Abstract
Calcium (Ca2+) influx through the plasma membrane store-operated Ca2+ channel ORAI1 is controlled by Ca2+ sensors of the stromal interaction molecule (STIM) family. STIM1 responds to endoplasmic reticulum (ER) Ca2+ store depletion by redistributing and activating ORAI1 from regions of the ER juxtaposed to the plasma membrane. Unlike STIM1, STIM2 can regulate ORAI1 in a store-dependent and store-independent manner, but the mechanism by which this is achieved is unknown. Here we find that STIM2 is translated from a highly conserved methionine residue and is directed to the ER by an incredibly long 101-amino acid signal peptide. We find that although the majority of the total STIM2 population resides on the ER membrane, a second population escapes ER targeting to accumulate as a full-length preprotein in the cytosol, signal peptide intact. Unlike STIM2, preSTIM2 localizes to the inner leaflet of the plasma membrane where it interacts with ORAI1 to regulate basal Ca2+ concentration and Ca2+-dependent gene transcription in a store-independent manner. Furthermore, a third protein comprising a fragment of the STIM2 signal peptide is released from the ER membrane into the cytosol where it regulates gene transcription in a Ca2+-independent manner. This study establishes a new model for STIM2-mediated regulation of ORAI1 in which two distinct proteins, STIM2 and preSTIM2, control store-dependent and store-independent modes of ORAI1 activation.
Keywords: Calcium, Calcium Channels, NFAT Transcription Factor, NF-κB Transcription Factor, Protein Translocation, ORAI1, SOCe, STIM1, STIM2, Basal Ca2+
Introduction
Calcium (Ca2+) entry through store-operated Ca2+ channels (SOCs)2 is essential for gene transcription-induced regulation of cell differentiation and function (1). Stromal interaction molecule 1 (STIM1) plays a critical role in activating the plasma membrane (PM)-localized SOCs of the ORAI family (2–4). Localized predominantly on the endoplasmic reticulum (ER), STIM1 spans the membrane with the N terminus confined to the ER lumen and the longer C terminus facing the cytosol (5). Control over STIM1 activation is mediated by ER Ca2+ store depletion; dissociation of Ca2+ from the N-terminal EF hand Ca2+ binding domain induces oligomerization, which drives STIM1 to precise locations on the ER juxtaposed to the PM (6). From there the cytosolic C terminus recruits ORAI1 by uncovering an integral activation domain (CRAC activation domain (CAD), also known as SOAR or OASF), which is also essential for channel gating (7–10). Ca2+ influx is terminated by store refilling, which returns STIM1 to its resting conformation (11). The essential involvement of STIM1 in regulating the activity of SOCs has been demonstrated by experimental manipulation of STIM1 levels; depletion of STIM1 abrogates store-operated Ca2+ entry (SOCe), overexpression increases SOCe (1, 12), and co-overexpression of STIM1 and ORAI1 results in massive Ca2+ influx that is almost entirely controlled by ER store depletion (4).
In contrast to STIM1, the function of its closely related homolog, STIM2 (13), is enigmatic. Despite almost complete structural conservation between STIM homologs, STIM2 depletion has little effect on SOCe (2, 14), and overexpression has more frequently been demonstrated to inhibit endogenous SOCe rather than to activate it (15–17). Co-overexpression of STIM2 and ORAI1 results in modest SOCe characterized by delayed kinetics and much reduced efficiency compared with STIM1, indicating that the ability of STIM2 to activate ORAI1 in a store-operated manner appears limited (16–19). STIM2 also activates ORAI1 in a store-independent manner, an interaction critical for maintaining basal intracellular Ca2+ homeostasis (17, 20). How STIM2 can simultaneously operate in a store-dependent and store-independent manner is not resolved, and no clear model has yet been presented to explain the functional differences between STIM1 and STIM2.
A recent study indicated that the ability of STIM2 to inhibit endogenous SOCe and delay the kinetics of ORAI1 activation could be transferred to STIM1 by chimeric exchange of a short N-terminal region (16). This region contains a 55–60-amino acid (aa) portion of the luminal domain and also the cleavable signal peptide responsible for ER targeting (13, 16). We hypothesized that differences in the size or structure of the STIM1 and STIM2 signal peptides could be responsible for the functional differences in these two proteins. Here we reveal that the STIM2 signal peptide is directly responsible for the production of three distinct STIM2-derived proteins. Each of these proteins regulates ORAI1 activation in a distinct manner, leading to combinatorial activation of gene transcription.
EXPERIMENTAL PROCEDURES
Reagents
Fura-2AM and Lipofectamine 2000 were from Invitrogen; DAPI, probenecid, carbachol, ionomycin, and phorbol 12-myristate 13-acetate (PMA) were from Sigma; thapsigargin was from EMD Bioscience and (Z-LL)2 ketone was from Merck. Recombinant Peptide N-glycosidase F was a kind gift from Shaun Lott (University of Auckland, New Zealand).
Plasmids and Cells
Construction of all plasmids used in this study are described in supplemental Fig. S5. pNFAT-SEAP and pNF-κB-SEAP were from Clontech, pGL3 Luc+ Control was from Promega, and ORAI1-HA was a kind gift from Jonathan Soboloff, Temple University School of Medicine. HEK293T cells (ATCC) were maintained in DMEM with 10% heat-inactivated FBS, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 2 mm l-glutamine (all from Invitrogen).
Molecular Biology
Standard molecular biology techniques were used for subcloning, high fidelity PCR, plasmid preparations, and DNA sequencing. Oligonucleotides were from Invitrogen. Site-directed mutagenesis was performed using the QuikChange Lightning Site-directed Mutagenesis kit (Stratagene). 5′-Rapid amplification of cDNA ends was performed with a SMART RACE kit (Clontech) using total RNA extracted from Danio rerio fins and the primers 5′-TGCCGAAGCGCCTCTAAACTGAAGC, 5′-TTGGTCTGGTGCTGCTTCATGTCCTC, and 5′-TGTGCTGGTCCTCTCGGTGGAGATT.
Transfections, Subfractionation, and Immunoblotting
HEK293T cells were transfected at 60–90% confluence with purified plasmid DNA (400 ng/24 well and 200 ng/96 well) complexed with Lipofectamine 2000 for 6 h. Cells were harvested 24–48 h post-transfection as described (13) or fractionated using a Subcellular Protein Fractionation kit (Pierce). Supernatants were collected from transfected cells 24 h after the addition of fresh media containing 0.1% FBS. For immunoprecipitation of endogenous STIM2 proteins, lysates from subconfluent HEK293T cells (3 × 108) harvested as above were immunoprecipitated with 50 μg of anti-STIM2-CT overnight at 4 °C. Immunoblotting and detection were performed essentially as described previously (12) using 4–12% NuPAGE Bis-Tris gels and a dry blotting system (Invitrogen). HRP activity was detected with ECL PlusTM (Amersham Biosciences). Anti-STIM1-CT, anti-STIM1-NT, anti-STIM2-CT, and anti-PAN STIM were used as described previously (13, 21). Anti-STIM2-IN (AnaSpec) was used at 1:500, anti-human IgG-HRP (Jackson Immunochemicals) was used at 1:40,000, and anti-FLAG (Sigma) and anti-HA (Berkeley Antibody Co., Inc.) were used at 1:1000. For detection of endogenous STIM2, secondary (anti-sheep IgG-Biotin; 1:2000) and tertiary (extrAvidin-AP; 1:300,000) antibodies were employed.
Deglycosylation
After immunoprecipitation, proteins were eluted from beads at 70 °C in 20 mm sodium phosphate buffer, pH 7.5, containing 0.1% SDS, 50 mm β-mercaptoethanol, and 1% Triton X-100 and deglycosylated with 50 μg/ml peptide N-glycosidase F or vehicle at 37 °C for 24 h.
