Abstract
Basophils mediate many of their biological functions by producing IL-4. However, it is unknown how the Il4 gene is regulated in basophils. Here, we report that CCAAT/enhancer-binding protein α (C/EBPα), a major myeloid transcription factor, was highly expressed in basophils. We show that C/EBPα selectively activated Il4 promoter-luciferase reporter gene transcription in response to IgE cross-linking, but C/EBPα did not activate other known Th2 or mast cell enhancers. We found that the PI3K pathway and calcineurin were essential in C/EBPα-driven Il4 promoter-luciferase gene transcription. Our mutation analyses revealed that C/EBPα drove Il4 promoter-luciferase activity depending on its DNA binding domain. Mutation of the C/EBPα-binding site in the Il4 promoter region abolished C/EBPα-driven Il4 promoter-luciferase activity. Our results further showed that a mutation in nuclear factor of activated T cells (NFAT)-binding sites in the Il4 promoter also negated C/EBPα-driven Il4 promoter-luciferase activity. Our study demonstrates that C/EBPα, in cooperation with NFAT, directly regulates Il4 gene transcription.
Keywords: Bone Marrow, Calcineurin, Cytokine, Gene Expression, PI 3-Kinase, C/EBPα, Il4 Gene, Basophils
Introduction
Basophils are a minor cell population, constituting less than 1% of peripheral blood and bone marrow cells. Basophils have been linked to allergic diseases because of their presence at the site of allergic inflammation. Basophils are found in the lung and sputum of allergic asthmatics, in the nasal mucosa and secretions of allergic rhinitis patients, in the skin lesions of atopic dermatitis patients during allergic late phase reactions, and in various other chronic allergic disease conditions (1). Many novel functions mediated by basophils have been uncovered in recent years. Basophils have been documented to play critical roles in initiating Th2-type immune responses to allergen challenges (2–4). Medzhitov and co-workers (3) reported that upon protease allergen sensitization basophils are activated and recruited into draining lymph nodes where they direct Th2 cell differentiation. More recently, basophils have been demonstrated to function as antigen-presenting cells in initiating Th2 responses (5–7), although two recent reports have challenged the role of basophils in antigen presentation and initiation of Th2 responses (8, 9). Mack and co-workers (10) revealed yet another novel function of basophils; they demonstrated that basophils help to maintain B cell memory responses because of their ability to capture free flowing antigens. The role of basophils in mediating effective memory responses has been verified by a recent report using genetically modified mice that have a 90% reduction in basophil number (9).
Basophils mediate many of their biological functions by producing IL-4 (3, 5–7, 10). Compared with Th2 cells, basophils are more robust IL-4-producing cells on a per cell basis. In the past decade, we have learned a great deal about how CD4+ Th2 cells regulate their Il4 gene transcription. The activation of STAT6 by IL-4 up-regulates GATA3, c-Maf, and JunB expression as naïve CD4+ T cells differentiate into Th2 cells (11–13). The up-regulated GATA3, c-Maf, and JunB together with NFAT5 and activator protein 1 (AP-1) are essential for Th2 cells to transcribe the Il4 gene. However, it is not known whether basophils use the same molecules as Th2 cells to regulate the Il4 gene.
Akashi and co-workers (14, 15) showed that the order of expression of GATA2, an essential transcription factor for early hematopoietic stem cell development (14), and C/EBPα, a required transcription factor for all myeloid cell differentiation (15), has been implicated as the deciding factor in determining the fate of basophils. If GATA2 expression precedes C/EBPα expression at the granulocyte-monocyte progenitor stage, then GATA2 together with C/EBPα will drive basophil differentiation. Conversely, if C/EBPα expression precedes GATA2 expression, then C/EBPα and GATA2 will drive eosinophil differentiation (14). However, it remains to be determined whether or not C/EBPα can directly regulate the Il4 gene along the basophil development pathway.
Regulatory regions of the Il4 gene that confer Th2-specific expression have been identified. Several non-coding regions of the Il4 gene that show hypersensitivity to DNase I digestion have been reported to be critical in Th2-specific Il4 gene expression. These regulatory regions include the intergenic (between the Il13 and the Il4 gene) DNase I-hypersensitive site (HS)/consensus non-coding sequence (CNS)-1, the Il4 promoter (HS1), the intronic enhancer (IE), the proximal 3′ enhancer (HS4), and the distal 3′ enhancer (HS5/CNS-2/HS5a). Among these regions, HS4 has been shown to act as a silencer for Il4 gene transcription (16).
Regulatory regions that confer basophil specific IL-4 expression are virtually unknown. Using the transgenic approach, Kubo and co-workers (17) tested the regulatory regions known for regulating Il4 gene expression in Th2 cells and showed that a 4-kb-long HS4 element together with a 5′ enhancer (−863 to −5448) and the Il4 promoter (−64 to −827) conferred basophil-specific GFP expression. This study, which shows that HS4, a defined silencer of the Il4 gene transcription in Th2 cells (18), is an enhancer of the Il4 gene transcription in basophils, also suggests that basophils might use a different set of regulatory regions and transcription factors to confer basophil-specific Il4 gene expression (17).
Here, we report that C/EBPα, an essential transcription factor for basophil development, can also drive Il4 promoter-luciferase expression in response to FcϵRI receptor cross-linking in a DNA-binding site-specific manner. We also show that C/EBPα was activated by signals generated via the PI3K pathway and the calcium signaling pathway.
EXPERIMENTAL PROCEDURES
Primary Basophil Preparation and Cell Culture
IL-3 (10 μg) was mixed with anti-IL-3 antibody (5 μg; MP2–8F8, BD Pharmingen) at room temperature for 1 min based on published methods (19, 20). The cytokine and antibody mixture (in 0.2–0.3 ml of PBS) was injected into mice via the lateral tail vein. Three days after injection, mice were killed, and bone marrow cells were collected. Primary basophils were purified from the IL-3 complex-treated bone marrow cells by positive selection using biotinylated anti-FcϵRIα antibody (MAR-1) and anti-biotin beads (Miltenyi Biotec Inc., Auburn, CA). We routinely obtained primary basophils with purity greater than 90%.
A rat basophilic leukemic (RBL) cell line was provided to us by Dr. Stephen Dreskin (Allergy and Clinic Immunology, University of Colorado School of Medicine, Denver, CO). IL-4-producing myeloid leukemia progenitor cells (32D.IL-4) were generated in our laboratory by retroviral insertion. 32D.IL-4 cells were grown in RPMI 1640 medium in the presence of G418 (500 ng/ml) and IL-3 (10 ng/ml) plus 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 5% CO2 and 37 °C.
