Abstract
The type I transmembrane protein with epidermal growth factor and two follistatin motifs 2 (TMEFF2) is expressed in brain and prostate and overexpressed in prostate cancer, but its role in this disease is unclear. Several studies have suggested that TMEFF2 plays a role in suppressing the growth and invasive potential of human cancer cells, whereas others suggest that the shed portion of TMEFF2, which lacks the cytoplasmic region, has a growth-promoting activity. Here we show that TMEFF2 has a dual mode of action. Ectopic expression of wild-type full-length TMEFF2 inhibits soft agar colony formation, cellular invasion, and migration and increases cellular sensitivity to apoptosis. However, expression of the ectodomain portion of TMEFF2 increases cell proliferation. Using affinity chromatography and mass spectrometry, we identify sarcosine dehydrogenase (SARDH), the enzyme that converts sarcosine to glycine, as a TMEFF2-interacting protein. Co-immunoprecipitation and immunofluorescence analysis confirms the interaction of SARDH with full-length TMEFF2. The ectodomain does not bind to SARDH. Moreover, expression of the full-length TMEFF2 but not the ectodomain results in a decreased level of sarcosine in the cells. These results suggest that the tumor suppressor activity of TMEFF2 requires the cytoplasmic/transmembrane portion of the protein and correlates with its ability to bind to SARDH and to modulate the level of sarcosine.
Keywords: Cancer Tumor Promoter, Cell Motility, Growth Factors, Sarcosine, Tumor Suppressor, TMEFF2, Prostate Cancer, Sarcosine Dehydrogenase
Introduction
The transmembrane protein with an epidermal growth factor and two follistatin motifs 2 (TMEFF2)3 is an evolutionarily conserved type I transmembrane protein expressed in the embryo (1, 2) and selectively in the adult brain and prostate (3–5). The extracellular (ecto-) domain can be cleaved from the membrane in an ADAM17/γ-secretase-dependent fashion (6, 7) and consists of an epidermal growth factor-like motif and two follistatin motifs. The cytoplasmic domain contains a potential G-protein activation motif (2). A critical role for this protein in tumorigenesis is suggested by the fact that it is up-regulated in a significant fraction of primary and metastatic prostate tumors (3–5, 8). In fact, ectopic expression or the addition of purified recombinant TMEFF2 ectodomain promotes neuronal cell survival (9), cell growth (6), and phosphorylation of erbB4 and ERK1/2 (2, 6). However, it has also been suggested that TMEFF2 functions as a tumor suppressor because ectopic expression of full-length TMEFF2 demonstrates in vitro anti-proliferative effects (4, 10) and suppresses tumor growth in vivo in nude mouse xenografts (10). Consistent with a tumor suppressor activity, Tmeff2 has been shown to be hypermethylated in a number of cancer types (Refs. 11–16 and references therein), and the Tmeff2 promoter is repressed by c-Myc (17).
Recently, sarcosine, a glycine derivative, was identified as a potential marker of prostate cancer progression (18). Sarcosine levels were highest in metastatic cancer, and in urine, its levels were higher in men with prostate cancer than in controls. Importantly, using cell lines, Sreekumar et al. (18) provided evidence that the enzymes involved in sarcosine metabolism act as regulators of cell invasion and therefore as potential therapeutic targets for prostate cancer. The addition of sarcosine or knockdown of sarcosine dehydrogenase (SARDH), the enzyme that converts sarcosine into glycine, in benign prostate epithelial cells enhances invasion. Conversely, lowering the levels of glycine N-methyltransferase, the enzyme that catalyzes the conversion of glycine into sarcosine, in DU145 cells reduces their invasiveness.
Here we present data to demonstrate that TMEFF2 interacts with SARDH and regulates the cellular levels of sarcosine. The data also indicate that there is an association between the ability of TMEFF2 to bind SARDH and modulate the level of sarcosine and its ability to act as a tumor suppressor. Further, this activity requires the transmembrane and/or cytoplasmic portion of the protein. Ectopic expression of TMEFF2 results in monolayer and anchorage-independent growth inhibition and decreased sarcosine-induced cellular motility. However, the TMEFF2 ectodomain fails to bind to SARDH and to modulate the cellular levels of sarcosine and reverts the tumor suppressor phenotype, demonstrating no effect on anchorage-independent growth and an increase in monolayer growth.
EXPERIMENTAL PROCEDURES
Proliferation Assay
Cells were seeded at 3,000–5,000 cells/well in 96-well plates. After incubation for the indicated times, MTT reagent (Sigma) was added at a concentration of 5 mg/ml in phenol red-free RPMI containing 1% FBS. Following a 3.5-h incubation at 37 °C, 200 μl of dimethyl sulfoxide (DMSO) (Sigma) was added to each well, and optical density was measured at 562 nm.
Apoptosis Assay
To induce apoptosis, 30,000 cells/well were plated into 6-well plates and treated with 2 or 3 μm staurosporine (Sigma) for 24 h. Cells were then washed with 1× PBS and stained with 1 μl of annexin V-FITC reagent (BioVision) and 1 μl of a 5 μg/ml propidium iodide solution (Invitrogen) to stain necrotic cells. Cells were analyzed by flow cytometry (FACScan, BD Biosciences).
Anchorage-independent Growth
2,500–3,500 cells were suspended in 0.35% agarose in DMEM containing 5% FBS. This suspension was overlaid onto a solidified layer of 0.4% agarose in a 60-mm plate. Fresh DMEM was maintained on top plates during a 14–21-day incubation, at which time the cells were stained with 0.005% crystal violet, photographed, and counted.
Migration Assays
Cell migration was assayed using cell culture Boyden chambers (BD Biosciences) and NIH 3T3 conditioned medium as chemoattractant. Following a 24-h incubation, cells adhering to the bottom of the membrane were fixed with 70% ethanol, stained with 0.1% crystal violet, and photographed. The inserts were then treated with 10% acetic acid, and absorbance was measured at 562 nm. Migration was also assessed by a wound-healing assay essentially as described (19). To analyze sarcosine-induced migration, the cells were grown in the presence of 50 μm sarcosine or alanine for a total of 96 h, and TMEFF2 expression was induced by the addition of doxycycline (250 ng/ml) for 48 h, before the wound was made.
Affinity Chromatography and Mass Spectrometry Analysis
TMEFF2-Myc-His was overexpressed in FreeStyle 293-F cells (Invitrogen) and captured on nickel-Sepharose 6 Fast Flow beads (GE Healthcare) using 20 mm imidazole-containing buffer. Beads were washed in 20 mm imidazole buffer, and TMEFF2-Myc-His complexes were eluted with 500–800 mm imidazole-containing buffer. Eluted proteins were resolved by SDS-PAGE on 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen) and stained with Imperial protein stain (Pierce). Bands of interest were excised and digested with trypsin (Sigma) and subjected to MALDI-mass spectrometry proteomic analysis using a Voyager DE Pro-MALDI-TOF mass spectrometer. Protein database searching was performed with Mascot peptide mass fingerprint (Matrix Science) against the human Swiss-Prot protein database.