Mass Spectrometry
Relevant bands were excised from Coomassie-stained gels, digested with trypsin, and interrogated by QSTAR XL Hybrid LS/MS/MS (Applied Biosystems). Peptides were identified using Mascot (Matrix Science) against an in-house data base.
Confocal Microscopy
Transfected cells were seeded onto poly-d-lysine-coated glass chamber slides (Nunc) at 6 h post-transfection. After 24–48 h, cells were fixed and immunostained with the indicated antibodies at 1:500 (anti-FLAG), 1:200 (anti-HA), and 1:100 (anti-Calnexin; Santa Cruz). Cells were imaged using an Olympus FV1000 confocal microscope equipped with FluoView 2.0b software.
Ca2+ Imaging
HEK293T cells in 96-well poly-d-lysine-coated, black-walled plates were transfected at 60–70% confluence as described above. 24 h after transfection cells were washed once in a HEPES-buffered salt solution (HBSS: 107 mm NaCl, 7.2 mm KCl, 1.2 mm MgCl2, 11.5 mm glucose, 2 mm CaCl2, and 10 mm HEPES with pH adjusted to 7.4 with NaOH) and loaded in HBSS containing 2 μm Fura-2 AM, 2.5 mm probenecid, and 1% BSA at room temperature (22 °C) in the dark for 30 min. Before [Ca2+]i were made, cells were washed thoroughly and incubated at room temperature for 20 min in the dark. Calcium measurements were performed on an EnVision multilabel plate reader (PerkinElmer Life Sciences) equipped with a high energy flash lamp and photomultiplier tube. The dye was alternately excited at 340 and 380 nm, and the emission wavelength was collected at 510 nm every 0.5 s. Agonists (carbachol, 100 μm; thapsigargin, 2 μm) were dispensed in a 20-μl volume at 100 μl/s into wells containing 100 μl of HBSS. Some assays required a brief (<5 min) incubation in nominally Ca2+-free solution (HBSS with no added Ca2+) before agonist dispense. Changes in intracellular calcium are represented as the ratio of Fura-2 fluorescence excited at 340 nm to that at 380 nm (F340/F380). In all cases, ratio values have been corrected for contributions by autofluorescence, which were measured in each well after treating cells with 10 μm ionomycin and 20 mm MnCl2. Data were manipulated in Excel, and statistical analysis was performed using one-way analysis of variance followed by Tukey's test, with p > 0.05 considered nonsignificant.
Reporter Gene Assays
HEK283T cells in 96-well poly-d-lysine-coated plates were transfected at 60% confluence as described above, except media contained 1% FBS. Plasmid DNA (200 ng/well) consisted of pGL3 Luc+ control, pSEAP reporter, and test plasmid at a 1:2:7 ratio. After 6 h, complexes were replaced with DMEM containing 0.1% FBS and 2 mm l-glutamine. 24 h after transfection, media were replaced with 10 ng/ml PMA or equivalent dilutions of DMSO in DMEM containing 0.1% FBS and 2 mm l-glutamine. After 10 h, secreted alkaline phosphatase (SEAP) activity in supernatants was measured by the Great EscAPe SEAP Chemiluminescence kit 2.0 (Clontech), and firefly luciferase was measured in cells by Bright-Glo Luciferase assay system (Promega). Chemiluminescence was detected on an EnVision Multilabel Plate Reader (PerkinElmer Life Sciences). Statistical analysis was performed using one-way analysis of variance followed by Tukey's test, with p > 0.05 considered nonsignificant.
Calmodulin (CaM) Binding Assay
Lysates from transfected HEK293T cells were prepared as above, except that EGTA and EDTA were omitted from the lysis buffer. CaM-agarose beads (Sigma) were pre-equilibrated in lysis buffer containing either 2 mm EDTA or 2 mm CaCl2. Equal quantities of lysate (to which either EGTA or CaCl2 was added to a final concentration of 2 mm) were incubated with pre-equilibrated CaM-agarose for 2 h at 4 °C. After washing in appropriate lysis buffer, proteins were eluted from the beads in SDS-PAGE sample buffer (Invitrogen) at 70 °C before immunoblotting analysis.
RESULTS
Met1–Gly101 Are Required for ER Targeting and Secretion of STIM2-Fc
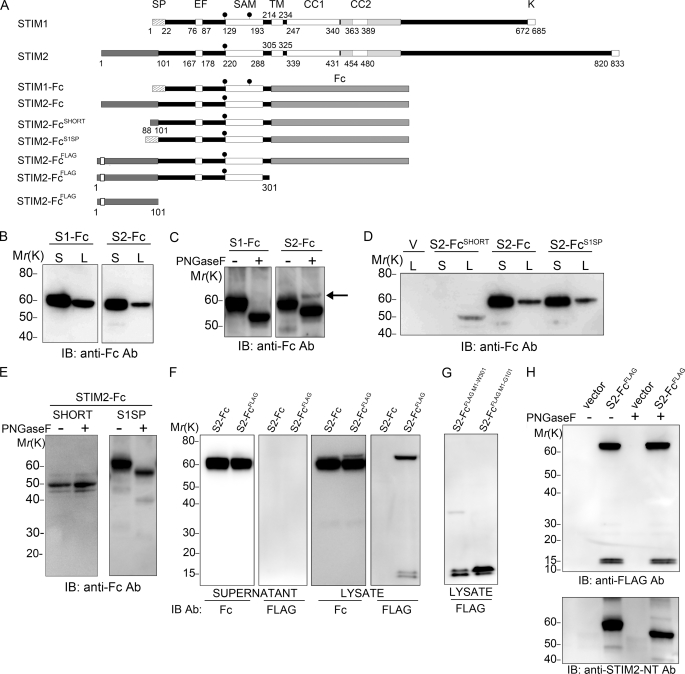
STIM1 is translocated across the ER membrane by a cleavable signal peptide at Met1-Ser22 (20, 21). The STIM2 signal peptide is less well defined; although the open reading frame extends 101 aa upstream of the endogenous signal peptidase cleavage site (Gly101-Cys102; Fig. 1A), our previous study suggested that translation likely started at a non-AUG codon around Leu88, implicating Leu88–Gly101 as the signal peptide sequence (13). To measure signal peptide functionality, we initially fused the STIM1 and STIM2 luminal domains with their cognate cleavable N-terminal region to the constant (Fc) region of human IgG (Fig. 1A), a strategy that, in mouse Stim1, results in glycosylation and secretion of the Fc fusion protein (22). Both STIM1-Fc and STIM2-Fc were glycosylated and secreted from HEK293T cells, indicating the presence of a functional signal peptide (Fig. 1, B and C). We consistently detected a small fraction of STIM2-Fc in lysates that was not glycosylated (Fig. 1C, arrow) and that was never detected in STIM1-Fc transfectants. To resolve the exact location of the STIM2 signal peptide, we removed Met1–Leu87 (STIM2-FcSHORT; Fig. 1A). STIM2-Fc secretion (Fig. 1D) and glycosylation (Fig. 1E, left panel) were abolished, indicating that Leu88–Gly101 alone was unable to function as a signal peptide. Several studies have utilized the STIM1 signal peptide to drive STIM2 expression in vitro (19, 20). Chimeric exchange of STIM2 residues Leu88–Cys102 with the STIM1 signal peptide (STIM2-FcS1SP) rescued STIM2-Fc glycosylation and secretion (Fig. 1, D and E), confirming that STIM2-Fc could be targeted to the ER by a short signal peptide. However, the STIM1 signal peptide could not rescue expression of the unglycosylated fraction seen in STIM2-Fc transfectants (Fig. 1E, right panel), suggesting that the STIM1 signal peptide differed in functionality to that of STIM2. Altogether, these results indicate that Leu88–Gly101 is not a bona fide signal peptide sequence and that sequences upstream of Leu88 are required for correct translation, ER targeting, and processing of STIM2-Fc.
