Abstract
Peptidoglycan recognition proteins (PGRPs) are involved in the recognition of pathogen-associated molecular patterns. The well known pathogen-associated molecular patterns include LPS from Gram-negative bacteria and lipoteichoic acid (LTA) from Gram-positive bacteria. In this work, the crystal structures of two complexes of the short form of camel PGRP (CPGRP-S) with LPS and LTA determined at 1.7- and 2.1-Å resolutions, respectively, are reported. Both compounds were held firmly inside the complex formed with four CPGRP-S molecules designated A, B, C, and D. The binding cleft is located at the interface of molecules C and D, which is extendable to the interface of molecules A and C. The interface of molecules A and B is tightly packed, whereas that of molecules B and D forms a wide channel. The hydrophilic moieties of these compounds occupy a common region, whereas hydrophobic chains interact with distinct regions in the binding site. The binding studies showed that CPGRP-S binds to LPS and LTA with affinities of 1.6 × 10−9 and 2.4 × 10−8 m, respectively. The flow cytometric studies showed that both LPS- and LTA-induced expression of the proinflammatory cytokines TNF-α and IL-6 was inhibited by CPGRP-S. The results of animal studies using mouse models indicated that both LPS- and LTA-induced mortality rates decreased drastically when CPGRP-S was administered. The recognition of both LPS and LTA, their high binding affinities for CPGRP-S, the significant decrease in the production of LPS- and LTA-induced TNF-α and IL-6, and the drastic reduction in the mortality rates in mice by CPGRP-S indicate its useful properties as an antibiotic agent.
Keywords: Crystal Structure, Innate Immunity, Lipopolysaccharide (LPS), Pathogen-associated Molecular Pattern (PAMP), Pattern Recognition Receptor, Lipoteichoic Acid (LTA), PGRP-S, Peptidoglycan (PGN)
Introduction
The innate immune system in animals provides the first line of defense against microbial infections (1). It works by an early sensing of the invading microorganisms through the recognition of molecular patterns present on the cell surface of bacteria that are absent in the host (2). Initially, the recognition was considered to be based on the binding of well known bacterial cell-surface molecules, peptidoglycans, by a class of innate immunity molecules. As a result, these proteins were called peptidoglycan recognition proteins (PGRPs).3 However, other commonly occurring pathogen-associated molecular patterns (PAMPs), such as LPS from Gram-negative bacteria (3) and lipoteichoic acid (LTA) from Gram-positive bacteria, were subsequently identified and included among the most commonly occurring PAMPs (4). Fortunately, these molecular patterns are not present on mammalian cell surfaces (5). The defense responses are generally activated when the microbial components are recognized by PGRPs, the pathogen sensors such as Toll-like receptors, Nod-like receptors, and double-stranded RNA sensors (6). Both LPS and LTA are glycol-lipids that are composed of an amphipathic lipid component and a hydrophilic polysaccharide core (7, 8). The excessive response to endotoxic LPS and LTA can lead to severe sepsis, which is a rapidly progressing inflammatory disease (9). The PGRPs are highly conserved protein molecules that may detect PAMPs from either Gram-negative or Gram-positive bacteria or both. The short form of PGRP isolated from the secretions of camel mammary glands (CPGRP-S) has been found to act potently against both Gram-negative and Gram-positive bacteria (10). We report here binding studies performed with CPGRP-S and two commonly occurring PAMPs, LPS and LTA, as well as detailed analyses of the three-dimensional structures of the two complexes of CPGRP-S with LPS (structure X) and with LTA (structure Y) determined at high resolutions. Real-time solution studies on the binding of LPS and LTA to CPGRP-S were carried out by surface plasmon resonance (SPR), which showed affinities at nanomolar concentrations for both compounds. The crystal structures of the complexes of CPGRP-S with LPS and LTA revealed the tight binding of both ligands. Studies were also carried out to determine the effects of CPGRP-S on LPS- and LTA-activated cultured peripheral blood mononuclear cells from healthy human volunteers using flow cytometry. The results indicate considerable reductions in the LPS- and LTA-induced expression of the proinflammatory cytokines TNF-α and IL-6 when CPGRP-S was added. Studies were further extended to evaluate the LPS- and LTA-induced effects in mouse models, in which the recoveries after introducing CPGRP-S were found to be significant.
EXPERIMENTAL PROCEDURES
Purification
Fresh samples of camel milk were obtained from the National Research Center on Camels (Bikaner, India). The procedures for purification of CPGRP-S have been described previously (11). The purity of the purified protein samples was checked by SDS-PAGE and MALDI-TOF mass spectrometry.
Fluorescence Spectroscopy
The fluorescence emission spectra were recorded on a Shimadzu FP-6200 spectrofluorometer using a 1-cm quartz cell in the wavelength range of 300–450 nm at an excitation wavelength of 280 nm at 298 K. The excitation and emission slits were both set at 5 nm with a scanning speed of 125 nm/min. The final concentrations of LPS and LTA used for the recording of emission spectra were 5, 10, 15, 20, and 25 μl in 10 mm HEPES (pH 8.0) from stock solutions of LPS and LTA prepared at 1 × 10−7 m in the same buffer while the CPGRP-S concentration was held constant at 1 × 10−9 m for all measurements. In both cases, the fluorescence effects of LPS and LTA were subtracted from the respective spectra of CPGRP-S in complex with LPS and LTA.
Binding Studies Using SPR
All SPR measurements were carried out using a Biacore 2000 system (Pharmacia Biosensor AB, Uppsala, Sweden) at 25 °C. CPGRP-S was immobilized onto a Biacore CM5 sensor chip using the amine coupling method until the SPR signal reached 1200 resonance units at a flow rate of 10 μl/min. The running buffer used was 10 mm HEPES-buffered saline (pH 7.4) containing EDTA and 0.005% Surfactant P20. Three different concentrations of the analytes LPS and LTA were passed through the CM5 chip to measure the binding of LPS and LTA to CPGRP-S. The dissociation of LPS and LTA from CPGRP-S was monitored by passing the buffer alone through the CM5 chip. The association (Kon) and dissociation (Koff) rate constants for LPS and LTA binding to CPGRP-S were calculated, and the values of the dissociation constants (Kd) were determined by the mass action relation Kd = Koff/Kon using BIAevaluation 3.0 software provided by the manufacturer.