Co-immunoprecipitation
HEK293T cells were transfected with the indicated constructs and lysed in cell lysis buffer (Cell Signaling). The lysate (50 μg) was incubated with antibody-coupled Dynabeads (Invitrogen) for 30 min at 4 °C. Beads were washed with PBS, and the immunoprecipitated proteins, remaining on the beads, were eluted in 50 mm glycine, pH 2.8, or by heating at 70 °C for 10 min in 50 mm glycine, pH 2.8, plus NuPAGE LDS sample buffer (Invitrogen), resolved by SDS-PAGE, and analyzed by Western blot using the appropriate primary horseradish peroxidase (HRP)-conjugated antibody. For co-immunoprecipitation analysis using LNCaP or 22Rv1 lysates, 100–250 μg of total protein was used, and elution of the target protein was in SDS sample buffer. To increase sensitivity, subsequent Western blot analysis was performed using the appropriate primary antibody and a light chain-specific secondary antibody (Jackson ImmunoResearch Laboratories).
Immunostaining and Live Imaging
Cells were seeded on poly-l-lysine- and laminin (Sigma)-coated coverslips, transfected with TMEFF2 and SARDH-Myc-His expression constructs, and incubated for 24–48 h. Cells were fixed with cold 4% paraformaldehyde in PBS (USB/Affymetrix), permeabilized with 0.1% Triton X-100 in PBS, and blocked with 5% normal goat serum in 0.1% PBS + 0.1% Tween-20. Samples were incubated with the indicated primary antibodies overnight at 4 °C and then with the corresponding goat anti-mouse FITC (Santa Cruz Biotechnology) or goat anti-rabbit Alexa Fluor 568 (Invitrogen) secondary antibody at room temperature and mounted with medium containing DAPI (Santa Cruz Biotechnology). Images were obtained with a Zeiss LSM 510 confocal microscope. For live cell protein imaging, HEK293T cells were seeded in collagen-coated glass bottom culture dishes (MatTek), transfected with SARDH-YFP and TMEFF2-CFP, and incubated for 24–48 h.
Sarcosine Assay
Cells were washed with PBS, lysed in sarcosine assay buffer (MBL International), and briefly sonicated. Insoluble material was removed by centrifugation at 4 °C. The cellular levels of sarcosine were measured in the supernatant using a sarcosine assay kit (MBL International), and the data were normalized to the level of total l-amino acid present in the same supernatant (l-amino acid quantification kit, MBL International).
Statistical Analyses
Data are expressed as mean ± S.D. Differences were analyzed by paired, two-tailed Student's t test. A p value ≤0.05 was considered statistically significant.
RESULTS
Increased Expression of TMEFF2 Inhibits Cell Growth in HEK293T Cells
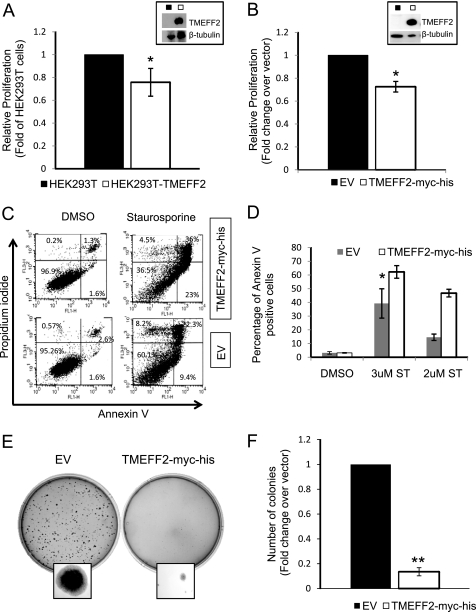
To investigate the function of TMEFF2 in tumorigenesis, we determined whether ectopic expression of this protein could affect cellular proliferation. HEK293T cells stably expressing untagged (TMEFF2-wt) or c-Myc-His-tagged (TMEFF2-Myc-His) TMEFF2 proteins, along with control cells transfected with empty vector or untransfected cells, were generated for this purpose (Table S1 and supplemental Fig. S1A). Overexpression of either untagged (Fig. 1A) or C-terminal c-Myc-His-tagged TMEFF2 (Fig. 1B) in HEK293T cells decreased cell numbers by 20–30% with respect to the untransfected cells or the cells transfected with the empty vector. The presence of the C-terminal c-Myc-His tag did not change the effect of TMEFF2 on cell growth. Therefore, subsequent experiments were done using the c-Myc-His-tagged form of the protein.
FIGURE 1.
TMEFF2 inhibits proliferation and anchorage-independent growth and sensitizes cells to apoptosis. A–F, stable expression of untagged (A) or c-Myc-His (B–F)-tagged form of TMEFF2 decreases proliferation of HEK293T cells (A and B), sensitizes the cell to an apoptotic stimulus (C and D), and inhibits anchorage-independent growth (E and F). Overexpression of TMEFF2 was confirmed by Western blot analysis (A and B, insets). The effect of TMEFF2 on growth (A and B) was determined using an MTT assay after 96 h of growth. The A562 at 96 h was normalized first to the value obtained at zero time (to correct for plating variability) and then to the value obtained for the parental cell line (HEK293T; A) or the cell line carrying the empty vector (EV) (B) as control. The effect of TMEFF2 on apoptosis of HEK293T cells (C and D) was determined in the presence of staurosporine or the vehicle, as a control, by analyzing the number of annexin V-positive cells and comparing it with the numbers obtained when expressing the empty vector. C and D, a representative image of the flow cytometry analysis (C) and percentage of apoptotic cells (D). E and F, a representative image showing anchorage-independent growth (E) and number of colonies formed by HEK293T cells stably expressing TMEFF2-Myc-His or the empty vector as a control (F) after 14 days of growth. Data shown are mean ± S.D. of at least three independent experiments with multiple replicates. Several clones were tested to rule out that the effects are due to the insertion site. *, p < 0.05, and **, p < 0.01.
To further characterize the nature of the alteration in proliferation rate, FACS analysis was used to investigate the effect of TMEFF2 in apoptosis and cell cycle progression. HEK293T cells stably transfected with TMEFF2-Myc-His or with the empty vector as a control were induced to undergo apoptosis with staurosporine, a protein kinase inhibitor that triggers both caspase-dependent and caspase-independent apoptotic pathways (20, 21). The presence of TMEFF2 in HEK293T cells had no effect on the number of apoptotic cells. However, it increased the sensitivity of the cells to staurosporine-induced apoptosis when compared with empty vector transfected cells (Fig. 1, C and D). TMEFF2 had a small effect on cell cycle, resulting in a slightly increased cell number in G1 (supplemental Fig. S1B). Supporting this observation, our preliminary array data indicate that overexpressing TMEFF2 results in increased expression of the cyclin-dependent kinase inhibitor p15 and decreased expression of cyclin E2 (supplemental Fig. S1C).
