Abstract
The crystal structure of NADPH-cytochrome P450 reductase (CYPOR) implies that a large domain movement is essential for electron transfer from NADPH via FAD and FMN to its redox partners. To test this hypothesis, a disulfide bond was engineered between residues Asp147 and Arg514 in the FMN and FAD domains, respectively. The cross-linked form of this mutant protein, designated 147CC514, exhibited a significant decrease in the rate of interflavin electron transfer and large (≥90%) decreases in rates of electron transfer to its redox partners, cytochrome c and cytochrome P450 2B4. Reduction of the disulfide bond restored the ability of the mutant to reduce its redox partners, demonstrating that a conformational change is essential for CYPOR function. The crystal structures of the mutant without and with NADP+ revealed that the two flavin domains are joined by a disulfide linkage and that the relative orientations of the two flavin rings are twisted ∼20° compared with the wild type, decreasing the surface contact area between the two flavin rings. Comparison of the structures without and with NADP+ shows movement of the Gly631–Asn635 loop. In the NADP+-free structure, the loop adopts a conformation that sterically hinders NADP(H) binding. The structure with NADP+ shows movement of the Gly631–Asn635 loop to a position that permits NADP(H) binding. Furthermore, comparison of these mutant and wild type structures strongly suggests that the Gly631–Asn635 loop movement controls NADPH binding and NADP+ release; this loop movement in turn facilitates the flavin domain movement, allowing electron transfer from FMN to the CYPOR redox partners.
Keywords: Crystal Structure, Cytochrome P450, Electron Transfer, Flavoproteins, Nicotinamide, Cytochrome P450 Reductase, Diflavin Enzyme, Hydride Transfer, Protein Domain Movement
Introduction
NADPH-cytochrome P450 oxidoreductase (CYPOR)5 is an ∼78-kDa diflavin microsomal protein that provides electrons to a variety of microsomal cytochromes P450 (P450) and heme oxygenase (reviewed in Ref. 1). It receives electrons from NADPH and transports them via FAD and FMN to its redox partners. FMN is the ultimate donor of electrons to acceptor proteins (2, 3). In humans, microsomal P450s catalyze the oxidation of a cornucopia of endogenous and exogenous substrates. P450s are responsible for the metabolism of at least one step in the biodegradation of the vast majority of drugs consumed by humans. P450s also metabolize the myriad of essential endogenous compounds, including steroids, vitamins, eicosanoids, and hormones (for review see Ref. 4). Although the liver harbors the highest concentration of P450s, they are found in almost every tissue and organ, including the brain and kidney, where their metabolites participate in regulating anxiety levels and blood pressure, respectively (5, 6). Because CYPOR is an essential electron donor to microsomal P450s, it is critical to the function of the large number of physiologic processes regulated by P450.
The electron transfer pathway in CYPOR begins with the obligate two electron donor, NADPH, which transfers a hydride ion to FAD, which in turn donates electrons to the FMN cofactor. The FMN hydroquinone then transfers electrons, one at a time, to its redox partners. An anionic convex surface surrounds the FMN, which is complementary both in charge and shape to the concave cationic CYPOR-docking surface on P450 (7–10). P450 receives electrons from CYPOR at two separate steps in its complex reaction mechanism. The ferric protein is reduced to the ferrous P450, which is then able to bind oxygen. A second electron is delivered to the oxyferrous P450, forming a peroxo species (Fe3+OO2−), which rapidly and irreversibly proceeds through the catalytic cycle to yield an oxidized substrate and a water molecule (for review see Ref. 11). The crystal structure of the wild type reductase reveals that the two flavins are in van der Waals contact, which accounts for the rapid electron transfer from FAD to FMN (7). However, in this structure, the nicotinamide moiety of NADP+ is blocked from interaction with FAD by Trp677, whose indole ring stacks against the re-face of the isoalloxazine ring of FAD. When Trp677 was deleted, the crystal structure of the mutant revealed that the nicotinamide moiety of NADP+ was now in van der Waals contact with the isoalloxazine ring with a tilt of ∼ 30° between the planes of the two rings (12). This result indicates that, in the wild type protein, Trp677 moves away from the isoalloxazine ring of FAD and allows the nicotinamide ring to interact with the flavin and donate a hydride ion to FAD. Thus, the structure of CYPOR provided a plausible structural explanation for the mechanism of electron transfer from NADPH to FAD and from FAD to FMN. Unfortunately, it was not possible to dock P450 to the closed conformation of CYPOR, and it was hypothesized that the FMN domain would rotate away from the FAD domain to dock with P450. Support for that hypothesis was obtained by crystallizing a mutant CYPOR in three distinct “open” conformations in which FAD and FMN were ∼30–60 Å apart. The mutant CYPOR lacked four amino acids (236TGEE239) in the linker connecting the FMN domain to the remainder of the protein. When provided with sufficient electrons, the open form of CYPOR, which exhibited slow interflavin electron transfer, was able to support catalysis by P450 (8). In this study, we report the crystal structure of a mutant CYPOR with an engineered disulfide bond between the FAD and FMN domains that is essentially incapable of supporting P450 activity unless the disulfide bond is reduced, demonstrating that CYPOR must undergo a large conformational change in order for the FMN domain to interact with P450.
EXPERIMENTAL PROCEDURES
Materials
Sodium dithionite, dithiothreitol (DTT), benzphetamine, superoxide dismutase (SOD), catalase (CAT), and horse heart cytochrome c were purchased from Sigma. Redistilled glycerol was purchased from Roche Diagnostics, and dilauroylphosphatidylcholine (DLPC) was from Doosan Serdary Research Laboratory (Toronto, Canada).
Generation of the 147CC514 Mutant CYPOR
The rat wild type CYPOR has seven cysteines at positions 136, 228, 363, 445, 472, 566, and 630 in the amino acid sequence. Prior to engineering a disulfide bond between the FAD and FMN domains, it was first necessary to remove the nonessential cysteines in CYPOR to prevent unwanted disulfide bond formation. A sequence alignment of CYPOR from yeast and mammalian sources indicated that only Cys630 was invariant and presumably essential. Mutation of Cys630 to alanine decreased the activity of CYPOR by 98%, thereby confirming its critical role in CYPOR function (13). Cys566, which is in contact with NADPH, was highly conserved but not invariant. The C566A mutant protein demonstrated full catalytic activity but exhibited a 2.5-fold increased KmNADPH (14). Accordingly, the six nonconserved and nonessential cysteines (positions 136, 228, 363, 445, 472, and 566) were mutated to residues chosen on the basis of their occurrence in CYPOR proteins of nonmammalian species, primarily yeast. The mutant protein lacking the six cysteines possessed a normal visible absorption spectrum and cyt c reductase activity (78 versus 73 units for the wild type; 1 unit = nmol of cyt c reduced per s/nmol of CYPOR), confirming that the six cysteines were not essential for the function of rat CYPOR.
Cloning and Site-directed Mutagenesis
Unless otherwise stated, all DNA manipulations were performed by standard procedures as described by Ausubel et al. (15). Mutagenesis of the full-length rat CYPOR was carried out by PCR as described previously (16). For all mutants, DNA sequencing confirmed the presence of the desired mutation and the absence of any other base changes. For preparation of mutants containing only a single cysteine mutation, DNA fragments coding for one of the following mutations, C136A, C228A, C363T, C445L, and C472T, were prepared by overlap extension PCR (17), digested with appropriate restriction enzymes, and cloned one at a time into the CYPOR expression plasmid C566A (14), which contains a lpp/lac hybrid promoter and an ompA signal peptide with eight extra amino acids at the N terminus of the expressed protein. The CYPOR mutant protein lacking six of the seven cysteines was prepared by restriction enzyme digestion and ligation of appropriate restriction fragments from the singly substituted plasmids into the C566A reductase expression plasmid. The supplemental Table S1 shows the primer pairs used to generate the mutations and restriction fragments used for cloning. Expression and purification indicated that the protein containing a single cysteine residue at position 630, designated Cys630-only, exhibited kinetic properties identical to that of the C566A enzyme, i.e. full catalytic activity and a 2.5-fold increased KmNADPH (14). For site-specific cross-linking mutant, the D147C and R514C substitutions were prepared by overlap extension PCR and cloning into the plasmid, pCys630-only, to generate the plasmid p147CC514.
Construction of the Plasmid Harboring the cDNA of the Truncated Form of the 147CC514 Mutant of Rat CYPOR
The truncated form of the disulfide-bridged CYPOR plasmid containing the residues 57–678 was generated from the plasmid p147CC514, which contains the full-length 147CC514 CYPOR, by excising a cDNA fragment encoding residues 86–678 using restriction sites BbvCI/HindIII. The mutant cDNA fragment was then cloned into the BbvCI/HindIII site of plasmid p57-678-CYPOR, which contains the truncated form of wild type CYPOR in a Novagen pET-23b vector (16). The overall procedure resulted in replacing a cDNA fragment coding for residues 86–678 of wild type CYPOR with a cDNA fragment (residues 86–678) coding for the 147CC514 mutant CYPOR.