Real Time PCR
Total RNA was isolated from untreated or treated primary basophils using an RNAqueousR-Micro® kit (Ambion Inc., Austin, TX) according to the manufacturer's instructions. cDNA was synthesized by reverse transcription. Real time PCR was performed in an ABI PRISMTM 7700 Sequence Detection System. The following primers were used: Il4: forward, 5′-ACAGGAGAAGGGACGCCAT-3′ and reverse, 5′-GAAGCCCTACAGACGAGCTCA-3′; C/EBPα: forward, 5′-AAAGCCAAGAAGTCGGTGGAC-3′ and reverse, 5′-CTTTATCTCGGCTCTTGCGC-3′; c-maf: forward, 5′-AGCAGTTGGTGACCATGTCG-3′ and reverse, 5′-TGGAGATCTCCTGCTTGAGG-3′; GATA1: forward, 5′-GCCTGTGGCTTGTATCACAAG-3′ and reverse, 5′ CCTCCGCCAGAGTGTTGTAGT-3′; GATA2: forward, 5′-GAATGGACAGAACCGGCC-3′ and reverse, 5′-AGGTGGTGGTTGTCGTCTGA-3′; GATA3: forward, 5′-CCTACCGGGTTCGGATGTAA-3′ and reverse, 5′-TTCACACACTCCCTGCCTTCT-3′; JunB: forward, 5′-CCTGTCTCTACACGACTACA-3′ and reverse, 5′-TTGAGGCTAGCTTCAGAGAT-3′; RBPJκ: forward, 5′-TCCACCAGCCTTACCTTCAC-3′ and reverse, 5′-AGTTAGGACACCACGGTTGC-3′; C/EBPδ: forward, 5′-GCTTTTCAGCCTGGACAGCC-3′ and reverse, 5′-TCGATGGCGCTCTCGTCGT-3′; C/EBPϵ: forward, 5′-GGACCTACTATGAGTGCGAGC-3′ and reverse, 5′-GCTGTTCTTCCCCAGACTCG-3′; and HPRT: forward, 5′-CTCATGGACTGATTATGGACAGGAC-3′ and reverse, 5′-GCAGGTCAGCAAAGAACTTATAGCC-3′. The cycling conditions were 95 °C for 10 min followed by 95 °C for 15 s and 60 °C for 1 min for 40 cycles. The amount of mRNA was expressed as the amount relative to that of HPRT (relative amounts = 2−ΔCT where ΔCT = CTSample − CTHPRT).
Plasmid Construction and Site-directed Mutagenesis
The full lengths of C/EBPα, C/EBPβ, and C/EBPϵ cDNAs were amplified from the cDNA of primary basophils by using the following primers: C/EBPα forward, 5′-AATAGATCTCCATGGAGTCGGCCGACTTC-3′ and C/EBPα reverse, 5′-AATGCGGCCGCCTCACGCGCAGTTGCCCATG-3′; C/EBPβ forward, 5′-AAATAGATCTATGCACCGCCTGCTGGCCTGG-3′ and C/EBPβ reverse, 5′-AAATGCGGCCGCCTAGCAGTGGCCCGCCGAGGC-3′; and C/EBPϵ forward, 5′-AAATAGATCTATGTCCCACGGGACCTACTAT-3′ and C/EBPϵ reverse, 5′-AAATGCGGCCGCTCAGCTGCAGCCCCCGACACC-3′. The PCR product was digested with BglII and NotI (restriction sites are underlined) and cloned into the retroviral expression vector MSCV2.2. C/EBPα deletion mutants were generated by PCR. The primers used were as follows: for ΔTE1 (deletion of transactivation domain 1): forward, 5′-AAATAGATCTCCATGTCTATAGACATCAGCGCCTAC-3′ and reverse, 5′-AATGCGGCCGCCTCACGCGCAGTTGCCCATG-3′; for ΔTE1–2: forward, 5′-AAATAGATCTATGTCCGCGGGGGCGCACGGG-3′ and reverse, 5′-AATGCGGCCGCCTCACGCGCAGTTGCCCATG-3′; for ΔTE1–3: forward, 5′-AAATAGATCTATGCACGCGTCTCCCGCGCACCTGG-3′ and reverse, 5′-AATGCGGCCGCCTCACGCGCAGTTGCCCATG-3′); and for ΔbZIP: forward, 5′-AATAGATCTCCATGGAGTCGGCCGACTTC-3′ and reverse, 5′-AAATGCGGCCGCTCAGGCACCGCTGCCACCGCCGCC-3′.
C/EBPα site-directed mutations of phosphorylation sites, the basic region, and the leucine zipper of the bZIP domain were generated by the overlapping PCR method (21). The primers used for preparing various mutants are listed in supplemental Table S1. All mutations were verified by sequencing.
Luciferase vectors Il4P110 and Il4P575 have been described earlier (22). Il4P-IE, HSS-Il4P, HSS-Il4P-IE, and Il4 minilocus were kindly provided by Dr. Richard A. Flavell (23). The HS4 fragment was amplified from an Il4 genomic fragment and cloned into Il4P575 to generate Il4P-HS4. PCR was used to make the Il4 promoter-luciferase deletion constructs. The primers used were as follows: Il4P reverse, 5′-AATTAAGCTTCAATAGCTCTGTGCCGTCAG-3′; Il4P187 forward, 5′-AATTGGTACCGTTTCATTTTCCAATTGGTCTG-3′; Il4P237 forward, 5′-AATTGGTACCTTTCCTATGCTGAAACTTTGTAG-3′; and Il4P287 forward, 5′-AATTGGTACCTGGCAACCCTACGCTGATAA-3′. The amplified PCR products were digested with KpnI and HindIII and cloned into the luciferase vector pGL3. Linker-scan mutations and binding site mutations of the Il4 promoter were also generated by the overlapping PCR method. The primers used are listed in supplemental Table S2. Constructs made were verified by sequencing.
Full-length cDNA for c-Jun, JunB, GATA1, GATA2, and c-Fos were ordered from Thermo Scientific; C/EBPδ, NFAT1, and NFAT2 were ordered from Addgene (24). Renilla vector PRL-SV40 ordered from Promega (Madison, WI) was used as an internal transfection control. All the plasmids used for transfection were confirmed by sequencing.
Cell Transfection, IgE Cross-linking, and Luciferase Measurement
RBL cells (1 × 107) were electroporated with 10 μg of luciferase plasmid, 0.5 μg of Renilla vector, and 10 μg of transcription factor expression vectors at 320 V and 950 microfarads using a Bio-Rad Gene Pulser II. For transfecting 32D.IL-4 cells, voltage and capacity were set at 350 V and 950 microfarads, respectively. Twenty hours after electroporation, the RBL cells were stimulated by IgE cross-linking with 1 μg/ml IgE (D8406, Sigma) and 1 μg/ml anti-IgE (R35–72, BD Pharmingen) for 6 h. 32D.IL-4 cells were stimulated with 50 ng/ml PMA and 1 μm ionomycin for 5 h. After stimulation, cells were collected, and luciferase activities were measured by an Infinite M1000® microplate reader (Tecan Systems, Inc., San Jose, CA) using the Dual-Luciferase reporter assay system (E1960, Promega). Transcription activity was expressed as the ratio of luciferase activity divided by Renilla activity.
Inhibitor Treatment and IL-4 ELISA Measurement
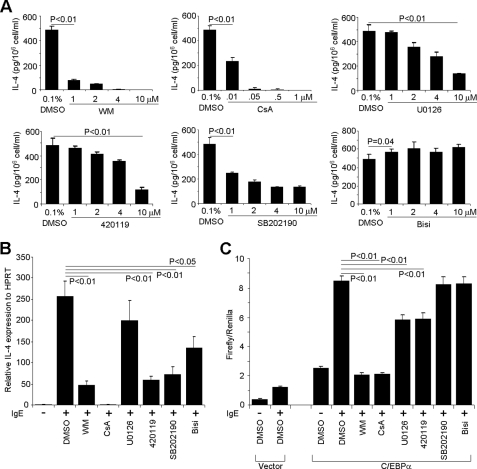
MEK inhibitor (U0126), JNK inhibitor II (catalog number 420119), protein kinase C inhibitor (bisindolylmaleimide I), p38 mitogen-activated protein kinase inhibitor (SB202190), calcineurin inhibitor (cyclosporin A), and PI3K inhibitor (wortmannin) were ordered from Calbiochem. Bone marrow basophils (1 × 106) and RBL cells were treated with inhibitors at the concentration indicated for 2 and 12 h, respectively, before IgE cross-linking. IL-4 protein was measured using an ELISA kit (catalog number 554390, BD Pharmingen).