To further investigate the tumor suppressor potential of TMEFF2, we assessed its ability to promote anchorage-independent growth using a soft agar growth assay. HEK293T cells stably expressing TMEFF2-Myc-His formed ∼5-fold fewer colonies, which were of smaller size than cells carrying the empty vector (Fig. 1, E and F). Thus, TMEFF2 suppresses the formation and the growth of HEK293T colonies in soft agar. Overexpression of TMEFF2 had no effect on the migration or invasion ability of HEK293T cells as measured using Boyden chambers (not shown).
TMEFF2 Inhibits Sarcosine-induced Cell Migration of Prostate Epithelial Cells
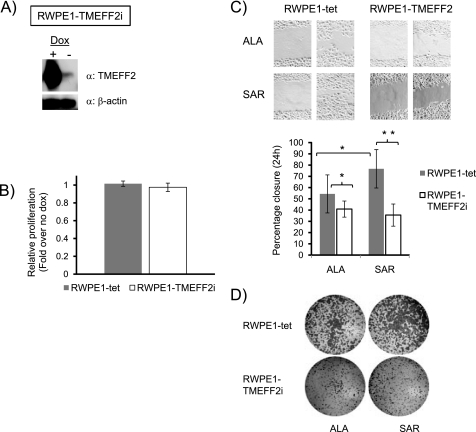
Because the expression of TMEFF2 is mainly restricted to brain and prostate (3–5), we sought to analyze the effect of TMEFF2 overexpression in prostate cells. We selected RWPE1 cells, derived from non-neoplastic human prostatic epithelial cells (22), which express very low levels of endogenous TMEFF2 as demonstrated by quantitative RT-PCR (not shown). Full-length TMEFF2 was introduced into the RWPE1 cells by retroviral gene transfer to generate an RWPE1 cell line that inducibly expresses TMEFF2 with the addition of doxycycline to the growth medium (RWPE1-TMEFF2i). Control cells were transduced with the transactivator construct only (RWPE1-tet). High level of expression of TMEFF2 in the RWPE1-TMEFF2i cell line upon the addition of doxycycline was demonstrated (Fig. 2A). To test whether TMEFF2 affects the growth rate of RWPE1 cells, RWPE1-TMEFF2i cells were grown in the absence (no TMEFF2 expression) and presence (TMEFF2 expression) of doxycycline, and the effect of TMEFF2 on the growth rate was determined. No significant effect of TMEFF2 on the growth rate of RWPE1 cells was observed when compared with the RWPE1-tet cells (Fig. 2B).
FIGURE 2.
TMEFF2 inhibits migration of RWPE cells. A, Western blot demonstrating the induction of TMEFF2 expression in response to doxycycline (Dox, 250 ng/ml) in the RWPE1-TMEFF2i cell line. β-Actin was used as loading control. B, the effect of TMEFF2 overexpression on the growth of RWPE1 cells was determined using an MTT assay after 96 h of growth. The A560 at 96 h was normalized first to the value obtained at zero time (to correct for plating variability) and then to the value obtained for same cells grown in the absence of doxycycline. C, the effect of TMEFF2 on migration was determined using a 24-h wound-healing assay. The cells were grown in the presence of 50 μm alanine (ALA) or sarcosine (SAR) and 250 ng/ml doxycycline to induce the expression of TMEFF2. The RWPE1-tet cell line was used as a control. A representative image (top) and quantification of the results (bottom) are shown. D, migration of cells from a random experimental repeat was also analyzed using Boyden chambers. Cells adhering to the bottom of the membrane were fixed, stained with crystal violet, and photographed. Single cell clones were analyzed and gave similar results. Data shown are mean ± S.D. of three independent experiments with multiple replicates. *, p < 0.05, and **, p < 0.01.
The addition of sarcosine to RWPE cells increases the migration and invasion ability of these cells (18).We therefore tested whether TMEFF2 can reverse the sarcosine-induced migration effect. Briefly, RWPE1-TMEFF2i cells were grown in the presence of sarcosine or alanine and doxycycline to induce TMEFF2 expression before their migration potential was analyzed using a wound-healing assay. The effect of TMEFF2 was investigated by comparing the migration of RWPE1-TMEFF2i cells with the migration ability of the control cell line, RWPE1-tet, both in the presence of doxycycline. The addition of sarcosine resulted in an increase in migration of the RWPE1-tet cells (Fig. 2C) when compared with cells grown in the presence of alanine. Overexpressing TMEFF2 in these cells blocked the increased migration associated with the addition of sarcosine (Fig. 2C). The addition of alanine also had a small effect on migration that was also reversed by TMEFF2 overexpression. Migration data obtained using Boyden chambers confirmed the results of the wound assay (Fig. 2D). These results suggest that TMEFF2 can block the intrinsic and the sarcosine-induced migration potential of RWPE1 cells. In addition, using a Boyden chamber invasion assay, we observed that TMEFF2 overexpression was also able to reverse the intrinsic and the sarcosine-induced invasion ability of the cells (supplemental Fig. S2). It is worth noting that although in HEK293T cells TMEFF2 negatively affects monolayer and anchorage-independent growth but has no effect on migration or invasion, the reverse seems to be true when TMEFF2 is overexpressed in RWPE cells, indicative of the cell line-specific effect of TMEFF2.
TMEFF2 Binds Specifically to Sarcosine Dehydrogenase
To gain insight into the molecular mechanisms of TMEFF2 action, we sought to search for candidate functional partner(s) of TMEFF2 by screening TMEFF2 affinity complexes using mass spectrometry. Freestyle 293-F cells were transfected with TMEFF2-Myc-His, and TMEFF2 complexes were purified using a histidine affinity column, resolved in a polyacrylamide gel, and subjected to MALDI-TOF/MS analysis. Binding to and elution of the TMEFF2 protein from the column were verified by Western blot (supplemental Fig. S3). To identify specific TMEFF2 interactors, we compared TMEFF2 and empty vector affinity eluates and chose those bands that were mainly represented only in the TMEFF2 affinity eluates. Furthermore, to qualify as a specific interactor, a protein had to be identified in at least two out of four independent TMEFF2 affinity/MS analysis. With six peptides displaying a probability-based Mowse score of 53 (23), one of the candidate proteins identified was SARDH. Additional searches provided up to nine different peptides corresponding to SARDH (Table 1).
TABLE 1.
