. (2003) Curr. Biol. 13, 315–320 [DOI] [PubMed] [Google Scholar]
- 28. Ventura J. J., Hübner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 21, 701–710 [DOI] [PubMed] [Google Scholar]
- 29. Kang Y. H., Lee S. J. (2008) Oncol. Rep. 20, 637–643 [PubMed] [Google Scholar]
- 30. Kang Y. H., Lee S. J. (2008) J. Cell. Physiol. 217, 23–33 [DOI] [PubMed] [Google Scholar]
- 31. Wagner E. F., Nebreda A. R. (2009) Nat. Rev. Cancer 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 32. Stolk J., Hiltermann T. J., Dijkman J. H., Verhoeven A. J. (1994) Am. J. Respir. Cell Mol. Biol. 11, 95–102 [DOI] [PubMed] [Google Scholar]
- 33. Ito Y., Mishra N. C., Yoshida K., Kharbanda S., Saxena S., Kufe D. (2001) Cell Death Differ. 8, 794–800 [DOI] [PubMed] [Google Scholar]
- 34. Dougherty C. J., Kubasiak L. A., Frazier D. P., Li H., Xiong W. C., Bishopric N. H., Webster K. A. (2004) FASEB J. 18, 1060–1070 [DOI] [PubMed] [Google Scholar]
- 35. Nemoto S., Takeda K., Yu Z. X., Ferrans V. J., Finkel T. (2000) Mol. Cell. Biol. 20, 7311–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soberanes S., Urich D., Baker C. M., Burgess Z., Chiarella S. E., Bell E. L., Ghio A. J., De Vizcaya-Ruiz A., Liu J., Ridge K. M., Kamp D. W., Chandel N. S., Schumacker P. T., Mutlu G. M., Budinger G. R. (2009) J. Biol. Chem. 284, 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao D., Qiao B., Ge Z., Yuan Y. (2005) J. Cell. Biochem. 96, 810–820 [DOI] [PubMed] [Google Scholar]
- 38. Pagliarini D. J., Dixon J. E. (2006) Trends Biochem. Sci. 31, 26–34 [DOI] [PubMed] [Google Scholar]
- 39. Soubannier V., McBride H. M. (2009) Biochim. Biophys. Acta 1793, 154–170 [DOI] [PubMed] [Google Scholar]
- 40. Garcia J., Han D., Sancheti H., Yap L. P., Kaplowitz N., Cadenas E. (2010) J. Biol. Chem. 285, 39646–39654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hurd T. R., Costa N. J., Dahm C. C., Beer S. M., Brown S. E., Filipovska A., Murphy M. P. (2005) Antioxid. Redox. Signal 7, 999–1010 [DOI] [PubMed] [Google Scholar]
- 42. Anuradha C. D., Kanno S., Hirano S. (2001) Free Radic. Biol. Med. 31, 367–373 [DOI] [PubMed] [Google Scholar]
- 43. Patel V. B., Spencer C. H., Young T. A., Lively M. O., Cunningham C. C. (2007) Free Radic. Biol. Med. 43, 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horbinski C., Chu C. T. (2005) Free Radic. Biol. Med. 38, 2–11 [DOI] [PubMed] [Google Scholar]
- 45. Koopman W. J., Nijtmans L. G., Dieteren C. E., Roestenberg P., Valsecchi F., Smeitink J. A., Willems P. H. (2010) Antioxid. Redox Signal. 12, 1431–1470 [DOI] [PubMed] [Google Scholar]
- 46. Grivennikova V. G., Vinogradov A. D. (2006) Biochim. Biophys. Acta 1757, 553–561 [DOI] [PubMed] [Google Scholar]
- 47. Reitzer L. J., Wice B. M., Kennell D. (1979) J. Biol. Chem. 254, 2669–2676 [PubMed] [Google Scholar]
- 48. Dickens M., Rogers J. S., Cavanagh J., Raitano A., Xia Z., Halpern J. R., Greenberg M. E., Sawyers C. L., Davis R. J. (1997) Science 277, 693–696 [DOI] [PubMed] [Google Scholar]
- 49. Kristiansen M., Hughes R., Patel P., Jacques T. S., Clark A. R., Ham J. (2010) J. Neurosci. 30, 10820–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.