Generation of Mouse Chimera
C57BL/6 mice were treated with 5-fluorouracil (5 mg/mouse) for 5 days. Bone marrow cells were harvested from the treated mice and cultured in the presence of IL-3 (20 ng/ml), IL-6 (50 ng/ml), and stem cell factor (50 ng/ml) for 1 day. The treated cells were infected with either GFP or C/EBPα virus by spin infection. Infected cells (2 × 106) were injected into irradiated C57BL/6 recipient mice (600 rads) intravenously. Mice were kept under pathogen-free conditions with water containing antibiotics. Four to 6 weeks later, GFP+ basophils were isolated from the bone marrow of the reconstituted mice by FACS sorting and IL-4 ELISA measurement was performed.
siRNA Transfection into Basophils
siRNA was ordered from Thermo Scientific. The control siRNA sequence is 5′-UGGUUUACAUGUCGACUAA-3′, and two specific siRNAs for targeting the sequences 5′-GAGCCGAGAUAAAGCCAAA-3′ and 5′-CCUGAGAGCUCCUUGGUCA-3′ were used for C/EBPα knockdown. Basophils were isolated and enriched from bone marrow of C57BL/6 mice that were injected with the IL-3 complex. The enriched basophils were cultured in the presence of IL-3 for 2 days and used for siRNA transfection by using a Nucleofector kit (Amaxa, Lonza Ltd.). Twenty-four hours later, the transfected basophils were assayed for IL-4 production by ELISA.
Western Blot Analysis
Purified basophils were used for protein extraction, and 50 μg of whole cell lysates were separated by 4–12% SDS-PAGE followed by blotting to PVDF membrane. The following antibodies were used: anti-C/EBPα (14ΑΑ, Santa Cruz Biotechnology), anti-C/EBPδ (M-17, Santa Cruz Biotechnology), and anti-C/EBPϵ (H-75, Santa Cruz Biotechnology). The blot was detected by enhanced chemiluminescent substrate (Thermo Scientific).
RESULTS
Basophils Might Use Different Transcription Factors from Those Used by Th2 Cells to Transcribe Il4 Gene
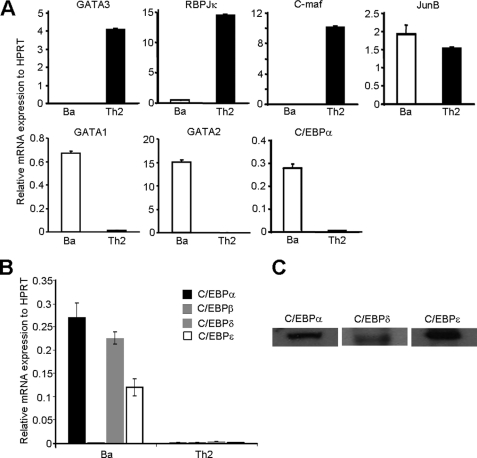
To search for potential transcription factors that might play critical roles in regulating Il4 gene transcription in basophils, we used the candidate gene approach. We compared mRNA expression of known Th2 transcription factors or known transcription factors that are pivotal in basophil development. We showed that GATA3, RBPJκ, and c-Maf were not detectable, whereas JunB, C/EBPα, GATA1, and GATA2 were all highly expressed in basophils (Fig. 1A). In addition to C/EBPα, the C/EBP family consists of several other isoforms. In fact, C/EBPβ has been shown to bind to both human and mouse Il4 promoter and to drive Il4 reporter gene expression in T cells (25–27). Thus, we further examined which of the other C/EBP isoforms is expressed in basophils and showed that C/EBPδ and C/EBPϵ, but not C/EBPβ, were also expressed in basophils (Fig. 1, B and C).
FIGURE 1.
mRNA expression analysis of known Th2 transcription factors in primary basophils and Th2 cells. A, primary basophils (Ba) were isolated by FACS sorting from the bone marrow of C57BL/6J mice. Th2 cells were prepared in vitro by culturing naïve CD4+ T cells in the presence of IL-4 as described previously (47). C/EBPα, c-Maf, JunB, RBPJκ, GATA1, GATA2, and GATA3 mRNAs were quantified by real time RT-PCR. The experiment was repeated at least three times with similar results. B, mRNA expression of C/EBPα, C/EBPβ, C/EBPδ, and C/EBPϵ in basophils and Th2 cells. C, Western blot analysis of C/EBPα, C/EBPδ, and C/EBPϵ in IL-3-expanded bone marrow basophils. The data shown represent mean ± S.D. of three independent experiments.
C/EBPα Drives Il4-Luciferase Gene Transcription
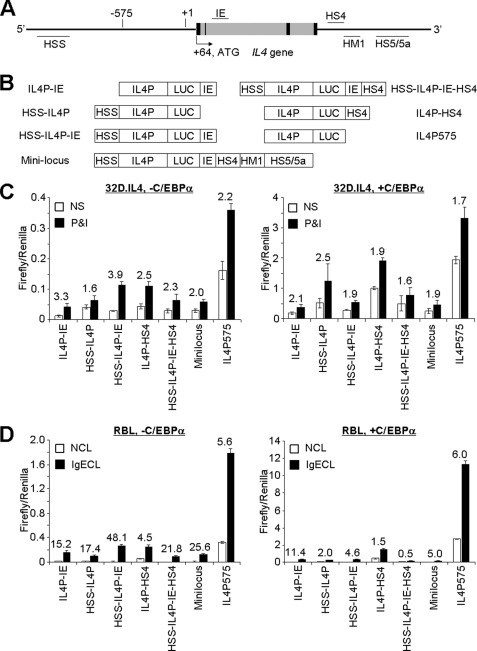
We first examined whether C/EBPα can activate or enhance Il4 gene transcription in myeloid cells. We co-transfected C/EBPα with various Il4 promoter-enhancer luciferase plasmids in murine 32D.IL-4 myeloid progenitor cells, which produced a large amount of IL-4 as the result of retroviral insertional mutagenesis (these cells produced around 5–10 ng/ml/106 cells, a >10-fold increase in IL-4 production compared with the parental 32D cell line). The Il4 promoter-enhancer luciferase used in this experiment included the regulatory regions that have been shown to promote Il4 gene transcription in CD4+ Th2 cells (IL4P and hypersensitive site S (HSS)), mast cells (IE), and basophils (HS4). We showed that C/EBPα drove the Il4 promoter-luciferase reporter gene transcription, but not that of other Il4 promoter-enhancer luciferase reporter genes, after the transfected cells were stimulated with PMA and ionomycin (Fig. 2C). On average, the C/EBPα-driven Il4 promoter-luciferase reporter gene activity was around 8-fold higher than the vector-driven luciferase activity.