Protein Expression and Purification
Expression of full-length 147CC514 was performed in freshly transformed competent C41 (DE3) Escherichia coli (18, 19), grown in 1 liter of LB medium (with an additional 5 g of yeast extract) containing 0.24 mm carbenicillin, 0.2% (w/v) glucose, and 5.3 nm riboflavin in a 2.8-liter Fernbach flask with shaking at 180 rpm and 35 °C. When the absorbance at 600 nm (A600) reached ∼2, the culture was cooled to 21 °C for 30 min, and isopropyl β-d-thiogalactopyranoside was added to a final concentration of 400 μm, and incubation at 21 °C was continued in the dark for ∼33 h with shaking at 140 rpm. Inclusion bodies were not observed at 21 °C. The cells were harvested when the A600 reached ∼5. The cells were pelleted at ∼3000 × g and subsequently resuspended in 0.1 m Tris acetate, pH 8.1, at 4 °C, 250 μm EDTA, and 30 μg/ml of lysozyme and left on ice for 30 min without shaking. Once again, the cells were pelleted at ∼3000 × g and resuspended in 50 mm Tris acetate, pH 8.1, at 4 °C, 0.1 mm EDTA, 10% glycerol (v/v). Two complete miniprotease inhibitor mixture tablets (Roche Diagnostics) were added. Cells were lysed by sonication, being careful to maintain the temperature at less than 12 °C. Cell debris was removed from the lysed cells by centrifugation at ∼6400 × g. The supernatant was then subjected to centrifugation at 100,000 × g for 45 min to pellet the “membrane fraction,” which was resuspended in buffer A (50 mm Tris acetate, pH 8.1, at 4 °C, 0.1 mm EDTA, 10% v/v glycerol) containing 1 complete miniprotease inhibitor mixture tablet (Roche Diagnostics). The resuspended membrane fraction was sonicated and subsequently solubilized by stirring at 4 °C at least 3 h to overnight in a solution with a final concentration of 0.3% (v/v) Triton X-100. The solubilized protein-containing solution was centrifuged at 100,000 × g for ∼45 min to remove unsolubilized material. The clarified supernatant was diluted 3-fold with buffer A and loaded onto a DEAE-Sepharose column (Sigma), which had been pre-equilibrated with buffer A plus 0.1% (v/v) Triton X-100. The column was washed with ∼4 column volumes of buffer A plus Triton X-100 and 0.15 m NaCl, until the absorbance of the eluate between 410 nm and 420 nm was less than 0.005. This wash removes unwanted heme proteins. The CYPOR protein was then eluted with a linear gradient between 0.15 and 0.5 m NaCl in the column washing solution. Fractions without a shoulder at 410–420 nm in the flavin spectrum were pooled and concentrated to 1/5 to 1/6 of their original volume in an Amicon YM30 concentrator. NaCl was added to increase the salt concentration by 0.1 m NaCl. The CYPOR-containing solution was loaded onto a Bio-Beads SM-2 column (152-3920, Bio-Rad), which had been pre-equilibrated in buffer B (100 mm potassium phosphate buffer, pH 7.4, 10% glycerol, 0.1 mm EDTA, and 0.2 μm FMN). The eluate was directly loaded onto an octyl-Sepharose column also pre-equilibrated with buffer B. The octyl-Sepharose column was washed with three column volumes of buffer B. The reductase was then eluted with buffer B plus 0.3% (v/v) Triton X-100. Fractions with greater than 1 μm reductase were pooled and concentrated with a YM30 membrane. The detergent was removed on a hydroxyapatite Bio-Gel HT column (Bio-Rad). The concentrated reductase was filtered through a 0.22-μm filter (Fisher) and stored in aliquots in 0.1 m potassium phosphate buffer and 15% glycerol at −80 °C. The purified protein gave a single band on an SDS-polyacrylamide gel at ∼78,000 daltons and a UV-visible absorption spectrum identical to that of the wild type protein. The CYPOR concentration was determined using an extinction coefficient of 21.4 mm−1 cm−1 at 454 nm. An average of 6 mg of pure full-length 147CC514 CYPOR was obtained from 1 liter of cell culture. DTT was not added during the purification of 147CC514. The reason was that when the reducing agent was added, a significant amount of the purified protein was isolated as a cross-linked homodimer.
The truncated Δ56 form of the rat 147CC514 mutant, in which the first 56 residues of CYPOR were truncated, was expressed in E. coli Rosetta gami (DE3) pLysS cells (Novagen). Transformation of the Rosetta-gami competent cells by the mutant plasmid was performed as described in the Novagen user protocol. The E. coli cells were grown overnight at 37 °C on Luria Broth (LB) agar plates containing 0.24 mm carbenicillin. A single colony was selected and used to inoculate 200 ml of LB medium containing 0.24 mm carbenicillin and grown overnight at 37 °C, with shaking at 250 rpm. 10 ml of this overnight culture were then used to inoculate 1 liter of LB medium containing 0.24 mm carbenicillin, 0.2% (w/v) glucose, 53 nm riboflavin, and 1% ethanol (v/v) in a 2.8-liter Fernbach flask. The cells were grown in the dark on an Innova shaker at 37 °C, with shaking at 200 rpm to an A600 of 2 (∼4 h), at which time isopropyl β-d-thiogalactopyranoside was added to a final concentration of 400 μm to initiate expression of the reductase. The cultures were incubated for an additional ∼64 h at 19 °C, with shaking at 130 rpm. The temperature at which expression occurs is critical. When the temperature at which expression occurred was lowered from 25 to 19 °C, the amount of protein included in the inclusion bodies decreased drastically, and the yield of properly folded protein improved by at least a factor of 3.
The cells expressing the truncated 147CC514 protein were sonicated in 50 mm Tris, pH 7.7, at room temperature (pH 8.05 at 4 °C). Triton X-100 was added to 0.1% (v/v). The mixture was stirred in the cold for ∼3 h. The addition of the low concentration of Triton X-100 increases the recovery of the reductase and facilitates the removal of contaminating heme proteins on the DEAE column. After stirring, the solution was centrifuged at 100,000 × g for 45 min. Subsequently, the supernatant was loaded onto a DEAE-Sepharose Fast Flow resin column pre-equilibrated with the Affinity buffer (50 mm Tris acetate, pH 7.7, at room temperature, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100 (v/v)). The column was washed with ∼3 column volumes of Affinity buffer containing 10 μm FMN and 0.15 m NaCl until the shoulder at 410–420 nm on the flavin spectrum disappeared. The enzyme was eluted with a NaCl gradient from 0.12 to 0.2 m in Affinity buffer. The fractions containing the greenish CYPOR protein were combined, concentrated, and subsequently loaded onto a 2′,5′-ADP-Sepharose column equilibrated with Affinity buffer and then washed with 50 mm potassium phosphate buffer containing 5 mm adenosine, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100 (v/v). Protein was eluted with 10 mm 2′-AMP in the buffer. The detergent was removed on a hydroxyapatite column, as in the purification of the full-length protein. The concentration of the purified 147CC514 protein was determined from its flavin content using an extinction coefficient of 21.4 mm−1cm−1 at 454 nm. P450 2B4 was expressed and purified as described previously (20). The concentration of P450 2B4 was determined spectrophotometrically using an extinction coefficient of ϵ450–490 nm of 91 mm−1 cm−1 for the absorbance difference between the ferrous P450-CO form and the ferrous form (21).
Activity Measurements
The ability of CYPOR to reduce cyt c and ferricyanide was measured under steady-state conditions at 30 °C in buffer containing 270 mm potassium phosphate, pH 7.7, as described previously (8, 18). Briefly, a solution containing 65 μm ferric cyt c and 10 nm CYPOR was incubated in a cuvette for 5 min at 30 °C. The reaction was started by addition of NADPH to a final concentration of 50 μm. The initial rate of cyt c reduction was followed at 550 nm. Reduction of ferricyanide was measured by preincubating 10 nm CYPOR with 500 μm oxidized ferricyanide for 5 min at 30 °C. The reaction was started by addition of NADPH to a final concentration of 100 μm. Rates of cyt c and ferricyanide reduction were determined using Δϵ = 21 mm−1 cm−1 at 550 nm and 1.02 mm−1cm−1 at 420 nm, respectively. Km and kcat values were obtained by fitting the data to the Michaelis-Menten equation. The steady-state activity of P450 2B4 for benzphetamine metabolism was determined by quantitating the amount of formaldehyde produced using Nash's reagent as described previously (9).
Measurement of 147CC514 Mutant Activity after Treatment with Dithiothreitol and Iodoacetamide
The disulfide cross-linked mutant or wild type CYPORs were treated in a glove box with a 500-fold excess of an anaerobic solution of freshly prepared dithiothreitol (DTT). The DTT was prepared in the glove box with anaerobic buffer and solid DTT. The mixture was then incubated at 4 °C in the glove box ([O2] = ∼2.5 ppm) overnight. Excess DTT was removed under anaerobic conditions from the reductases using a PD-10 (G-25) desalting column (Amersham Biosciences) that had been pre-equilibrated with anaerobic 270 mm potassium phosphate buffer, pH 7.7. The DTT-treated CYPOR sample was divided into two equal parts. One-half was further treated with 30 mm iodoacetamide for 30 min at 22 °C. The iodoacetamide (CH2ICONH2) alkylates the free thiols and prevents reformation of the disulfide bond when the mutant protein is assayed under aerobic conditions. The excess iodoacetamide was also removed on a PD-10 column under anaerobic conditions. In some experiments, the iodoacetamide was added to the mutant that had been treated overnight with DTT before the DTT had been removed. The mixture was incubated at 22 °C for 30 min, at which point both the DTT and iodoacetamide were removed on the PD-10 column. This experiment demonstrated that the two thiol groups of DTT did not interfere with the reaction of iodoacetamide and the CYPOR thiol groups. As a control, wild type CYPOR was treated in a similar manner.
To measure cyt c reductase activity of the DTT-treated protein, a solution containing 270 mm potassium phosphate buffer and a final concentration of 65 μm cyt c and 50 μm NADPH was incubated in a cuvette for 5 min at 30 °C in a Cary 300 UV-visible spectrophotometer (Varian). The treated CYPOR (DTT-only or DTT + iodoacetamide) was then incubated under aerobic conditions from 0 to 300 s before being added to the assay mixture at a final concentration of 10 nm. The initial rate of cyt c reduction was followed at 550 nm, as described above (8, 18). This experiment measured how rapidly the disulfide bond reformed upon exposure to oxygen and the ability of iodoacetamide to block reformation of the disulfide linkage.
The steady-state activity of purified P450 2B4 with mutant and wild type CYPOR was measured using benzphetamine as the substrate. The wild type and 147CC514 CYPOR were treated with DTT alone or both DTT and iodoacetamide, as described above, and the reagents were removed under anaerobic conditions on a PD-10 column. The reduction of cyt c by CYPOR does not require oxygen, and complex formation between CYPOR and cyt c occurs rapidly. In contrast, the ability of the full-length wild type and mutant CYPOR to catalyze substrate oxidation by full-length P450 has a requirement for oxygen and a lag of ∼5 min for maximal complex formation between CYPOR and P450. P450, DLPC, and CYPOR were preincubated in room air in 100 mm potassium buffer at 30 °C for 0–30 min to allow complex formation between P450 and CYPOR. After incubating for the indicated amount of time, the reaction was started by addition of benzphetamine and NADPH. Final concentrations of reagents were P450 0.2 μm, CYPOR 0.2 μm, DLPC 24 μm, benzphetamine 1 mm, NADPH 0.5 mm in 100 mm potassium phosphate buffer pH 7.4. The resulting P450 assay solution was incubated for 6 min, with shaking at 30 °C. The reaction was stopped by addition of trifluoroacetic acid (TFA) to a final concentration of 7% (v/v). Formaldehyde, the product of benzphetamine metabolism, was quantified spectrophotometrically with Nash's reagent as described previously (9).
Measurement of Activity of DTT or DTT/Iodoacetamide-treated 147CC514 CYPOR in the Presence of SOD and CAT
The reduction of cyt c and the metabolism of benzphetamine by the DTT- and DTT/iodoacetamide-treated wild type and 147CC514 mutant in the presence of SOD and CAT were measured, as described above. The initial rate of cyt c reduction was followed at 550 nm at 30 °C. After 10 s of reaction, either SOD or CAT or both SOD and CAT were added to the reaction mixture. The final concentrations of the reagents in the reaction mixture were 65 μm cyt c, 50 μm NADPH, 10 nm CYPOR, 750 units of SOD/ml, 1000 units of CAT/ml, in 1 ml of 270 mm potassium phosphate buffer. The rate of cyt c reduction was calculated from rates of the reaction before and after the addition of SOD, CAT, or SOD/CAT.