Isolation of Peripheral Blood Mononuclear Cells from Peripheral Blood
In this study, a total of seven peripheral blood samples from healthy adults were analyzed. All peripheral blood samples were collected in heparinized vials and immediately processed for analysis of cytokine production. All samples were obtained with the approval of the local ethical committee after informed consent had been given by the donor. Mononuclear cells were isolated from heparinized blood by Ficoll-Hypaque gradient centrifugation and suspended in complete RPMI 1640 medium (Caisson Laboratories, Logan, UT) supplemented with 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal calf serum. The viability of cells was measured by trypan blue dye exclusion and was >97%. These cells were used for in vitro culture.
Cell Culture and Flow Cytometry
We performed in vitro stimulation of freshly isolated peripheral blood mononuclear cells to measured cytokine production with LPS (Escherichia coli, serotype O55:B5) and LTA (Staphylococcus aureus) from Sigma. These cells were cultured under five different experimental conditions: (i) medium alone, (ii) 10 μg/ml LPS, (iii) 10 μg/ml LPS + 5 μg/ml PGRP, (iv) 10 μg/ml LTA, and (v) 10 μg/ml LPS + 5 μg/ml PGRP in the presence of 10 μg/ml brefeldin A, a Golgi transport inhibitor (Sigma). After 24 h, the cultured cells were washed and surface-stained with anti-CD3 antibody (BD Biosciences), followed by intracellular staining for TNF-α and IL-6 (BD Biosciences) Stained cells were run in a BD FACSCalibur (BD Biosciences) and subsequently analyzed using FlowJo software (TreeStar Inc., Ashland, OR).
LPS-induced Septic Shock in Mice
8-week-old mice (weighing 25–30 g) were obtained from and housed in our institution's central animal facility. The mice were divided into three groups of eight each. The LPS group of mice were injected intraperitoneally with a lethal dose of LPS (30 mg/kg) only, and the LPS/PGRP group of mice were first given LPS (30 mg/kg) and then PGRP (10 mg/kg) immediately after the LPS injection. The third group of mice were used as controls and were injected with sterile saline solution only. The percent survival of the mice was analyzed using GraphPad Prism 4.0, and p values were calculated using the log-rank t test. The experimental procedure was approved by the Ethics Committee of the All India Institute of Medical Sciences and was performed in accordance with institutional animal care guidelines.
Crystallization
Freshly purified samples of protein were dissolved in buffer containing 50 mm Tris-HCl (pH 8.0) to a concentration of 15 mg/ml. 10 μl of the protein solution was mixed with an equal volume of a reservoir solution of 10% PEG-3350 containing 0.2 m potassium sodium tartrate and vortexed to make it homogeneous. 10-μl drops were set up for the hanging drop vapor diffusion method against the above reservoir solution. The crystals grew to dimensions of 0.4 × 0.3 × 0.3 mm3 in ∼2 weeks. The freshly grown crystals were soaked for 48 h in reservoir buffer containing LPS or LTA at 5 mg/ml.
X-ray Intensity Data Collection and Processing
The crystals of CPGRP-S soaked in the solutions of LPS and LTA were stabilized by the addition of 30% PEG-3350 and 0.2 m potassium sodium tartrate for data collection at low temperature. The crystals were mounted in nylon loops and flash-frozen in liquid nitrogen at 100 K. Two data sets, one each for crystals of the complexes of CPGRP-S with LPS and LTA, were collected using the Department of Biotechnology-sponsored MX beamline BM14 at ESRF (Grenoble, France) at a wavelength (λ) of 0.98 Å on a 165-mm MarCCD detector. The data were processed with AUTOMAR and SCALEPACK from the HKL package (12). The overall completeness of data for crystals of the complex of CPGRP-S with LPS was 100% to 1.7-Å resolution, and that with LTA was 95% to 2.1-Å resolution. The results of the data collection are given in Table 1.
TABLE 1.
Data collection and refinement statistics for the structures of the complexes of CPGRP-S with LPS and LTA
The values in parentheses correspond to the values in the highest resolution shell. r.m.s.d., root mean square deviation.
| PGRP-S + LPS | PGRP-S + LTA | |
|---|---|---|
| Protein Data Bank code | 3MU9 | 3O4K |
| Space group | I222 | I222 |
| Unit cell dimensions (Å) | a = 87.1, b = 100.8, c = 161.9 | a = 88.1, b = 101.5, c = 162.8 |
| No. of molecules in unit cell | 32 | 32 |
| Resolution range (Å) | 32.13–1.7 | 47.86–2.1 |
| Total number of measured reflections | 642,952 | 425,791 |
| No. of unique reflections | 78,352 | 40,418 |
| Rsym (%)a | 4.3 (39.1) | 9.0 (36.2) |
| I/σ(I) | 18.5 (2.1) | 30.0 (2.7) |
| Overall completeness of data (30.0–1.7) (%) | 99.9 (99.6) | 94.8 (88.0) |
| B-factor (Wilson plot; Å2) | 22.7 | 30.5 |
| Rcryst (%)b | 21.8 | 23.4 |
| Rfree (%) | 23.2 | 26.1 |
| Protein atoms | 5348 | 5348 |
| Water oxygen atoms | 613 | 523 |
| Atoms of tartrate | 10 | 10 |
| Atoms of glycerol | 6 | |
| Atoms of ligands | 47 | 54 |
| r.m.s.d. in bond lengths (Å) | 0.007 | 0.02 |
| r.m.s.d. in bond angles | 1.6° | 1.9° |
| r.m.s.d. in torsion angles | 23.9° | 15.7° |
| Mean B-factor for main chain atoms (Å2) | 26.4 | 39.5 |
| Mean B-factor for side chain atoms and waters (Å2) | 28.9 | 43.7 |
| Mean B-factor for all atoms (Å2) | 29.1 | 41.8 |
| Ramachandran φ,ψ map | ||
| Residues in most favored regions (%) | 88.9 | 89.7 |
| Residues in additionally allowed regions (%) | 11.1 | 10.3 |
a Rsym = Σhkl;Σi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIii(hkl).
b Rcryst = Σhkl;|Fo(hkl) − Fc(hkl)|/Σhkl|Fo(hkl)|, where Fo and Fc are the observed and calculated structure factors, respectively.