FIGURE 2.
C/EBPα activates proximal region of Il4 promoter. A, a schematic representation of regulatory elements of the Il4 gene. The map of DNase I-HSS and IE region as indicated. Black and gray boxes indicate the exon and intron of the IL4 gene, respectively. B, Il4-luciferase constructs used in this experiment. C, transfection of 32D.IL-4 cells with the Il4-luciferase constructs as indicated. The transfected cells were stimulated with or without (not stimulated is abbreviated as NS) PMA (50 ng/ml) and ionomycin (1 μm) (P&I) for 5 h. D, transfection of RBL cells. The transfected cells were cross-linked with or without (NCL) IgE and anti-IgE (IgECL) for 6 h. The number above the bars indicates the relative -fold of induction by stimulation. The mean ± S.D. value indicates one experiment with triplicate samples. Data represent three independent experiments with similar results.
The 32D.IL-4 cells did not express FcϵRI receptor. To test whether C/EBPα could drive the Il4 promoter-luciferase reporter gene transcription in response to IgE cross-linking (a more physiological stimulation), we used an RBL cell line. RBL cells have been shown to produce IL-4 in response to IgE cross-linking (28). We showed that C/EBPα directed the Il4 promoter-luciferase reporter gene transcription in a similar manner in response to IgE cross-linking (Fig. 2, C and D).
Although the DNA binding sequences of CEBP isoforms are highly conservative, the transactivation domains of CEBP isoforms differ significantly. To test whether other C/EBP isoforms can also activate Il4 gene expression, we performed co-transfection of C/EBPs with Il4 promoter-luciferase in RBL cells. As shown in Fig. 3A, C/EBPδ activated Il4 promoter-luciferase expression albeit less potently compared with C/EBPα. C/EBPϵ also weakly activated Il4 promoter-luciferase expression. C/EBPβ failed to activate Il4 promoter-luciferase expression.
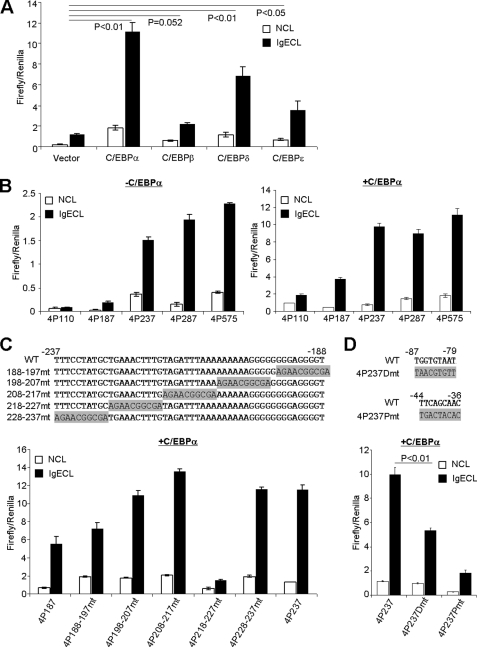
FIGURE 3.
−218 to −227 region and C/EBPα-binding sites are critical for C/EBPα-driven Il4 promoter-luciferase activity. A, RBL cells were co-transfected with Il4P575 luciferase plasmid and C/EBPα, C/EBPβ, C/EBPδ, or C/EBPϵ. B, RBL cells were transfected with or without C/EBPα plasmid. C, RBL cells were transfected with a series of linker-scan mutants (depicted) together with C/EBPα co-transfection. D, RBL cells were transfected with Il4 promoter-luciferase plasmid containing a distal C/EBPα-binding site mutation (4P237Dmt) and a proximal site mutation (4p237Pmt). The mean ± S.D. value indicates one experiment with triplicate samples. The results represent three independent experiments with similar results. NCL, no cross-linking with IgE; IgECL, cross-linking with IgE.
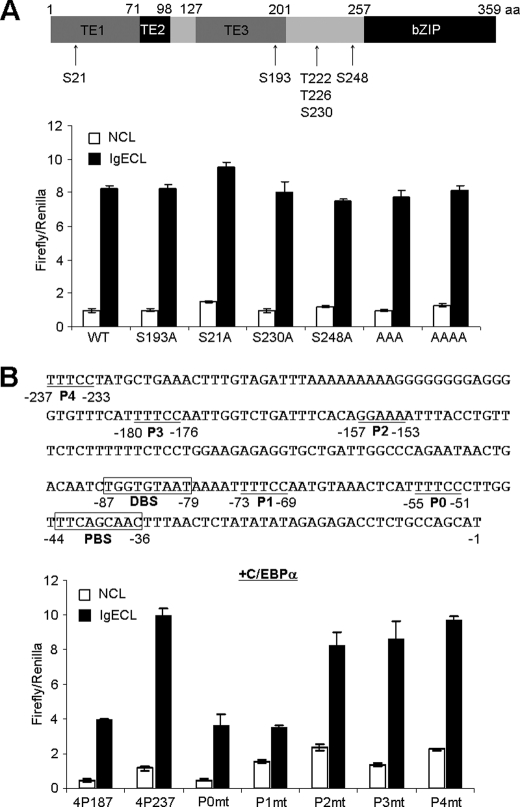
Next, we performed deletion mutation analysis to demonstrate that the −237 to +1 region of the Il4 promoter was crucial in mediating the C/EBPα-driven luciferase transcription (Fig. 3B) and was a potentially important element in the response to IgE cross-linking located in the −237 to −187 region (Fig. 3B, left). Using the linker-scan mutation method (29), we narrowed the region that was critical in enhancing C/EBPα-driven Il4 promoter-luciferase gene transcription to −227 to −218 (Fig. 3C). Surprisingly, the region did not contain a C/EBPα-binding site. To determine whether C/EBPα-binding sites are imperative in mediating Il4 reporter gene transcription, we mutated a previously identified distal C/EBPβ-binding site (TGGTGTAT→TAACGTGTT; located in the −87 to −79 region) and found that the mutation reduced the C/EBPα-driven luciferase gene transcription by 50% (Fig. 3D). Mutation of the proximal C/EBPα-binding site (TTCAGCAAC→TGACTACAC; located in the −44 to −36 region) almost abolished the C/EBPα-driven luciferase expression (Fig. 3D).