FIGURE 1.
Met1-Gly101 are required for ER targeting and secretion of STIM2-Fc. A, shown are the domain structure of wild type STIM1 and STIM2 proteins (5) and constructs used. SP, signal peptide; EF, EF hand domain; SAM, sterile α motif; TM, transmembrane domain; CC1 and CC2, coiled-coil 1 and 2; gray shading surrounding CC2, CAD (7); K, lysine-rich polybasic domain; filled circles, glycosylation site. B–H, HEK293T cells were transfected with the indicated constructs, and lysates (L) or supernatants (S) were analyzed by immunoblotting (IB) with the indicated antibodies (Ab). B, STIM1-Fc (S1-Fc) and STIM2-Fc (S2-Fc) are secreted. C, STIM1-Fc and STIM2-Fc are glycosylated, and a peptide N-glycosidase F (PNGase F)-resistant fraction (arrow) is present in STIM2-Fc, but not STIM1-Fc, lysates. D, STIM2 residues Leu88–Gly101 do not mediate secretion of STIM2-Fc, unlike the STIM1 signal peptide. E, proteins produced by STIM2-FcSHORT are not glycosylated (left panel). The STIM1 signal peptide rescues glycosylation, but not the production of the peptide N-glycosidase F-resistant fraction (right panel). F, production of FLAG-tagged proteins in STIM2-FcFLAG lysates but not in supernatants is shown. G, the 15-kDa doublet is derived from the N terminus. H, FLAG-tagged proteins are not glycosylated.
Met1–Gly101 is highly conserved in STIM2 orthologs from placental mammals but not in lower vertebrates (supplemental Fig. S1A), suggesting it may have functional significance. To examine whether Met1 was the STIM2 translation start site, a FLAG tag was inserted after Pro9 in STIM2-Fc, creating STIM2-FcFLAG. STIM2-Fc secretion was not hampered by the tag, which had clearly been removed before secretion (Fig. 1F, first and second panels). STIM2-Fc production in lysates was also identical in cells derived from tagged and untagged constructs (Fig. 1F, third panel), except that the unglycosylated fraction identified in Fig. 1C appeared slightly larger, at 70 kDa, in STIM2-FcFLAG transfectants and was specifically detected by anti-FLAG antibodies (Fig. 1F, fourth panel). Furthermore, the anti-FLAG antibody also detected a doublet at 15 kDa in STIM2-FcFLAG-transfected cells (Fig. 1F, fourth panel). An identical 15-kDa doublet was produced from a construct comprising Met1–Trp301, or indeed only Met1–Gly101 (Fig. 1G), indicating that the doublet was a cleavage fragment derived from the N terminus. Both the FLAG-reactive 70-kDa band and the 15-kDa doublet were unglycosylated (Fig. 1H), indicating neither accessed the ER lumen. To identify the proteins, we analyzed 60- and 70-kDa STIM2-Fc proteins and the 15-kDa doublet by mass spectrometry (supplemental Fig. S1, B–D). Peptides recovered from secreted 60-kDa STIM2-Fc were derived exclusively from residues C-terminal to the signal peptidase cleavage site (supplemental Fig. S1B) unlike those recovered from 70-kDa FLAG-reactive STIM2-Fc, which additionally spanned Met1–Gly101 (supplemental Fig. S1C). From the 15-kDa doublet, a single peptide was recovered comprising Gly45–Arg57 (supplemental Fig. S1D). Altogether, the above data indicate that STIM2 is translated from Met1 and that Met1–Gly101 is cleaved before secretion of mature STIM2-Fc.
Met1-Gly101 Is the STIM2 Signal Peptide, and a Fragment Is Cleaved and Released to the Cytosol
The above data suggested that Met1–Gly101 represented an extraordinarily long signal peptide. This hypothesis was confirmed by the introduction of mutations around the signal peptidase cleavage site (Arg99/Gly101) that inhibited signal peptide cleavage, abolishing STIM2-Fc secretion and production of the 15 kDa doublet (supplemental Fig. S2A). We next considered whether further cleavage of the signal peptide resulted in its appearance as a doublet, as some signal peptides are sequentially processed first by signal peptidase then by signal peptide peptidases, resulting in release of signal peptide fragments into the cytosol (23). The signal peptide peptidase inhibitor (Z-LL)2 ketone (24) specifically inhibited production of the lower band of the 15-kDa doublet (supplemental Fig. S2B), and subfractionation of STIM2-FcFLAG-transfected cells confirmed that the lower band was released into the cytosol (supplemental Fig. S2C). Finally, we created a series of signal peptide fragments (SPFs) that implicated residues between Leu88 and Leu91 as the likely signal peptide peptidase cleavage site (supplemental Fig. S2, D and E).
Altogether, the data show that residues Met1–Gly101 represent the STIM2 signal peptide, which is cleaved by signal peptidase after productive ER translocation of STIM2-Fc. Further processing by signal peptide peptidases results in release of an 88–91-aa SPF into the cytosol. Moreover, the STIM2 signal peptide is unable to translocate all STIM2-Fc precursors to the ER with 100% efficiency, as a stable proportion of STIM2-Fc preproteins remain in the cytosol, signal peptide intact and unglycosylated. We have named this cytosolic population preSTIM2-Fc.
STIM2 Biosynthesis Is Identical to That of STIM2-Fc
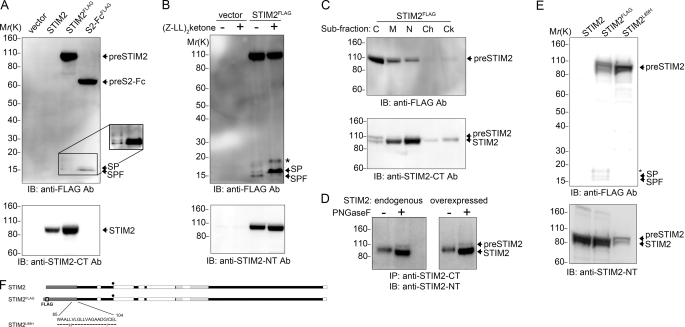
It was important to confirm that STIM2 biosynthesis mirrored that of STIM2-Fc. We placed a FLAG tag within wild type STIM2 in an identical position to that of STIM2-FcFLAG (Fig. 2). Like STIM2-Fc, expression of 100-kDa STIM2 was unaffected by the FLAG tag (Fig. 2A, lower panel). Anti-FLAG antibodies detected a 110-kDa STIM2 preprotein (preSTIM2) and a 15-kDa doublet comprising the STIM2 signal peptide and the SPF (Fig. 2A, top panel and supplemental Fig. S3, A and B). The abundance of the signal peptide and SPF was consistently lower when derived from the STIM2FLAG construct. An additional doublet at 17 kDa, also derived from the signal peptide, was variably produced in STIM2FLAG-transfected cells (Fig. 2A, inset and supplemental Fig. S3C); this appeared dependent on the presence of the STIM2 C terminus (supplemental Fig. S3, D and E) but was not investigated further. Signal peptide and SPF cleavage proceeded in a sequential manner, identical to that determined for STIM2-Fc (Fig. 2B and supplemental Fig. S3F). Cell fractionation confirmed the predominantly cytosolic localization of preSTIM2 (Fig. 2C, upper panel), unlike STIM2, which resided principally in ER membrane-associated fractions (Fig. 2C, lower panel). To determine whether endogenous STIM2 was processed in the same manner as exogenously expressed STIM2FLAG, we immunoprecipitated endogenous STIM2 from HEK293T cells. Deglycosylation revealed the presence of an endogenous preSTIM2 population (Fig. 2D, left panel) similar to that observed upon expression of STIM2FLAG (Fig. 2D, right panel). Together, these data confirm that endogenous STIM2 is translated and processed in an identical manner to that of STIM2-Fc and exists concomitantly in three distinct forms: ER resident STIM2, cytosolic preSTIM2, and a cytosolic SPF.