Structure Determination and Refinement
The structures of both complexes of CPGRP-S with LPS and LTA were determined with the molecular replacement method using the structure of native CPGRP-S (Protein Data Bank code 3C2X) (11) as the search model. The initial models were improved by repeated manual model building using program O (13) and Coot (14). The structures were refined with REFMAC 5.5 (15). The tight main chain and side chain non-crystallographic symmetry restraints between four crystallographically independent molecules (A, B, C, and D) were used in the refinement. The 2Fo − Fc and Fo − Fc electron density maps were calculated to adjust the protein chain in the electron density. After several rounds of model rebuilding and intermittent cycles of refinement, Rcryst factors dropped to 0.282 and 0.308, respectively. Group temperature factor (B) refinement was used with further model adjustments, yielding Rcryst factors of 25.3 and 27.8% for the structures with LPS and LTA, respectively. The tight non-crystallographic restraints and B-factor grouping were removed in the subsequent refinement cycles. Both 2Fo − Fc and Fo − Fc Fourier maps computed at this stage showed characteristic electron densities at 2.5σ in the presence of the ligands LPS and LTA at the interface of molecules C and D in the two structures (Fig. 1). The ligand atoms were added to the model in the further refinement cycles with isotropic B-factors. At this stage, the water molecules were also added in the subsequent refinement cycles. The final models of the complexes of CPGRP-S with LPS comprises four crystallographically independent protein molecules (A, B, C, and D), each having residues 1–171 and a bound LPS molecule. The positions of 613 water oxygen atoms and one tartrate molecule were obtained. The crystal structure of CPGRP-S with LTA comprises four protein molecules, one bound LTA molecule, 523 water oxygen atoms, and one tartrate molecule. The final Rcryst and Rfree factors for the structure of the complex with LPS are 21.8 and 23.2%, respectively, whereas the corresponding values for the structure of the complex with LTA are 23.4 and 26.1%, respectively.
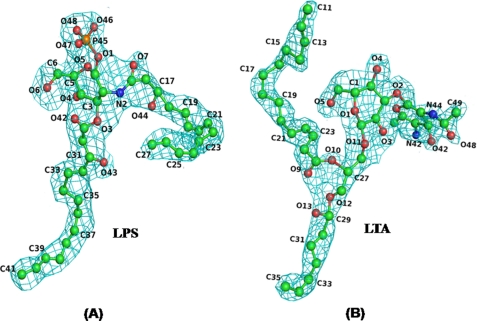
FIGURE 1.
Difference Fo − Fc electron density maps for the ligands LPS (A) and LTA (B) computed at the stages at which Rcryst factors were 25.3 and 27.8%, respectively, before introducing water oxygen atoms into the structures. The contour levels are drawn at 2.5σ. The atom numbers in ligands are also indicated.
Statistical Analysis
The statistical significance of results was determined using SigmaPlot and Prism 4.0 software (GraphPad Software, La Jolla, CA).
RESULTS
Fluorescence Spectroscopic Analysis
Fluorescence emission spectroscopy was used to determine the interactions of LPS and LTA with CPGRP-S, from which the binding constants were calculated. The intrinsic fluorescence of CPGRP-S when excited at 280 nm is due to the presence of four tryptophan residues, Trp-C66, Trp-C98, Trp-D66, and Trp-D98, and six tyrosine residues, Tyr-C59, Tyr-C71, Tyr-C160, Tyr-D59, Tyr-D71, and Tyr-D160, which were found to be associated with the binding cleft. Increasing concentrations of LPS and LTA with a fixed concentration of CPGRP-S showed that the fluorescence maxima of CPGRP-S at 352 nm were quenched upon the binding of LPS (supplemental Fig. S1A, panel a) and LTA (supplemental Fig. S1B, panel a). This indicates that both LPS and LTA are strong quenchers and hence must have strong affinities for CPGRP-S involving several hydrogen bonds, highlighting the changed polarity of the emitting fluorophore environment of the binding region. The observed fluorescence data were also used for calculating the resultant fluorescence quenching coefficient: Q = (F0 − F)/F0, where F represents the fluorescence intensities in the presence of LPS and LTA and F0 is the intensity in the absence of LPS and LTA. The values of Q (percent) were plotted against the concentrations of the ligands (supplemental Fig. S1, A, panel b, and B, panel b). The R2 values, indicating the goodness of the fit of the curves, were obtained using SigmaPlot 8.0 (16) and were 0.96 and 0.93, respectively. The error bars on the experimental points were estimated from the average of values that were obtained by repeating each experiment at least five times. The approximate values of the binding constants calculated using the binding equation described by Scatchard (see Ref. 17) as represented by dissociation constants (Kd) were 2.4 × 10−9 and 2.6 × 10−8 m, respectively.
SPR Spectroscopic Analysis
The binding studies on CPGRP-S with LPS and LTA were carried out using real-time SPR spectroscopy. The SPR sensograms showing the association and dissociation curves for LPS and LTA with immobilized CPGRP-S are given in supplemental Fig. S2. The global fitting of the primary data to a Langmuir 1:1 association model using the BIAevaluation 3.0 software package provided Kd values of 1.6 × 10−9 and 2.4 × 10−8 m for LPS and LTA, respectively.