The steady-state activity of P450 2B4 for benzphetamine metabolism was also determined in the presence of SOD or CAT. P450, DLPC, and the various preparations of 147CC514 were incubated for 5 min at 30 °C, and then a solution containing benzphetamine and SOD (and/or CAT) was added to the protein mixture. The reaction was started by addition of NADPH. The final concentrations of reagents in the assay mixture were as follows: P450, 0.2 μm; CYPOR, 0.2 μm; DLPC, 24 μm; benzphetamine, 1 mm; NADPH, 300 μm; 750 units of SOD/ml; and 1000 units of CAT/ml in 100 mm potassium phosphate buffer, pH 7.4. Formaldehyde, the product of benzphetamine metabolism, was measured spectrophotometrically with Nash's reagent as described above.
Titration of 147CC514 by NADPH under Anaerobic Conditions
To ensure that the mutation did not modify the redox potential of the flavins, we measured the spectrum of the disulfide cross-linked mutant protein during titration with NADPH under anaerobic conditions. The samples were made oxygen-free by repeated evacuation and flushing with nitrogen and argon. The reaction mixture was then incubated overnight at 4 °C in an anaerobic Belle Technology glove box (Hi-Tech, Salisbury, UK) to ensure that all oxygen was removed. The titrant (NADPH) was prepared in anaerobic buffer, and its concentration was calculated using an extinction coefficient of 6.22 mm−1 cm−1 at 340 nm. The reaction mixtures contained 100 mm potassium phosphate buffer, pH 7.4, and ∼19 μm of protein in which both flavins were oxidized. All manipulations of the titrator assembly were performed in a glove box to minimize contamination with oxygen. Following the addition of each aliquot of NADPH, the reaction mixture was permitted to equilibrate at room temperature until no further absorbance change occurred.
Kinetics of Reduction of 147CC514 by NADPH
All the experiments were performed at 30 °C under anaerobic conditions using a Hi-Tech SF61DX2 stopped-flow spectrophotometer housed in an anaerobic Belle Technology glove box (Hi-Tech) as described previously (8). Before being introduced into the glove box, the buffer and sample solutions were deoxygenated by three cycles of purging with argon and then applying a vacuum for 1 min. The samples were then incubated overnight at 4 °C in the glove box to minimize the amount of oxygen in the reaction mixtures. For kinetic measurements, one syringe of the apparatus contained a solution of 20 μm of the oxidized flavoprotein in 100 mm potassium phosphate, pH 7.4, 15% glycerol (v/v), and the other syringe contained either 20 or 200 μm NADPH. Equal volumes of the two solutions were mixed, and the absorbance changes at 452 nm (flavin reduction) and 585 nm (semiquinone formation) were recorded. Rate constants and amplitudes were obtained by fitting the absorbance change at a particular wavelength with multiple exponential functions using KinetAsyst2 software (Hi-Tech).
Kinetics of Reduction of cyt c under Single Turnover Conditions
Electron transfer from the two electron-reduced 147CC514 variant to ferric cyt c was studied under single-turnover conditions using a High-Tech SF61DX2 stopped-flow spectrophotometer housed in an anaerobic Belle Technology glove box (Hi-Tech) at 30 °C. A 10 μm solution of the mutant protein was preincubated with 1 m eq of NADPH for 20 min in the anaerobic glove box. The two electron-reduced mutant protein was then loaded in syringe 1 of the stopped flow, whereas syringe 2 contained an anaerobic solution of 100 μm of ferric cyt c. The reaction was started by rapid mixing of an equal volume from both syringes. The reaction was monitored at 550 nm, and the kinetic traces were fitted using a standard exponential equation. The Δϵ 550 nm for cyt c reduction was 21.1 mm−1cm−1.
Crystallization, Data Collection, and Model Building
Crystals were grown using the hanging drop method (22) by mixing 2 μl of 15 mg/ml solution of the truncated form of 147CC514 and 2 μl of reservoir solution containing 100 mm HEPES, pH 7.2, 150 mm MgCl2, and 17% polyethyleneglycol (PEG) 3350. Prior to all crystallization setups, purified protein was treated with 2 and 20 times molar excess of FMN and NADP+, respectively, followed by several cycles of concentration/dilution to remove excess cofactors. The protein solution was concentrated to a final concentration of 15 mg/ml in 50 mm HEPES, pH 7.5. The crystals were either directly flash-frozen after briefly being soaked in a cryosolution (mother liquor plus 13% additional PEG 3350) (NADP+-free crystals) or soaked in the same well solution plus 5 mm DTT and 2 mm NADP+ for 30 min prior to treating with the cryosolution and freezing in liquid nitrogen (disulfide-reduced, NADP+-bound crystals). All data were collected using an R-AXIS IV2+ detector system with a Micromax 007 generator. Data processing was done using the program HKL2000 (23). The crystals belong to the space group P212121 with one molecule/asymmetric unit.
Crystal Structure Determination
The initial phases were determined by the molecular replacement method using Phaser (24) with the FAD domain and the FMN domain of the rat CYPOR structure (7) (PDB code 1AMO) as the search models. The final Phaser solution has the rotation function Z scores/translation function Z scores/log likelihood gain (RFZ/TRZ/LLG) of 15.9/30.4/1022 and 11.5/44.0/2290 for the FAD and FMN domain sequentially. Because all the cofactors were excluded from the search model, the correctness of the solution was further confirmed by the strong electron densities corresponding to the bound FAD and FMN at their expected positions. Further refinements were carried out using CNS (25) and manual model adjustments with using COOT (26). Data collection and the refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Crystals | 147CC514, PDB code 3OJW | 147C/C514(NADP+ soaked), PDB code 3OJX | |
|---|---|---|---|
| Data collection | |||
| Resolution | 30-2.20 Å (2.28-2.20 Å)a | 30-2.5 Å (2.59-2.50 Å) | |
| No. of measured reflections | 268,640 | 104,504 | |
| No. of unique reflections | 33,491 | 23,362 | |
| Completeness | 97.0% (96.1%) | 99.6% (99.7%) | |
| Redundancy | 8.0 (8.0) | 4.5 (4.1) | |
| 〈I/σ(I)〉 | 27.5 (4.9) | 16.3 (3.0) | |
| Unit cell | |||
| a | 65.8 Å | 65.4 Å | |
| b | 73.0 Å | 73.3 Å | |
| c | 138.2 Å | 137.5 Å | |
| Space group | P212121 | P 212121 | |
| Rsym | 0.080 (0.423) | 0.084 (0.475) | |
| No. of molecules in asymmetric units | 1 | 1 | |
| Vm | 2.37 Å3/Da | 2.35 Å3/Da | |
| Refinement | |||
| Resolution | 30-2.20 Å | 30-2.50 Å | |
| Rcrystal | 20.52% | 20.9% | |
| Rfree | 25.2% | 26.7% | |
| Deviations from ideal geometry for bond length/bond angle | 0.006 Å/1.2° | 0.007 Å /1.3° | |
| No. of water molecules | 216 | 70 | |
| B-factor analysis | |||
| Wilson B | 36.7 Å2 | 48.7 Å2 | |
| Protein | 37.6 Å2 | 40.3 Å2 | |
| Water molecules | 36.4 Å2 | 29.5 Å2 | |
| Flavins/NADP+ | 30.4/ | 28.9/51.8 | |
| Ramachandran analysis | |||
| Most favored | 90.1% | 88.5% | |
| Allowed | 9.3% | 11.0% | |
| Generously allowed | 0.6% | 0.6% | |
| Disallowed | 0.0% | 0.0% | |
a Numbers in the parentheses are values for the highest resolution shell.
RESULTS
Examination of the crystal structure of the wild type NADP+-bound CYPOR ((7) PDB code 1AMO) identified two salt bridges between the FMN and FAD domains, Glu179–Lys664 and Asp147–Arg514. The Asp147–Arg514 salt bridge was chosen for mutation to cysteines. The other pair (Glu179–Lys664) was located closer to the surface of the closed CYPOR molecule and was therefore considered more likely to form undesirable inter-molecular disulfide linkages. The mutated protein was expressed in either E. coli C41 strain (for full-length CYPOR) or a Rosetta-gami strain (for the truncated Δ56 form) that carried inactivating mutations in the thioredoxin and glutaredoxin reductases. The resulting variant protein has an engineered disulfide linkage (Cys147 and Cys514) in addition to the catalytic cysteine (Cys630). Six native cysteines (at positions 136, 228, 363, 445, 472, and 566) out of seven have been removed. This variant protein is referred to as 147CC514.
Altered Positioning of Flavin Domains in Cross-linked 147CC514
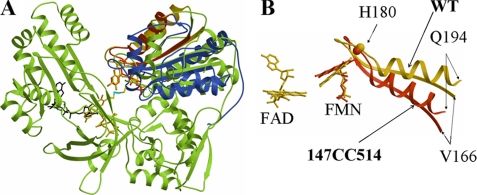
To verify that the 147CC514 protein as purified from E. coli contained a disulfide bond and to determine whether any structural changes accompanied the disulfide linkage, the crystal structure of the Δ56 form of the double mutant protein (C147CC514) was determined. The crystal structure of the mutant protein determined at 2.2 Å resolution indeed showed the cross-linked disulfide linkage. Fig. 1A shows the electron density in the vicinity of Cys147 and Cys514 clearly showing the disulfide bond between the two cysteines. The overall structures of each domain of the mutant are essentially the same as those of wild type CYPOR (PDB code 1AMO (7)) with root mean square values of 0.34 and 0.72 Å for the FMN and FAD domain, respectively (Fig. 2A). However, the relative positions of the two domains in the cross-linked structure are different from the wild type structure (Fig. 2A). In the 147CC514 structure, the entire FMN domain is slightly pulled toward the FAD domain and is twisted about 20°, with a pivot point located near the His180 Cα of the polypeptide (Fig. 2B), which is located about 5 Å away from the O2 and O4 atoms of the isoalloxazine ring of FMN. In the cross-linked 147CC514 structure, the distance between the two 7-CH3 groups of FAD and FMN is 5.6 Å and that of 8-CH3 methyl groups is 5.1 Å, both of which are over 1 Å longer than the corresponding distances of the wild type structure (4.4 and 4.0 Å, respectively). However, the distance between 7-CH3 of FMN and 8-CH3 of FAD in the mutant structure is 4.1 Å, whereas that in the wild type structure is 3.9 Å. Thus, the alignment of the two flavin rings in the mutant structure is also twisted by about 20°. The longer distances between the corresponding methyl groups of the two flavin rings (i.e. 7CH3-7CH3 and 8CH3-8CH3 distances), decrease in surface contact area, and the twist between the two flavin rings are consistent with the ∼46-fold poorer electron transfer rate between the two flavins in the mutant compared with that of wild type (78.7 versus 1.7 s−1, in Table 2). The relative orientations of the flavins are shown in Fig. 2B.
