Abstract
The relative activity of the AKT kinase has been demonstrated to be a major determinant of sensitivity of tumor cells to mammalian target of rapamycin (mTOR) complex 1 inhibitors. Our previous studies have shown that the multifunctional RNA-binding protein heterogeneous nuclear ribonucleoprotein (hnRNP) A1 regulates a salvage pathway facilitating internal ribosome entry site (IRES)-dependent mRNA translation of critical cellular determinants in an AKT-dependent manner following mTOR inhibitor exposure. This pathway functions by stimulating IRES-dependent translation in cells with relatively quiescent AKT, resulting in resistance to rapamycin. However, the pathway is repressed in cells with elevated AKT activity, rendering them sensitive to rapamycin-induced G1 arrest as a result of the inhibition of global eIF-4E-mediated translation. AKT phosphorylation of hnRNP A1 at serine 199 has been demonstrated to inhibit IRES-mediated translation initiation. Here we describe a phosphomimetic mutant of hnRNP A1 (S199E) that is capable of binding both the cyclin D1 and c-MYC IRES RNAs in vitro but lacks nucleic acid annealing activity, resulting in inhibition of IRES function in dicistronic mRNA reporter assays. Utilizing cells in which AKT is conditionally active, we demonstrate that overexpression of this mutant renders quiescent AKT-containing cells sensitive to rapamycin in vitro and in xenografts. We also demonstrate that activated AKT is strongly correlated with elevated Ser(P)199-hnRNP A1 levels in a panel of 22 glioblastomas. These data demonstrate that the phosphorylation status of hnRNP A1 serine 199 regulates the AKT-dependent sensitivity of cells to rapamycin and functionally links IRES-transacting factor annealing activity to cellular responses to mTOR complex 1 inhibition.
Keywords: Akt PKB, mTOR, mTOR Complex (mTORC), Mutant, Protein Phosphorylation
Introduction
A broad range of tumor types have been reported to exhibit hypersensitivity to mTORC12 inhibition with rapalogs depending on their degree of AKT activation (1–3). The cells that have elevated AKT activity as a result of dysregulated PI3K activity, AKT gene amplification, or a loss of PTEN display markedly increased G1 arrest following rapamycin exposure relative to cells having quiescent AKT (2, 3). Our previous studies have demonstrated that this differential sensitivity can be explained, in part, by continued IRES-initiated mRNA translation of cyclin D1 and c-MYC in the face of mTOR inhibition mediated by the ITAF hnRNP A1 (4). We have also demonstrated that direct phosphorylation of the ITAF hnRNP A1 on serine 199 by AKT regulates differential cyclin D1 and c-MYC IRES activity (5).
The ability of IRES-mediated protein synthesis to contribute to aberrant gene expression in cancer and during integrated cell stress responses is well documented (6–8); however, the processes regulating IRES function are poorly defined. Cellular IRESs require ITAFs to recruit the 40 S small ribosomal subunit leading to the formation of a competent preinitiation complex (9). Some ITAFs have been shown to directly interact with components of the ribosome to facilitate IRES-mediated initiation (10–13). However, these factors may also contribute to cellular IRES activities by promoting the formation of critical RNA-RNA interactions required for the formation of a productive IRES (14, 15).
The multi-functional RNA-binding protein hnRNP A1 has several established roles in mRNA metabolism (16). hnRNP A1 binds nascent pre-mRNAs in a sequence-specific manner and is known to promote RNA annealing (17–19). hnRNP A1 is also known to be involved in the export of mature transcripts from the nucleus, as well as in mRNA turnover and both cap-dependent and IRES-mediated translation (20–23). Although primarily a nuclear protein, hnRNP A1 shuttles continually between the nucleus and the cytoplasm. This shuttling activity is dependent on ongoing RNA polymerase II transcription and the integrity of a 38-amino acid C-terminal domain (M9 domain) (24).
Previously, we demonstrated that in IRES reporter assays utilizing translation competent cell extracts, the phosphorylation of hnRNP A1 at serine 199 specifically governed cyclin D1 and c-MYC IRES activities (5). To understand how this phosphorylation event may regulate the biochemical activities of hnRNP A1 and to further explore whether this particular phosphorylation event is critical and sufficient for AKT-dependent hypersensitivity to mTORC1 inhibition, we examined a substitution mutant of hnRNP A1. Additionally, because AKT activity is known to broadly affect many signaling pathways including MAPK signaling (25, 26), which is known to influence IRES-dependent translation initiation, we were interested in identifying mutants of hnRNP A1 that would circumvent hnRNP A1-independent effects of AKT on IRES activity.
In the present study, we describe a phosphomimetic mutant of the ITAF hnRNP A1 (S199E), which is able to bind to the cyclin D1 and c-MYC IRESs normally but is deficient in nucleic acid annealing activity. The mutant inhibits IRES activity in vitro, and overexpression of this mutant in cells inhibits cyclin D1 and c-MYC IRES activity in an AKT-dependent manner. Ectopic expression of the mutant also confers rapamycin hypersensitivity to quiescent AKT-containing cells both in culture and in xenograft experiments. Moreover, in primary glioblastoma samples, elevated levels of serine 473-phosphorylated AKT directly correlated with high levels of serine 199-phosphorylated hnRNP A1, supporting its applicability as a predictive biomarker for mTORC1 inhibitor therapies.
EXPERIMENTAL PROCEDURES
Cell Lines, Constructs, and Transfections
The glioblastoma line LN229 was obtained from ATCC (Manassas, VA), and mouse embryonic fibroblasts (MEFs) were generously provided by Dr. Hong Wu (Department of Molecular and Medical Pharmacology, UCLA). These lines were transfected with a myristoylated AKT-estrogen receptor ligand-binding domain fusion (myr-AKT-MER) cloned into pTracer-SV40 and stably expressing clones isolated (2). Control lines were transfected with empty vector (EV). Constructs expressing native and S199E mutated full-length hnRNP A1 as GST fusions, cloned into pcDNA3.1, have been described previously (5). The GFP-tagged hnRNP A1 construct was a gift from Claudio Sette (Department of Cell Biology, University of Rome Tor Vergata, Rome, Italy) (27). This construct was then subjected to site-directed mutagenesis to introduce the S199E mutation using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Transfections were performed using X-treme GENE Q2 transfection reagent (Roche Applied Science) and then cultured in the presence of G418. The dicistronic reporter plasmid pRF contains the Renilla and firefly luciferase ORFs separated by an intercistronic region and has been described (4). pRCD1 and pRmycF contain the minimal cyclin D1 and c-MYC IRES sequences, respectively, cloned into the intercistronic region of pRF (4).