FIGURE 2.
STIM2 biosynthesis is identical to STIM2-Fc and is modified by mutations in the signal peptide. A–E, HEK293T cells were transfected with the indicated constructs, and samples were analyzed by immunoblotting (IB) with the indicated antibodies (Ab). A, STIM2FLAG is processed in an identical manner to that of STIM2-FcFLAG. Inset, shown is enhancement of the boxed area. B, the SPF (lower band) is produced by cleavage of the signal peptide (SP; upper band). The asterisk (*) indicates an additional 17-kDa band. C, preSTIM2 is enriched in the cytosolic fraction, whereas STIM2 is predominantly ER membrane-associated. C, cytosol; M, membrane; N, soluble nuclear; Ch, chromatin; Ck, cytoskeletal. D, production of endogenous preSTIM2 in HEK293T cells (left panel) is identical to that from STIM2FLAG-expressing cells (right panel). E, the L89H mutation enhances preSTIM2 expression (top panel) and abolishes signal peptide (SP), SPF (top panel), and STIM2 expression (bottom panel). F, shown are the constructs used in Fig. 2.
Signal Peptide Targeting Efficiency Is Modified by Substitution of Residues within Its Membrane Spanning Domain
In a previous study, we found that Leu89 mutations abrogate STIM2 expression (13). Leu89 is located within the membrane-spanning domain of the STIM2 signal peptide, and we hypothesized that the mutation, rather than inhibiting translation, may actually affect signal peptide function. We recreated the L89H mutation in STIM2FLAG to create STIM2L89H (Fig. 2, E and F). Little or no STIM2 was produced from STIM2L89H (Fig. 2E, lower panel) consistent with our previous study. Signal peptide and SPF production were also abolished by the L89H mutation, whereas preSTIM2 expression was enhanced (Fig. 2E, top panel). Like preSTIM2 derived from STIM2FLAG, preSTIM2 derived from STIM2L89H was not glycosylated (supplemental Fig. S3F) and localized predominantly to the cytosolic fraction (supplemental Fig. S3G). Together, these data show that the L89H mutation reduces signal peptide ER targeting efficiency, resulting in enhanced cytosolic accumulation of preSTIM2 while abrogating ER-localized STIM2 expression.
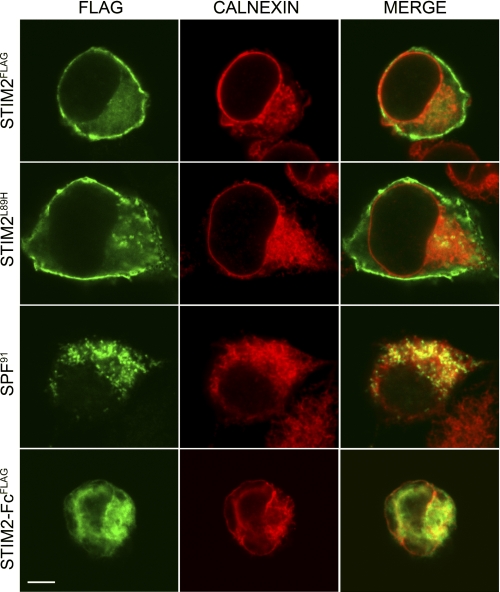
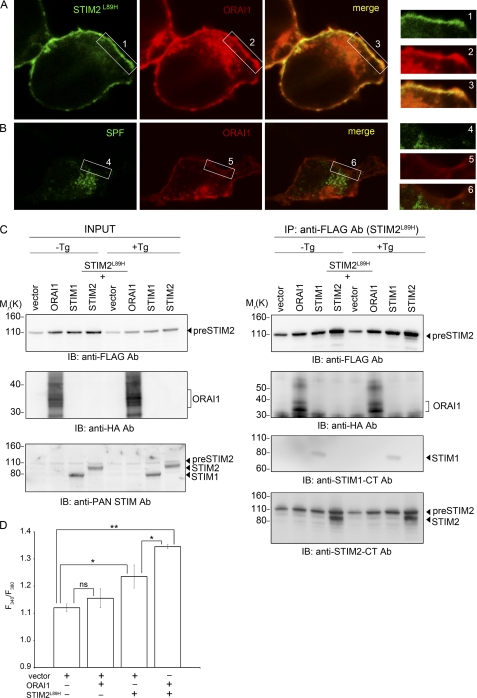
preSTIM2 Accumulates at the Plasma Membrane
We used immunolocalization to confirm the cellular location of preSTIM2 and the SPF (Fig. 3). In STIM2FLAG-transfected cells, where anti-FLAG antibodies simultaneously detected preSTIM2, the signal peptide, and the SPF, cells exhibited a punctate cytosolic staining pattern and clear perimembrane association that was distinct from the ER marker calnexin. Membrane association was not observed in STIM2-FcFLAG-transfected cells, which was predominantly localized within the ER and the cytosol. In STIM2L89H-transfected cells, where preSTIM2 expression could be analyzed in isolation, striking plasma membrane localization was observed. In contrast, the SPF gathered in punctate structures both on the ER and within the cytosol. Altogether, we concluded that preSTIM2 displays impressive targeting to the inner leaflet of the plasma membrane, dependent on the C terminus, whereas the SPF is distributed in the cytosol.
FIGURE 3.
preSTIM2 and the STIM2 SPF are localized in the cytosol. HEK293T cells transfected with STIM2FLAG, STIM2L89H, SPF91, or STIM2-FcFLAG were co-stained with anti-FLAG (green) to visualize FLAG-tagged proteins and anti-calnexin (red) to visualize the ER and imaged by confocal microscopy. All images are from the middle of the cell. Scale bar, 10 μm.
preSTIM2 Increases Basal Cytosolic Ca2+ Levels and Reduces SOCe
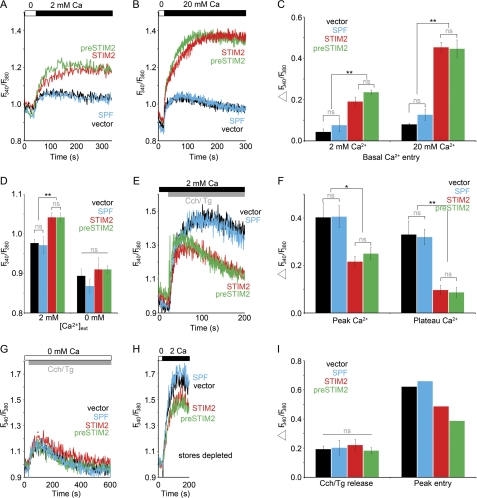
The identification of cytosolic preSTIM2 and SPF populations suggested that these proteins might regulate Ca2+ influx independent of ER Ca2+ store depletion. In store-replete cells briefly incubated in nominally Ca2+-free conditions, re-addition of Ca2+ results in Ca2+ influx, indicative of the basal activity of SOCs (16). STIM2 and preSTIM2 both increased basal Ca2+ influx to the same extent, the level of which was significantly higher than that of vector-transfected cells, and was dependent on [Ca2+]ext (Fig. 4, A–C). In contrast, SPF91 was without effect (Fig. 4, A–C). STIM2 and preSTIM2 transfectants additionally had a significantly higher resting [Ca2+]i than vector- or SPF91-transfected cells, which were dependent on extracellular Ca2+ (Fig. 4D).