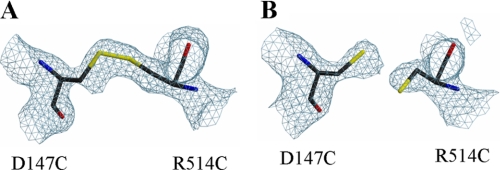
FIGURE 1.
Fo − Fc omit maps in the vicinity of Cys147 and Cys514 contoured at 3.6 σ level. A, disulfide linkage is clearly shown between Cys147 and Cys514 in the structure of 147CC514, when the purified protein was crystallized. B, reduced sulfhydryls shown in the binary structure of the 147CC514 crystal soaked with 5 mm DTT and 2 mm NADP+ for 30 min, referred to as 147C/C514-NADP+.
FIGURE 2.
A, comparison of the structures of 147CC514 (blue) and wild type CYPOR (green). The structures are overlaid with only their FAD domains superimposed. For clarity, only the wild type FAD domain is shown. The FAD, FMN (both brown), and NADP+ (black) of wild type CYPOR as well as FMN (red) of 147CC514 are shown as a stick model. The disulfide bond between D147C and R514C is shown in cyan. The entire FMN domain in 147CC514 is twisted about 20° with respect to the FAD domain at a pivot point near the His180 Cα atom in the strand-loop-helix containing Val166 to Gln194 (highlighted with gold (wild type) or red (147CC514). B, enlarged view of FAD and FMN along with the strand-loop-helix. The color scheme is the same as in A.
TABLE 2.
Kinetics of reduction of CYPOR by 1 and 10 molar equivalents of NADPH at 30 °C
See under “Experimental Procedures” for experimental details. k represents the rate constant, and A represents the phase amplitude.
| Reductase | λ nm | A1 | k1 | A2 | k2 | A3 | k3 |
|---|---|---|---|---|---|---|---|
| % | s−1 | % | s−1 | % | s−1 | ||
| 1 Equivalent | |||||||
| Wild type | 452 | 79 | 75 | 21 | 6.1 | ||
| 585 | 62 | 79 | 38 | 6.2 | |||
| 147CC514 | 452 | 25 | 64 | 43 | 12 | 32 | 0.8 |
| 585 | 39 | 1.7 | 61 | 0.2 | |||
| 10 Equivalents | |||||||
| Wild type | 452 | 73 | 76 | 27 | 9.1 | ||
| 585 | 91 | 11 (decay)a | |||||
| 147CC514 | 452 | 43 | 52 | 17 | 2.5 | 40 | 0.7 |
| 585 | 2.4 | 0.02 (decay)a | |||||
a Data represent a decrease in absorbance due to formation of the four electron-reduced CYPOR.
147CC514 Exhibits Decreased NADP+ Binding and Alterations in the Conformation of the NADP+-binding Site
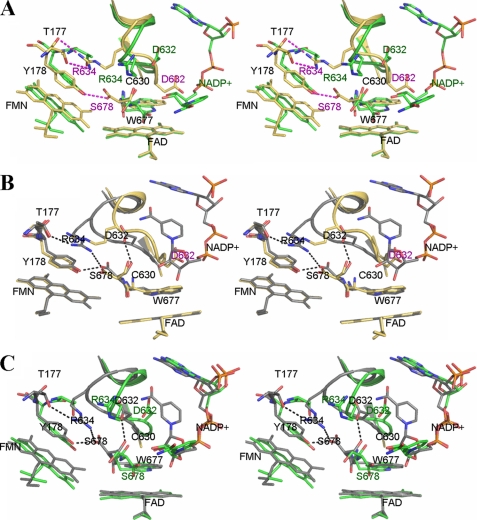
Although the mutant protein was treated with excess NADP+ prior to the crystallization setup, as with crystallization of other CYPOR proteins (7, 12), no bound NADP+ was found in the crystal structure. This was totally unexpected, because all the CYPOR structures solved so far, except the structure of yeast-human hybrid CYPOR (27), contained either NADP+ or the 2′,5′-ADP moiety of NADP+ (7, 8, 12, 28) and indicated that the cross-linked mutant has a lower affinity for NADP+ than wild type CYPOR (PDB code 1AMO). For structural comparison, the triple mutant S457A/C630A/D675N structure (PDB code 1JA1) was of particular interest. Although the mutant was catalytically inactive, its structure was almost identical to that of the wild type structure with a root mean square deviation of 0.54 Å for all visible 615 Cα atoms. In addition, the triple mutant structure was of a higher resolution (1.8 versus 2.5 Å), and most importantly the ribityl-nicotinamide moiety of NADP+ in the triple mutant structure was less disordered than the wild type structure (estimated occupancy of >60% compared with that of ∼30% in wild type), so that the entire NADP+ molecule could be modeled in one of the two molecules in the asymmetric unit. For these reasons, the better ordered triple mutant structure, referred to as “wild type” (wild type is the triple mutant S457A/C630A/D675N of CYPOR), was used for structural comparisons together with the wild type structure.
Fig. 3A shows a comparison of the vicinity of the NADP+-binding site of the structures of 147CC514 (NADP+-free form) and the NADP+-bound triple mutant (PDB code 1JA1). In the cross-linked structure, the loop containing residues Gly631–Asn635 has moved by 2–4 Å with respect to the NADP+-bound wild type 1JA1 structure (2.4 Å at the Asp632 Cα and 3.8 Å at the Ala633 Cα atom) such that the Asp632 side chain is within van der Waals contact with the wild type pyrophosphate ribose moiety. This relocation of the Gly631–Asn635 loop (hereafter referred to as Asp632 loop) also resulted in formation of a hydrogen bond between the guanidine group of Arg634 and the hydroxyl of Thr177. In addition, the orientation of Trp677 in the 147CC514 structure was also different from that of wild type and 1JA1. As in the wild type structure, the indole ring of Trp677 and the isoalloxazine ring of FAD were coplanar. However, the orientation of the indole ring was different in the 147CC514 mutant such that the ring overlap was more extensive suggesting a stronger interaction between the FAD and indole rings in the 147CC514 structure. It is reasonable, when NADP(H) was not bound to the enzyme, to have the strongest interaction between Trp677 and FAD by maximizing the π-π overlap between the two ring systems. Interestingly, the Asp632 loop conformation and the orientation of the Trp677 indole ring observed in 147CC514 were very similar to those found in the structure of yeast-human hybrid CYPOR, which also lacks NADP+ (27). Fig. 4A depicts an overlay of the structures of 147CC514 and the yeast-human hybrid CYPOR in the vicinity of the NADP+-binding site. In both structures, the Asp632 carboxylate was less than 2 Å away from the ribose moiety (steric hindrance) and was less than 3.5 Å away from the pyrophosphate group of NADP+ (both steric hindrance and electrostatic repulsion), thus preventing NADP+ from binding to the enzyme in the conformation observed in the wild type structure. In addition, it should be noted that Asp632 was conserved in all known diflavin-containing enzymes, including CYPOR, P450 BM3, nitric-oxide synthase isozymes, and methionine synthase reductase. Taken together, the conformation of the Asp632 loop and the orientation of the Trp677 indole ring observed in the structure of NADP+-free 147CC514 most likely represents the structure of wild type CYPOR when NADP(H) was not bound.
FIGURE 3.
Stereo overlays of the structures of wild type (PDB code 1JA1) (green), 147CC514 (gold), and the binary complex of reduced 147C/C514-NADP+ (gray). For clarity, only the isoalloxazine ring-ribityl portions of flavin cofactors are shown. A, comparison of wild type (green) and 147CC514 (gold with red labels). The NADP+ structure is that found in the wild type structure, because the cross-linked 147CC514 structure lacks bound NADP+. Residues labeled in black are those superimposed in both structures. The loop containing residues Gly631–Asn635 (Asp632 loop) in the structure of 147CC514 is extended down toward where the indole ring of Trp677 lies. This loop movement results in the guanidine group of Arg634 making hydrogen bonds with both the side chain and main chain amide nitrogens of Thr177. Furthermore, the side chain of Asp632 flips down and occupies the space where the ribityl-pyrophosphate moiety of NADP+ would ordinarily bind, causing steric hindrance as well as charge repulsion between the NADP+ pyrophosphate and Asp632 carboxylate, preventing NADP+ from binding to 147CC514. B, overlay of the structure of NADP+-free 147CC514 (gold) and that of NADP+ bound disulfide-reduced 147CC514 (gray). Upon binding of NADP+, the Asp632 loop of 147C/C514 is pushed further toward the FMN domain by as much as 6 Å for the Ala633 Cα atom and 4 Å for the Asp632 Cα, making room for NADP+ to bind. Arg634 makes a salt bridge with the C-terminal carboxylate of Ser678. As shown in A, Asp632 of 147CC514, which is located in the middle of the Asp632 loop, may impair NADP+ binding by both steric hindrance and charge repulsion. C, comparison between wild type (green) and 147C/C514-NADP+ (gray). Both the Asp632 loop and NADP+ conformations are different between these two structures; an extended NADP+ conformation is found in the wild type structure (green carbons with red oxygen and phosphorus atoms), although a more compact, folded conformation is observed in the mutant structure (gray carbons).
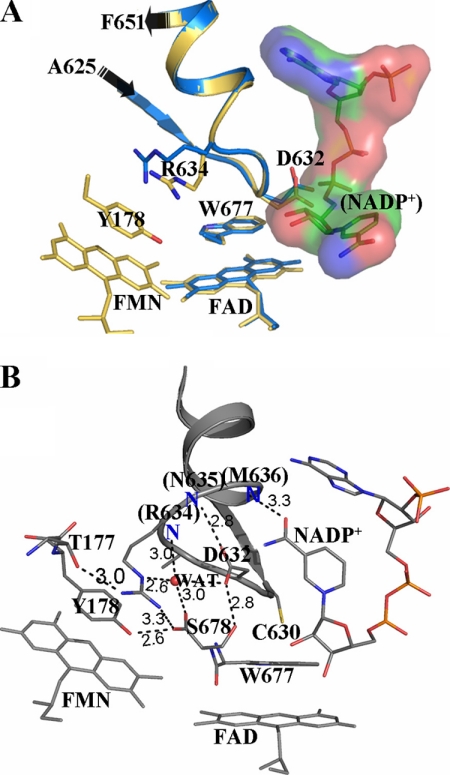
FIGURE 4.
A, superposition of the structures of 147CC514 (gold) and the yeast-human hybrid CYPOR (PDB code 1JFO; blue). The conformation of the strand-loop-helix containing Ala625–Phe651 and the orientation of the indole ring of Trp677 in the two structures are almost identical, strongly suggesting that the observed structure of 147CC514 is indeed the structure of wild type CYPOR in the absence of bound NADP+. B, structure of 147C/C514-NADP+ in the vicinity of the NADP+-binding site. The bound NADP+ adopts a folded conformation similar to the conformation seen free in solution (29). The amide group of NADP+ forms a hydrogen bond with the main chain amide nitrogen of Met636 (note: the occupancy of the nicotinamide-ribityl moiety is estimated to be ∼60%). A hydrogen bonding network involving a water molecule stabilizes the Asp632 loop conformation. Positions of the main chain amide nitrogen atoms in the Asp632 loop are marked with a blue N.