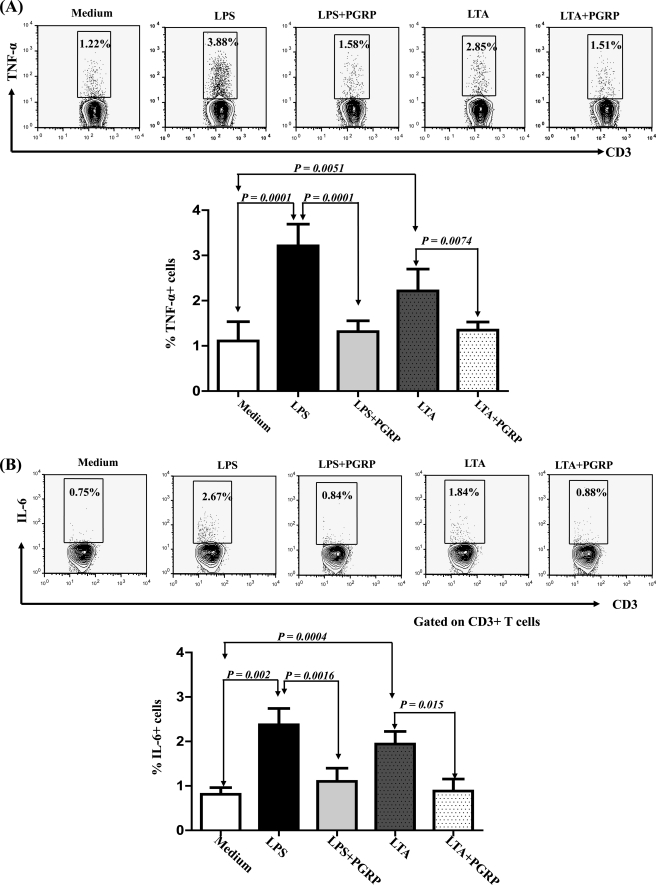
Inhibition of LPS- and LTA-induced Expression of TNF-α and IL-6 by CPGRP-S
It is well known that LPS and LTA induce the production of proinflammatory cytokines because they are ubiquitous cell-surface components of Gram-negative and Gram-positive bacteria. They participate in the pathogenesis of sepsis (18) by producing copious amounts of TNF-α and IL-6 predominantly in macrophages and T cells (19, 20). The levels of TNF-α were increased by ∼3.5-fold (Fig. 2A). Treatment with 1 μg/ml CPGRP-S significantly reduced LPS-induced TNF-α biosynthesis, whereas treatment with 5 μg/ml CPGRP-S drastically reduced LPS-augmented TNF-α production by CD3+ T cells. However, concentrations of CPGRP-S higher than 5 μg/ml did not cause further reduction in TNF-α levels in CD3+ T cells. Therefore, in subsequent experiments, 5 μg/ml CPGRP-S was used as the standard concentration for inhibition of LPS- and LTΑ-induced production of TNF-α and IL-6. T lymphocytes were treated with 10 μg/ml LPS and LTΑ, which increased the levels of TNF-α by 3.5- and 2.5-fold, respectively, compared with treatment with medium alone. The increased levels of TNF-α were almost completely abolished (>90% reduction) when the T cells were incubated with 5 μg/ml CPGRP-S along with 10 μg/ml LPS/LTΑ (Fig. 2B). Similarly, 2.3- and 2-fold increases in IL-6 levels were observed when 10 μg/ml LPS and LTΑ were administered, respectively. These were reduced by >90% with 5 μg/ml CPGRP-S. These data clearly indicate that CPGRP-S neutralizes effectively the proinflammatory effects of LPS and LTA in vitro presumably by blocking the availability of LPS and LTA to various PAMP receptors (CD14, Toll-like receptors, and CD6) expressed on T cells, which are known to recognize LPS/LTA (19, 20).
FIGURE 2.
Inhibition of LPS- and LTA-induced cytokine production in human T cells by CPGRP-S. Freshly isolated peripheral blood mononuclear cells derived from peripheral blood of healthy donors were cultured for 24 h with Golgi body transport blocker in the presence and absence of various doses of LPS/LTA and CPGRP-S. Cultured cells were washed and surface-stained with anti-CD3 antibody, followed by intracellular staining for TNF-α and IL-6. A, representative FACS plot showing percent production of TNF-α on gated CD3+ T cells and bar diagram, with error bars representing the mean ± S.D. calculated using data from six individual experiments. B, representative FACS plot showing percent production of IL-6 on gated CD3+ T cells and bar diagram for six individual experiments.
Survival Rate of Mice after LPS-induced Septic Shock
The survival graph (supplemental Fig. S3) obtained for the mice shows that nothing happened to the mice until 6 h after LPS injection. However, after the next 6 h, the survival rate dropped significantly, and at the end of 24 h, all of the mice that received a lethal dose of LPS died. On the other hand, the mice that received LPS and CPGRP-S showed a >75% survival rate. This is a significant improvement, indicating the therapeutic potential of CPGRP-S against LPS-induced septic shock.