Recombinant Proteins, Antibodies, and Reagents
Full-length native and S199E hnRNP A1-GST, which had been cloned into pGEX-2T, were expressed in Escherichia coli and purified as described previously (5). Antibodies were from the following sources. Anti-Ser(P)473)-AKT, anti-AKT, anti-Thr(P)389-S6K, anti-S6K, anti-mTOR, anti-cyclin D1, anti-MYC, anti-GST, anti-ERK, and anti-Thr(P)202/Tyr(P)204-ERK were from Cell Signaling (Danvers, MA); actin antibody was from Sigma. Anti-hnRNP A1 was from Abcam. Anti-Ser(P)199-hnRNP A1 antibody was generated in rabbits immunized with the phosphorylated peptide SQRGRSGpSGNFGGGR (where pS represents phosphoserine) and subsequently affinity-purified (5). The Ser(P)473-AKT and Thr(P)202/Tyr(P)204-ERK blocking peptides were obtained from Cell Signaling, and control peptides were synthesized by GenScript (Piscataway, NJ). Rapamycin and CCI-779 were obtained from LC Laboratories (Woburn, MA), and 4-hydroxy-tamoxifen was from Sigma. Rapamycin was used at a concentration of 100 μm for 24 h for all treatments.
Dicistronic IRES Reporter Assays
The indicated dicistronic reporters were co-transfected into cells with pSVβ-galactosidase to normalize for transfection efficiency as described previously (5). The cells were harvested 18 h after transfection, and Renilla, firefly, and β-galactosidase activities were determined (Dual-Glo luciferase and β-galactosidase assay systems; Promega).
Filter Binding Assays and ELISA
The indicated amounts of GST-hnRNP A1 and GST-S199E-hnRNP A1 were added to in vitro transcribed 32P-labeled RNAs corresponding to either the cyclin D1 or c-MYC IRESs in separate reactions in a volume of 10 ml in buffer containing 5 mm HEPES (pH 7.6), 30 mm KCl, 2 mm MgCl2, 200 mm DTT, 4% glycerol, and 10 ng of yeast tRNA for 10 min at room temperature (5). 8 μl of each binding reaction was applied to nitrocellullose membranes on a slot blot apparatus (Minifold II; Schleicher & Schuell). The membranes were washed and dried, and signals were quantified using a PhosphorImager. Binding curves of three independent experiments were fitted by using SigmaPlot to determine the apparent dissociation constants. ELISAs were performed as described (28). Briefly, in vitro transcribed biotinylated cyclin D1 or c-MYC IRES RNAs in binding buffer containing 25 mm HEPES (pH 7.5), 500 μm EGTA, 100 mm NaCl, 500 μm DTT, 4 mm MgCl2, 20 mm KCl, 0.05% Nonidet P-40, 0.5 mg/ml yeast tRNA, 0.05 mg/ml poly(A) RNA, 0.125 mg/ml BSA, 0.4 mm vanadyl ribonucleoside complexes, and 80 units/ml RNasin were mixed with various concentrations of recombinant hnRNP A1 S199E and immediately added to ELISA plates containing adsorbed native hnRNP A1. Following a 30-min incubation at room temperature, the plates were washed, and the amount of bound RNA was quantified via colorimetry by the addition of alkaline phosphatase-streptavidin and the substrate p-nitrophenyl phosphate. Absorbances were determined using a microplate reader using a 405-nm filter.
Cell Cycle Analysis
Cell cycle distributions were determined by propidium iodide staining of cells, followed by flow cytometry as described previously (2).
In Vitro Translation of Dicistronic mRNA Reporters
The dicistronic plasmids were linearized using BamHI and capped RNA transcribed in vitro (mMESSAGE T7 transcription kit; Ambion). Capped RNA transcripts were used to program extracts of LN229 cells as described (5).
hnRNP A1 in Vitro Annealing Assays
Reactions were performed as described previously by Kumar and Wilson (29). Briefly, a 19-mer oligonucleotide 5′-ACGGCCAGTGCCAAGCTTG-3′ complementary to positions 6280–6298 on M13amp18(+) strand DNA was used as template in the annealing reactions. The oligonucleotide was 5′-end labeled and mixed with equimolar amounts (0.2 nm) of M13mp18 single-stranded DNA in the presence of native or S199E hnRNP A1 as indicated. Annealing reactions were allowed to proceed at 25 °C for 5 min and subsequently electrophoresed, and the gels were exposed to film.
Immunohistochemistry of Primary Glioblastoma Samples
Flash-frozen normal brain and glioblastoma samples were obtained from the Cooperative Human Tissue Network, National Cancer Institute (Western Division, Vanderbilt University Medical Center) under an institutional review board-approved protocol. Each glioblastoma sample was histopathologically reviewed, and those containing greater than 95% tumor were utilized in this analysis. Sections of paraffin-embedded tumors on slides were processed for immunohistochemistry and scored on a scale of 0–2 (0, no staining; 1, mild intensity staining; and 2, strong staining). Staining scores of 1 and 2 were considered positive. Correlation analysis used the Spearman nonparametric correlation test. Differences were considered significant when p < 0.05. For additional multivariate analysis, we used a logistic regression model. The samples were also homogenized in radioimmune precipitation assay (lysis) buffer using a Polytron homogenizer (Fisher) to generate protein extracts for Western blot analysis.
Xenograft Studies
Male SCID mice were injected subcutaneously with single cell suspensions of the LN229EV and LN229AKT-MER as described previously (30). AKT activity was induced by intraperitoneal maintenance injections of 4-hydroxy-tamoxifen (4OHT) in peanut oil. Tumor growth was measured, and mice were randomized to CCI-779 versus vehicle when tumors reached 200 mm3. Treatment (eight mice/experimental group) was given via intraperitoneal injection for five consecutive days. Tumor growth was assessed on day 8 or 12 after initiation of CCI-779 treatment. Tumor volume was determined using the formula L × W2 × 0.5, where L is the longest length, and W is the shortest length. IC50 was determined by extrapolation of plots of percent growth inhibition by CCI-779 versus log concentration. Statistical analysis was done with Student's t test and analysis of variance models using Systat 13 (Systat Software, Chicago, IL).