Nicotinamide Binding Is Regulated by the Asp632 Loop
When preformed crystals of 147CC514 were soaked in a solution containing NADP+, the crystals cracked. Attempts to crystallize the thiol form of the 147CC514 protein were not successful. However, when preformed crystals of cross-linked 147CC514 were soaked in a solution containing DTT, a data set could be obtained at ∼3.3 Å resolution. The resulting structure showed that the disulfide bond was reduced and that the overall structure was identical to the structure of the oxidized 147CC514 (data not shown). However, by soaking the crystals of cross-linked 147CC514 in a solution containing both DTT and NADP+, the disulfide bond in the protein was reduced (Fig. 1B), and NADP+ was bound to the protein. A comparison of the oxidized NADP+-free structure (147CC514) with disulfide-reduced NADP+-bound structure (referred to as 147C/C514-NADP+) is shown in Fig. 3B. The overall structures are essentially identical (root mean square of 0.47 Å), except in the vicinity of the NADP+-binding site. The major difference between the two structures again lies in the loop containing Gly631 and Asn635 (Asp632 loop). Upon binding of NADP+, the loop swings back toward the FMN domain by about as much as 6 Å at the Ala633 Cα atom, resulting in the Asp632 side chain flipping away from the NADP+-binding site to provide room for NADP+ to bind. The binding mode of NADP+ is different from that found in the wild type structure (Fig. 3C). The molecule adopts a compact, folded conformation in contrast to the more extended conformation found in the wild type structure or in the structure of the mutant CYPOR in which the last two C-terminal residues (Trp677 and Ser678) were truncated (referred to as W677X) (12). In the W677X structure (PDB code 1JA0), the ribityl-nicotinamide moiety of NADP+ rotates, and the nicotinamide ring is stacked on the re-side of the flavin ring poised to transfer hydride ion to the N5 atom of FAD. However, in the structure of 147C/C514-NADP+, although the AMP-PPi portion of NADP+ binds to the enzyme in the same manner as that observed in the structures of wild type and W677X, the nicotinamide moiety adopts a very different conformation. The ribityl-nicotinamide ring portion folds toward the adenine ring, and the nicotinamide group makes a relatively weak hydrogen bond with the main chain amide nitrogen of Met636 (Fig. 4B), which is reminiscent of the conformation of NADP+ free in solution (29). The binary structure of 147C/C514-NADP+ observed here is most likely the structure of CYPOR immediately after binding NADPH or just prior to release of the bound NADP+ cofactor. Thus, the Asp632 loop movement and the change of the Trp677 indole ring orientation may facilitate the binding and release of NADPH/NADP+.
CYPOR Redox Properties Are Unaffected by Cross-linking
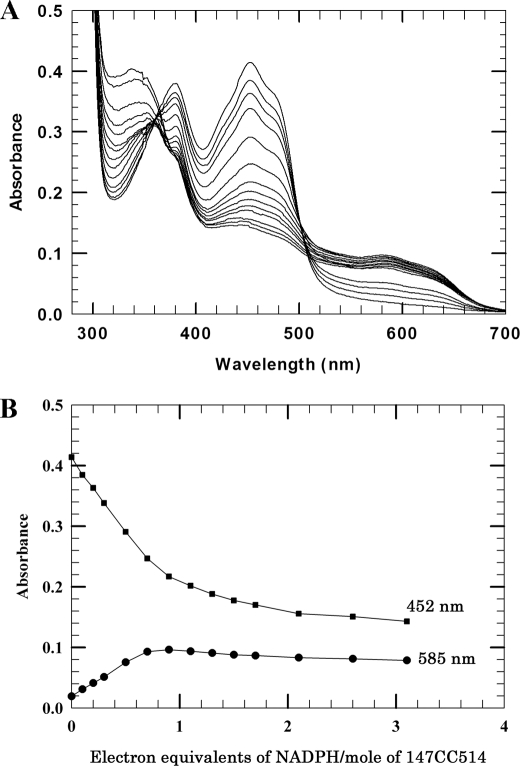
The steady-state oxidation-reduction properties of FMN and FAD in the 147CC514 mutant were investigated by titration with NADPH under anaerobic conditions. Reduction with DTT or dithionite will reduce the disulfide bond. As shown in Fig. 5A, titration of the cross-linked 147CC514 enzyme with NADPH proceeded in a manner identical to that reported for wild type CYPOR (30). Addition of slightly less than one electron equivalent of NADPH/mol of fully flavin-oxidized 147CC514 enzyme produced the spectrum of the air-stable blue semiquinone (FAD/FMNH•), characterized by an absorbance decrease at 452 nm and an absorbance increase at 585 nm, with isosbestic points at 363 and 502 nm, (Fig. 5). Further addition of NADPH led to continued bleaching at 452 and 502 nm, corresponding to formation of FMNH2 and the three electron-reduced CYPOR (FADH•/FMNH2). Low concentrations of NADPH (E = −320 mV) were unable to fully reduce the FAD (E is for FADH•/FADH2 = −365 mV) of the enzyme. As a result, addition of NADPH to the three electron-reduced enzyme produced minimal absorbance changes in the flavin spectra (Fig. 5B) and an increase at 340 nm, which represents an accumulation of NADPH (Fig. 5A). This demonstrates that the potential of the FADH•/FADH2 couple, like wild type CYPOR, is lower than that of the NADP+/NADPH couple. Apparently the introduction of a disulfide bond between residues 147 and 514 did not significantly modify the redox potential of the flavins despite the 20° rotation of the FMN domain and FMN with respect to the FAD domain and FAD (Fig. 2) The remainder of the 147CC514 polypeptide arrangement surrounding the FAD ring, especially in the vicinity of the redox active N5 atom, is completely conserved as in wild type. The lack of a significant redox potential change is consistent with crystallographic data showing minimal changes in the environment of the FAD.
FIGURE 5.
Titration of the 147CC514 variant with NADPH under anaerobic conditions. The titration was performed as described under “Experimental Procedures.” CYPOR concentration was 19 μm. A, spectral changes during the titration. B, absorbance changes at 452 and 585 nm.
Interflavin Electron Transfer Is Impaired in the Cross-linked 147CC514 Protein
To examine electron transfer from either NADPH to FAD or FADH2 to FMN, we investigated the kinetics of reduction of the mutant by both equimolar and a 10-fold excess of NADPH. The kinetics of both the reduction of FAD by NADPH and interflavin electron transfer are impaired. The absorbance changes during the anaerobic reduction of the mutant CYPOR were monitored at 452 nm, the wavelength associated with the flavin reduction, and at 585 nm, which reflects blue semiquinone formation as a result of electron transfer from FAD to FMN (2). In the wild type protein, the reduction of the flavins is biphasic with an initial fast phase at k1 ∼75 s−1 (∼79% of the total absorbance change at 452 nm) and a slower second phase (k = ∼6 s−1), likely reflecting reduction by a second molecule of NADPH. Semiquinone formation, monitored at 585 nm, occurred with a rate constant, k = ∼79 s−1, essentially identical to the rate of flavin reduction observed at 452 nm. The t½ of the electron transfer is ∼0.2 s. The disemiquinone resulting from intramolecular electron transfer from FADH2 hydroquinone produces two forms of the reductase at an ∼1:2 ratio (FADH•/FMNH• and FAD/FMNH2) at equilibrium (3). Note that in the wild type protein, the reduction of FAD by NADPH, and the intramolecular electron transfer from FADH2 to FMN, followed at 452 and 585 nm, respectively, are rapid and indistinguishable under our experimental conditions.
In the case of the 147CC514 mutant, the kinetic traces at 452 nm are triphasic compared with biphasic for the wild type protein. As shown in Table 2, the initial phase, which represents 25% of the total absorbance change versus 79% in the wild type, proceeds with a rate constant of k1 ∼63.6 s−1, is almost as fast as the wild type. This is followed by two other phases, an intermediate phase with a k2 ∼12 s−1 (∼43%) and a slow phase with k3 ∼0.75 s−1 (32%). The rate constant of the first two phases in the mutant are slightly decreased compared with the first two rate constants in the wild type protein. What the third phase represents is not known with certainty. One possibility is that it may reflect the time required for movement of the Asp632 loop from a sterically obstructing position (Fig. 3A) to a nonobstructing position in the NADP+-binding site (Figs. 3B and 4B). The kinetics of electron transfer from FAD to FMN as measured by semiquinone formation at 585 nm are slower in the mutant, with rate constants of 1.7 and 0.2 s−1 versus 79 and 6 s−1 in the wild type (Table 2). These data clearly indicate that, in the cross-linked variant, electron transfer from FADH2 to FMN is markedly impeded, presumably because of the 1 Å greater separation of the flavins and the ∼20° rotation of their isoalloxazine rings, resulting in the decreased suface contact area between the two rings.
The kinetics of the initial phase of reduction of wild type CYPOR at 452 nm in the presence of a 10-fold molar excess of NADPH (k1 = 76 s−1, amplitude of phase = 73%) are not significantly different from those observed in the presence of 1 m eq of NADPH (Table 2). However, in the 147CC514 mutant, the kinetics observed at 452 nm are triphasic, with the rate constant of the fast initial phase decreased 32%, although the amplitude is only 43 versus 73% in the wild type protein. The absorbance changes at 452 nm represent the reduction of FAD, initially by a single molecule of NADPH. Following transfer of two electrons from FADH2 to form FMNH2/FAD CYPOR, a second molecule of the two electron donor, NADPH, reacts with the FMNH2/FAD form of the reductase more slowly, presumably because of a diminished thermodynamic driving force and a lower concentration of FAD-containing enzyme (3). In the presence of excess NADPH, more (43 versus 25%) of the mutant FAD is reduced in the fast phase, indicating that there is an impediment to binding of NADPH in the mutant protein. This conclusion is consistent with the 4.8-fold increased Ki for NADP+ in the C566A mutant (14). However, the rate constant for the third phase ∼0.7 s−1 and its amplitude is similar in the presence of both 1 and 10 molar eq of NADPH, indicating it is not dependent on NADPH concentration. As stated above, one possibility is that this rate constant may reflect the time required for rotation of Asp632 to a nonobstructing position in the NADPH/NADP+-binding site.