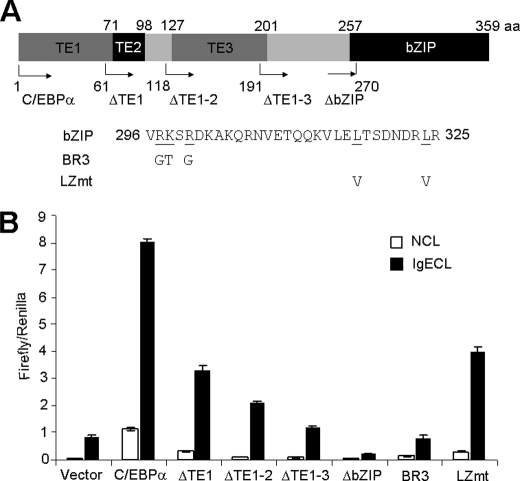
C/EBPα protein contains three transcription activation domains (TE1–3) and a basic leucine zipper domain (Fig. 4A). To determine which domain(s) is essential for driving the Il4 gene transcription, we performed deletion mutation analysis and found that C/EBPα lacking TE1 lost about half of its activity, C/EBPα lacking TE1–2 lost about two-thirds of its activity, and C/EBPα lacking all three TE domains completely failed to drive the Il4 gene transcription (Fig. 4B). These results support that C/EBPα is likely to directly interact with RNA polymerase II. Mutants lacking the bZIP domain were not only completely unable to initiate the Il4 gene transcription, but they also abolished the basal levels of Il4 gene transcription driven by endogenous transcription factor serving as a dominant negative mutant (Fig. 4B). The bZIP domain can be further divided into the DNA binding domain and the leucine zipper domain, which is needed for interaction with other transcription factors, such as PU.1, GATA1, RUNX-1, and p21waf1/cip1 (30). We showed that mutation in Arg-297, Lys-298, and Arg-300 (collectively, these mutants are called BR3 (31)) eliminated C/EBPα-driven transcription activity, whereas deletion of the leucine zipper domain only reduced C/EBPα-driven transcription activity by half. Taken together, these results demonstrate that C/EBPα directly regulates Il4 gene transcription.
FIGURE 4.
Activation domain and DNA binding domain of C/EBPα contribute to C/EBPα-driven Il4 promoter-luciferase activity. A, a schematic presentation of the various domains of the C/EBPα protein. ΔTE1, ΔTE1–2, ΔTE1–3, and ΔbZIP indicate that transactivation domain TE1, TE1 and TE2, TE1 to TE3, and bZIP domains were deleted, respectively. BR3 indicates a C/EBPα DNA binding domain mutation with substitution from Arg-297/Lys-298/Arg-300 to Gly-297/Thr-298/Gly-300, respectively. LZmt indicates a C/EBPα leucine zipper domain mutation with substitution from Leu-317/Leu-324 to Val-317/Val-324. B, RBL cells were transfected with various C/EBPα mutants as indicated. The mean ± S.D. value indicates one experiment with triplicate sample. The results represent three independent experiments with similar results. NCL, no cross-linking with IgE; IgECL, cross-linking with IgE.
PI3K Signaling and Calcium Signaling Pathways Are Required for C/EBPα-driven Il4 Gene Transcription
The PKC/MAPK pathway, calcium signaling pathway, and PI3K pathway have been shown to be critical in regulating mast cell degranulation and de novo cytokine production (32, 33). However, the FcϵRI signaling pathways in basophils have not been studied extensively. To determine which signaling pathway is involved in FcϵRI activation-induced IL-4 production by basophils, we injected mice with the IL-3 and anti-IL-3 antibody complex, a method that expands the number of basophils in the bone marrow by 10-fold in 3 days (20). We used the treated bone marrow without further enriching basophils because we found that basophils were the only cell type in the bone marrow that produced IL-4 (data not shown). IL-3 complex-treated bone marrow cells were treated with various inhibitors. We showed that PI3K (both wortmannin and LY294002; supplemental Fig. S1), MEK, JNK, the calcium signaling pathway, and the p38 signaling pathway were critical to IL-4 production by bone marrow basophils after IgE stimulation (Fig. 5A). Identical findings were obtained using the RBL cell line (Fig. 5B).
FIGURE 5.
PI3K pathway and calcineurin are responsible for C/EBPα-driven Il4 promoter-luciferase activity. A, IL-3 complex-treated bone marrow cells (1 × 106) were treated with wortmannin (WM; PI3K inhibitor), cyclosporin A (CsA; calcineurin inhibitor), U0126 (MEK inhibitor), 420019 (JNK inhibitor II), SB202190 (p38 mitogen-activated protein kinase inhibitor), or bisindolylmaleimide I (Bisi; protein kinase C inhibitor) at the indicated concentration for 2 h followed by IgE cross-linking for 6 h. IL-4 production was measured by ELISA. B, inhibitor effects on IL-4 expression in RBL cells. RBL cells (1 × 106) were treated with 10 μm wortmannin, 0.5 μm cyclosporin A, 10 μm U0126, 10 μm 420119, 10 μm SB202190, or 10 μm bisindolylmaleimide I for 12 h before IgE cross-linking for 6 h. IL-4 mRNA was quantified by real time RT-PCR. C, effects of various inhibitors on C/EBPα-driven Il4 promoter-luciferase activity. RBL cells (1 × 107) were transfected with Il4P575 luciferase plasmid (10 μg) and Renilla luciferase plasmid (0.5 μg) plus C/EBPα expression plasmid (10 μg) or vector control (10 μg). After recovery for 12 h, the transfected cells were treated with the indicated inhibitors at the same concentrations as in B for another 12 h. The transfected cells were stimulated with IgE cross-linking. The results represent three independent experiments with similar results.
To further determine which of PI3K, MEK, JNK, calcium signaling pathway, and p38 signaling pathway are essential in C/EBPα-mediated IL-4 production, we examined whether C/EBPα could drive the Il4 promoter-luciferase gene transcription in the presence of various inhibitors. We showed that only the calcium signaling pathway and the PI3K inhibitor nearly abolished the C/EBPα-driven Il4 promoter-luciferase gene transcription, whereas other inhibitors did not show any significant effects. These results demonstrate that the calcium signaling pathway and the PI3K signaling pathway are imperative in the C/EBPα-driven IL-4 production (Fig. 5C). This conclusion is consistent with a recent report (34).
Post-translational modifications of C/EBPα often occur as the result of signaling activation and have been shown to regulate C/EBPα functions (30). Timchenko and co-workers (35) demonstrated that PI3K signaling activated a phosphatase, pp2a, which in turn dephosphorylated C/EBPα at Ser-193. Dephosphorylation at Ser-193 leads to the loss of cyclin-dependent kinases and E2F hepatocytes and thus the loss of C/EBPα-mediated growth inhibition (35). To examine whether dephosphorylation at Ser-193 could affect the C/EBPα-driven Il4 promoter-luciferase activity, we co-transfected RBL cells with various C/EBPα serine/threonine mutants and found that a mutation at Ser-193 did not reduce the ability of C/EBPα to direct Il4 gene transcription (Fig. 6A). Mutation at Ser-21 has been reported to be imperative in detecting ERK signaling (36, 37); Thr-222, Thr-226, and Ser-230 are phosphorylated by GSK3 signaling (38); and Ser-248 is phosphorylated by RAS signaling (39). A single mutation at Ser-21, Ser-230, or Ser-248 did not show any significant effects on C/EBPα-driven Il4 promoter-luciferase activity. Triple mutations at Thr-222, Thr-226, and Ser-230 and even quadruple mutations at Thr-222, Thr-226, Ser-230, and Ser-247 also did not change C/EBPα-driven Il4 promoter-luciferase activity (Fig. 6A).
FIGURE 6.
NFAT-binding site is crucial for C/EBPα-driven Il4 promoter-luciferase activity. A, effects of serine/threonine of C/EBPα on Il4 promoter-luciferase activity. RBL cells were transfected with Il4P575 luciferase plasmid and the various mutants indicated. B, effects of NFAT-binding site mutation on C/EBPα-driven Il4 promoter-luciferase activity. P0 to P4 indicate NFAT-binding sites, and DBS and PBS indicate C/EBPα distal and proximal binding sites, respectively. The results represent two independent experiments with similar results. NCL, no cross-linking with IgE; IgECL, cross-linking with IgE.