Sarcosine dehydrogenase peptides identified by mass spectrometry analysis of TMEFF2 affinity complexes
| Start to end | m/z | Peptide sequence |
|---|---|---|
| 167–173 | 804.0 | RLMSLGK |
| 174–188 | 1,588.8 | AYGVESHVLSPAETK |
| 189–222 | 3,771.0 | TLYPLMNVDDLYGTLYVPHDGTMDPAGTCTTLAR |
| 228–241 | 1,514.4 | GAQVIENCPVTGIR |
| 242–251 | 1,250.1 | VWTDDFGVRR |
| 286–300 | 1,737.8 | VPLVAMHHAYVVTER |
| 312–320 | 1,076.2 | DHDASVYLR |
| 467–484 | 2,101.4 | NYSVVFPHDEPLAGRNMR |
| 566–587 | 2,398.8 | GAAAVFDMSYFGKFYLVGLDAR |
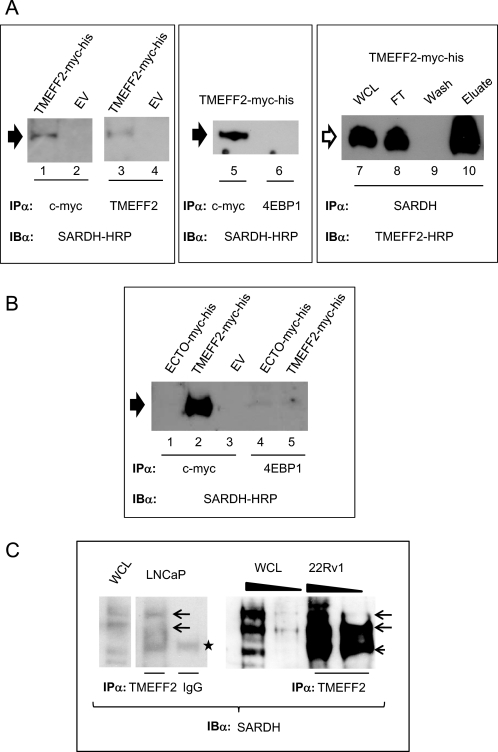
An interaction between TMEFF2 and SARDH was confirmed by co-immunoprecipitation (co-IP) analysis (Fig. 3). A plasmid expressing SARDH from a CMV promoter (pCMV-SARDH) was transiently transfected into HEK293T cells stably expressing the TMEFF2-Myc-His protein or the empty vector, and anti-TMEFF2 immunocomplexes were analyzed for the presence of SARDH by Western blotting. SARDH was clearly detected by the SARDH antibody in the TMEFF2 immunoprecipitates from cells expressing the TMEFF2-Myc-His construct (Fig. 3A, lanes 1 and 3), but not the empty vector (Fig. 3A, lanes 2 and 4), demonstrating that the presence of TMEFF2 is required to detect SARDH. Two different antibodies, c-Myc and a TMEFF2-specific antibody, were used to immunoprecipitate the TMEFF2-Myc-His protein with the same results (Fig. 3A, compare lanes 1 and 2 with lanes 3 and 4). However, when the TMEFF2-specific antibody and strong elution conditions were used, we observed an additional higher molecular weight band specific for SARDH also present in the whole cell lysates (supplemental Fig. S3). The same band was also observed when the co-IP experiments were conducted with endogenous proteins (see below). As negative controls, two isotype-matched, anti-4EBP1 (Fig. 3A, lane 6) and anti-eIF2 (not shown) antibodies were utilized for the IP step. SARDH was not co-immunoprecipitated when either one of these antibodies was used for the IP step, demonstrating that the interaction between SARDH and TMEFF2 is specific. Reciprocal co-IP experiments using anti-SARDH antibodies for immunoprecipitation and TMEFF2 antibodies for Western blot were also performed (Fig. 3A, lanes 7–10) and confirmed the TMEFF2-SARDH interaction (Fig. 3A, lane 10).
FIGURE 3.
TMEFF2 interacts with sarcosine dehydrogenase. A, TMEFF2 associates with SARDH in cells. Cell lysates from HEK293T cells overexpressing SARDH and TMEFF2-Myc-His or the empty vector (EV) as a control were immunoprecipitated with the indicated antibodies (IPα) anti-c-Myc, anti-TMEFF2, anti-SARDH, or anti-4EBP1 (control) and immunoblotted (IBα) with anti-SARDH or anti-TMEFF2 HRP-conjugated antibodies. The size of the band corresponding to SARDH (black arrowhead) lies between the 55–70-kDa marker. The size of the band corresponding to TMEFF2 (empty arrowhead) lies between the 40–55-kDa marker. WCL, whole cell lysate; FT, flow-through. B, the TMEFF2 ectodomain (ECTO) fails to associate with SARDH in cells. A cell lysate from HEK293T cells overexpressing SARDH and ECTO-Myc-His was immunoprecipitated with anti-Myc or anti-4EBP1 (control) and immunoblotted with anti-SARDH-HRP antibody. SARDH is indicated with a black arrowhead. C, association of endogenous TMEFF2 and SARDH proteins. Cell lysates from LNCaP or 22Rv1 cells were immunoprecipitated with anti-TMEFF2 antibody or IgG as a control and immunoblotted with anti-SARDH antibody. The wedge shape indicates increasing amount of lysate used (150–250 μg of total protein). The arrows indicate the position of the major SARDH bands (between the 55–70- and 70–100-kDa markers). The small arrowhead indicates a band with a migration similar to the IgG heavy chain (star).
To further validate the specificity of the TMEFF2-SARDH interaction, we performed co-IP studies as described above with cells that ectopically express the TMEFF2 ectodomain (ECTO-Myc-His; supplemental Fig. S3 for schematic). The results indicate that SARDH was not present in the co-immunoprecipitates obtained from the ECTO-Myc-His-expressing cells (Fig. 3B, lanes 1–5). The ECTO-Myc-His protein is readily secreted into the medium (see below), but it is also abundant in the cellular lysates and able to bind to the nickel or antibody affinity columns (supplemental Fig. S3), ruling out that the lack of interaction reflects the absence of intracellular protein or failure to bind to the columns.
To better establish the physiological relevance of the above results, the TMEFF2-SARDH interaction was also analyzed in two prostate cancer cells, LNCaP and 22RV1, known to express some endogenous TMEFF2. Co-IP studies indicated that SARDH is present in the TMEFF2 co-immunoprecipitates obtained using LNCaP or 22Rv1 cell lysates and therefore that the interaction occurs and is detectable even with the low endogenous levels of these proteins (Fig. 3C). Polyclonal rabbit IgG was used as a negative control. The ability of endogenous TMEFF2 to bind to the antibody affinity column was verified (supplemental Fig. S3). Collectively, these results indicate that TMEFF2 specifically interacts with SARDH and that the presence of the transmembrane and/or cytoplasmic domains is essential for this interaction.