Overall Structures of the Complexes of CPGRP-S with LPS and LTA
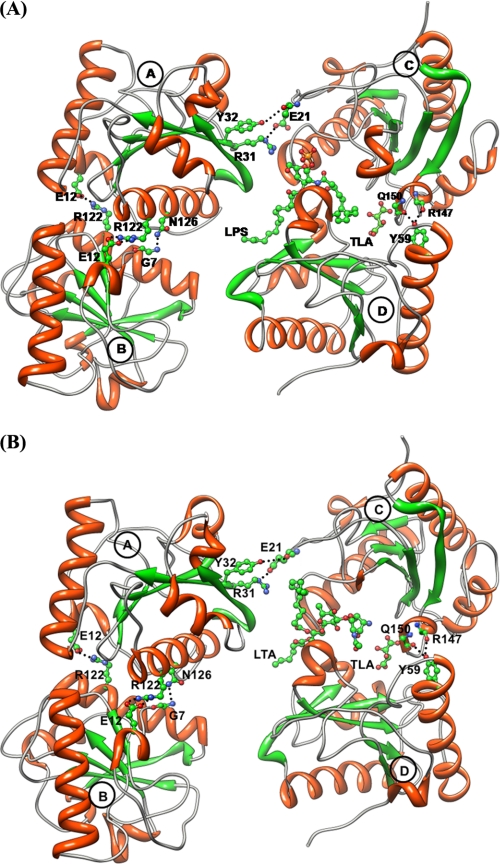
The refined structures of the complexes of CPGRP-S with LPS (Protein Data Bank code 3MU9) and LTA (code 3O4K) consist of four protein molecules with one ligand molecule in each structure (Fig. 3). The structure of unliganded CPGRP-S (11) shows that it forms a stable tetrameric complex in which the interface between molecules A and B is tightly packed, whereas the interfaces between molecules A and C and molecules C and D are partially closed, covering nearly one-third portions of the interfaces on the distal side. The inner parts of these interfaces form a cleft-like structure consisting of features favorable for intermolecular interactions (Fig. 4). In contrast, the interface between molecules B and D lacks intermolecular interactions and forms a channel presumably to allow the diffusion of ligands, as it connects the deeply situated cleft to the surface. In structure 3MU9, LPS is bound in the cleft primarily at the C-D interface, and one of its two hydrocarbon chains extends into the diffusion channel. The aromatic and hydrophilic parts of the LTA molecule occupy a similar region of the cleft at the C-D interface as occupied by the aromatic and hydrophilic moieties of LPS. However, unlike LPS, one of its two hydrocarbon chains is observed in the binding space provided by the A-C interface. In the case of LPS, the region of the A-C interface is empty. The second hydrocarbon arm of LTA is placed in the B-D interface along the surface of molecule D. The positions of 721 water molecules were determined in structure 3MU9, whereas the coordinates of 560 water oxygen atoms were obtained in structure 3O4K. The tetrameric association of molecules A, B, C, and D of CPGRP-S is stabilized through direct intermolecular contacts between molecules A and B, A and C, and C and D, whereas the interface between molecules B and D is devoid of direct protein-protein intermolecular interactions. The B-D interface with approximate dimensions of 36 Å (length) and 9 Å (width) is filled by bulk solvent molecules.
FIGURE 3.
Binding of the ligands LPS (A) and LTA (B) to the CPGRP-S tetramer. A few key interactions between molecules A and B, A and C, and C and D are shown. Dotted lines indicate hydrogen bonds.
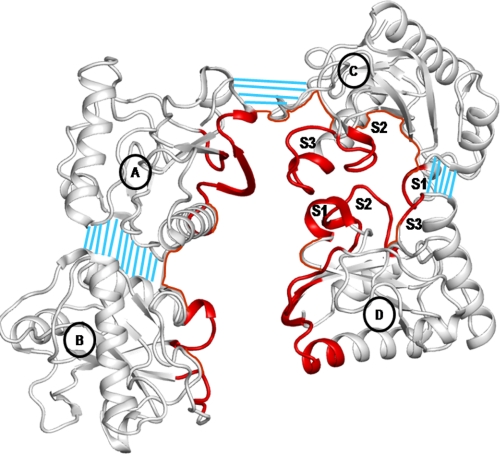
FIGURE 4.
The ligand-binding cleft in the CPGRP-S tetrameric complex is shown in red. Three characteristic loops, S1, S2, and S3, are indicated at the C-D interface. The intermolecular contact regions are indicated by blue grid lines.
Molecular Structure
The superimpositions of Cα positions of the four crystallographically independent molecules of CPGRP-S on each other in structures 3MU9 and 3O4K show a maximum root mean square shift of 0.6 Å, indicating that the backbone folding of all four molecules is identical. Therefore, in the subsequent discussion on the molecular structure, only a single protein molecule of CPGRP-S will be described. The molecular architecture of CPGRP-S described previously includes a central β-sheet surrounded by three major α-helices. The molecular structure is stabilized by three disulfide linkages, Cys-6–Cys-130, Cys-22–Cys-67, and Cys-43–Cys-49. The structure of CPGRP-S in complex is identical to that of the unbound protein, with a root mean square shift of 0.7 Å for the Cα traces. The corresponding value upon comparison with truncated human PGRP-S (HPGRP-S) for 170 residues is 0.9 Å.
Molecular Association and Formation of the Binding Site for PAMPs
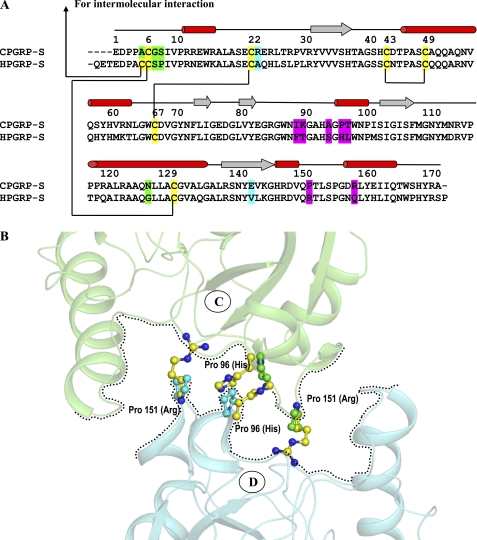
Four CPGRP-S molecules (A, B, C, and D) associate in an asymmetrical tetrameric assembly. A novel binding site is formed mainly at the interface of molecules C and D, with an extension into the interface of molecules A and C (Fig. 4). The molecular pairs of C-D and A-C are held together by several hydrogen bond interactions and van der Waals contacts on the distal part of the interface. In contrast, molecules A and B form a tightly packed structure with more than one dozen hydrogen bonds and at least two dozen van der Waals contacts with distances of <4.2 Å (21, 22). The tightly formed dimer of molecules A and B is shifted by ∼10 Å with respect to a relatively less tightly formed dimer of molecules C and D in the direction perpendicular to the axis of the interface of molecules C and D. The total length of the dimer in the direction perpendicular to the dimeric interface is ∼54 Å. The two dimers are inclined inwardly at ∼30°. The A-B dimer interacts with the C-D dimer through the A-C interface, forming several intermolecular contacts. The cleft between molecules C and D is formed with multiple binding subsites. The total binding space of the cleft includes three-fourths of the area of the C-D interface, nearly half of the region of the A-C interface, and the central core of the tetramer. The cleft is connected to the surface through a channel formed between molecules B and D. The cleft contains features compatible with a wide range of PAMPs. Such an arrangement has not been observed in other PGRPs (23–28). The closely related HPGRP-S has been reported in the monomeric form. However, the first nine residues from the N terminus are not part of the structure. This also includes Cys-8, which has been suggested to facilitate the formation of a covalently linked dimer of HPGRP-S (29, 30). Interestingly, an interface similar to that of molecules C and D of CPGRP-S cannot be formed in HPGRP-S because of incompatibility of critical residues, where Pro-96 and Pro-151 in CPGRP-S are replaced with His-96 and Arg-151 (Fig. 5A). Pro-96 and Pro-151 from molecules C and D define the lines of the C-D interface, whereas the corresponding residues, His-96 and Arg-151, in the human counterpart disrupt the interfacial arrangement (Fig. 5B). These residues in the corresponding domain of PGRPs from other species are not conserved (supplemental Fig. S4), indicating that this might be a unique feature of CPGRP-S. In general, proline residues at the interface promote protein-protein association (31).