Our calcineurin inhibitor experiment prompted us to first explore the possibility that calcineurin might directly dephosphorylate C/EBPα, leading to its activation in a way that resembles how calcineurin dephosphorylates NFAT. Nevertheless, initially we failed to find the typical calcineurin-binding motif (PXIXIT (40)) that exists in NFAT. Next, we observed that C/EBPα is mostly located inside the nucleolus already, and IgE cross-linking did not change its cellular location (data not shown). Thus, we hypothesized that C/EBPα might cooperate with NFAT, a well characterized calcineurin substrate, to drive the Il4 promoter-luciferase transcription. We mutated five NFAT-binding sites (P0 to P4) in the Il4 promoter and found that mutations in the P0 and P1 sites abolished the C/EBPα-driven Il4 promoter-luciferase activity (Fig. 6B). Thus, our data demonstrate that NFAT is crucial for C/EBPα-driven Il4 gene transcription.
C/EBPα Does Not Cooperate with GATA or JunB to Transcribe Il4 Gene
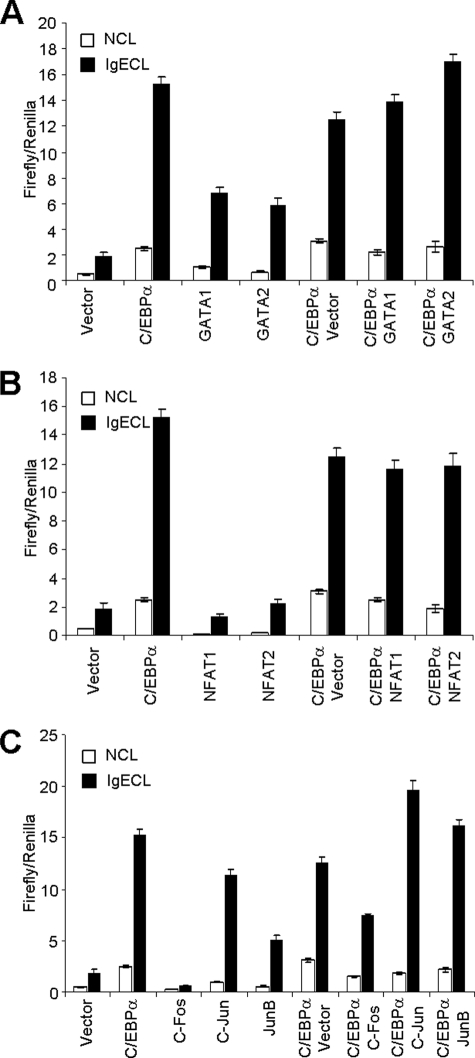
Both GATA1 and GATA2 have been reported to bind to the IE of the Il4 gene (41). JunB was also reported to synergize with c-Maf to promote Th2 cytokine gene transcription (13). Our expression analysis of known Th2 transcription factors demonstrated that GATA2 and JunB were highly expressed in primary basophils (Fig. 1). To determine whether C/EBPα cooperates with GATAT1, GATA2, or JunB, we co-transfected the Il4 promoter-luciferase reporter plasmid and C/EBPα plasmid with GATA1, GATA2, NFAT1–2, or JunB. We did not find any synergistic effect between C/EBPα and GATA1 or between C/EBPα and GATA2 (Fig. 7A). Surprisingly, given that the NFAT-binding site was required for C/EBPα-driven Il4 promoter-luciferase activity, no additive effects were found between C/EBPα and NFAT1 or between C/EBPα and NFAT2 (Fig. 7B). Nor did we find cooperation between C/EBPα and JunB or JunC (Fig. 7C). Repeatedly, we found that c-Fos inhibited the C/EBPα-driven Il4 promoter-luciferase activity (Fig. 7C).
FIGURE 7.
C/EBPα does not cooperate with other known Il4-promoting transcription factors to drive Il4 promoter-luciferase activity. A, effects of GATA1 and GATA2 on C/EBPα-driven Il4 promoter-luciferase activity. B, effects of NFAT1 and NFAT2 on C/EBPα-driven Il4 promoter-luciferase activity. C, effects of c-Fos, c-Jun, and JunB on C/EBPα-driven Il4 promoter-luciferase activity. RBL cells were transfected with Il4P575 luciferase plasmid and various expression plasmids as indicated. The results represent three independent experiments with similar results. NCL, no cross-linking with IgE; IgECL, cross-linking with IgE.
C/EBPα Is Critical for Primary Basophils to Express IL-4
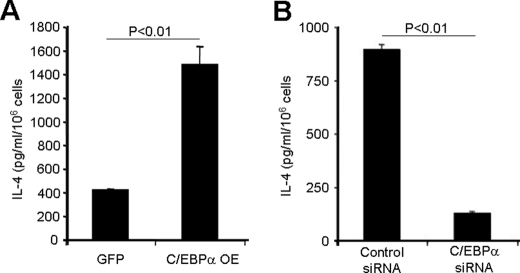
To test whether C/EBPα was capable of transcribing the Il4 gene in primary basophils, we overexpressed C/EBPα in bone marrow stem cells by retroviral infection and used the infected stem cells to generate a mouse radiation chimera. We found that C/EBPα overexpression in basophils resulted in increased IL-4 production (Fig. 8A).
FIGURE 8.
C/EBPα is critical for basophils to produce IL-4. A, GFP+ basophils were isolated from the bone marrow of the reconstituted mice with either C/EBPα or GFP virus-infected bone marrow by FACS sorting. The sorted cells were adjusted to a concentration of 106 cells/ml and stimulated with PMA and ionomycin overnight. OE, overexpression. B, purified IL-3-expanded basophils were transfected, using a Nucleofector kit, with 300 nm control siRNA or C/EBPα-specific siRNAs mixture, and 24 h later, the cells were stimulated with PMA and ionomycin overnight. Supernatants were used for IL-4 measurement by ELISA.
C/EBPα deficiency results in embryonic lethality (42), and C/EBPα-deleted granulocyte-monocyte progenitors failed to differentiate into basophils (15). In our previous work, we have shown that IL-3 together with anti-IL-3 antibody induced an 8–10-fold expansion of basophil lineage-restricted progenitors and basophils (20). We transfected the C/EBPα-specific siRNA into IL-3-expanded basophil precursors and basophils. The result showed that the C/EBPα-knocked down basophils produced much less IL-4 than did control basophils (Fig. 8B). These results demonstrate that C/EBPα plays a critical role in regulating the Il4 gene expression in basophils.
DISCUSSION
In this study, we investigated whether or not basophils use different transcription machinery to drive Il4 gene transcription. We demonstrate that the key Th2 transcription factors, such as GATA3, c-Maf, and RBPJκ, were not expressed in freshly isolated primary basophils. C/EBPα, -δ, and -ϵ along with GATA1, GATA2, and JunB were highly expressed in the freshly isolated primary basophils (Fig. 1). We demonstrate that C/EBPα overexpression was able to drive Il4 promoter-luciferase gene transcription in myeloid IL-4-producing cells in response to IgE-induced activation (Figs. 2 and 3).