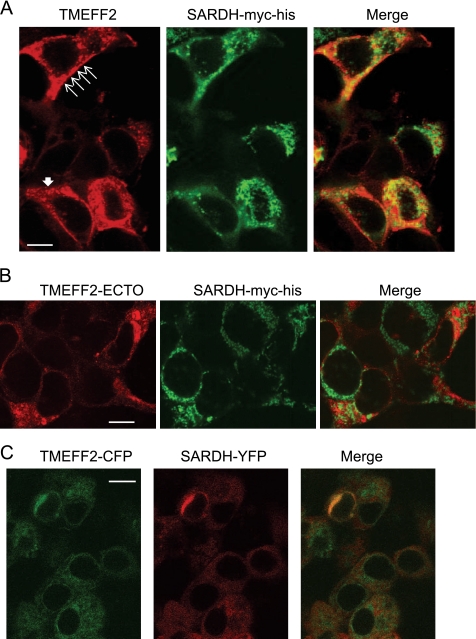
TMEFF2 Co-localizes with Sarcosine Dehydrogenase
The possibility that TMEFF2 and SARDH can temporarily localize to the same cellular compartment was examined by immunofluorescence co-localization studies. HEK293T cells ectopically expressing TMEFF2 were transiently transfected with a tagged SARDH-Myc-His construct and incubated with polyclonal TMEFF2 antibody (to detect TMEFF2) and monoclonal Myc antibody (to detect SARDH). The localization of the proteins was subsequently visualized by confocal microscopy using Alexa Fluor 568- and FITC-conjugated secondary antibodies. Cells expressing TMEFF2 exhibited fluorescence concentrated at the plasma membrane but also in the cytoplasm in a punctate pattern (Fig. 4A and supplemental Fig. S4). Confirming published observations, SARDH mainly localized to the mitochondria (36), with less fluorescence detected in the cytoplasm (Fig. 4A and supplemental Fig. S4). Co-localization of the proteins in the cytoplasm was evidenced by overlapping fluorescence signals (Fig. 4A and supplemental Fig. S4, yellow). As anticipated based on the results from the co-IP analysis, the ectodomain of TMEFF2 did not demonstrate co-localization with SARDH (Fig. 4B).
FIGURE 4.
TMEFF2 co-localizes with sarcosine dehydrogenase. A, HEK293T cells ectopically expressing TMEFF2 and SARDH-Myc-His were fixed and stained with anti-TMEFF2 and anti-c-Myc (to detect SARDH) antibodies. Thin and thick filled arrows point to the membranous and vesicular localization of TMEFF2, respectively. B, HEK293T cells ectopically expressing the TMEFF2 ectodomain (ECTO) and SARDH-Myc-His were treated and processed for immunofluorescent staining as in A. C, in vivo co-localization of TMEFF2 and SARDH. HEK293T cells were transfected with TMEFF2-CFP and SARDH-YFP fusion constructs, and living cells were observed 24 h later. CFP fluorescence is shown as green signal, YFP fluorescence is shown as red signal, and co-localization of CFP and YFP is illustrated by yellow signal. The confocal images in each panel are representative of more than 40 fields observed over four or five different experiments. Scale bars represent 10 μm.
Co-localization of TMEFF2 and SARDH was also examined in living cells. For this purpose, TMEFF2 and SARDH were C-terminally tagged with cyan (CFP) and yellow fluorescent proteins (YFP) to generate TMEFF2-CFP and SARDH-YFP, respectively. The resulting fusion proteins were expressed in HEK293T cells, and the localization of the fluorescent proteins was imaged by confocal microscopy (Fig. 4C and supplemental Fig. S4). Interestingly, when TMEFF2-CFP and SARDH-YFP were co-expressed, they co-localize in an area surrounding the nuclear envelope that could correspond to the Golgi apparatus.
Co-localization of endogenously expressed proteins was examined using LNCaP and 22Rv1 cells as described above and in Cytospin preparations of cell suspensions. As observed with the TMEFF2-overexpressing cells, co-localization of the proteins in the cytoplasm was evidenced by overlapping fluorescence signals (supplemental Fig. S4, yellow).
Overexpression of the Full-length TMEFF2 Results in Decreased Cellular Sarcosine Levels
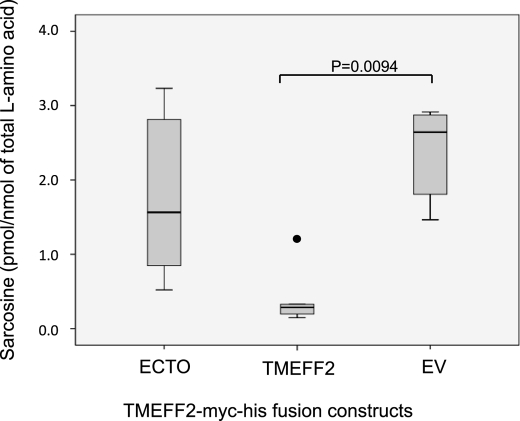
SARDH catalyzes sarcosine to glycine conversion and, consequently, siRNA to SARDH results in increased sarcosine levels. Interestingly, this also results in an increase in the invasion potential of the cells, linking sarcosine metabolism with tumorigenesis (18). Because TMEFF2 interacts with SARDH, we hypothesized that TMEFF2 modulates SARDH activity to promote changes in cell growth and/or invasion. We analyzed changes in sarcosine levels in response to TMEFF2 overexpression. Lysates were prepared from HEK293T cells stably transfected with the TMEFF2-Myc-His and ECTO-Myc-His expression constructs or with the empty vector as a control, and sarcosine levels were determined in the resulting cell lines. Overexpression of the ectodomain (ECTO-Myc-His) does not affect sarcosine levels (Fig. 5). However, overexpression of TMEFF2 resulted in a significant decrease in the amount of sarcosine with respect to the lysates expressing the empty vector control (Fig. 5), whereas having no effect on SARDH expression as measured by Western blot (supplemental Fig. S5). Expression of TMEFF2 also resulted in a small but significant decrease in the amount of sarcosine in RWPE1 cells (supplemental Fig. S5) that could account for the observed effect of TMEFF2 on the migration and invasion properties of this cell line. Although it is possible that the observed decrease in sarcosine levels is due to an indirect effect of TMEFF2, the physical interaction between SARDH and TMEFF2 demonstrated above suggests that the reduction in sarcosine may be due to an increase in SARDH activity mediated by its interaction with TMEFF2. A small but detectable effect of TMEFF2 on the activity of commercially available purified Pseudomonas sp. SARDH enzyme further supports this possibility (supplemental Fig. S5).