RESULTS
Conditional Regulation of Rapamycin Sensitivity and IRES Function by a myr-AKT-MER Fusion Protein
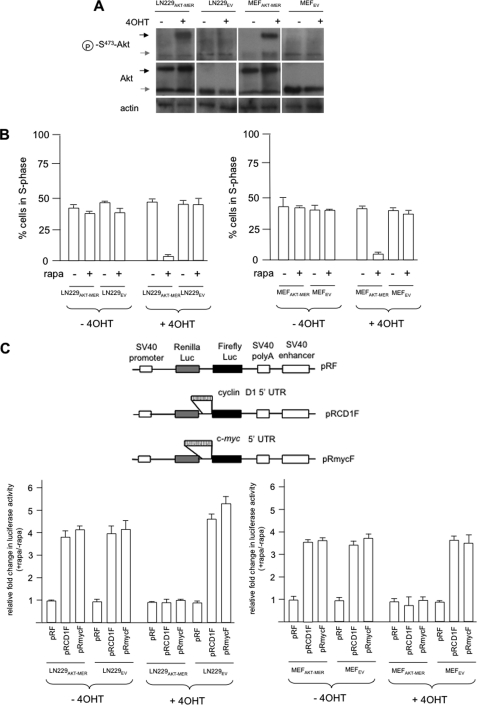
To investigate AKT-dependent rapamycin sensitivity, our previous studies have utilized a panel of isogenic cell lines in which AKT activity had been modulated via stable expression of a myristoylated constitutively active allele of AKT or via ectopic expression of PTEN in PTEN-null lines. To determine whether we could conditionally regulate rapamycin sensitivity in an AKT-dependent manner, we engineered either LN229 glioblastoma cells (PTEN wt) or MEFs to express myr-AKT-MER, which is a fusion protein consisting of the active form of AKT fused to the ligand-binding domain of the estrogen receptor (MER). In both of these cell lines, myr-AKT-MER (subsequently referred to as AKT-MER) was expressed at levels comparable with endogenous AKT (Fig. 1A). The activity of the AKT-MER fusion protein has been demonstrated to be induced following exposure to the MER ligand 4OHT and inactive in its absence (31). To determine whether expression of the AKT-MER fusion conferred differential sensitivity to rapamycin in these cells, we exposed LN229AKT-MER, LN229EV, MEFAKT-MER, and MEFEV cells to 4OHT in the absence or presence of rapamycin and determined the relative cell cycle distributions by flow cytometry. As shown in Fig. 1B, both the LN229 and MEF paired lines were relatively insensitive to rapamycin under these conditions; however, upon stimulation of AKT activity by 4OHT, both LN229AKT-MER and MEFAKT-MER cells were markedly sensitive to rapamycin and displayed significant reductions in S phase cell numbers as compared with controls. To determine whether conditional activation of AKT activity would also lead to differential cyclin D1 and c-MYC IRES activity as demonstrated in our previous studies (4), we transiently transfected LN229AKT-MER, LN229EV, MEFAKT-MER, and MEFEV with the indicated dicistronic reporter constructs (Fig. 1C, top panel) containing either the cyclin D1 or c-MYC IRESs within the intercistronic regions of the constructs. The relative amount of firefly luciferase activity is indicative of IRES-mediated protein synthesis directed by either the cyclin D1 or c-MYC IRESs, whereas Renilla luciferase activity is a readout of cap-dependent initiation (4). As shown in Fig. 1C, in both the LN229 and MEF paired cell lines, expressing the myr-AKT-MER fusion stimulation of AKT activity strongly inhibited cyclin D1 and c-MYC IRES activity in LN229AKT-MER and MEFAKT-MER cells following rapamycin treatment, consistent with our previous studies showing that AKT activity negatively regulates cyclin D1 and c-MYC IRES function (4). Renilla luciferase activity was not significantly affected by differential AKT activity but was reduced by ∼70–90% following rapamycin treatment compared with values obtained in its absence consistent with the effects of the drug on eIF-4E-mediated initiation (data not shown). Finally, we also examined the steady-state levels of various proteins (supplemental Fig. S1), including cyclin D1 and c-MYC, in these lines following AKT induction and exposure to rapamycin. As demonstrated previously, these lines exhibited the AKT-dependent alterations in cyclin D1 and c-MYC protein levels following rapamycin treatment and reflected the changes in IRES-mediated protein synthesis. These data demonstrate that the relative activity of the myr-AKT-MER fusion governs cyclin D1 and c-MYC IRES activities and rapamycin sensitivity in these lines.
FIGURE 1.
Conditionally active AKT regulates rapamycin hypersensitivity and cyclin D1 and c-MYC IRES activities. A, immunoblot analysis of LN229 and MEF cells expressing the myr-AKT-MER fusion protein or EV control transfectants. The indicated cells were treated with the ligand 4OHT (1 μm, 24 h), and the extracts were prepared and separated by SDS-PAGE. The blots were incubated with anti-AKT, anti-Ser(P)473-AKT, and anti-β-actin antibodies. The black and gray arrowheads represent the myr-AKT-MER and endogenous AKT, respectively. B, LN229AKT-MER and LN229EV (left panel) and MEFAKT-MER and MEFEV (right panel) were treated in the presence or absence of 4OHT and rapamycin as shown and subjected to propidium iodide staining followed by flow cytometry. The means ± S.D. are shown for three independent experiments. C, AKT-dependent differential cyclin D1 and c-MYC IRES activities in myr-AKT-MER-expressing cell lines following 4OHT and rapamycin treatment as shown (LN229AKT-MER and LN229EV, left bottom panel; MEFAKT-MER and MEFEV, right bottom panel). The top panel depicts schematic diagrams of the dicistronic vectors. The relative fold change in firefly luciferase activity is shown as compared with activities obtained in the absence of rapamycin and normalized to values obtained for pRF in each cell line. The means ± S.D. are shown for three independent experiments.
The hnRNP A1 S199E Mutant Does Not Support IRES Activity in Vitro
Initially we investigated whether the phosphomimetic mutant would support cyclin D1 and c-MYC IRES activity in vitro. Translation-competent cell extracts were prepared from cells in which hnRNP A1 had been knocked down via RNA interference (supplemental Fig. S2A) and programmed with in vitro transcribed dicistronic mRNAs containing either the cyclin D1 or c-MYC IRES within the intercistronic region. siRNA-mediated knockdown of hnRNP A1 resulted in undetectable levels as determined by immunoblotting, relative to a nontargeting scrambled sequence siRNA that had no appreciable effect on expression. The knockdown was also specific for hnRNP A1 because actin levels were unchanged in cells following treatment with the targeting siRNAs. The reduction in hnRNP A1 expression also did not significantly affect the ability of these extracts to support cap-dependent protein synthesis of exogenous Renilla mRNAs (supplemental Fig. S2B). These extracts were then supplemented with a negative control ITAF, glyceraldehyde-3-phosphate dehydrogenase, which binds the HAV IRES, native hnRNP A1, or hnRNP A1 S199E. The integrity and purity of the recombinant hnRNP A1 proteins were confirmed (supplemental Fig. S2C). As can be seen in Fig. 2, adding recombinant hnRNP A1 markedly stimulated both cyclin D1 (left panel) or c-MYC (right panel) in vitro IRES activity, whereas the addition of the hnRNP S199E mutant or the negative control glyceraldehyde-3-phosphate dehydrogenase was unable to stimulate IRES activity. These data demonstrate that the addition of native hnRNP A1 is sufficient to support IRES activity and strongly suggest that the siRNA-mediated knockdown does not have apparent off target effects, while also confirming that the inhibition of IRES activity in these extracts is due to a direct effect of hnRNP A1. Additionally, these data demonstrate that the phosphomimetic mutant is unable to support cyclin D1 or c-MYC IRES activity in these cell extracts.