FIGURE 5.
A, sequence alignment of CPGRP-S and HPGRP-S is shown with elements of secondary structures on top. Cysteine residues are highlighted in yellow. The important residues at the A-B (green) and C-D (red) interfaces are compared with the corresponding residues in HPGRP-S. The differences indicate incompatibilities in HPGRP-S for making an oligomer as observed in CPGRP-S. B, the formation of a PAMPs-binding cleft at the interface of molecules C and D is hampered in HPGRP-S because four proline residues (Pro-C96, Pro-C151, Pro-D96, and Pro-D151) at the C-D interface in CPGRP-S are replaced with His-96 and Arg-151 in HPGRP-S.
Structure of the Complex of PGRP-S with LPS
The structure of the complex of CPGRP-S with LPS (CPGRP-S·LPS) was determined at 1.7-Å resolution. When the Cα traces of molecules A, B, C, and D from the structure of the CPGRP-S·LPS complex were superimposed on those of unbound CPGRP-S, the root mean square shifts were found to be <0.7 Å, indicating that the binding of LPS to CPGRP-S did not perturb the conformation of the protein chain. However, the orientations of several side chains in the regions where LPS bound to CPGRP-S were altered considerably. The protein segments consisting of residues 59–66 (designated subsite S1), 93–98 (designated subsite S2), and 149–153 (designated subsite S3) belonging to molecules C and D are the major contributors to the binding cleft at the C-D interface. These subsites of molecules C and D are arranged in the reverse order. If the order in molecule C is S1, S2, and S3, it is S3, S2, and S1 in molecule D. These segments form outwardly protruding loops and generate a zigzag interface, as the opposing loops from molecules C and D are slightly shifted with respect to each other. The length of the cleft as measured along the C-D interface is ∼24 Å. Although highly irregular with deep cleavages on either side of the central line of the interface, the approximate width of the binding cleft is nearly 8.5 Å. The cleft opens into the center of tetramer, whereas it is closed from the opposite side. The binding space of the cleft is extendable into the A-C interface, which is ∼10 Å in length. The hydrophilic center of the cleft is situated in the proximity of subsites S2. In the CPGRP-S·LPS complex, the hydrophilic moiety of LPS is held at this site while the two hydrophobic chains are pulled in opposite directions, where one arm occupies the space on the distal side of the cleft, and the second arm is expelled into the B-D interface along the surface of molecule D. The LPS molecule makes extensive contacts with protein atoms, forming at least two dozen hydrogen bonds and a similarly large number of van der Waals interactions. The prominent residues that contribute to the hydrogen bonding are Arg-A170, Trp-C66, Arg-C85, Lys-C90, Gly-C91, Ala-C92, His-C93, Asn-C99, Thr-D97, Asp-D98, Val-D149, and Gln-D150 (supplemental Table S1). The observed interactions indicate that Lys-90, Asn-99, and Arg-170 play an important role in the recognition of PAMPs.
Structure of the Complex of PGRP-S with LTA
The structure of the CPGRP-S·LTA complex was determined at 2.1-Å resolution. When the Cα traces of CPGRP-S·LTA were superimposed on the Cα traces of unbound CPGRP-S (11), root mean square shifts of 0.8 Å or less were obtained, indicating that the binding of LTA did not disturb the backbone conformation. However, the orientations of some of the side chains at the interfaces of molecules C and D and molecules A and C changed appreciably. Notable conformational differences were observed in the side chains of Glu-C21, Trp-C66, Pro-C96, Glu-D24, Pro-D96, Trp-D98, Gln-D150, Pro-D151, and Glu-A142. The hydrophilic moiety of LTA occupies an extra space in addition to the site occupied by LPS. The additional space includes part of the body of molecule D, which is in the proximity of Gly-D95, Pro-D96, Asn-D99, and His-D146. On the other hand, LTA does not occupy a particular space occupied by LPS in molecule C, which is in the proximity of Lys-C90, Gly-C91, Asn-C99, and Pro-C100. The two hydrophobic arms of LTA bifurcate at the mouth of the cleft of the C-D interface in the core of the tetramer. The two chains move in opposite directions toward the A-C and B-D interfaces. The LTA molecule forms a number of hydrogen bonds and a large number of hydrophobic interactions with protein atoms (supplemental Table S2). In this structure, the unique hydrogen bond interactions are provided by Asn-A140, Gly-D95, and Glu-D150.