FIGURE 5.
TMEFF2 affects the levels of cellular sarcosine. Overexpression of TMEFF2 significantly reduces the levels of sarcosine in HEK293T cells. Circles on the box plot indicate outliers. Data shown are the result of six different experiments with multiple replicates.
The Ectodomain Region of TMEFF2 Acts as a Ligand to Promote Increased Cell Growth
Based on our findings, we also hypothesized that because of the inability of the TMEFF2 ectodomain to interact with SARDH and to affect the levels of cellular sarcosine, it would not demonstrate a tumor suppressor phenotype. We therefore expressed the ectodomain form of TMEFF2 (ECTO-Myc-His) in HEK293T cells and analyzed its effect on cell growth. As reported previously (6), overexpression of the ectodomain resulted in increased monolayer growth when compared with empty vector transfected cells (Fig. 6A). In contrast to full-length TMEFF2, overexpression of the ectodomain did not have any effect on anchorage-independent growth in soft agar or cellular invasion (data not shown).
FIGURE 6.
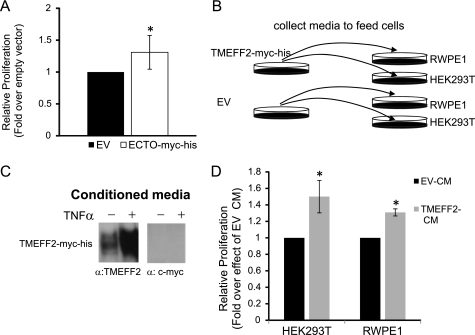
The TMEFF2 ectodomain promotes cell growth. A, the effect of the TMEFF2 ectodomain on growth was determined in HEK293T cells stably transfected with the ECTO-Myc-His construct using an MTT assay. After 96 h of growth, the A562 was measured and normalized first to the value obtained at zero time and then to the value obtained for the cell line carrying the empty vector (EV) as control. B, schematics of the experiment used to determine the effect of the secreted ectodomain on cell growth (shown in D). C, detection of ectodomain sequences in the conditioned medium using the specified antibodies. Because the ectodomain region is produced by TMEFF2 shedding from the membrane, it is not detected by the c-Myc antibody to the C-terminal region. Specificity was further confirmed by the addition of TNFα to induce shedding. (See supplemental Fig. S6.) D, effect of the secreted ectodomain on the growth of HEK293T and RWPE1 cells was determined by MTT assay after 48 h of growth on conditioned medium obtained from cells expressing TMEFF2-Myc-His (TMEFF2 CM) or the empty vector as control (EV-CM). The A562 at 48 h was normalized first to the value obtained at zero time and then to the value obtained for the same cell line grown in the empty vector conditioned medium. Data shown are mean ± S.D. of three independent experiments with multiple replicates. *, p < 0.05.
The ectodomain region expressed throughout these experiments corresponds essentially to the naturally shed ectodomain of the TMEFF2 protein. Because it lacks a transmembrane domain, it is directly secreted and can be detected in the conditioned medium (supplemental Fig. S6). It was therefore likely that the observed effect on monolayer growth was due to the secreted form of the ectodomain acting as a ligand from the outside of the cell. To examine this possibility, we determined the effect that conditioned medium collected from cell cultures overexpressing full-length TMEFF2 had on the growth of two different cell lines, HEK293T and RWPE1 (Fig. 6, B–D). Exponentially growing HEK293T cells transfected with the TMEFF2-Myc-His construct were starved for 24 h, and the conditioned medium was collected and supplemented with 0.4% FBS. The presence of TMEFF2 sequences in the conditioned medium was analyzed by Western blot using antibodies against the TMEFF2 ectodomain or against the c-Myc tag (Fig. 6C). The collected conditioned medium was used to replace the growth medium of cultures of HEK293T and RWPE1 cells, and the growth rate of these cells was measured at different time intervals after the medium replacement. As a control, conditioned medium collected from vector transfected HEK293T cultures was used. The addition of conditioned medium from the TMEFF2-Myc-His-overexpressing cultures resulted in a significant growth increase of HEK293T and RWPE1 cells (Fig. 6D). Similar results were observed when conditioned medium from ECTO-Myc-His-overexpressing cells was used to feed the HEK293T or RWPE1 cell cultures (supplemental Fig. S6). These results suggest that the ectodomain may act as a ligand to promote increased growth rate of HEK293T and RWPE1 cells.
DISCUSSION
Deregulated expression of TMEFF2 has been documented in a variety of tumor types (3, 8, 11–16, 25, and references therein). However, the relationship of TMEFF2 to the biology of tumor development or suppression and the molecular bases of these activities remain unknown. The present study reveals a novel functional and physical interaction between TMEFF2 and SARDH, an enzyme involved in sarcosine metabolism, and characterizes the role of this interaction in the tumorigenic activity of TMEFF2. We demonstrate that TMEFF2 expression results in a decrease in the level of cellular sarcosine and that this effect correlates with its ability to act as a tumor suppressor.
SARDH is responsible for conversion of sarcosine to glycine and is therefore one of the regulators of sarcosine levels in the cell. Because sarcosine is formed as a result of glycine methylation, the decrease in sarcosine levels that we observed with TMEFF2 overexpression could be due to an effect on the global cellular amino acid metabolism and/or methylation activity or to an effect on the activity of the SARDH enzyme. Our data indicating that TMEFF2 physically interacts with SARDH favor the latter possibility. This interpretation bestows an active role for sarcosine in tumorigenesis consistent with data indicating that in cell culture, the addition of sarcosine promotes cell invasion of prostate epithelial cells (18). Our results demonstrate that the ability of TMEFF2 to decrease the cellular sarcosine levels correlates with its function. Expression of full-length TMEFF2 decreases cellular sarcosine levels and leads to a tumor suppressor phenotype, whereas expression of the ectodomain does not alter sarcosine levels and leads to reversion of the tumorigenic phenotype. These results also suggest that the effect of TMEFF2 on sarcosine levels requires the presence of a transmembrane domain and/or the cytoplasmic tail and could possibly be mediated by the G-protein-activating domain present in this region.
As shown under “Results,” the ectodomain promotes cellular proliferation, confirming previous reports (6, 9). Cleavage of the extracellular domain of TMEFF2 is induced by proinflammatory cytokines and regulated by ADAM17 (6, 7) and, as proposed (6), this effect could contribute to explain the opposing results described for TMEFF2, such as growth suppression dependent on full-length TMEFF2, its ability to modulate sarcosine levels, and proliferation dependent on regulated release of the ectodomain, which is unable to modulate sarcosine levels. Interestingly, a soluble isoform of TMEFF2 has also been described (25), and although its role is unknown, we predict that it will not modulate sarcosine levels. The identification of proteins with both pro-oncogenic and anti-oncogenic activities has been previously described, emphasizing the complexity of cellular events that occur during tumorigenesis (26–30). Several mechanisms account for the switch in oncogenic activity including the cellular context, the type of tumor, the activation of different pathways, or the presence of different isoforms with opposing roles (26–30).