FIGURE 2.
Recombinant hnRNP A1 S199E does not support IRES activity in vitro. Translation-competent extracts were prepared from LN229 cells in which hnRNP A1 expression had been knocked down via siRNA exposure, and the indicated proteins were added to the extracts prior to programming the extracts with either in vitro transcribed dicistronic cyclin D1 (left panel) or dicistronic c-MYC (right panel) reporter mRNAs. Translations were performed at 30 °C for 40 min. An irrelevant ITAF, GAPDH was added as a negative control. Renilla (black bars) and firefly (open bars) luciferase activities were determined and normalized to values obtained for extracts alone. The means ± S.D. are shown for three independent experiments.
Phosphomimetic Mutation of hnRNP A1 Ser199 Does Not Impede IRES RNA Binding and Can Compete with Native hnRNP A1
To begin to address the mechanism by which the hnRNP A1 S199E mutant inhibited IRES activity, we initially sought to determine whether this mutant would bind the cyclin D1 and c-MYC IRESs normally. As shown in Fig. 3 (A and B), the hnRNP A1 mutant bound to both the cyclin D1 and c-MYC IRES RNAs, respectively, with equilibrium dissociation constants (Kd) that were comparable with native hnRNP A1 (∼200 nm) as determined in filter binding assays. These values were consistent with our previous apparent Kd measurements (5) as well as those obtained for other hnRNP A1-IRES RNA interactions (23). Based on these findings, we also wanted to determine whether the hnRNP A1 S199E mutant could compete with native hnRNP A1 for cyclin D1 and c-MYC IRES binding. To investigate this, we utilized an ELISA-based RNA binding assay (28). In this assay, hnRNP A1 was affixed to the ELISA well and subsequently allowed to bind biotinylated cyclin D1 or c-MYC IRES RNAs and hnRNP A1 S199E. Cyclin D1 or c-MYC IRES RNAs that bound the mutant are inaccessible for hnRNP A1 binding and are removed during washing procedures. The amount of retained RNA was quantified using streptavidin-conjugated alkaline phosphatase and a colorimetric substrate. As shown in Fig. 3 (C and D), there was a concentration-dependent reduction of hnRNP A1 binding to the cyclin D1 and c-MYC IRESs, respectively, in the presence of the hnRNP A1 S199E mutant. An ITAF that binds specifically to the HAV IRES, GAPDH, was included as a negative control and did not display significant IRES RNA binding (32, 33). These data taken together suggest that the phosphomimetic mutant of hnRNP A1 S199E is competent to bind both the cyclin D1 and c-MYC IRES RNAs and additionally can compete for binding with native hnRNP A1.
FIGURE 3.
Phosphomimetic hnRNP A1 S199E mutant in vitro IRES RNA binding characteristics and ability to compete with native hnRNP A1 for IRES binding. A and B, hnRNP A1 (closed circles) and S199E mutant (open circles) binding curves for the cyclin D1 (165 nucleotides) (A) and c-MYC (233 nucleotides) (B) IRES RNAs. C and D, mutant hnRNP A1 (open circles) competes with native protein for binding to cyclin D1 (C) or c-MYC (D) IRES RNAs. Native hnRNP A1 was adsorbed to the ELISA well and the binding reaction initiated by the addition of biotinylated IRES RNA and mutant hnRNP A1. IRES RNA binding was determined via colorimetry. The irrelevant ITAF, GAPDH (closed circles in C and D, was included as a negative control and did not bind either the cyclin D1 or c-MYC IRES RNAs. The results are expressed as percentages of the value obtained in the absence of competitor. The means ± S.D. are shown (n = 3).
hnRNP A1 S199E Mutation Suppresses Strand Annealing Activity
To further investigate a possible mechanistic basis for the reduced IRES activities observed, we determined whether the S199E mutation affected the ability of hnRNP A1 to promote strand annealing. hnRNP A1 is known to facilitate strong RNA or DNA strand annealing comparably (19, 29), and a critical function of this property may be to provide required structural elements that favor the formation of a productive IRES conformation capable of ribosome recruitment. Additionally, in vitro phosphorylation of serine 199 of hnRNP A1 has been reported to inhibit its native annealing activity (18). As shown in Fig. 4A, native hnRNP A1 efficiently promoted reannealing of single-stranded M13 mp18 plus strand DNA and a base pair complementary 19-mer oligonucleotide in a concentration-specific manner in an in vitro annealing assay. However, under identical experimental conditions, the S199E mutation completely abolished the reannealing activity of hnRNP A1.
FIGURE 4.
hnRNP S199E mutant displays reduced annealing activity. A, recombinant native or S199E hnRNP A1 was purified and analyzed for reannealing activity. The amount of protein used in the annealing reactions is shown above the panel, and the migration positions of the indicated species are also displayed. B and C, subcellular localization of GFP-tagged native (B) and S199E hnRNP A1 (C) in LN229 cells following the indicated treatments. The cells were transfected with GFP-hnRNP A1 or GFP-hnRNP A1 S199E plasmids and treated with 10 nm rapamycin (rap) for 24 h and GFP visualized using fluorescence microscopy.
Because hnRNP A1 also has demonstrated roles in mRNA trafficking, which may play a role in the regulation of IRES activity (34), we examined whether the mutant hnRNP A1 exhibited altered localization patterns relative to the native protein. As shown in Fig. 4 (B and C), native GFP-hnRNP A1 showed similar localization patterns as compared with GFP-S199E-hnRNP A1 under basal and post-rapamycin-treated conditions, being primarily localized to the nucleus and perinuclear regions under basal conditions and found mostly in the cytoplasm with a high degree of membrane-association following rapamycin exposure (35). These GFP fusions were expressed and remained intact prior to and following rapamycin treatments (supplemental Fig. S3A). Although it is likely that hnRNP A1 redistribution following rapamycin exposure plays a role in IRES-mediated translation, these data taken together strongly suggest that the S199E mutation in hnRNPA1 affects its ITAF function via suppressing RNA annealing activity.