FIGURE 4.
preSTIM2 regulates Ca2+ dynamics in HEK293T cells. A–I, [Ca2+]i measurements in HEK293T cells transiently transfected (24 h) with empty vector (black), STIM2FLAG (red), STIM2L89H (green), or SPF91 (blue) are shown. A and B, basal Ca2+ entry measurements in 2 mm (A) or 20 mm (B) Ca2+ are shown. Traces are the mean for n = 3 (A) or n = 4 (B) individual wells per construct. C is a summary of peak basal Ca2+ entry from A and B; data are the mean ± S.D. for n = 3 (A) and n = 4 (B) individual wells. Data represent six individual experiments. D, shown are resting [Ca2+]i measurements in 0 or 2 mm [Ca2+]ext. Data are the mean ± S.E. for n = 4 individual wells per construct and are representative of 2 individual experiments. E, Ca2+ influx measurements in 2 mm [Ca2+]ext are shown. Cch, carbachol; Tg, thapsigargin. F, a summary of peak and plateau [Ca2+]i from E; data are the mean ± S.E. for n = 4 (vector, STIM2FLAG, STIM2L89H) or n = 3 (SPF91) individual wells. Data represent three individual experiments. G, ER store release measurements in 0 [Ca2+]ext is shown. H, SOCe measurements in store-depleted cells are shown. I, shown is a summary of peak ER store release and SOCe from G and H; data are the mean ± S.D. for n = 4 (vector and SPF91) and n = 2 (STIM2FLAG and STIM2L89H) individual wells. Data represent four individual experiments. ns, not significant; *, p < 0.05; **, p < 0.01.
A defining feature of STIM2 is that it reduces SOCe when overexpressed (15–17). STIM2 significantly reduced the SOCe-mediated plateau phase in HEK cells activated in the presence of 2 mm [Ca2+]ext (Fig. 4, E and F). preSTIM2 was as effective as STIM2, whereas the SPF91 had no effect (Fig. 4, E and F). This could not be attributed to differences in ER Ca2+ store content or prior store depletion, as the magnitude of Ca2+ release from ER stores of cells transfected with STIM2, preSTIM2, or SPF91 was identical to the vector control (Fig. 4, G and I). As a second measure of SOCe, we analyzed Ca2+ entry after store depletion in Ca2+-free medium. STIM2 reduced the magnitude of SOCe in store-depleted cells, and preSTIM2, but not SPF91, was as effective as STIM2 (Fig. 4, H and I). These data indicate that preSTIM2, but not the SPF, is able to mimic the effects of STIM2 expression on Ca2+ dynamics in HEK cells, regulating basal Ca2+ levels, basal Ca2+ influx, and SOCe.
preSTIM2 and ORAI1 Interact to Regulate Basal Ca2+ Levels
STIM2 and cytoplasmically localized STIM1 and STIM2 C-terminal fragments can elevate basal Ca2+ concentration via a store-independent association with ORAI1 (7, 8, 25–27). To determine whether preSTIM2 regulates basal Ca2+ levels through ORAI1, we co-expressed STIM2L89H with HA-tagged ORAI1. Plasma membrane expression of preSTIM2 overlapped significantly with that of ORAI1-HA (Fig. 5A), in contrast to the SPF91 (Fig. 5B). To determine whether preSTIM2 and ORAI1-HA interacted biochemically, pulldown assays were conducted with co-transfected cells with and without prior store depletion. After immunoprecipitation of preSTIM2 (Fig. 5C, top panel), a strong interaction with ORAI1-HA (Fig. 5C, second panel) and STIM2 (Fig. 5C, bottom panel) was detected. A weak interaction with STIM1 was also detected (Fig. 5C, third panel). These interactions were not enhanced by store depletion (Fig. 5C, +Tg). Reciprocal immunoprecipitations showed that ORAI1 could co-immunoprecipitate preSTIM2 (data not shown). Finally, we measured basal Ca2+ influx in cells co-overexpressing STIM2L89H and ORAI1-HA. Expression of ORAI1 alone did not elevate a basal Ca2+ leak in HEK cells, unlike preSTIM2 (Fig. 5D). However, co-overexpression significantly elevated basal Ca2+ influx above that of preSTIM2 alone (Fig. 5D). Altogether, these data indicate that preSTIM2 associates with plasma membrane ORAI1 in a store-independent manner to regulate basal Ca2+ levels.
FIGURE 5.
preSTIM2 and ORAI1 synergize to regulate constitutive Ca2+ entry. Co-localization of preSTIM2 and ORAI1-HA (A) but not SPF91 and ORAI1-HA (B) is shown. C, interactions of preSTIM2 with STIM2, STIM1, and ORAI1-HA are stable in store replete (−Tg) and store-depleted (+Tg) HEK293T cells. Note how the overexpression level of preSTIM2 is very similar to that of endogenous STIM2 (INPUT, bottom panel). D, basal Ca2+ entry measurements after the addition of 20 mm [Ca2+]ext are increased in cells co-expressing preSTIM2 and ORAI1-HA. Data are the mean peak Ca2+ entry ± S.D. for n = 2 wells and are representative of three individual experiments. ns, not significant; *, p < 0.05; **, p < 0.01. Ab, antibody; IP, immunoprecipitation; IB, immunoblot.
preSTIM2 and the STIM2 SPF Regulate Gene Expression
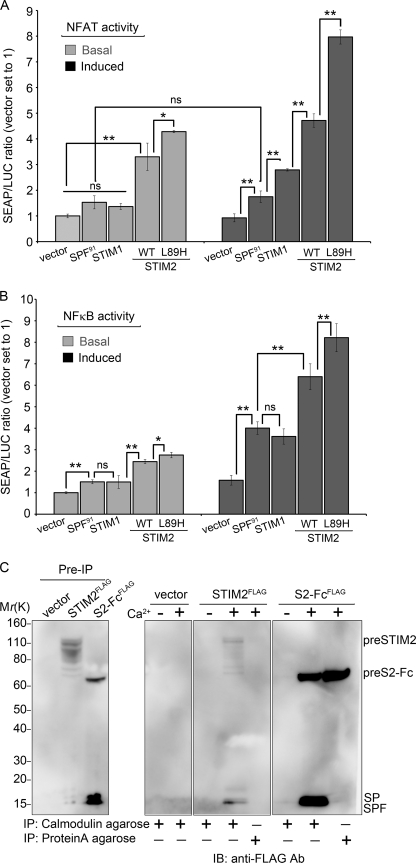
To determine whether the elevation of [Ca2+]i by preSTIM2 was translated into a cellular response, we monitored induction of NFAT and NF-κB-dependent SEAP reporter gene activity. NFAT-dependent transcription requires sustained elevation of [Ca2+]i combined with a phorbol ester to activate PKC. Accordingly, PMA in conjunction with transfected STIM1 C-terminal fragments, which constitutively elevate resting [Ca2+]i, activate NFAT significantly (7). Therefore, we compared SEAP production in transfected cells in the absence and presence of PMA (Fig. 6A). Under basal conditions, NFAT-SEAP production was enhanced 3-fold by STIM2 and 4-fold by preSTIM2 expression, increasing to 5- and 8-fold, respectively, upon PMA stimulation. In comparison, STIM1 had no effect on basal NFAT-SEAP activity and moderately (3-fold) enhanced PMA-stimulated activity, consistent with a previous study (7). SPF91 had no significant effect on either basal or PMA-induced NFAT-SEAP activity.