As shown in Table 2, the kinetics at 585 nm were biphasic in both the wild type and 147CC514 mutant and consistent with a decreased rate of interflavin electron transfer. The rapid absorbance increase at 585 nm (k = 91 s−1 versus 2.4), which reflects disemiquinone formation, was followed by a subsequent monophasic decay of absorbance (k = 10.8 s−1 versus 0.017) for the wild type and mutant, respectively. The rates of interflavin electron transfer obtained with excess NADPH are comparable with those observed with a 1 m eq amount of NADPH (Table 2). Following the initial increase in semiquinone formation, the absorbance decays, indicating further reduction of reductase because of the transfer of a second hydride from NADPH. Indeed, it was proposed that the second hydride transfer is limited by the dissociation of NADP+ (3). The delivery of the second hydride ion by NADPH to the mutant was 635-fold slower than in the wild type (11 versus 0.02/s) (Table 2), presumably primarily because electron transfer from FAD to FMN is delayed. The FAD remains reduced and cannot accept a second hydride ion. In the case of the mutant, all of the kinetic data are consistent with the fact that by cross-linking the FAD domain to the FMN domain of the reductase, we severely perturbed the intramolecular electron transfer, while moderately decreasing the rate of hydride transfer from NADPH to FAD.
Electron Transfer to cyt c and P450 Is Severely Impaired by Cross-linked 147CC514
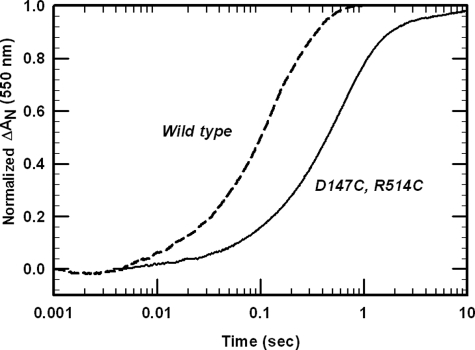
Two acidic clusters of amino acids, Asp207–209 and Glu213, Glu214, and Asp215, are known to be involved in interprotein binding with basic residues on cyt c and subsequent electron transfer (31). In the cross-linked mutant, these residues are not readily accessible to cyt c. To gain further insight into the interactions of cyt c with the mutant CYPOR, we have investigated the ability of the 147CC514 protein to reduce cyt c under single turnover conditions (Table 3). In these experiments, the wild type and the cross-linked mutant were stoichiometrically reduced to the two electron-reduced state by a molar equivalent of NADPH. Under anaerobic conditions, the two electron-reduced state of wild type CYPOR exists as a mixture of ∼70% (FAD and FMNH2), which is catalytically competent, and ∼30% of the enzyme in the disemiquinone form (FMNH• and FADH•), which rapidly interconverts to the catalytically competent (FAD and FMNH2) form (3). Multiple turnovers do not occur because only the FMNH2 form of rabbit and rat wild type CYPOR is able to reduce cyt c (2, 30, 32).
TABLE 3.
Kinetics of oxidation of the two electron-reduced wild type and 147CC514 mutant CYPOR by a 10-fold excess of cyt c
The reduction of cyt c was measured at 30 °C as described under “Experiment Procedures,” using a 10-fold molar excess of cyt c versus CYPOR. The phase amplitude is in parentheses.
| Reductase | kfast | kslow |
|---|---|---|
| s−1 | s−1 | |
| Wild type | 8.7 ± 1.2 (89%) | 1.19 ± 2.5 (11%) |
| 147CC514 | 2.6 ± 0.3 (63%) | 0.65 ± 0.005 (37%) |
As shown in Fig. 6, the reduction of cyt c was initiated by the rapid mixing of two electron-reduced CYPOR and a 10-fold excess of ferric cyt c. The reduction of a molar equivalent of cyt c by wild type enzyme shows biphasic kinetics with a k1 of 8.7 s−1, accounting for ∼89% of the absorbance changes (Table 3). This result is comparable with that obtained with human CYPOR (k = 12 ± 0.4 s−1) (33). Electron transfer from the 147CC514 mutant occurs four times more slowly (Table 3 and Fig. 6). This low rate constant for electron transfer from FMNH2 to an excess of cyt c is consistent with the hypothesis that the interaction surface for electron transfer to cyt c has been decreased, but not totally eliminated, because the FMN domain is not free to dissociate from the FAD domain and expose its acidic convex surface surrounding FMN. The data also suggest that the interaction surface for cyt c on the reductase is not solely restricted to the negatively charged surface surrounding FMN.
FIGURE 6.
Reduction of cyt c by the two electron-reduced wild type and 147CC514 proteins. The reaction was conducted as under “Experimental Procedures” with a 10-fold molar excess of cyt c over the reductase. The absorbance change is normalized to similar concentrations.
Steady-state Kinetics of 147CC514
Table 4 shows the kinetic constants of the mutant CYPOR under steady-state conditions compared with the wild type protein. The kcat(Vmax) of the mutant was decreased ∼20-fold for cyt c and ∼3.5-fold for Fe(CN)63−. The decreases in Km for the substrates cyt c and Fe(CN)63− are consistent with a ping-pong mechanism and a decrease in the rate of the reductive half-reaction (34, 35), due to decreased rates of electron transfer from NADPH to FAD and FAD to FMN. The KmNADPH was increased 2.5-fold in C566A CYPOR mutant (14). In addition, the structure suggests a modest impairment of NADPH binding. The relative contributions of each of these factors to the increased KmNADPH observed in the 147CC514 mutant cannot be ascertained at this time.
TABLE 4.
Kinetic constants of the wild type and mutant CYPOR with NADPH, cyt c, and ferricyanide under steady state conditions
The kinetics of cyt c and Fe(CN)63− reduction were measured at 30 °C as described under “Experimental Procedures.”
| Reductase | Fe3+ cyt c |
Fe(CN)63− |
NADPH, Km | ||
|---|---|---|---|---|---|
| kcata | Km | kcat | Km | ||
| μm | μm | μm | |||
| Wild type | 65.6 ± 7.6 | 15.8 ± 0.6 | 85 ± 14 | 31.1 ± 4 | 5.4 ± 0.4 |
| 147CC514 | 3.5 ± 0.4 | 1.9 ± 0.5 | 25.4 ± 4.6 | 19.5 ± 2.5 | 11.3 ± 1.9 |
a The kcat value with cyt c and Fe(CN)63− is reported in units of nmol of product reduced per s/nmol CYPOR.
Restoration of P450 and cyt c Activity following Reduction of the 147CC514 Mutant Disulfide Bond
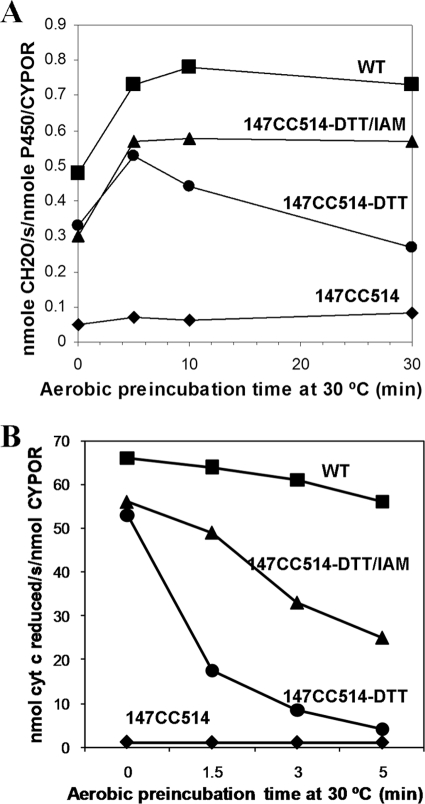
To test the hypothesis that inhibition of domain movement by cross-linking was responsible for decreased catalytic activity, the ability of the thiol and the disulfide forms of 147CC514 to support the P450-catalyzed metabolism of benzphetamine to formaldehyde and reduction of cyt c was investigated. The data in Table 5 and Fig. 7 demonstrate that reduction of the disulfide bond of the cross-linked mutant CYPOR with DTT under anaerobic conditions reduces the disulfide bond and restores 88% of the cyt c reductase activity and 57% of the P450 activity. When the DTT-treated mutant protein was also treated with iodoacetamide, which alkylates the free thiols and prevents reformation of the disulfide bond, 69% of the P450 activity could be recovered.
TABLE 5.
Activity of wild type and 147CC514 CYPOR with cyt c and P450 before and after treatment with DTT alone or with DTT and iodoacetamide
The experiments were performed as discussed under “Experiment Procedures.” The DTT and iodoacetamide (IAM) were removed from the 147CC514 protein prior to the test of its activity.
| Protein | Nanomoles of cyt c reduced per s/nmol CYPOR |
Nanomoles of CH2O produced per s/nmol P450-CYPOR |
||||
|---|---|---|---|---|---|---|
| −DTT | +DTT | +DTT/IAM | −DTT | +DTT | +DTT/IAM | |
| Wild type CYPOR | 62 ± 6.2 | 68 ± 5.0 | 62 ± 4.5 | 0.84 ± 0.11 | 0.88 ± 0.11 | 0.83 ± 0.13 |
| Full-length 147CC514 | 2.31 ± 0.35 | 60 ± 4.5 | 54 ± 6.0 | 0.09 ± 0.01 | 0.50 ± 0.09 | 0.57 ± 0.08 |
FIGURE 7.
Rapid reoxidation of DTT-reduced 147CC514. A, effect of aerobic preincubation on activity of the mutant protein with P450. B, effect of aerobic incubation of disulfide cross-linked DTT and both DTT- and iodoacetamide-treated mutant CYPOR on the ability of the protein to reduce cyt c. Note the rapid loss of activity of the DTT-treated protein. The data suggest iodoacetamide treatment does not alkylate all free thiols. The reactions were conducted as described under “Experimental Procedures.”
As shown in Fig. 7, disulfide bond reformation occurs rapidly under aerobic conditions in seconds to minutes after DTT treatment; iodoacetamide partially prevents the reformation of the disulfide bond and inactivation of the 147CC514 protein. Initially, the activities of the DTT- and DTT/iodoacetamide-treated CYPOR were similar with both cyt c and P450 (Fig. 7). However, with aerobic incubation, activities with both cytochromes declined, presumably because of reformation of the inactive cross-linked protein. Iodoacetamide slowed the decline of activity with cyt c and prevented the activity decline with P450. Taken together, these data demonstrate that restoration of the ability of the FAD and FMN domains to undergo conformational changes through reduction of the disulfide bond restores the ability to rapidly reduce the redox partners P450 and cyt c. Conversely, reoxidation of the disulfide abrogates electron transfer to cyt c and P450.