Expression of hnRNP A1 S199E Inhibits Cyclin D1 and c-MYC IRES Activity in Cells
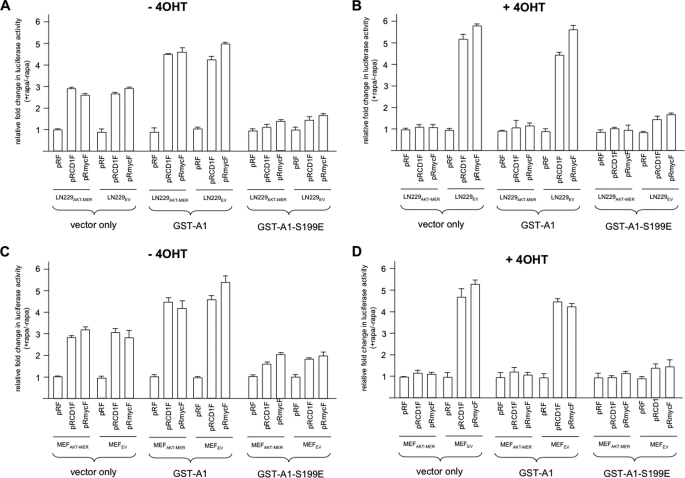
We subsequently determined whether forced expression of the hnRNP A1 S199E mutant would have effects on AKT-dependent cyclin D1 and c-MYC IRES activity in cells. The conditionally active AKT LN229 and MEF cell line pairs were stably transfected with an empty mammalian expression vector alone (pCDNA3.1+), the vector containing a full-length GST-tagged hnRNP A1, or the vector containing GST-tagged hnRNP A1 S199E. G418-resistant clones were isolated and screened for stable expression of the respective transgene (supplemental Fig. S3B). We then tested whether rapamycin-stimulated cyclin D1 or c-MYC IRES activity was influenced in cells expressing the hnRNP A1 S199E mutant by transient transfection of the indicated lines in Fig. 5 with dicistronic mRNA reporter constructs as before. As shown in Fig. 5A, in the absence of 4OHT, rapamycin stimulated cyclin D1 and c-MYC IRES activity in both LN229AKT-MER and LN229EV cells transfected with the vector only control or GST-A1. A modestly higher degree of rapamycin-stimulated cyclin D1 and c-MYC IRES activity was observed in GST-A1-expressing cells consistent with the ability of A1 to facilitate IRES activity. However, in LN229AKT-MER and LN229EV cells, which expressed the GST-A1-S199E mutant, both cyclin D1 and c-MYC rapamycin-stimulated IRES activity was blunted. In the presence of the AKT inducer 4OHT (Fig. 5B), LN229EV (quiescent AKT-containing cells) cells exhibited a 5–6-fold stimulation of rapamycin-induced cyclin D1 and c-MYC IRES activity in vector only or GST-A1-expressing cells, whereas in LN229AKT-MER cells (active AKT-containing cells) little or no stimulation of cyclin D1 or c-MYC IRES activity, relative to control pRF transfected cells, was observed following rapamycin exposure. In LN229EV cells expressing the phosphomimetic hnRNP A1 mutant (GST-A1-S199E), however, a marked reduction in rapamycin-stimulated cyclin D1 and c-MYC IRES activity was observed as compared with identically treated LN229AKT-MER cells expressing GST-A1-S199E. Similar results were obtained in the MEFAKT-MER and MEFEV paired lines shown in Fig. 5 (C and D). Again, in the absence of 4OHT, rapamycin exposure led to a significant stimulation of IRES activity in MEFAKT-MER and MEFEV cells transfected with the vector only or GST-A1, which was inhibited in lines expressing the GST-A1-S199E mutant (Fig. 5C). In the presence of 4OHT, MEFEV (quiescent AKT-containing cell) displayed significant rapamycin-stimulated cyclin D1 and c-MYC IRES activity relative to MEFAKT-MER cells, consistent with the AKT-dependent regulation of IRES activity. However, in MEFEV cells expressing the GST-A1-S199E mutant, a marked reduction in rapamycin-stimulated cyclin D1 and c-MYC IRES activity was observed (Fig. 5D). These results demonstrate that forced expression of the phosphomimetic hnRNP A1 S199E mutant is sufficient to inhibit rapamycin-stimulated and AKT-dependent cyclin D1 and c-MYC IRES activity and suggest that the mutant protein has significant dominant negative effects on IRES activity.
FIGURE 5.
The hnRNP A1 S199E mutant inhibits IRES activity in cells. Rapamycin stimulated IRES activity is inhibited in LN229AKT-MER, LN229EV, MEFAKT-MER, and MEFEV cells expressing the hnRNP A1 S199E mutant. The indicated cell lines expressing empty vector, GST-hnRNP A1 (GST-A1), or GST-hnRNP A1 S199E were transfected with the dicistronic reporter plasmids shown in the absence or presence of rapamycin. A, without 4OHT; B, with 4OHT. C and D are identical to A and B, respectively, except using the MEFAKT-MER and MEFEV lines as indicated. Relative fold change in firefly activity is shown as compared with activities obtained in the absence of rapamycin and normalized to values obtained for pRF in each cell lines. The means ± S.D. are shown (n = 3).
Ectopic Expression of the Phosphomimetic Mutant of hnRNP A1 S199E Confers Rapamycin Sensitivity
To address whether forced expression of the phosphomimetic hnRNP A1 mutant was sufficient to result in rapamycin sensitivity of resistant cells with relatively quiescent AKT, we treated the conditionally active AKT LN229 and MEF paired lines expressing the GST-hnRNP A1-S199E mutant with rapamycin and determined cell cycle distributions by flow cytometry as before. As shown in Fig. 6A, LN229AKT-MER and LN229EV cells, in the absence of 4OHT, which expressed vector only sequences or GST-A1, were relatively resistant to rapamycin. However, both LN229AKT-MER and LN229EV cells expressing the GST-hnRNP A1-S199A mutant were markedly sensitive to rapamycin. We then examined the sensitivity of these cells in the context of differential AKT activity. As shown in Fig. 6B, in the presence of 4OHT, LN229AKT-MER (active AKT-containing) cells either expressing the vector only or GST-A1 demonstrated marked sensitivity to rapamycin, whereas LN229EV (quiescent AKT-containing cells) expressing vector only and GST-A1 were relatively resistant. In LN299AKT-MER and LN229EV cells stably expressing the GST-hnRNP A1-S199E mutant, however, the AKT-dependent differential sensitivity of the cells was abrogated, because both active and quiescent AKT-containing lines were equally sensitive to rapamycin. Similar rapamycin sensitivities were observed in the MEFAKT-MER and MEFEV paired lines, which expressed the phosphomimetic hnRNP A1 mutant as shown in Fig. 6 (C and D). In the absence of 4OHT, both MEFAKT-MER and MEFEV lines expressing either vector sequences only or GST-hnRNP A1 were relatively resistant to rapamycin. MEFAKT-MER and MEFEV cell lines expressing the GST-hnRNP A1-S199E mutant were sensitive to rapamycin as observed in the LN229 transfectants. In MEFAKT-MER and MEFEV cells treated with 4OHT to differentially activate AKT and stably expressing vector or GST-hnRNP A1, we observed AKT-dependent rapamycin sensitivity, which was abolished in these lines expressing the GST-hnRNP A1-S199E mutant (Fig. 6D). Collectively, these data demonstrate that overexpression of the phosphomimetic mutant of hnRNP A1 is sufficient to confer rapamycin sensitivity and strongly suggest that this phosphorylation event is capable of regulating the cellular response to mTORC1 inhibitors.
FIGURE 6.