FIGURE 6.
preSTIM2 and the STIM2 SPF regulate gene transcription. A and B, shown are basal- and PMA-induced NFAT-SEAP (A) or NF-κB-SEAP (B) reporter gene activity in HEK293T cells transiently transfected with indicated constructs. Data represent the mean ± S.D. for n = 3 individual wells and are representative of four (NFAT) and five (NF-κB) experiments. WT, wild type. ns, not significant; *, p < 0.05; **, p < 0.01. C, preSTIM2 and the SPF specifically interact with Ca2+/calmodulin. Slight degradation of preSTIM2 is visible in this experiment. Identical results were obtained in two individual experiments. IP, immunoprecipitation; IB, immunoblot.
NF-κB-dependent transcription is activated by PMA alone and is enhanced by [Ca2+]i (28). Under basal conditions, NF-κB-SEAP production was enhanced 2.3-fold by STIM2 and 2.5-fold by preSTIM2, increasing to 6- and 7.5-fold, respectively, upon PMA stimulation (Fig. 6B). STIM1 had no significant effect on basal NF-κB-SEAP activity but increased PMA-mediated activity 3.2-fold. Surprisingly, SPF91 had a small, but significant effect on basal NF-κB-SEAP activity and increased PMA-mediated activity 3.6-fold over background.
SPF91 was unable to alter basal [Ca2+]i, basal Ca2+ influx, or SOCe. A search for potential regulatory motifs uncovered a conserved CaM binding site between Trp76 and Leu91 (supplemental Fig. S4). The SPF immunoprecipitated with CaM-agarose beads strongly in the presence of 2 mm Ca2+ and weakly in Ca2+ free conditions, whether derived from STIM2FLAG or STIM2-FcFLAG (Fig. 6C). preSTIM2-Fc and preSTIM2 also interacted with CaM-agarose in the presence of Ca2+. Protein A-agarose immunoprecipitated preSTIM2-Fc via the Fc tag but not preSTIM2 or the SPF, confirming that preSTIM2 and SPF interactions were CaM-specific. Altogether, the data show that regulation of basal Ca2+ influx by preSTIM2 supports NFAT- and NF-κB-mediated transcription, whereas the STIM2 SPF enhances NF-κB-mediated gene transcription in a Ca2+-independent manner.
DISCUSSION
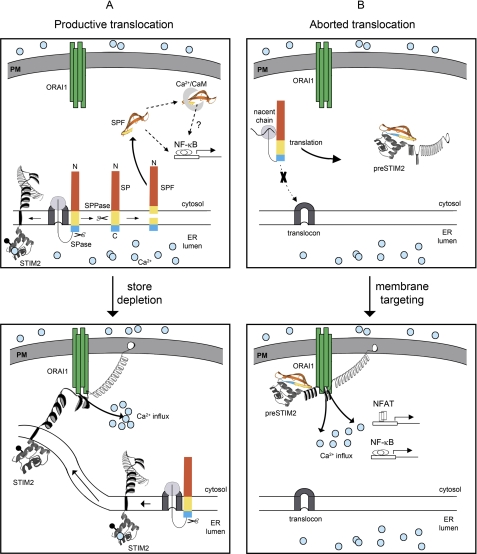
Our study has revealed for the first time that STIM2 exists in at least three distinct functional forms that reside in the ER and in the cytosol. The major population is the widely investigated ER-membrane resident STIM2 that we find is produced after cleavage of the 101-aa STIM2 signal peptide following translocation across the ER membrane. We find a second minor population, preSTIM2, escapes ER targeting to remain signal peptide intact within the cytosol. preSTIM2 stably associates with the plasma membrane, where it regulates basal Ca2+ concentration in a store-independent manner. A third population is synthesized after cleavage of the STIM2 signal peptide into a ∼91-aa SPF. Released into the cytosol, the SPF regulates NF-κB transcription. We make the novel finding that production of these functional populations is dependent on the endogenous STIM2 signal peptide, which is extraordinary long. Our data reveal that translation begins at a methionine residue highly conserved in mammals, producing a 101-aa signal peptide of unprecedented length. Unable to mediate ER targeting and translocation with 100% efficiency, the STIM2 signal peptide controls the ratio of STIM2 versus preSTIM2 populations. Previously, we hypothesized that STIM2 was translated from a non-AUG codon at Leu88 or Leu89, as the majority of STIM2 expression is abrogated after mutation of either residue (13). Here we find that, rather than aborting translation, Leu89 mutations modify signal peptide function and instead skew the preSTIM2:STIM2 ratio in favor of the low abundance preSTIM2 population.
Under endogenous conditions, preSTIM2 comprises a minor proportion of “total” STIM2, estimated to be ∼2–10%. Although small, cytosolic populations derived from similarly inefficient signal peptides, such as calreticulin (2–14%) or prion protein (5–15%), have significant physiological or pathological relevance (29, 30). Indeed, from its location at the inner leaflet of the plasma membrane, preSTIM2 constitutively interacts with ORAI1 to regulate basal Ca2+ concentration independent of ER Ca2+ store depletion, much like cytosolically expressed STIM1 and STIM2 C-terminal domain fragments (7–9, 25, 26, 31, 32). Although the exact mechanisms of targeting and activation remain to be determined, our data suggest a role for the C-terminal polybasic domain (33) in driving plasma membrane targeting. The cytosolic STIM1 C terminus is recruited to sites of PM ORAI1 expression via the CAD domain, which alone is sufficient for binding to, and gating ORAI1 (7–9). This same region in preSTIM2 is >80% identical to CAD (7) and, in the low Ca2+ environment of the cytosol, may be sufficiently exposed in preSTIM2 to recruit and gate adjacent ORAI1 channels. The preSTIM2 N terminus may also play a regulatory role in ORAI1 activation as co-expression of ORAI1 with preSTIM2 did not promote cell death in this study, unlike co-expression with the STIM2 C terminus (27) or the STIM1 CAD (7). Conceivably, this could be achieved through binding of CaM to the preSTIM2 N terminus, as CaM is a known inhibitor of STIM2-mediated, store-independent ORAI1 activation (17).
When SOCe is activated, preSTIM2 partially reduces the amplitude of Ca2+ entry, like wild type STIM2 (15) and cytosolic STIM1 C-terminal fragments (31, 32). Under these conditions, interactions between preSTIM2 and ORAI1, like those between ORAI and the STIM C terminus (31), remain stable. Together, these results suggest that preSTIM2 may partially block access to ORAI1 when SOCe is activated. The exact nature of STIM2-mediated inhibition of SOCe is controversial. This study supports our previous report (15) and that of others (16, 17), which suggest that STIM2 is a specific inhibitor of endogenous SOCe. These studies have in common their use of constructs containing the endogenous STIM2 signal peptide. In contrast, studies using the STIM1 signal peptide to drive STIM2 expression (19, 20, 34) have suggested that STIM2-mediated inhibition is rather nonspecific, inhibiting SOCe only at high expression levels or after a delayed period of overexpression (19, 20). These differences may be reconciled by our demonstration that the STIM1 signal peptide cannot entirely substitute for that of STIM2. Constructs containing the STIM1 signal peptide would efficiently express ER-localized STIM2 to the exclusion of preSTIM2 and the SPF, suggesting that differences in preSTIM2 expression between studies may have resulted in different outcomes.