DISCUSSION
Although the C-terminal domain of ∼165 amino acid residues known as the PGRP domain is conserved in various PGRPs (21–26), structural comparison shows a significant structural diversity, indicating a considerable functional variability. Because the capacity for the recognition of a variety of PAMPs is dependent on the versatility of the binding site, the novelty in the formation of binding sites in various PGRPs is of particular significance. The crystal structures of PGRP-LB, PGRP-SA, PGRP-LC, and PGRP-LE from Drosophila and the C-terminal peptidoglycan-binding domain of PGRP-IαC and PGRP-IβC and cloned truncated PGRP-S from human represent different structural units of PGRPs. The first complete crystal structure of a mammalian PGRP belonging to the S class is that of CPGRP-S (11). The sequence alignment of CPGRP-S and HPGRP-S shows an overall sequence identity of ∼75%, whereas the segment with the first 30 residues from the N terminus shows <50% sequence identity. HPGRP-S has three extra residues (Gln-1–Glu-2–Thr-3) on the N terminus with an extra cysteine at position 8, which corresponds to Ala-5 in CPGRP-S. The reported structure of HPGRP-S is as a monomer in which the first nine residues, including Cys-8, were not observed. However, it has been reported in the literature that HPGRP-S forms a disulfide-linked dimer through Cys-8 (29, 30). In contrast, CPGRP-S forms a noncovalent association with four molecules, resulting in the formation of a novel PAMP-binding site that is situated deep inside the asymmetric homotetramer. In contrast, in HPGRP-S, due to the indicated covalent linkage and additional incompatible residues at the corresponding faces of molecules C and D, the structure is unlikely to be a tetramer, which is similar to CPGRP-S. Thus, CPGRP-S is the only structure determined so far in which a tetrameric complex giving rise to a versatile binding site has been observed. It has been found to be capable of binding to a wide range of PAMPs with high specificities and potencies. The structures of the complexes of CPGRP-S with two important PAMPs, LPS from Gram-negative bacteria and LTA from Gram-positive, have clearly shown the potential of the binding site, which is capable of recognizing a wide range of microorganisms. The solution studies using fluorescence spectroscopy and observations with SPR showed that CPGRP-S binds to both LPS and LTA with nearly similar affinities as indicated by Kd values of 1.6 × 10−9 and 2.4 × 10−8 m, respectively. As revealed by the structures of two complexes of CPGRP-S with LPS and LTA, the hydrophilic moieties of the two ligands occupy an overlapping space as well as distinct non-overlapping regions to accommodate their non-similar moieties. This happens because the binding cleft offers a variety of features. As observed from the structure of CPGRP-S·LPS, the distinct interactions with the hydrophilic moiety of LPS are provided by Lys-C90, Ala-C92, Asn-C99, and Arg-A170, whereas the residues that interact with LTA are Thr-D97, Asn-C99, Gln-D150, and Asn-A140. In both cases, the interactions are optimally achieved, but the residues involved in the interactions are not identical. The predominant interactions with LPS are provided by molecule C, whereas molecule D is the major contributor for the interactions with LTA. Similarly, parts of the hydrocarbon chains of LPS and LTA are placed in identical regions, whereas other parts are aligned differently but with nearly similar numbers of attractive van der Waals contacts. In the structure of CPGRP-S·LPS, one of the two hydrocarbon arms is adjusted in the remaining space at the C-D interface, where van der Waals interactions are provided by Trp-D66, Ala-D92, His-D93, Gly-D95, Pro-C96, Gln-C150, and Pro-C151. The second hydrophobic arm of LPS lies in the B-D interface along the surface of molecule D. This moiety of LPS is stabilized by a number of van der Waals contacts provided by Trp-D98, Asp-D148, Val-D149, and Glu-D150.
In the case of LTA, neither of its two hydrocarbon arms could be accommodated in the cleft at the C-D interface due to stereochemical incompatibilities. Instead, both chains are placed outside the C-D interface at the junction near the core of the tetrameric complex. One chain is stabilized in the A-C interface through van der Waals contacts provided by Arg-A31, Glu-A142, Lys-A144, Arg-A170, Glu-C24, Cys-C67, Lys-C90, and Ala-C92. The interface of molecules A and C is empty in the structure of CPGRP-S·LPS. The second chain of LTA is placed in the opposite direction into the B-D interface along molecule D. This is stabilized by van der Waals interactions provided by Thr-D97, Trp-D98, Val-D149, and Ala-D171. Thus, the two chemically and structurally distinct PAMPs, LPS and LTA, from Gram-negative and Gram-positive bacteria, respectively, are recognized equally well by CPGRP-S. The formation of such a versatile binding site is the result of the association of four molecules of CPGRP-S, which is the only example among the proteins of the PGRP-S family. The flow cytometric studies confirmed that CPGRP-S suppressed the LPS- and LTA-induced biosynthesis of the proinflammatory cytokines TNF-α and IL-6 by >90% in both T lymphocytes and monocytes. The in vivo experiments carried out using mouse models showed that lethal doses of LPS led to the death of all mice in 24 h. However, if CPGRP-S was injected soon after the administration of LPS, a survival rate of >80% was observed.
In general, the specific binding clefts on protein surfaces are present to interact with compounds having stereochemically favorable features. Because the structures of various PAMPs differ considerably, it is unlikely that a single binding site could accommodate them with such high affinities and specificities. If a single binding site has to perform such diverse binding roles, it could do so only at moderate potencies. To produce high potencies for a variety of ligands, the binding site must be equipped with multiple features that are hard to generate in a single protein molecule. On the other hand, binding studies have shown that CPGRP-S binds to various PAMPs with high affinities. The structure showed that it is because of the formation of the tetrameric complex of CPGRP-S, which allowed the construction of a versatile ligand-binding site. The observed binding site at the interface of four protein molecules with multiple features specific to different PAMPs makes the structure of CPGRP-S a potent binder. The interaction site is buried deep inside the tetramer, which is connected through a broad and long channel with favorable features to attract the PAMPs. Indeed, this is an excellent example of designing a highly potent binding site by using multimeric complexes of protein molecules. Evolutionarily, it appears to be of utmost significance in the case of camels because of the extreme conditions of their habitat as well as their unusual body metabolism. It is also well known that their immunological makeup with single-chain IgG molecules carries a compromised immune system. Furthermore, unlike in other species, the other supporting antibacterial proteins, such as lactoperoxidase and lysozyme C, are available only at very low concentrations in the mammary secretions of camels (10). Therefore, these very useful properties of CPGRP-S may be of great therapeutic value as an antibiotic to protect mammary glands and newborns from bacterial infection. It should also be mentioned that mastitis, a rampant infection in domesticated milking animals, is rarely reported in camels. This useful antibacterial property of CPGRP-S can also be exploited as a common antibiotic agent for humans. This is particularly significant as there is an alarming rise in the incidence of bacterial resistance to known antibiotics. This also brings the amino acid sequence of CPGRP-S into focus, particularly the presence of Pro-96 and Pro-151 at one of the interfaces and the absence of three N-terminal residues and Cys-8 compared with HPGRP-S. In the PGRP-S family, such a homotetrameric complex has been observed only for CPGRP-S.