The effect of SOD and catalase on the activity of the different CYPOR preparations (cross-linked, DTT-treated, and DTT/iodoacetamide-treated) was examined (Table 6). In the presence of NADPH and oxygen, we and others have shown that wild type CYPOR produces superoxide, which dismutates to hydrogen peroxide (36, 37). Because superoxide and hydrogen peroxide are known to reduce cyt c, it was expected that the dismutase and catalase would decrease cyt c reductase activity of wild type CYPOR. Table 6 demonstrates that SOD and catalase do indeed decrease wild type cyt c reductase activity by ∼30%. Cyt c reductase activities of the 147CC514 proteins treated with DTT or DTT/iodoacetamide were diminished by 13 and 24%, respectively, although the activity of the cross-linked 147CC514 mutants were unaffected. These results indicate that the cross-linked mutant does not form a large amount of reactive oxygen species, which is consistent with its slower reduction by NADPH (Table 2).
TABLE 6.
Effect of SOD and CAT on the activity of 147CC514
CYPOR had been pretreated with DTT alone or with both DTT and iodoacetamide (IAM). DTT and DTT/iodoacetamide were removed prior to assay. Experimental details and controls are described under “Experimental Procedures.”
| Reductase | Nanomoles of cyt c reduced per s/nmol CYPOR | Nanomoles of CH2O/s/nmol P450-CYPOR |
|---|---|---|
| WT CPR | 62.0 ± 6.2 | 0.73 ± 0.06 |
| WT CPR + SOD | 41.0 ± 3.9 | 0.83 ± 0.10 |
| WT CPR + CAT | 46.0 ± 3.2 | 0.70 ± 0.09 |
| WT CPR + SOD + CAT | 46.0 ± 7.0 | |
| 147CC514 | 1.1 ± 0.1 | 0.07 ± 0.005 |
| 147CC514 + SOD | 1.0 ± 0.1 | 0.06 ± 0.008 |
| 147CC514 + CAT | 1.0 ± 0.1 | 0.01 ± 0.01 |
| 147CC514-DTT | 53.0 ± 6.0 | 0.53 ± 0.01 |
| 147CC514-DTT + SOD | 46.0 ± 5.2 | 0.62 ± 0.08 |
| 147CC514-DTT + CAT | 46.0 ± 5.1 | 0.47 ± 0.06 |
| 147CC514-DTT-IAM | 55.0 ± 3.2 | 0.57 ± 0.03 |
| 147CC514-DTT-IAM + SOD | 42.0 ± 7.0 | 0.64 ± 0.05 |
| 147CC514-DTT-IAM + CAT | 42.0 ± 4.0 | 0.56 ± 0.04 |
The effect of SOD and catalase on the metabolism of benzphetamine by P450 was examined (Table 6). SOD produced a slight, but not statistically significant, enhancement of P450-mediated benzphetamine metabolism. Because of the extremely low activity with 147CC514, the effect of SOD and catalase could not be accurately evaluated. However, the experiments with SOD and catalase do not alter our conclusion that a large conformational change of CYPOR is essential for electron transfer from the FMN domain to its redox partners.
DISCUSSION
Requirement for Domain Movements in Electron Transfer
The FMN domain of CYPOR is of particular interest as it is the domain that interacts with P450. Structural and site-directed mutagenesis experiments have been able to identify a potential binding site for cyt c and P450 on the FMN domain. The acidic clusters, Asp207–Asp208–Asp209 and Glu213–Glu214–Asp215, which encircle the FMN on the convex surface of the FMN domain, have been shown to be a region that interacts with both cyt c and P450 (31, 38). However, those residues are not accessible to the redox partners in the closed conformation of the wild type protein (7). This suggests that the FMN and FAD domains must alternate between an open and closed conformation in order for the FMN domain to shuttle electrons from FAD to its redox partners. In fact, a rotation of at least ∼90° of the FMN domain relative to the FAD domain is necessary to be able to dock the acidic convex surface of the FMN domain at its known binding site on the concave basic surface of P450 near to where the heme comes closest to the surface (9). Because the 10 residues extending from Gly232 to Arg243 were mobile in the closed conformation of CYPOR, in which the FAD and FMN were in contact, it was hypothesized that this linker, which joins the FMN domain to the remainder of the protein, might serve to allow the FMN domain to rotate into a position suitable for docking with P450 in a conformation appropriate for physiologic electron transfer (7). The structure of a mutant with four amino acids deleted from this linker was crystallized in three different open conformations in which the FAD and FMN were separated by 30–60 Å (8). The least extended of the three conformations could be docked with P450 2B4 in a manner that satisfied known mutagenesis constraints and in which the heme and flavin cofactors were ∼12 Å apart. This distance is known to support rapid electron transfer between redox coenzymes in proteins (39, 40). To demonstrate that a rotation of the FMN domain away from the FAD domain was indeed essential for CYPOR to transfer electrons from FAD to P450, a disulfide bond was engineered between the FAD and FMN domains of the protein.
The disulfide bond has been considered to be one of the major structural factors restricting protein motions (41–44). Previous studies have shown that introduction of artificial disulfide bonds into a protein molecule enhanced protein stability by lowering the entropy and increasing enthalpic stabilization of the folded state, indicating that a disulfide bond perturbs and/or restricts the protein fluctuations (45–49). It has become a useful tool for studying the importance of the motion and dynamics of a protein during catalysis. It allows us to stabilize and isolate protein conformers that are usually in equilibrium in solution and most of the time impossible to observe. Therefore, we decided to use this strategy to trap the reductase in a closed conformation by an appropriately engineered disulfide bond between the FMN and the FAD domains of CYPOR. If the hypothesis that the FMN and FAD domains must separate to be capable of donating electrons to P450 is correct, the disulfide bond cross-linked CYPOR will be compromised in its ability to transfer electrons to P450.
NADPH-supported microsomal P450 monooxygenase activity requires a specific interaction between P450 and CYPOR. Complex formation has been reported to occur via complementary charged residues and hydrophobic interactions (1, 9). Although mutagenesis studies have been able to identify a binding site for CYPOR on P450, the binding site on CYPOR for P450 is less well characterized. Nevertheless, it is well established that the concave, proximal basic surface of P450 receives the electrons from the FMN cofactor, which is surrounded by negatively charged acid residues on a convex surface (7, 31). Mutagenesis of Asp208 in the three aspartic acid cluster (Asp207–Asp209) diminishes catalysis by P450. In the cross-linked 147CC514 mutant, these residues are not accessible to the P450. If indeed the FMN domain of CYPOR is required to swing back and forth between the FAD domain and the P450 during electron transfer, the cross-linked mutant should not be able to reduce P450. Consistent with this hypothesis, the activity of the cross-linked mutant with P450 was reduced by 90%, electron transfer from FMNH2 to cyt c was inhibited by 96%, and electron transfer from FMNH2 to cyt P450 was decreased to below our detection limits. The observation of restoration of reductase activity upon cleavage of the disulfide bond demonstrates that a large conformational change in CYPOR is required for efficient electron transfer to P450. Various biochemical, structural, and site-directed mutagenesis data support this conclusion (7–9, 50–52). The cross-linked form of the protein is able to reduce Fe(CN)63−, consistent with the known ability of this substrate to accept electrons from FAD. The ability of the cross-linked protein to reduce cyt c, albeit at greatly diminished rates, is indicative of the less stringent structural requirements, with the possibility of multiple binding sites, previously proposed for this electron acceptor. However, it appears that domain movement is more essential for electron transfer to P450, with a 99% decrease in benzphetamine demethylase activity observed, when peroxide reduction of P450 was inhibited by catalase.
Conformational Changes Regulating NADPH Binding and NADP+ Releasing
In addition to the previously known structures of NADP+-bound CYPOR (PDB codes 1AMO/1JA1 and 1JA0, observed in the structure of the mutant in which the last two C-terminal residues (Trp677 and Ser678) were truncated (7, 12)), this study now provides a structure of the NADP+-free form of the protein as well as one with a different NADP+-binding mode. These structures allow us to envision a scenario for NADPH binding and NADP+ release.
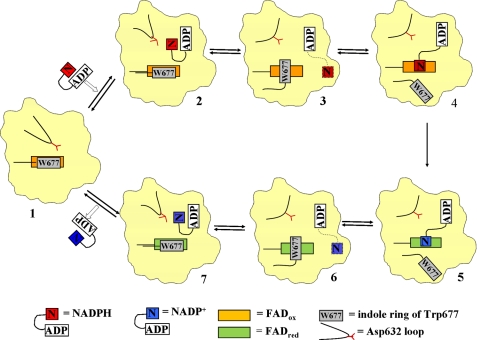
Fig. 8 shows a schematic representation of NADPH binding and NADP+ release for hydride transfer during the catalytic cycle of CYPOR. Upon binding to the enzyme (Fig. 8, state 1), the NADPH cofactor adopts a compact, folded conformation, in which the nicotinamide and adenine rings are almost parallel, as observed in the NADP+-bound form of 147C/C514 (state 2 of Fig. 8; Figs. 3 and 4B) and also found free in solution (29). Interactions of the 2′-AMP-pyrophosphate portion of NADP+ with Arg567, Arg597, and Lys602 anchor the cofactor, and the ribityl-nicotinamide moiety binds at the Asp632, as shown in Fig. 4B. Subsequently, the ribityl-nicotinamide moiety is displaced by the movement of the Asp632 loop and is searching for a binding site near FAD for efficient hydride transfer. The ribityl-nicotinamide adopts an extended conformation observed in wild type CYPOR structure (Fig. 3A, green molecule; Fig. 8, state 3). The indole ring of Trp677 moves away, and the nicotinamide ring binds to the re-face of the FAD ring as observed in the structure of the W677X mutant, 1JA0 (Fig. 8, state 4) (12). The reduced planar nicotinamide ring binds to the oxidized planar FAD ring, such that the N5 atom of FAD and the C4 atom of the nicotinamide are appropriately aligned for an efficient hydride transfer (as seen in 1JA0) (Fig. 8, state 5). Once the hydride ion transfer occurs, both the nicotinamide and flavin rings would lose aromatization and planarity, weakening the π-π interaction between the two ring systems, which in turn facilitates the displacement of the nicotinamide ring by Trp677 from the re-side of the FAD ring, leading to the disordered conformation of the extended NADP+ as observed in wild type CYPOR, 1AMO, and 1JA1 (Fig. 3, A and C, and state 6 of Fig. 8). The Asp632 loop moves away from the cofactor-binding site, allowing NADP+ to form the folded conformation ready to dissociate from the enzyme (Fig. 3B and state 7 of Fig. 8) The Asp632 loop now moves in closer to the binding site of the 2′,5′-ribityl-pyrophosphate moiety of NADP+. At this loop conformation, the carboxylate of Asp632 occupies the sterically obstructing position, which weakens the affinity of the pyrophosphate ribose moiety for the enzyme, resulting in the dissociation of NADP+ from CYPOR, and the enzyme returns to the NADP+-free structure (Fig. 4A, and state 1 in Fig. 8).
FIGURE 8.