Similar to TMEFF2, TMEFF1, the only other known member of the TMEFF family, demonstrates different activities depending on the presence of the cytoplasmic tail and its anchorage to the membrane. In Xenopus, TMEFF1 inhibits TGFβ signaling by blocking the nodal co-receptor Cripto (31). The follistatin and EGF motifs contribute to this effect; however, anchorage to the membrane is also essential for this function. Conversely, TMEFF1 blocking of BMP2-mediated signaling requires the cytoplasmic domain of the protein, whereas deletion of either the follistatin or the EGF motifs does not interfere with this function (32). A function for the TMEFF1 ectodomain has not been described. TMEFF1 and TMEFF2 grossly differ in their tissue distribution. TMEFF1 is more widely distributed than TMEFF2, and even in brain, where both proteins are expressed, they exhibit distinct distribution patterns (33). Nevertheless, their high level of sequence similarity suggests that TMEFF1 and TMEFF2 could be playing similar roles in different tissues and/or developmental stages.
Using confocal microscopy, we have observed TMEFF2 localization to the membrane but also to the cytoplasm, where it appears in a punctate pattern. This vesicle-like immunoreactivity has also been described in neurons (34, 35), and it has been proposed to correspond to vesicles translocating newly synthesized TMEFF2 to the cell surface and/or cleaved TMEFF2 to the nucleus. SARDH has been described essentially as a mitochondrial enzyme (36); however, our results suggest that it can also be found in the cytosol and/or the Golgi apparatus, where it could be interacting with TMEFF2 during trafficking. We predict that this interaction modifies the activity of SARDH and, ultimately, the mitochondrial and/or cytosolic levels of sarcosine. Several mitochondrial enzymes have been described as tumor suppressors, for example, succinate dehydrogenase and fumarase hydratase (37) or a mitochondrial form of the sirtuin deacetylases, SIRT3 (38). Similar to these enzymes, SARDH should be considered a tumor suppressor because its inactivation leads to sarcosine accumulation and increased invasion. Interestingly, the level of SARDH protein has been reported to decrease in hepatocellular carcinoma (39). By binding to SARDH, TMEFF2 could be modulating the activity of SARDH and therefore the cellular level of sarcosine, suggesting that it may function as a “secondary” tumor suppressor. Whether the tumor suppressor activity of TMEFF2 depends entirely on SARDH or on additional interacting protein/signaling pathways is currently under investigation.
The results presented here indicate that the role of TMEFF2 in tumorigenesis correlates, at least partially, with its ability to bind to SARDH and modulate the cellular levels of sarcosine. However, how does sarcosine promote tumor invasion? Sarcosine is an endogenous amino acid with several important biological functions: (i) it participates in one-carbon metabolism essential for protein and nucleotide synthesis and DNA methylation (40), and (ii) it is a competitive inhibitor of the type I glycine transporter (GlyT1), a glycine transporter found in brain (41). It is therefore possible that the addition of exogenous sarcosine and its subsequent metabolism affect the methylation and/or synthetic ability of the cell; alternatively, sarcosine may have a yet unidentified role, for example, similar to its role on GlyT1, as an agonist/antagonist of a factor involved in tumorigenesis. This last possibility is supported by the fact that blocking sarcosine metabolism also promotes cellular invasion due to sarcosine accumulation.
Increasing evidence links sarcosine metabolism with disease state, including cancer (prostate, liver) and brain disease. Although the phenotypic expression of altered sarcosine metabolism is pleiotropic in several cell types, these cell type- and isoform-specific differences suggest that a broad range of biologic responses is mediated by TMEFF2 and sarcosine. Because TMEFF2 is also differentially expressed in brain, it is reasonable to hypothesize that its role there may also be related to sarcosine metabolism. In fact, TMEFF2 was identified in a genome-wide association study as a factor involved in schizophrenia (42). Whether the role of TMEFF2 in schizophrenia is related to its ability to modulate the level of sarcosine and therefore the activity of GlyT1 is not yet certain, but sarcosine is currently being investigated as a treatment for schizophrenia (24). The complex biology of TMEFF2 offers insight into metabolic regulation of cancer and perhaps other disease states. The contribution of different TMEFF2 isoforms to this biology and the regulation of their expression remain to be defined.
Supplementary Material
Acknowledgments
We thank M. Harris and S. Balagate for excellent technical assistance. We are grateful to M.-H. Lee for critical reading of the manuscript, Dr. Q. Lu for providing the RWPE cell line, and Dr. Q. Wu for help with biostatistics.
This work was supported in part by contributions from the Division of Research and Graduate Studies at East Carolina University and the Brody Brothers Foundation fund (to M. J. R.-E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S6, and supplemental Experimental Procedures.
- TMEFF2
- transmembrane protein with epidermal growth factor and two follistatin motifs 2
- SARDH
- sarcosine dehydrogenase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation.