Overexpressing the hnRNP A1 S199E mutant confers sensitivity to TORC1 inhibition. The indicated lines were stably transfected with vector alone, GST-hnRNP A1, and GST-hnRNP A1 S199E and treated without or with rapamycin (rapa) as shown. The percentage of cells in the S phase of the cell cycle was subsequently determined on propidium iodide-stained cells by flow cytometry. A, without 4OHT; B, with 4OHT in LN229AKT-MER and LN229EV cells. C and D are identical to A and B, respectively, except using the MEFAKT-MER and MEFEV lines. The means ± S.D. are shown (n = 3).
The hnRNP A1 S199E Mutant Confers mTORC1 Inhibitor Sensitivity in GBM Xenografts
To determine whether the phosphomimetic hnRNP A1 mutant would have effects on rapamycin sensitivity in vivo, we utilized the conditionally active AKT LN299AKT-MER and LN229EV cell line pair stably expressing the mutant hnRNP A1 in a series of murine xenograft studies. Mice were injected with either LN229AKT-MER or LN229EV lines expressing either vector, GST-hnRNP A1, or the phosphomimetic mutant GST-hnRNP A1-S199E pretreated with or without 4OHT. Mice injected with 4OHT-treated cells were given daily intraperitoneal maintenance injections of 4OHT. Upon tumors reaching a mean volume of 200 mm3, all of the mice were additionally given CCI-779 (vehicle, 0.01, 0.1, and 4 mg/kg) for 5 consecutive days and at 8 and 12 days after initiation of treatment tumor growth was assessed. As shown in Fig. 7, CCI-779 inhibited growth of all the cell lines in a dose-dependent manner. In the absence of 4OHT, LN229AKT-MER and LN229EV tumors expressing vector sequences or GST-hnRNP A1 were relatively refractory to CCI-779 except at the highest dose. However, LN229AKT-MER and LN229EV tumors that expressed the phosphomimetic hnRNP A1 S199E mutant showed marked inhibition of tumor growth in response to CCI-779 consistent with the previous data with cell lines (Fig. 6). In mice in which 4OHT was administered, LN229AKT-MER that expressed vector sequences or GST-hnRNP A1 was sensitive to CCI-779 and demonstrated significant inhibition of tumor growth at all concentrations tested. In contrast, LN229EV cells expressing either vector only sequences or GST-A1 were relatively resistant to CCI-779, corroborating previous studies demonstrating AKT-dependent sensitivity to mTORC1 inhibition (2). In LN229AKT-MER and LN229EV tumors that expressed the GST-hnRNP A1-S199A mutant, however, marked CCI-779 sensitivity was observed in both lines. These results support our previous experiments and demonstrate that overexpression of the GST-hnRNP A1-S199E mutant results in mTORC1 inhibitor sensitivity in tumor cell xenografts.
FIGURE 7.
The hnRNP A1 S199E mutant confers TORC1 inhibitor sensitivity in GBM xenografts. Inhibition of tumor growth by CCI-779 in xenografts of LN229AKT-MER and LN229EV cells overexpressing the hnRNP A1 mutant in SCID mice. The mice were treated with CCI-779 for 5 days with the indicated doses of drug and tumor growth assessed at day 8 (D8) or day 12 (D12) after initial treatment. The mice were treated with 4OHT as indicated via daily maintenance injections of 4OHT. The horizontal bars indicate 50% reduction in control tumor growth (vehicle-treated). The data represent tumor volumes (means ± S.D. of three experiments).
Primary GBM Tumors Containing Elevated AKT Activity Display High Levels of Phosphorylated Ser199 hnRNP A1
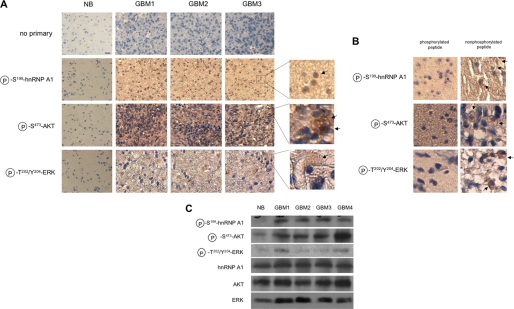
To determine whether the signaling relationship between AKT and hnRNP A1 could be detected in clinical samples, we evaluated a panel of 22 GBM samples determined previously to harbor elevated levels of Ser473-phosphorylated AKT (30). We analyzed these samples for Ser(P)199 hnRNP A1 (predominantly localized in nuclei) ands Ser(P)473 AKT and Thr(P)202/Tyr(P)204 ERK kinase using phosphorylation-specific antibodies via immunohistochemical analysis shown in Fig. 8A. Total protein levels for AKT, ERK, and hnRNP A1 were also determined, and no significant differences were found relative to normal brain expression levels (data not shown), consistent with the results of other studies (36, 37). The specificity of staining was also confirmed in a series of antibody blocking experiments in which samples were stained either with antibodies preabsorbed with phosphorylated or nonphosphorylated peptides (Fig. 8B). Specific staining was only observed in samples stained with antibodies preabsorbed with the relevant nonphosphorylated peptides. In statistical analysis of the scoring data, Ser(P)199 hnRNP A1 was significantly correlated with Ser(P)473 AKT phosphorylation (p = 0.0005) but not with Thr(P)202/Tyr(P)204 ERK activation (p = 0.61). We then also fit a regression model with Ser(P)199 hnRNP A1 as the outcome and Ser(P)473 AKT and Thr(P)202/Tyr(P)204 ERK as co-variates. A significant independent association was identified between Ser(P)199 hnRNP A1 and Ser(P)473 AKT (p = 0.0015), whereas the association between Ser(P)199 hnRNP A1 and Thr(P)202/Tyr(P)204 was not significant (p = 0.78). To corroborate these results, we also performed immunoblotting experiments on those tumors where sufficient material was available and was scored positive for elevated levels of Ser473-phosphorylated AKT by immunohistochemistry. As shown in Fig. 8C, tumor samples that displayed elevated levels of Ser473-phosphorylated AKT also contained high levels of Ser199-phosphorylated hnRNP A1. The results from these analyses are consistent with the known ability of AKT to phosphorylate hnRNP A1 at serine 199 as well as the evident inability of hnRNP A1 to be phosphorylated via ERK on this residue.
FIGURE 8.
GBM displaying elevated levels of AKT activity harbor high levels of phosphorylated Ser199 hnRNP A1. A, representative immunohistochemical analysis of Ser(P)199-hnRNP A1, activated AKT, and activated ERK in a panel of 22 human glioblastoma samples. The sections were prepared and stained with antibodies to the indicated proteins followed by an appropriate secondary antibody. The sections were subsequently incubated with streptavidin-horseradish peroxidase and developed with 3,3′-diaminobenzidine reagent. Magnified images of the indicated regions are shown to the right. NB, normal brain. The arrows indicate regions of significant staining. Bar, 10 μm. B, glioblastoma samples stained with the indicated antibodies following preabsorption with the relevant reacting phosphorylated peptide (left panels) or corresponding nonphosphorylated peptide (right panels). The arrows indicate areas of significant staining in samples stained with antibodies preabsorbed with the corresponding nonphosphorylated peptides. C, representative immunoblot analysis of glioblastoma samples displaying elevated Ser(P)473-AKT levels. Normal brain and tumor lysates were analyzed using antibodies against the indicated proteins.