Interestingly, the known regulatory regions identified in Th2 cells and mast cells did not show enhancer activities in response to FcϵRI receptor cross-linking in two of the cell lines used, suggesting that myeloid IL-4-producing cells might use different enhancer elements to enhance Il4 gene transcription. This notion is likely because different transcription factors used by basophils might bind to different parts of the Il4 gene. Different regulatory regions that could show more efficient enhancer activity might actually explain why, on a per cell basis, basophils are more robust IL-4-producing cells than Th2 cells. However, using data obtained from the 32D.IL-4 cell line and the RBL cell line to draw such conclusions might be somewhat premature because there is not sufficient information available on the regulatory region of basophils. Kubo and co-workers (17) report that a 4-kb-long HS4 element together with a 5′ enhancer (−863 to −5448) and the Il4 promoter (−64 to −827) conferred basophil-specific GFP expression. Our data did not show that this HS4 region could enhance C/EBPα-driven Il4 promoter-luciferase gene transcription in either of the IL-4-producing myeloid cell lines we used (Fig. 2). Future work designed to study the accessibility of the Il4 locus in primary basophils (such as DNA hypersensitivity mapping and histone modifications) will enhance our understanding of the regulatory regions that enhance Il4 gene transcription in basophils.
The key elements within the proximate Il4 promoter in Th2 cells and mast cells have been mapped within the −100 to −28 region of the Il4 promoter. There are two major composite sites termed P0 (−55 to −51) and P1 (−73 to −69), which contain an NFAT-binding site and an adjacent AP-1-binding site (43). An additional regulatory site within the P0 element is called the Maf recognition element half-site to which c-Maf binds (44). JunB has been shown to bind to the AP-1 site of the P1 element (13). C/EBPβ has also been demonstrated to bind to the sequence that has an overlap with the AP-1 site of the P1 element and the Maf recognition element half-site (25, 45). We found that the C/EBPβ sites were also important in the C/EBPα-driven Il4 promoter-luciferase gene transcription in response to FcϵR1 receptor cross-linking. We demonstrate that the NFAT-binding site inside the P0 element is critical for C/EBPα-driven Il4 promoter-luciferase gene transcription. Our analyses further revealed an additional site (−227 to −218) that is important for C/EBPα-driven Il4 promoter-luciferase gene transcription. Based on these results, we propose that multiple factorial transcription machinery, consisting of C/EBPα, NFAT, and at least one additional unidentified co-transcription factor, carries out the proximate Il4 promoter-driven gene transcription.
C/EBPα could directly or indirectly detect signals trigged by activation of the FcϵRI receptor. Site-directed mutagenesis analysis of serine/threonine residues did not reveal any evidence to support the notion that C/EBPα could sense signals from the PI3K pathway. Dephosphorylation of serine 193 of C/EBPα is PI3K-dependent (35). However, we did not find that mutation at Ser-193 affected C/EBPα-driven Il4 promoter-luciferase gene transcription. Surprisingly, mutation at other reported serine/threonine residues that have been shown to be critical in transducing signals carried by several major signaling pathways did not show significant effects on the C/EBPα-driven Il4 promoter-luciferase gene transcription (Fig. 6 and Refs. 30 and 38). For example, mutation at Ser-21, a serine residue that becomes phosphorylated after ERK activation (37), also did not change the C/EBPα-driven Il4 promoter-luciferase gene transcription. Nevertheless, our data do not rule out that C/EBPα could sense signals from the PI3K pathway via other unidentified serine/threonine residues.
On the other hand, we found that the calcium signaling pathway indirectly affected C/EBPα-driven Il4 promoter-luciferase gene transcription in response to FcϵRI receptor cross-linking. We demonstrate that the calcineurin inhibitor abolished the C/EBPα-driven Il4 promoter-luciferase gene transcription, and the NFAT-binding site mutant dramatically affected C/EBPα-driven Il4 promoter-luciferase gene transcription.
C/EBPα has been shown to be essential in all myeloid cell development; it plays a non-redundant role in basophil development (14). It has been demonstrated that induction of other C/EBP isoforms in myeloid cells is C/EBPα-dependent (46). The lack of other C/EBP isoforms in the absence of C/EBPα might explain why knockdown of C/EBPα in the IL-3-expanded basophil progenitors resulted in a reduction in IL-4 production. Together, our analyses lead us to conclude that C/EBPα, in cooperation with NFAT, directly regulates the Il4 gene in response to FcϵRI receptor activation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Rich Flavell of Yale University for providing the Il4 promoter-luciferase constructs; Dr. Timchenko of Baylor College of Medicine for providing Ser-193 mutant; Dr. C Sutherland of University of Dundee for providing C/EBPα triple mutants at Thr-222, Thr-226, and Ser-230; Dr. Stephen Dreskin of Colorado University for providing the RBL cell line; and Dr. Anthony Gerber of National Jewish Health for allowing us to use the luminometer. We thank J. D. Williams for technique assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AI05917 and RO1 AI068083 (to H. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
- NFAT
- nuclear factor of activated T cells
- CEBP
- CCAAT/enhancer-binding protein
- FcϵRI
- Fcϵ receptor I
- HS
- hypersensitive site
- HSS
- hypersensitive site S
- CNS
- consensus non-coding sequence
- IE
- intronic enhancer
- RBL
- rat basophilic leukemic
- HPRT
- hypoxanthine phosphoribosyltransferase
- bZIP
- basic leucine zipper
- PMA
- phorbol 12-myristate 13-acetate
- AP-1
- activator protein 1
- RBPJκ
- recombination signal binding protein for immunoglobulin κ J region.