Supplementary Material
This work was supported by the Departments of Science and Technology and Biotechnology, Ministry of Science and Technology, Government of India (New Delhi).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
- PGRP
- peptidoglycan recognition protein
- PAMP
- pathogen-associated molecular pattern
- LTA
- lipoteichoic acid
- CPGRP-S
- camel PGRP-short
- SPR
- surface plasmon resonance
- HPGRP-S
- human PGRP-S.
REFERENCES
- 1. Hoffmann J. A., Reichhart J. M. (2002) Nat. Immunol. 3, 121–126 [DOI] [PubMed] [Google Scholar]
- 2. Medzhitov R., Janeway C. A., Jr. (1998) Curr. Opin. Immunol. 10, 12–15 [DOI] [PubMed] [Google Scholar]
- 3. Beutler B., Rietschel E. T. (2003) Nat. Rev. Immunol. 3, 169–176 [DOI] [PubMed] [Google Scholar]
- 4. Ginsburg I. (2002) Lancet Infect. Dis. 2, 171–179 [DOI] [PubMed] [Google Scholar]
- 5. Janeway C. A., Jr. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 [DOI] [PubMed] [Google Scholar]
- 6. Fritz J. H., Ferrero R. L., Philpott D. J., Girardin S. E. (2006) Nat. Immunol. 7, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 7. Raetz C. R. (1990) Annu. Rev. Biochem. 59, 129–170 [DOI] [PubMed] [Google Scholar]
- 8. Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., Pinsky M. R. (2001) Crit. Care Med. 29, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 10. Kappeler S. R., Heuberger C., Farah Z., Puhan Z. (2004) J. Dairy Sci. 87, 2660–2668 [DOI] [PubMed] [Google Scholar]
- 11. Sharma P., Singh N., Sinha M., Sharma S., Perbandt M., Betzel C., Kaur P., Srinivasan A., Singh T. P. (2008) J. Mol. Biol. 378, 923–932 [DOI] [PubMed] [Google Scholar]
- 12. Otwinowsk Z., Minor W. (1996) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 13. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 14. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 16. Charland M. B. (1995) SigmaPlot for Scientists, Version 8.0, Wm. C. Brown Communications, Inc., Dubuque, IA [Google Scholar]
- 17. Hu Y. J., Ou-Yang Y., Zhang Y., Liu Y. (2010) Protein J. 29, 234–241 [DOI] [PubMed] [Google Scholar]
- 18. Wang J. E., Dahle M. K., McDonald M., Foster S. J., Aasen A. O., Thiemermann C. (2003) Shock 20, 402–414 [DOI] [PubMed] [Google Scholar]
- 19. Sarrias M. R., Farnós M., Mota R., Sánchez-Barbero F., Ibáñez A., Gimferrer I., Vera J., Fenutría R., Casals C., Yélamos J., Lozano F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komai-Koma M., Gilchrist D. S., Xu D. (2009) Eur. J. Immunol. 39, 1564–1572 [DOI] [PubMed] [Google Scholar]
- 21. Pauling L. (1960) The Nature of the Chemical Bond and the Structure of Molecules and Crystals: An Introduction to Modern Structural Chemistry, 3rd Ed., Cornell University Press, Ithaca, NY [Google Scholar]
- 22. Kitaigorodskii A. I. (1955) Organic Chemistry Crystallography, Consultants Bureau, New York [Google Scholar]
- 23. Guan R., Wang Q., Sundberg E. J., Mariuzza R. A. (2005) J. Mol. Biol. 347, 683–691 [DOI] [PubMed] [Google Scholar]
- 24. Kim M. S., Byun M., Oh B. H. (2003) Nat. Immunol. 4, 787–793 [DOI] [PubMed] [Google Scholar]
- 25. Reiser J. B., Teyton L., Wilson I. A. (2004) J. Mol. Biol. 340, 909–917 [DOI] [PubMed] [Google Scholar]
- 26. Chang C. I., Chelliah Y., Borek D., Mengin-Lecreulx D., Deisenhofer J. (2006) Science 311, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 27. Lim J. H., Kim M. S., Kim H. E., Yano T., Oshima Y., Aggarwal K., Goldman W. E., Silverman N., Kurata S., Oh B. H. (2006) J. Biol. Chem. 281, 8286–8295 [DOI] [PubMed] [Google Scholar]
- 28. Guan R., Brown P. H., Swaminathan C. P., Roychowdhury A., Boons G. J., Mariuzza R. A. (2006) Protein Sci. 15, 1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., van der Fits L., Voerman J. S., Melief M. J., Laman J. D., Wang M., Wang H., Wang M., Li X., Walls C. D., Gupta D., Dziarski R. (2005) Biochim. Biophys. Acta 1752, 34–46 [DOI] [PubMed] [Google Scholar]
- 30. Lu X., Wang M., Qi J., Wang H., Li X., Gupta D., Dziarski R. (2006) J. Biol. Chem. 281, 5895–5907 [DOI] [PubMed] [Google Scholar]
- 31. Kini R. M., Evans H. J. (1995) Biochem. Biophys. Res. Commun. 212, 1115–1124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.