DISCUSSION
Our previous studies have indicted the multi-functional RNA-binding protein hnRNP A1 as a critical factor in the AKT-dependent regulation of cyclin D1 and c-MYC IRES activity (5). Serine 199 phosphorylation of hnRNP A1 by AKT inhibits its ability to promote IRES-mediated translation initiation, and this phosphorylation event also confers rapamycin hypersensitivity. In this report, we have identified a phosphomimetic mutant of hnRNP A1, which we demonstrate is specifically deficient in nucleic acid annealing activity, resulting in markedly reduced ITAF function in vivo. Forced expression of this mutant abrogated both cyclin D1 and c-MYC IRES activity in quiescent AKT-containing cells and resulted in rapamycin sensitivity. We have proposed a model in which hnRNP A1 is constitutively associated with the cyclin D1 and c-MYC IRESs and promotes translation initiation of these determinants in the face of mTORC1 inhibition in cells harboring relatively quiescent AKT activity. However, in cells with elevated AKT activity, hnRNP A1 is phosphorylated at serine 199, rendering it unable to act as an ITAF and promote protein synthesis, resulting in rapamycin hypersensitivity.
There is significant experimental evidence supporting the role of secondary and tertiary structure generated by RNA-RNA interactions for the function of cellular and viral IRESs (14, 15). It has also been proposed that ITAFs may function as RNA chaperones that promote the formation of a competent IRES structure that can facilitate ribosomal small subunit binding (8, 12). An important biochemical activity of hnRNP A1 is its ability to enhance the renaturation of single-stranded nucleic acids in vitro (18, 19, 29). A possible implication for this activity in IRES-mediated initiation is that it may promote both localized RNA secondary structure formation as well as long range intramolecular RNA-RNA interactions critical for IRES activity. The observation that the S199E mutant of hnRNP A1 appears to be specifically deficient in annealing activity and also displays dominant negative ITAF function supports this concept. It is also conceivable that hnRNP A1-mediated annealing may additionally regulate the activity of a microRNA involved in cyclin D1 and c-MYC IRES-dependent initiation.
Previous in vitro data have shown that phosphorylation of hnRNP A1 at serine 199 is sufficient to inhibit annealing activity (18, 19). Additionally, circular dichroism spectroscopic analyses (19) demonstrated a marked change in secondary structure of hnRNP A1 upon phosphorylation of this residue, consistent with the generation of a more ordered structure in its C-terminal domain. This mostly disordered domain confers nucleic acid binding cooperativity to hnRNP A1 molecules and is required for annealing activity. Although the precise mechanism of strand annealing is unknown, it is tempting to speculate that the phosphomimetic mutant may also exhibit structural alterations mimicking the in vitro phosphorylation of hnRNP A1. However, our RNA binding data demonstrated that the S199E mutant displayed normal cooperative IRES-RNA binding similar to native hnRNP A1, suggesting that the RNA binding and annealing properties of hnRNP A1 are separable. Further study of this mutant may provide mechanistic insights into hnRNP A1-based strand annealing activity.
Our previous studies utilizing translation-competent cell extracts revealed that AKT negatively regulates cyclin D1 and c-MYC IRES activity in vitro. The present data utilizing the hnRNP A1 S199E phosphomimetic mutant in vivo closely mimic and support our in vitro findings. The simplest mechanism by which the mutant exerts its dominant negative effects in cells likely involves competing with endogenous hnRNP A1 for cyclin D1 and c-MYC IRESs, rendering them inactive, resulting in rapamycin sensitivity in quiescent AKT-containing cells. The RNA binding ELISA experiments utilizing native hnRNP A1 and the phosphomimetic mutant support this notion (Fig. 3, C and D). However, it is also possible that in cells the phosphomimetic mutant may additionally affect interactions among endogenous hnRNP A1 molecules as to alter the RNA-strand annealing properties of hnRNP A1, resulting in a IRES structure that is not formed properly and is markedly less efficient or nonfunctional.
The observation that rapamycin induces a dramatic relocalization of both native and the mutant hnRNP A1 (Fig. 4, B and C) is consistent with reports demonstrating cytoplasmic accumulation of this factor following stress (38). The subcellular localization of ITAFs may participate in the regulation of IRES activity (34). However, the current data suggest that hnRNP A1 can negatively or positively influence IRES-mediated translation initiation depending on the specific mRNA. Cytoplasmic localization of hnRNP A1 was demonstrated to promote human rhinovirus-2 IRES activity, whereas it was found to repress IRES-dependent initiation of the APAF-1 transcript (39). We envision a model in which hnRNP A1 accumulation in the cytoplasm may enhance cyclin D1 and c-MYC IRES activity and in which phosphorylation at serine 199 via AKT inactivates the RNA annealing function of this ITAF. However, our previous studies utilizing a dominant negative shuttling-defective mutant of hnRNP A1, which predominantly remains nuclearly localized, demonstrated that cells expressing this mutant exhibited partially reduced cyclin D1 and c-MYC IRES activities following rapamycin exposure (5), suggesting a role for hnRNP A1 trafficking in its ITAF function. It is possible that both the subcellular localization and annealing properties of hnRNP A1 are subject to regulation and that each contributes to the overall ability of hnRNP A1 to support IRES activity. Additional studies are being performed to address the mechanisms by which rapamycin induces hnRNP A1 redistribution to the cytoplasm and what role this may play in IRES-dependent initiation.
In summary, we have characterized a phosphomimetic mutant of hnRNP A1 that is specifically deficient in single-strand annealing. Cells overexpressing this mutant demonstrated reduced AKT-dependent cyclin D1 and c-MYC IRES activities in response to mTORC1 inhibition, resulting in increased sensitivity of quiescent AKT-containing cells to rapamycin in vitro and in xenograft experiments. Furthermore, in a panel of glioblastomas, elevated AKT activity directly correlated with increased Ser199 hnRNP A1 phosphorylation. Therefore, monitoring this phosphorylation event may have utility as a surrogate biomarker to stratify those tumors that may be most sensitive to mTORC1 inhibitor therapy.
Supplementary Material
Acknowledgments
We thank Hong Wu, Simona Pedrotti, and Claudio Sette for cell lines and reagents. We also are grateful to Johnathan Lee and YiJiang Shi for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA111448 (to A. L.) and R01CA109312 (to J. G.). This work was also supported by funds from the Department of Veterans Affairs and the Department of Defense.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- mTORC1
- mTOR complex 1
- mTOR
- mammalian target of rapamycin
- ITAF
- IRES-transacting factor
- IRES
- internal ribosome entry site
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- GBM
- glioblastoma
- MEF
- mouse embryo fibroblast
- CCI-779
- cell cycle inhibitor 779
- 4OHT
- 4-hydroxy-tamoxifen
- EV
- empty vector.