Schematic illustration of NADPH binding to and NADP+ release from CYPOR. For clarity, only the FAD/NADP(H)-binding domain of CYPOR is shown. NADPH enters oxidized CYPOR (state 1, modeled after the structure of 147CC514, which lacks bound NADP(H)). NADPH initially binds to the enzyme in a folded conformation, such that the adenine ring and the nicotinamide ring almost stack on each other. Upon binding of NADPH, the Asp632 loop of CYPOR moves away to allow NADPH to bind (state 2, modeled from the structure of 147C/C514-NADP+). Once the NADPH binds, the Asp632 loop moves in and displaces the nicotinamide ring, resulting in the ribityl-nicotinamide moiety extending to search for the proper binding site for hydride transfer, while keeping the ADP-PPi anchor at the arginine site. The indole ring of Trp677 rotates to be ready to move away from the flavin ring (state 3, the structure of wild type; PDB code 1JA1). The indole ring moves away to make room for the nicotinamide ring to bind at the re-side of the isoalloxazine ring (state 4, modeled after the structure of the mutant W677X, in which the two penultimate residues of CYPOR, Trp677 and Ser678, were truncated, PDB code 1JA0). Hydride transfer occurs; FAD is reduced, and NADPH becomes NADP+ (state 5, from PDB code 1JA0). Once the flavin is reduced and the nicotinamide is oxidized, the nicotinamide ring moves out and the indole ring returns to the re-face of the FAD ring (state 6, PDB code 1JA1). As the Asp632 loop moves away allowing for the nicotinamide ring to fold back, NADP+ adopts the folded conformation poised to dissociate from the enzyme (state 7, the structure of 147C/C514-NADP+). The Asp632 loop moves back closer to where the NADP+ pyrophosphate-2′-AMP lies, causing steric hindrance as well as electrostatic repulsion, resulting in dissociation of the cofactor from CYPOR. The enzyme now returns to state 1 and the cycle repeats.
The linkage between the large scale domain movement and the movement of the Asp632 loop provides a mechanism by which electron transfer is coupled to cofactor binding and release. Cofactor binding triggers movement of Trp677, followed by hydride transfer and FAD reduction. These steps must occur with the enzyme in the closed position for electron transfer to proceed immediately to FMN. However, the domain movement necessary to accommodate cyt c or P450 must be tightly coupled to electron transfer to prevent reaction with oxygen and production of superoxide. It is likely that this motion is coupled with movement of the Asp632 loop to facilitate NADP+ release. Details of the mechanism through which large scale domain motions are coupled to movements of loops and/or individual amino acids remain to be established.
Supplementary Material
Acknowledgment
The assistance of Launa Wakenhut in preparation of the manuscript is greatly appreciated.
This work was supported, in whole or in part, by National Institutes of Health Grants CA22484 (to A. L. S. and C. B. K.), GM35533 (to L. W.), and GM52682 (to J.-J. P. K.). This work was also supported by a Veterans Affairs merit review grant (to L. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
The atomic coordinates and structure factors (codes 3OJW and 3OJX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CYPOR
- NADPH-cytochrome P450 reductase
- P450
- cytochrome P450
- DLPC
- dilauroyl l-3-phosphatidylcholine
- cyt c
- cytochrome c
- SOD
- superoxide dismutase
- CAT
- catalase
- PDB
- Protein Data Bank.
REFERENCES
- 1. Paine M. J., Scrutton N. S., Munro A. W., Gutierrez A., Roberts G. C., Wolf C. R. (2005) in Cytochrome P450 (Ortiz de Montellano P. R. ed) 3rd Ed., pp. 115–148, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 2. Vermilion J. L., Ballou D. P., Massey V., Coon M. J. (1981) J. Biol. Chem. 256, 266–277 [PubMed] [Google Scholar]
- 3. Oprian D. D., Coon M. J. (1982) J. Biol. Chem. 257, 8935–8944 [PubMed] [Google Scholar]
- 4. Guengrich F. (2005) in Cytochrome P450 (Ortiz de Montellano P.R. ed) 3rd Ed., pp. 377–463, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 5. Belelli D., Lambert J. J. (2005) Nat. Rev. Neurosci. 6, 565–575 [DOI] [PubMed] [Google Scholar]
- 6. Gainer J. V., Lipkowitz M. S., Yu C., Waterman M. R., Dawson E. P., Capdevila J. H., Brown N. J. and AASK Study Group (2008) J. Am. Soc. Nephrol. 19, 1606–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang M., Roberts D. L., Paschke R., Shea T. M., Masters B. S., Kim J. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamdane D., Xia C., Im S. C., Zhang H., Kim J. J., Waskell L. (2009) J. Biol. Chem. 284, 11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bridges A., Gruenke L., Chang Y. T., Vakser I. A., Loew G., Waskell L. (1998) J. Biol. Chem. 273, 17036–17049 [DOI] [PubMed] [Google Scholar]
- 10. Scott E. E., White M. A., He Y. A., Johnson E. F., Stout C. D., Halpert J. R. (2004) J. Biol. Chem. 279, 27294–27301 [DOI] [PubMed] [Google Scholar]
- 11. Ortiz de Montellano P. R. (2010) Chem. Rev. 110, 932–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hubbard P. A., Shen A. L., Paschke R., Kasper C. B., Kim J. J. (2001) J. Biol. Chem. 276, 29163–29170 [DOI] [PubMed] [Google Scholar]
- 13. Shen A. L., Sem D. S., Kasper C. B. (1999) J. Biol. Chem. 274, 5391–5398 [DOI] [PubMed] [Google Scholar]
- 14. Shen A. L., Christensen M. J., Kasper C. B. (1991) J. Biol. Chem. 266, 19976–19980 [PubMed] [Google Scholar]
- 15. Ausubel F. M., Brent R., Kingston R. E., Moore D. E., Seidman J. G., Smith J. A., Struhl K., Albright L. M., Coen D. M., Varki A. (eds) (1997) Current Protocols in Molecular Biology, pp. 3.0.1–3.19.8 and 8.0.1–8.5.10, John Wiley & Sons, Inc., New York [Google Scholar]
- 16. Shen A. L., Kasper C. B. (2000) J. Biol. Chem. 275, 41087–41091 [DOI] [PubMed] [Google Scholar]
- 17. Higuchi R., Krummel B., Saiki R. K. (1988) Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen A. L., Porter T. D., Wilson T. E., Kasper C. B. (1989) J. Biol. Chem. 264, 7584–7589 [PubMed] [Google Scholar]
- 19. Miroux B., Walker J. E. (1996) J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 20. Saribas A. S., Gruenke L., Waskell L. (2001) Protein Expr. Purif. 21, 303–309 [DOI] [PubMed] [Google Scholar]
- 21. Omura T., Sato R. (1964) J. Biol. Chem. 239, 2379–2385 [PubMed] [Google Scholar]
- 22. McPherson A. (1999) Crystallization of Biological Macromolecules, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Otwinowski Z., Minor W., Getzoff D. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kuntstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Acta Crystallogr D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Aigrain L., Pompon D., Moréra S., Truan G. (2009) EMBO Rep. 10, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lamb D. C., Kim Y., Yermalitskaya L. V., Yermalitsky V. N., Lepesheva G. I., Kelly S. L., Waterman M. R., Podust L. M. (2006) Structure 14, 51–61 [DOI] [PubMed] [Google Scholar]
- 29. Hull R. V., Conger P. S., 3rd, Hoobler R. J. (2001) Biophys. Chem. 90, 9–16 [DOI] [PubMed] [Google Scholar]
- 30. Iyanagi T., Makino N., Mason H. S. (1974) Biochemistry 13, 1701–1710 [DOI] [PubMed] [Google Scholar]
- 31. Shen A. L., Kasper C. B. (1995) J. Biol. Chem. 270, 27475–27480 [DOI] [PubMed] [Google Scholar]
- 32. Masters B. S., Kamin H., Gibson Q. H., Williams C. H., Jr. (1965) J. Biol. Chem. 240, 921–931 [PubMed] [Google Scholar]
- 33. Gutierrez A., Lian L. Y., Wolf C. R., Scrutton N. S., Roberts G. C. (2001) Biochemistry 40, 1964–1975 [DOI] [PubMed] [Google Scholar]
- 34. Matthews R. G. (1991) in Flavins and Flavoproteins (Curti B., Ronchi G., Zanetti G. eds) pp. 593–597, Walter de Gruyter, Inc., New York [Google Scholar]
- 35. Sem D. S., Kasper C. B. (1994) Biochemistry 33, 12012–12021 [DOI] [PubMed] [Google Scholar]
- 36. Gruenke L. D., Konopka K., Cadieu M., Waskell L. (1995) J. Biol. Chem. 270, 24707–24718 [DOI] [PubMed] [Google Scholar]
- 37. Kuthan H., Ullrich V., Estabrook R. W. (1982) Biochem. J. 203, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nisimoto Y. (1986) J. Biol. Chem. 261, 14232–14239 [PubMed] [Google Scholar]
- 39. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]
- 40. Page C. C., Moser C. C., Dutton P. L. (2003) Curr. Opin. Chem. Biol. 7, 551–556 [DOI] [PubMed] [Google Scholar]
- 41. Careaga C. L., Falke J. J. (1992) Biophys. J. 62, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Careaga C. L., Falke J. J. (1992) J. Mol. Biol. 226, 1219–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daggett V., Levitt M. (1993) Annu. Rev. Biophys. Biomol. Struct. 22, 353–380 [DOI] [PubMed] [Google Scholar]
- 44. Jeng M. F., Dyson H. J. (1995) Biochemistry 34, 611–619 [DOI] [PubMed] [Google Scholar]
- 45. Perry L. J., Wetzel R. (1984) Science 226, 555–557 [DOI] [PubMed] [Google Scholar]
- 46. Pantoliano M. W., Ladner R. C., Bryan P. N., Rollence M. L., Wood J. F., Poulos T. L. (1987) Biochemistry 26, 2077–2082 [DOI] [PubMed] [Google Scholar]
- 47. Matsumura M., Signor G., Matthews B. W. (1989) Nature 342, 291–293 [DOI] [PubMed] [Google Scholar]
- 48. Kanaya S., Katsuda C., Kimura S., Nakai T., Kitakuni E., Nakamura H., Katayanagi K., Morikawa K., Ikehara M. (1991) J. Biol. Chem. 266, 6038–6044 [PubMed] [Google Scholar]
- 49. Gusev N. B., Grabarek Z., Gergely J. (1991) J. Biol. Chem. 266, 16622–16626 [PubMed] [Google Scholar]
- 50. Gutierrez A., Paine M., Wolf C. R., Scrutton N. S., Roberts G. C. (2002) Biochemistry 41, 4626–4637 [DOI] [PubMed] [Google Scholar]
- 51. Ellis J., Gutierrez A., Barsukov I. L., Huang W. C., Grossmann J. G., Roberts G. C. (2009) J. Biol. Chem. 284, 36628–36637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hay S., Brenner S., Khara B., Quinn A. M., Rigby S. E., Scrutton N. S. (2010) J. Am. Chem. Soc. 132, 9738–9745 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.