REFERENCES
- 1. Arock M., Schneider E., Boissan M., Tricottet V., Dy M. (2002) J. Leukoc. Biol. 71, 557–564 [PubMed] [Google Scholar]
- 2. Luccioli S., Brody D. T., Hasan S., Keane-Myers A., Prussin C., Metcalfe D. D. (2002) J. Allergy Clin. Immunol. 110, 117–124 [DOI] [PubMed] [Google Scholar]
- 3. Sokol C. L., Barton G. M., Farr A. G., Medzhitov R. (2008) Nat. Immunol. 9, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Obata K., Mukai K., Tsujimura Y., Ishiwata K., Kawano Y., Minegishi Y., Watanabe N., Karasuyama H. (2007) Blood 110, 913–920 [DOI] [PubMed] [Google Scholar]
- 5. Sokol C. L., Chu N. Q., Yu S., Nish S. A., Laufer T. M., Medzhitov R. (2009) Nat. Immunol. 10, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perrigoue J. G., Saenz S. A., Siracusa M. C., Allenspach E. J., Taylor B. C., Giacomin P. R., Nair M. G., Du Y., Zaph C., van Rooijen N., Comeau M. R., Pearce E. J., Laufer T. M., Artis D. (2009) Nat. Immunol. 10, 697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshimoto T., Yasuda K., Tanaka H., Nakahira M., Imai Y., Fujimori Y., Nakanishi K. (2009) Nat. Immunol. 10, 706–712 [DOI] [PubMed] [Google Scholar]
- 8. Hammad H., Plantinga M., Deswarte K., Pouliot P., Willart M. A., Kool M., Muskens F., Lambrecht B. N. (2010) J. Exp. Med. 207, 2097–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohnmacht C., Schwartz C., Panzer M., Schiedewitz I., Naumann R., Voehringer D. (2010) Immunity 33, 364–374 [DOI] [PubMed] [Google Scholar]
- 10. Denzel A., Maus U. A., Rodriguez Gomez M., Moll C., Niedermeier M., Winter C., Maus R., Hollingshead S., Briles D. E., Kunz-Schughart L. A., Talke Y., Mack M. (2008) Nat. Immunol. 9, 733–742 [DOI] [PubMed] [Google Scholar]
- 11. Kim J. I., Ho I. C., Grusby M. J., Glimcher L. H. (1999) Immunity 10, 745–751 [DOI] [PubMed] [Google Scholar]
- 12. Zheng W., Flavell R. A. (1997) Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 13. Li B., Tournier C., Davis R. J., Flavell R. A. (1999) EMBO J. 18, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwasaki H., Mizuno S., Arinobu Y., Ozawa H., Mori Y., Shigematsu H., Takatsu K., Tenen D. G., Akashi K. (2006) Genes Dev. 20, 3010–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M. L., Dayaram T., Owens B. M., Shigematsu H., Levantini E., Huettner C. S., Lekstrom-Himes J. A., Akashi K., Tenen D. G. (2004) Immunity 21, 853–863 [DOI] [PubMed] [Google Scholar]
- 16. Ansel K. M., Djuretic I., Tanasa B., Rao A. (2006) Annu. Rev. Immunol. 24, 607–656 [DOI] [PubMed] [Google Scholar]
- 17. Yagi R., Tanaka S., Motomura Y., Kubo M. (2007) Mol. Cell. Biol. 27, 8087–8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ansel K. M., Greenwald R. J., Agarwal S., Bassing C. H., Monticelli S., Interlandi J., Djuretic I. M., Lee D. U., Sharpe A. H., Alt F. W., Rao A. (2004) Nat. Immunol. 5, 1251–1259 [DOI] [PubMed] [Google Scholar]
- 19. Finkelman F. D., Madden K. B., Morris S. C., Holmes J. M., Boiani N., Katona I. M., Maliszewski C. R. (1993) J. Immunol. 151, 1235–1244 [PubMed] [Google Scholar]
- 20. Ohmori K., Luo Y., Jia Y., Nishida J., Wang Z., Bunting K. D., Wang D., Huang H. (2009) J. Immunol. 182, 2835–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urban A., Neukirchen S., Jaeger K. E. (1997) Nucleic Acids Res. 25, 2227–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang H., Hu-Li J., Chen H., Ben-Sasson S. Z., Paul W. E. (1997) J. Immunol. 159, 3731–3738 [PubMed] [Google Scholar]
- 23. Lee G. R., Fields P. E., Flavell R. A. (2001) Immunity 14, 447–459 [DOI] [PubMed] [Google Scholar]
- 24. Monticelli S., Rao A. (2002) Eur. J. Immunol. 32, 2971–2978 [DOI] [PubMed] [Google Scholar]
- 25. Berberich-Siebelt F., Klein-Hessling S., Hepping N., Santner-Nanan B., Lindemann D., Schimpl A., Berberich I., Serfling E. (2000) Eur. J. Immunol. 30, 2576–2585 [DOI] [PubMed] [Google Scholar]
- 26. Davydov I. V., Krammer P. H., Li-Weber M. (1995) J. Immunol. 155, 5273–5279 [PubMed] [Google Scholar]
- 27. Li-Weber M., Krammer P. H. (2003) Nat. Rev. Immunol. 3, 534–543 [DOI] [PubMed] [Google Scholar]
- 28. Erickson J., Kane P., Goldstein B., Holowka D., Baird B. (1986) Mol. Immunol. 23, 769–781 [DOI] [PubMed] [Google Scholar]
- 29. Rooney J. W., Hoey T., Glimcher L. H. (1995) Immunity 2, 473–483 [DOI] [PubMed] [Google Scholar]
- 30. Khanna-Gupta A. (2008) Blood Cells Mol. Dis. 41, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paz-Priel I., Cai D. H., Wang D., Kowalski J., Blackford A., Liu H., Heckman C. A., Gombart A. F., Koeffler H. P., Boxer L. M., Friedman A. D. (2005) Mol. Cancer Res. 3, 585–596 [DOI] [PubMed] [Google Scholar]
- 32. Gilfillan A. M., Tkaczyk C. (2006) Nat. Rev. Immunol. 6, 218–230 [DOI] [PubMed] [Google Scholar]
- 33. Kalesnikoff J., Huber M., Lam V., Damen J. E., Zhang J., Siraganian R. P., Krystal G. (2001) Immunity 14, 801–811 [DOI] [PubMed] [Google Scholar]
- 34. Kuroda E., Antignano F., Ho V. W., Hughes M. R., Ruschmann J., Lam V., Kawakami T., Kerr W. G., McNagny K. M., Sly L. M., Krystal G. (2011) J. Immunol. 186, 323–332 [DOI] [PubMed] [Google Scholar]
- 35. Wang G. L., Iakova P., Wilde M., Awad S., Timchenko N. A. (2004) Genes Dev. 18, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Radomska H. S., Bassères D. S., Zheng R., Zhang P., Dayaram T., Yamamoto Y., Sternberg D. W., Lokker N., Giese N. A., Bohlander S. K., Schnittger S., Delmotte M. H., Davis R. J., Small D., Hiddemann W., Gilliland D. G., Tenen D. G. (2006) J. Exp. Med. 203, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ross S. E., Radomska H. S., Wu B., Zhang P., Winnay J. N., Bajnok L., Wright W. S., Schaufele F., Tenen D. G., MacDougald O. A. (2004) Mol. Cell. Biol. 24, 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu H. K., Perrier S., Lipina C., Finlay D., McLauchlan H., Hastie C. J., Hundal H. S., Sutherland C. (2006) BMC Mol. Biol. 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Behre G., Singh S. M., Liu H., Bortolin L. T., Christopeit M., Radomska H. S., Rangatia J., Hiddemann W., Friedman A. D., Tenen D. G. (2002) J. Biol. Chem. 277, 26293–26299 [DOI] [PubMed] [Google Scholar]
- 40. Mulero M. C., Aubareda A., Orzáez M., Messeguer J., Serrano-Candelas E., Martínez-Hoyer S., Messeguer A., Pérez-Payá E., Pérez-Riba M. (2009) J. Biol. Chem. 284, 9394–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henkel G., Brown M. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7737–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang P., Iwama A., Datta M. W., Darlington G. J., Link D. C., Tenen D. G. (1998) J. Exp. Med. 188, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss D. L., Brown M. A. (2001) Immunol. Rev. 179, 35–47 [DOI] [PubMed] [Google Scholar]
- 44. Ho I. C., Hodge M. R., Rooney J. W., Glimcher L. H. (1996) Cell 85, 973–983 [DOI] [PubMed] [Google Scholar]
- 45. Li-Weber M., Salgame P., Hu C., Davydov I. V., Laur O., Klevenz S., Krammer P. H. (1998) J. Immunol. 161, 1380–1389 [PubMed] [Google Scholar]
- 46. Scott L. M., Civin C. I., Rorth P., Friedman A. D. (1992) Blood 80, 1725–1735 [PubMed] [Google Scholar]
- 47. Zhang Y., Apilado R., Coleman J., Ben-Sasson S., Tsang S., Hu-Li J., Paul W. E., Huang H. (2001) J. Exp. Med. 194, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.