REFERENCES
- 1. Shi Y., Gera J., Hu L., Hsu J. H., Bookstein R., Li W., Lichtenstein A. (2002) Cancer Res. 62, 5027–5034 [PubMed] [Google Scholar]
- 2. Gera J. F., Mellinghoff I. K., Shi Y., Rettig M. B., Tran C., Hsu J. H., Sawyers C. L., Lichtenstein A. K. (2004) J. Biol. Chem. 279, 2737–2746 [DOI] [PubMed] [Google Scholar]
- 3. Neshat M. S., Mellinghoff I. K., Tran C., Stiles B., Thomas G., Petersen R., Frost P., Gibbons J. J., Wu H., Sawyers C. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y., Sharma A., Wu H., Lichtenstein A., Gera J. (2005) J. Biol. Chem. 280, 10964–10973 [DOI] [PubMed] [Google Scholar]
- 5. Jo O. D., Martin J., Bernath A., Masri J., Lichtenstein A., Gera J. (2008) J. Biol. Chem. 283, 23274–23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holcík M. (2004) Curr. Cancer Drug Targets 4, 299–311 [DOI] [PubMed] [Google Scholar]
- 7. Spriggs K. A., Stoneley M., Bushell M., Willis A. E. (2008) Biol. Cell 100, 27–38 [DOI] [PubMed] [Google Scholar]
- 8. Le Quesne J. P., Spriggs K. A., Bushell M., Willis A. E. (2010) J. Pathol. 220, 140–151 [DOI] [PubMed] [Google Scholar]
- 9. Hellen C. U., Sarnow P. (2001) Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]
- 10. Fleischer T. C., Weaver C. M., McAfee K. J., Jennings J. L., Link A. J. (2006) Genes Dev. 20, 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Semler B. L., Waterman M. L. (2008) Trends Microbiol. 16, 1–5 [DOI] [PubMed] [Google Scholar]
- 12. Stoneley M., Willis A. E. (2004) Oncogene 23, 3200–3207 [DOI] [PubMed] [Google Scholar]
- 13. Sammons M. A., Antons A. K., Bendjennat M., Udd B., Krahe R., Link A. J. (2010) PLoS One 5, e9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mitchell S. A., Spriggs K. A., Coldwell M. J., Jackson R. J., Willis A. E. (2003) Mol. Cell 11, 757–771 [DOI] [PubMed] [Google Scholar]
- 15. Pickering B. M., Mitchell S. A., Spriggs K. A., Stoneley M., Willis A. E. (2004) Mol. Cell Biol. 24, 5595–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dreyfuss G., Kim V. N., Kataoka N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 17. Burd C. G., Dreyfuss G. (1994) Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 18. Cobianchi F., Calvio C., Stoppini M., Buvoli M., Riva S. (1993) Nucleic Acids Res. 21, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Idriss H., Kumar A., Casas-Finet J. R., Guo H., Damuni Z., Wilson S. H. (1994) Biochemistry 33, 11382–11390 [DOI] [PubMed] [Google Scholar]
- 20. Hamilton B. J., Nagy E., Malter J. S., Arrick B. A., Rigby W. F. (1993) J. Biol. Chem. 268, 8881–8887 [PubMed] [Google Scholar]
- 21. Henics T., Sanfridson A., Hamilton B. J., Nagy E., Rigby W. F. (1994) J. Biol. Chem. 269, 5377–5383 [PubMed] [Google Scholar]
- 22. Svitkin Y. V., Ovchinnikov L. P., Dreyfuss G., Sonenberg N. (1996) EMBO J. 15, 7147–7155 [PMC free article] [PubMed] [Google Scholar]
- 23. Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., Vagner S. (2005) J. Biol. Chem. 280, 4144–4153 [DOI] [PubMed] [Google Scholar]
- 24. Piñol-Roma S., Dreyfuss G. (1991) Science 253, 312–314 [DOI] [PubMed] [Google Scholar]
- 25. Zimmermann S., Moelling K. (1999) Science 286, 1741–1744 [DOI] [PubMed] [Google Scholar]
- 26. Gratton J. P., Morales-Ruiz M., Kureishi Y., Fulton D., Walsh K., Sessa W. C. (2001) J. Biol. Chem. 276, 30359–30365 [DOI] [PubMed] [Google Scholar]
- 27. Pedrotti S., Bielli P., Paronetto M. P., Ciccosanti F., Fimia G. M., Stamm S., Manley J. L., Sette C. (2010) EMBO J. 29, 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. King P. H. (2000) Nucleic Acids Res. 28, E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar A., Wilson S. H. (1990) Biochemistry 29, 10717–10722 [DOI] [PubMed] [Google Scholar]
- 30. Masri J., Bernath A., Martin J., Jo O. D., Vartanian R., Funk A., Gera J. (2007) Cancer Res. 67, 11712–11720 [DOI] [PubMed] [Google Scholar]
- 31. Kohn A. D., Barthel A., Kovacina K. S., Boge A., Wallach B., Summers S. A., Birnbaum M. J., Scott P. H., Lawrence J. C., Jr., Roth R. A. (1998) J. Biol. Chem. 273, 11937–11943 [DOI] [PubMed] [Google Scholar]
- 32. Schultz D. E., Hardin C. C., Lemon S. M. (1996) J. Biol. Chem. 271, 14134–14142 [DOI] [PubMed] [Google Scholar]
- 33. Yi M., Schultz D. E., Lemon S. M. (2000) J. Virol. 74, 6459–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewis S. M., Holcik M. (2008) Oncogene 27, 1033–1035 [DOI] [PubMed] [Google Scholar]
- 35. Siomi H., Dreyfuss G. (1995) J. Cell Biol. 129, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choe G., Horvath S., Cloughesy T. F., Crosby K., Seligson D., Palotie A., Inge L., Smith B. L., Sawyers C. L., Mischel P. S. (2003) Cancer Res. 63, 2742–2746 [PubMed] [Google Scholar]
- 37. Chakravarti A., Zhai G., Suzuki Y., Sarkesh S., Black P. M., Muzikansky A., Loeffler J. S. (2004) J. Clin. Oncol. 22, 1926–1933 [DOI] [PubMed] [Google Scholar]
- 38. Cammas A., Lewis S. M., Vagner S., Holcik M. (2008) Biochem. Pharmacol. 76, 1395–1403 [DOI] [PubMed] [Google Scholar]
- 39. Cammas A., Pileur F., Bonnal S., Lewis S. M., Lévêque N., Holcik M., Vagner S. (2007) Mol. Biol. Cell 18, 5048–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.