REFERENCES
- 1. Heanue T. A., Pachnis V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6919–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uchida T., Wada K., Akamatsu T., Yonezawa M., Noguchi H., Mizoguchi A., Kasuga M., Sakamoto C. (1999) Biochem. Biophys. Res. Commun. 266, 593–602 [DOI] [PubMed] [Google Scholar]
- 3. Afar D. E., Bhaskar V., Ibsen E., Breinberg D., Henshall S. M., Kench J. G., Drobnjak M., Powers R., Wong M., Evangelista F., O'Hara C., Powers D., DuBridge R. B., Caras I., Winter R., Anderson T., Solvason N., Stricker P. D., Cordon-Cardo C., Scher H. I., Grygiel J. J., Sutherland R. L., Murray R., Ramakrishnan V., Law D. A. (2004) Mol. Cancer. Ther. 3, 921–932 [PubMed] [Google Scholar]
- 4. Gery S., Sawyers C. L., Agus D. B., Said J. W., Koeffler H. P. (2002) Oncogene 21, 4739–4746 [DOI] [PubMed] [Google Scholar]
- 5. Glynne-Jones E., Harper M. E., Seery L. T., James R., Anglin I., Morgan H. E., Taylor K. M., Gee J. M., Nicholson R. I. (2001) Int. J. Cancer. 94, 178–184 [DOI] [PubMed] [Google Scholar]
- 6. Ali N., Knaüper V. (2007) J. Biol. Chem. 282, 37378–37388 [DOI] [PubMed] [Google Scholar]
- 7. Lin H., Wada K., Yonezawa M., Shinoki K., Akamatsu T., Tsukui T., Sakamoto C. (2003) Life Sci. 73, 1617–1627 [DOI] [PubMed] [Google Scholar]
- 8. Mohler J. L., Morris T. L., Ford O. H., 3rd, Alvey R. F., Sakamoto C., Gregory C. W. (2002) Prostate 51, 247–255 [DOI] [PubMed] [Google Scholar]
- 9. Horie M., Mitsumoto Y., Kyushiki H., Kanemoto N., Watanabe A., Taniguchi Y., Nishino N., Okamoto T., Kondo M., Mori T., Noguchi K., Nakamura Y., Takahashi E., Tanigami A. (2000) Genomics 67, 146–152 [DOI] [PubMed] [Google Scholar]
- 10. Elahi A., Zhang L., Yeatman T. J., Gery S., Sebti S., Shibata D. (2008) Int. J. Cancer. 122, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 11. Brücher B. L., Geddert H., Langner C., Höfler H., Fink U., Siewert J. R., Sarbia M. (2006) Int. J. Cancer. 119, 1298–1302 [DOI] [PubMed] [Google Scholar]
- 12. Gonzalo V., Lozano J. J., Muñoz J., Balaguer F., Pellisé M., Rodríguez de Miguel C., Andreu M., Jover R., Llor X., Giráldez M. D., Ocaña T., Serradesanferm A., Alonso-Espinaco V., Jimeno M., Cuatrecasas M., Sendino O., Castellví-Bel S., Castells A. (2010) PLoS One. 5, e8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hellwinkel O. J., Kedia M., Isbarn H., Budäus L., Friedrich M. G. (2008) BJU Int. 101, 753–757 [DOI] [PubMed] [Google Scholar]
- 14. Ivanauskas A., Hoffmann J., Jonaitis L. V., Markelis R., Juozaityte E., Kupcinskas L., Lofton-Day C., Röcken C., Malfertheiner P. (2008) Dig. Liver Dis. 40, 920–926 [DOI] [PubMed] [Google Scholar]
- 15. Suzuki M., Shigematsu H., Shames D. S., Sunaga N., Takahashi T., Shivapurkar N., Iizasa T., Frenkel E. P., Minna J. D., Fujisawa T., Gazdar A. F. (2005) Br. J. Cancer. 93, 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Tsunoda S., Smith E., De Young N. J., Wang X., Tian Z. Q., Liu J. F., Jamieson G. G., Drew P. A. (2009) Oncol. Rep. 21, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 17. Gery S., Koeffler H. P. (2003) J. Mol. Biol. 328, 977–983 [DOI] [PubMed] [Google Scholar]
- 18. Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 19. Rodriguez L. G., Wu X., Guan J. L. (2005) Methods Mol. Biol. 294, 23–29 [DOI] [PubMed] [Google Scholar]
- 20. Belmokhtar C. A., Hillion J., Ségal-Bendirdjian E. (2001) Oncogene 20, 3354–3362 [DOI] [PubMed] [Google Scholar]
- 21. Zhang X. D., Gillespie S. K., Hersey P. (2004) Mol. Cancer. Ther. 3, 187–197 [PubMed] [Google Scholar]
- 22. Bello D., Webber M. M., Kleinman H. K., Wartinger D. D., Rhim J. S. (1997) Carcinogenesis 18, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 23. Pappin D. J., Hojrup P., Bleasby A. J. (1993) Curr. Biol. 3, 327–332 [DOI] [PubMed] [Google Scholar]
- 24. Javitt D. C. (2009) Curr. Opin. Drug Discov. Devel. 12, 468–478 [PubMed] [Google Scholar]
- 25. Quayle S. N., Sadar M. D. (2006) Genomics 87, 633–637 [DOI] [PubMed] [Google Scholar]
- 26. Genander M., Halford M. M., Xu N. J., Eriksson M., Yu Z., Qiu Z., Martling A., Greicius G., Thakar S., Catchpole T., Chumley M. J., Zdunek S., Wang C., Holm T., Goff S. P., Pettersson S., Pestell R. G., Henkemeyer M., Frisén J. (2009) Cell 139, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang Y. K., Schiff R., Ko L., Wang T., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Cancer Res. 68, 7887–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006) Cell Death Differ. 13, 962–972 [DOI] [PubMed] [Google Scholar]
- 29. Niu Y., Altuwaijri S., Lai K. P., Wu C. T., Ricke W. A., Messing E. M., Yao J., Yeh S., Chang C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12182–12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan D., Zhu Q., Luo K. (2009) EMBO J. 28, 3500–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harms P. W., Chang C. (2003) Genes Dev. 17, 2624–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang C., Eggen B. J., Weinstein D. C., Brivanlou A. H. (2003) Dev. Biol. 255, 1–11 [DOI] [PubMed] [Google Scholar]
- 33. Kanemoto N., Horie M., Omori K., Nishino N., Kondo M., Noguchi K., Tanigami A. (2001) Brain Res. Mol. Brain Res. 86, 48–55 [DOI] [PubMed] [Google Scholar]
- 34. Siegel D. A., Huang M. K., Becker S. F. (2002) Int. J. Dev. Neurosci. 20, 373–389 [DOI] [PubMed] [Google Scholar]
- 35. Siegel D. A., Davies P., Dobrenis K., Huang M. (2006) J. Neurochem. 98, 34–44 [DOI] [PubMed] [Google Scholar]
- 36. Bergeron F., Otto A., Blache P., Day R., Denoroy L., Brandsch R., Bataille D. (1998) Eur. J. Biochem. 257, 556–561 [DOI] [PubMed] [Google Scholar]
- 37. King A., Selak M. A., Gottlieb E. (2006) Oncogene 25, 4675–4682 [DOI] [PubMed] [Google Scholar]
- 38. Kim H. S., Patel K., Muldoon-Jacobs K., Bisht K. S., Aykin-Burns N., Pennington J. D., van der Meer R., Nguyen P., Savage J., Owens K. M., Vassilopoulos A., Ozden O., Park S. H., Singh K. K., Abdulkadir S. A., Spitz D. R., Deng C. X., Gius D. (2010) Cancer. Cell. 17, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lim S. O., Park S. J., Kim W., Park S. G., Kim H. J., Kim Y. I., Sohn T. S., Noh J. H., Jung G. (2002) Biochem. Biophys. Res. Commun. 291, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 40. Stover P. J. (2009) J. Nutr. 139, 2402–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H. X., Hyrc K., Thio L. L. (2009) J. Physiol. 587, 3207–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Belouchi A., Raelson J. V., Bradley W. E., Paquin B., Fournier H., Croteau P., Paquin N., Dubois D., Bruat V., Van eerdewegh P., Segal J., Little R. D., Keith T. (September 18, 2008) World Intellectual Property Organization Patent WO/2008/112177 (PCT/US2008/003125)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.