A major finding of this study is that the increase in basal Ca2+ concentration mediated by preSTIM2 was sufficient to increase basal and PMA-induced activity of NFAT and NF-κB. Indeed, preSTIM2 was more active than wild type STIM2 despite being expressed at levels many times less than that of the total STIM2 population. This would suggest that a significant proportion of endogenous STIM2 transcriptional activity is mediated by the preSTIM2 population. In support, we found preSTIM2 to be highly active when overexpressed at levels approaching that of endogenous STIM2. By comparison, STIM1 was much less efficient than preSTIM2 or indeed the total STIM2 population in the absence of a Ca2+ signal. Although SOCe is a major mechanism for Ca2+-induced regulation of NFAT nuclear localization and gene transcription (14), Stim2-deficient T cells also display marked defects in NFAT-dependent signaling despite comparatively little change to the magnitude of endogenous SOCe (14). This combined with our data suggests that preSTIM2 plays a major role among the total STIM2 population in stabilizing NFAT transcription in T cells and is consistent with studies demonstrating that changes in basal Ca2+ concentration lead to altered NFAT signaling (35). This study has for the first time demonstrated the effectiveness by which STIM2, like STIM1 (36–38), additionally regulates NF-κB-mediated gene transcription. STIM1 and STIM2 cooperation is readily apparent in mice with a T cell-specific deletion in both Stim1 and Stim2, double deficiency leading specifically to a decrease in the number and function of thymic regulatory T cells (nTreg) (14). Emerging studies now demonstrate that nTreg development is critically dependent on NF-κB-mediated transcription (39–43), whereas NFAT appears to be required for correct function (43–46). Although speculative, it is possible that combined STIM1- and STIM2-mediated regulation of NF-κB and NFAT pathways are required for the development and function of nTreg cells in vivo.
Remarkably, the STIM2 signal peptide functions to augment NF-κB-mediated gene transcription after its release as a ∼91-aa SPF into the cytosol. Only four eukaryotic signal peptides have a demonstrable post targeting function after processing by ER signal peptide peptidases (47–51). Although the STIM2 SPF was unable to regulate basal Ca2+ or SOCe, it nevertheless productively interacted with Ca2+/CaM and proved as efficient as STIM1 at stimulating the activity of NF-κB-dependent gene transcription. Although the specific mechanisms underlying these effects are not yet clear, several SPFs and a number of endogenous peptides of a comparable size to the STIM2 SPF bind to Ca2+/CaM and act to inhibit a number of CaM-mediated processes (52–56). Conceivably, Ca2+/CaM may be required to facilitate SPF-mediated transcriptional activity as CaM acts via multiple mechanisms to regulate NF-κB transcription, both positively (57, 58) and negatively (58–60). Because there are multiple potential mechanisms, understanding how the SPF affects CaM activity and NF-κB activation must await additional studies.
In conclusion, our data challenge the prevailing theory regarding STIM2-mediated control of basal Ca2+ homeostasis and SOCe (16, 17, 19, 20, 61). We suggest a new model whereby basal Ca2+ and SOCe are predominantly controlled in an individual fashion by two distinct proteins, preSTIM2 and STIM2, rather than a single ER-localized STIM2 population (Fig. 7). Whether interactions between preSTIM2 and STIM2 are required to facilitate these processes will be an important area of future research. We demonstrate here that several of the functional differences between STIM1 and STIM2 are due to concomitant expression of multiple STIM2-derived populations. In the future, it will be essential to discriminate between these populations to more clearly understand the diverse effects of this unique protein on cell function. More broadly, the STIM2 signal peptide increased dramatically in length post-duplication of the single invertebrate STIM into vertebrate paralogs STIM1 and STIM2. Subcellular re-localization of paralogs within protein families is increasingly recognized as a mechanism for the functional diversification of duplicated genes (62, 63). Our study illustrates for the first time that this can be achieved by modifications to signal peptide structure and function.
FIGURE 7.
A model for STIM2 biogenesis, regulation, and activity. A, mature STIM2 and the SPF are produced after productive translocation of nacent STIM2 polypeptides through the ER translocon. Released into the cytosol, the SPF interacts with Ca2+-loaded CaM and drives NF-κB-mediated transcription. A possible link between CaM binding and NF-κB transcription is indicated. Upon store depletion, ER-localized STIM2 is activated, redistributing on the ER membrane to regions near ORAI1 to promote SOCe. B, aborted translocation promotes preSTIM2 accumulation within the cytosol, signal peptide intact and unglycosylated. Membrane targeting of preSTIM2, likely via the C-terminal polybasic domain, assists functional coupling between preSTIM2 and ORAI1. The resulting Ca2+ influx regulates basal Ca2+ concentration, and NFAT-, and NF-κB-mediated transcription and is independent of ER store depletion. For clarity, the nucleus is not shown. PM, plasma membrane; SP, signal peptide (red, N region; yellow, H-region; blue, C region); SPase, signal peptidase; SPPase, signal peptide peptidase; blue circles, Ca2+; filled black circles, glycosylation site.
Supplementary Material
Acknowledgments
We thank the University of Auckland Centre of Genomics and Proteomics and Biomedical Imaging Research Unit for sequencing and confocal microscopy, Shaun Lott, Keith Hudson, and Jonathan Soboloff for reagents, and John Taylor and Nigel Birch for comments on the manuscript.
This work was supported by the Health Research Council, Auckland Medical Research Foundation, and Lottery Health in New Zealand.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HQ444754.
- SOC
- store operated Ca2+ channel
- STIM
- stromal interaction molecule
- ER
- endoplasmic reticulum
- PM
- plasma membrane
- CAD
- CRAC activation domain
- SOCe
- store-operated Ca2+ entry
- aa
- amino acid
- NFAT
- nuclear factor of activated T-cells
- NF-κB
- nuclear factor κB
- SEAP
- secreted alkaline phosphatase
- Luc+
- luciferase
- Fc
- fragment crystallizable
- SPF
- signal peptide fragment
- PMA
- phorbol 12-myristate 13-acetate
- CaM
- calmodulin
- nTreg
- thymic/natural regulatory T-cells
- HBSS
- HEPES-buffered salt solution
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Johnstone L. S., Graham S. J., Dziadek M. A. (2010) J. Cell. Mol. Med. 14, 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vig M., Beck A., Billingsley J. M., Lis A., Parvez S., Peinelt C., Koomoa D. L., Soboloff J., Gill D. L., Fleig A., Kinet J. P., Penner R. (2006) Curr. Biol. 16, 2073–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. (2006) Nature 443, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dziadek M. A., Johnstone L. S. (2007) Cell Calcium 42, 123–132 [DOI] [PubMed] [Google Scholar]
- 6. Baba Y., Hayashi K., Fujii Y., Mizushima A., Watarai H., Wakamori M., Numaga T., Mori Y., Iino M., Hikida M., Kurosaki T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16704–16709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. A. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korzeniowski M. K., Manjarrés I. M., Varnai P., Balla T. (2010) Sci. Signal. 3, ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malli R., Naghdi S., Romanin C., Graier W. F. (2008) J. Cell Sci. 121, 3133–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graham S. J., Black M. J., Soboloff J., Gill D. L., Dziadek M. A., Johnstone L. S. (2009) Differentiation 77, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams R. T., Manji S. S., Parker N. J., Hancock M. S., Van Stekelenburg L., Eid J. P., Senior P. V., Kazenwadel J. S., Shandala T., Saint R., Smith P. J., Dziadek M. A. (2001) Biochem. J. 357, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh-Hora M., Yamashita M., Hogan P. G., Sharma S., Lamperti E., Chung W., Prakriya M., Feske S., Rao A. (2008) Nat. Immunol. 9, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soboloff J., Spassova M. A., Hewavitharana T., He L. P., Xu W., Johnstone L. S., Dziadek M. A., Gill D. L. (2006) Curr. Biol. 16, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 16. Zhou Y., Mancarella S., Wang Y., Yue C., Ritchie M., Gill D. L., Soboloff J. (2009) J. Biol. Chem. 284, 19164–19168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parvez S., Beck A., Peinelt C., Soboloff J., Lis A., Monteilh-Zoller M., Gill D. L., Fleig A., Penner R. (2008) FASEB J. 22, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wedel B., Boyles R. R., Putney J. W., Jr., Bird G. S. (2007) J. Physiol. 579, 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bird G. S., Hwang S. Y., Smyth J. T., Fukushima M., Boyles R. R., Putney J. W., Jr. (2009) Curr. Biol. 19, 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brandman O., Liou J., Park W. S., Meyer T. (2007) Cell. 131, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manji S. S., Parker N. J., Williams R. T., van Stekelenburg L., Pearson R. B., Dziadek M., Smith P. J. (2000) Biochim. Biophys. Acta 1481, 147–155 [DOI] [PubMed] [Google Scholar]
- 22. Oritani K., Kincade P. W. (1996) J. Cell Biol. 134, 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lemberg M. K., Martoglio B. (2004) FEBS Lett. 564, 213–218 [DOI] [PubMed] [Google Scholar]
- 24. Weihofen A., Lemberg M. K., Ploegh H. L., Bogyo M., Martoglio B. (2000) J. Biol. Chem. 275, 30951–30956 [DOI] [PubMed] [Google Scholar]
- 25. Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S., Worley P. F. (2006) Nat. Cell Biol. 8, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 26. Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Deng X., Zhou Y., Hendron E., Mancarella S., Ritchie M. F., Tang X. D., Baba Y., Kurosaki T., Mori Y., Soboloff J., Gill D. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mattila P. S., Ullman K. S., Fiering S., Emmel E. A., McCutcheon M., Crabtree G. R., Herzenberg L. A. (1990) EMBO J. 9, 4425–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rane N. S., Yonkovich J. L., Hegde R. S. (2004) EMBO J. 23, 4550–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaffer K. L., Sharma A., Snapp E. L., Hegde R. S. (2005) Dev. Cell. 9, 545–554 [DOI] [PubMed] [Google Scholar]
- 31. Penna A., Demuro A., Yeromin A. V., Zhang S. L., Safrina O., Parker I., Cahalan M. D. (2008) Nature 456, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. (2008) J. Biol. Chem. 283, 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ercan E., Momburg F., Engel U., Temmerman K., Nickel W., Seedorf M. (2009) Traffic 10, 1802–1818 [DOI] [PubMed] [Google Scholar]
- 34. Darbellay B., Arnaudeau S., Ceroni D., Bader C. R., Konig S., Bernheim L. (2010) J. Biol. Chem. 285, 22437–22447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lipskaia L., Hulot J. S., Lompré A. M. (2009) Pflugers Arch. 457, 673–685 [DOI] [PubMed] [Google Scholar]
- 36. Baba Y., Nishida K., Fujii Y., Hirano T., Hikida M., Kurosaki T. (2008) Nat. Immunol. 9, 81–88 [DOI] [PubMed] [Google Scholar]
- 37. Pani B., Ong H. L., Brazer S. C., Liu X., Rauser K., Singh B. B., Ambudkar I. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20087–20092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oh-hora M., Rao A. (2008) Curr. Opin. Immunol. 20, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Isomura I., Palmer S., Grumont R. J., Bunting K., Hoyne G., Wilkinson N., Banerjee A., Proietto A., Gugasyan R., Wu L., Li W., McNally A., Steptoe R. J., Thomas R., Shannon M. F., Gerondakis S. (2009) J. Exp. Med. 206, 3001–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vang K. B., Yang J., Pagán A. J., Li L. X., Wang J., Green J. M., Beg A. A., Farrar M. A. (2010) J. Immunol. 184, 4074–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deenick E. K., Elford A. R., Pellegrini M., Hall H., Mak T. W., Ohashi P. S. (2010) Eur. J. Immunol. 40, 677–681 [DOI] [PubMed] [Google Scholar]
- 42. Visekruna A., Huber M., Hellhund A., Bothur E., Reinhard K., Bollig N., Schmidt N., Joeris T., Lohoff M., Steinhoff U. (2010) Eur. J. Immunol. 40, 671–676 [DOI] [PubMed] [Google Scholar]
- 43. Zheng Y., Josefowicz S., Chaudhry A., Peng X. P., Forbush K., Rudensky A. Y. (2010) Nature 463, 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bopp T., Palmetshofer A., Serfling E., Heib V., Schmitt S., Richter C., Klein M., Schild H., Schmitt E., Stassen M. (2005) J. Exp. Med. 201, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A. D., Stroud J. C., Bates D. L., Guo L., Han A., Ziegler S. F., Mathis D., Benoist C., Chen L., Rao A. (2006) Cell. 126, 375–387 [DOI] [PubMed] [Google Scholar]
- 46. Müller M. R., Rao A. (2010) Nat. Rev. Immunol. 10, 645–656 [DOI] [PubMed] [Google Scholar]
- 47. Golde T. E., Wolfe M. S., Greenbaum D. C. (2009) Semin. Cell Dev. Biol. 20, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sato T., Nyborg A. C., Iwata N., Diehl T. S., Saido T. C., Golde T. E., Wolfe M. S. (2006) Biochemistry 45, 8649–8656 [DOI] [PubMed] [Google Scholar]
- 49. Narayanan S., Sato T., Wolfe M. S. (2007) J. Biol. Chem. 282, 20172–20179 [DOI] [PubMed] [Google Scholar]
- 50. Hegde R. S., Bernstein H. D. (2006) Trends Biochem. Sci. 31, 563–571 [DOI] [PubMed] [Google Scholar]
- 51. Kilic A., Klose S., Dobberstein B., Knust E., Kapp K. (2010) Eur. J. Cell Biol. 89, 449–461 [DOI] [PubMed] [Google Scholar]
- 52. Martoglio B., Graf R., Dobberstein B. (1997) EMBO J. 16, 6636–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mariotti M., De Benedictis L., Avon E., Maier J. A. (2000) J. Biol. Chem. 275, 24047–24051 [DOI] [PubMed] [Google Scholar]
- 54. Ballabio E., Mariotti M., De Benedictis L., Maier J. A. (2004) Cell. Mol. Life Sci. 61, 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanazawa Y., Makino M., Morishima Y., Yamada K., Nabeshima T., Shirasaki Y. (2008) Neuroscience. 154, 473–481 [DOI] [PubMed] [Google Scholar]
- 56. Johanson R. A., Sarau H. M., Foley J. J., Slemmon J. R. (2000) J. Neurosci. 20, 2860–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hughes K., Edin S., Antonsson A., Grundström T. (2001) J. Biol. Chem. 276, 36008–36013 [DOI] [PubMed] [Google Scholar]
- 58. Bae J. S., Jang M. K., Hong S., An W. G., Choi Y. H., Kim H. D., Cheong J. (2003) Biochem. Biophys. Res. Commun. 305, 1094–1098 [DOI] [PubMed] [Google Scholar]
- 59. Antonsson A., Hughes K., Edin S., Grundström T. (2003) Mol. Cell. Biol. 23, 1418–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Edin S., Oruganti S. R., Grundström C., Grundström T. (2010) Mol. Immunol. 47, 2057–2064 [DOI] [PubMed] [Google Scholar]
- 61. Berna-Erro A., Braun A., Kraft R., Kleinschnitz C., Schuhmann M. K., Stegner D., Wultsch T., Eilers J., Meuth S. G., Stoll G., Nieswandt B. (2009) Sci. Signal. 2, ra67. [DOI] [PubMed] [Google Scholar]
- 62. Byun-McKay S. A., Geeta R. (2007) Trends Ecol. Evol. 22, 338–344 [DOI] [PubMed] [Google Scholar]
- 63. Rosso L., Marques A. C., Weier M., Lambert N., Lambot M. A., Vanderhaeghen P., Kaessmann H. (2008) PLoS Biol. 6, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.