Abstract
Autoantibodies against astrocyte water channel aquaporin-4 (AQP4) are highly specific for the neuroinflammatory disease neuromyelitis optica (NMO). We measured the binding of NMO autoantibodies to AQP4 in human astrocyte-derived U87MG cells expressing M1 and/or M23 AQP4, or M23 mutants that do not form orthogonal array of particles (OAPs). Binding affinity was quantified by two-color fluorescence ratio imaging of cells stained with NMO serum or a recombinant monoclonal NMO autoantibody (NMO-rAb), together with a C terminus anti-AQP4 antibody. NMO-rAb titrations showed binding with dissociation constants down to 44 ± 7 nm. Different NMO-rAbs and NMO patient sera showed a wide variation in NMO-IgG binding to M1 versus M23 AQP4. Differences in binding affinity rather than stoichiometry accounted for M1 versus M23 binding specificity, with consistently greater affinity of NMO-IgG binding to M23 than M1 AQP4. Binding and OAP measurements in cells expressing different M1:M23 ratios or AQP4 mutants indicated that the differential binding of NMO-IgG to M1 versus M23 was due to OAP assembly rather than to differences in the M1 versus M23 N termini. Purified Fab fragments of NMO-IgG showed similar patterns of AQP4 isoform binding, indicating that structural changes in the AQP4 epitope upon array assembly, and not bivalent cross-linking of whole IgG, result in the greater binding affinity to OAPs. Our study establishes a quantitative assay of NMO-IgG binding to AQP4 and indicates remarkable, OAP-dependent heterogeneity in NMO autoantibody binding specificity.
Keywords: Antibodies, Biophysics, Fluorescence, Microscopic Imaging, Water Channel, AQP4, NMO, Aquaporin, Neuromyelitis Optica
Introduction
A defining feature of the neuroinflammatory demyelinating disease neuromyelitis optica (NMO)2 is the presence of serum autoantibodies (NMO-IgG) against aquaporin-4 (AQP4), a water channel expressed in astrocytes throughout the central nervous system (1). The presence of NMO-IgG is specific for NMO, and in some reports serum NMO-IgG titers correlate with NMO disease activity (2, 3). Studies in rodents suggest that NMO-IgG is pathogenic in NMO. Human NMO-IgG produces many features of NMO disease in rats with preexisting experimental autoimmune encephalomyelitis (4, 5) or pretreated with complete Freund's adjuvant (6) and in naïve mice when injected together with human complement (7). These animals develop characteristic NMO lesions with neuroinflammation, perivascular deposition of activated complement, demyelination, and loss of astrocyte glial fibrillary acidic protein and AQP4 immunoreactivity.
The target of the NMO autoantibody, AQP4, is involved in water balance in brain (8, 9) and spinal cord (10), sensory signal transduction (11, 12) and neuroexcitatory phenomena including seizure activity (13) and cortical spreading depression (14), and astrocyte migration and glial scarring (15, 16). AQP4 is expressed in astrocytes as two major isoforms: a long (M1) isoform with translation initiation at Met-1, and a shorter (M23) isoform with translation initiation at Met-23 (17–20). M23 AQP4 assembles in membranes as regular square arrays called orthogonal arrays of particles (OAPs), which were originally seen by freeze-fracture electron microscopy (21, 22). OAP formation by M23 results from tetramer-tetramer interactions involving residues just downstream of Met-23 at its cytoplasmic N terminus, whereas residues in M1 AQP4 just upstream of Met-23 disrupt this interaction (23). Although M1 does not form OAPs on its own, it can co-assemble with M23 in heterotetramers that limit OAP size (24–27). The biological significance of OAP formation by AQP4 remains unknown, with speculated functions including cell-cell adhesion, enhanced AQP4 water permeability, and AQP4 polarization to astrocyte end-feet.
Our study here is focused on the AQP4 binding specificity of NMO autoantibodies to the M1 versus M23 isoforms of AQP4 and to OAP- versus non-OAP-associated AQP4. A prior report that analyzed NMO sera concluded that OAPs are the exclusive target of NMO-IgG (28). However, this conclusion cannot be correct because the clinical serum assay for serum anti-AQP4 autoantibody uses M1 AQP4 (29), which does not form OAPs, and we (4, 26) and others (30) reported strong binding of some NMO autoantibodies to cells expressing only M1 AQP4. Although a recent study showed that clinical sensitivity of the NMO-IgG binding assay could be improved by using M23-expressing cells (31), there have been no quantitative studies of NMO-IgG AQP4 specificity, nor has the absolute affinity of NMO-IgG binding to AQP4 been measured.
We used quantitative ratio imaging to measure NMO autoantibody binding to AQP4 in which NMO-IgG binding, as revealed by a fluorescent secondary antibody, was normalized to total AQP4 protein using an antibody directed against the AQP4 C terminus. For these studies we identified a human astrocyte-derived cell line that expressed AQP4 in a plasma membrane pattern after transfection. Our strategy to assess independently the AQP4 isoform and OAP specificities of NMO-IgG binding was to express M1 and M23 AQP4 in different ratios, or an M23 mutant containing an OAP-disrupting, single-amino acid substitution in its N terminus. Measurements were made on serum samples from NMO patients, as well as purified monoclonal antibodies generated by recombinant technology from cloned sequences derived from plasma cells in the cerebrospinal fluid of an NMO patient. Studies using monoclonal NMO antibodies allowed, for the first time, the measurement of absolute binding affinities of NMO-IgG to AQP4. Studies using OAP-deficient mutants of M23 AQP4 and various heterotetramer-forming mixed AQP4 isoforms indicated enhanced NMO-IgG binding to AQP4 in OAPs. Studies comparing whole NMO-IgG with purified Fab fragments suggested a molecular basis for the enhanced NMO-IgG binding to OAP-assembled AQP4.
EXPERIMENTAL PROCEDURES
DNA Constructs, Cell Culture, and Transfection
DNA constructs encoding full-length human AQP4 (M1 and M23 isoforms) were generated by PCR amplification using whole brain cDNA as template. For some studies a Myc epitope (NH2-EQKLISEEDL-COOH) was inserted in the second extracellular loop by PCR amplification using the nontagged constructs as template. Mutants of M1 and M23 were generated by PCR amplification using either tagged or nontagged templates. All PCR fragments were ligated into mammalian expression vector pcDNA3.1 and fully sequenced. U87MG cell cultures (American Type Culture Collection HTB-14) were maintained at 37 °C in 5% CO2/95% air in Eagle's minimum essential medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were grown on glass coverslips and transfected with DNA in antibiotic-free medium using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Stable AQP4-expressing clones were selected following enrichment in Geneticin (Invitrogen) and plating in 96-well plates at very low density.
NMO Patient Sera and Recombinant AQP4 Autoantibodies
NMO serum was obtained from four NMO-IgG-seropositive individuals who met the revised diagnostic criteria for clinical disease (29). Control (non-NMO) human serum was purchased from the University of California San Francisco cell culture facility. Recombinant monoclonal NMO antibodies were generated from clonally expanded cerebrospinal fluid plasma blasts as described previously (4). Heavy and light chain constructs were co-transfected into HEK293 cells, the supernatant harvested, centrifuged to remove any cells and debris, and incubated overnight with protein A-Sepharose (Sigma-Aldrich) at 4 °C. The rAb was eluted in 0.1 m glycine/1 m NaCl (pH 3.0) and adjusted to pH 7.5 with 0.1 m Tris-HCl, pH 8.0. Recombinant IgG was subsequently exchanged and concentrated in PBS containing 0.1% protease-free bovine serum albumin using Ultracel YM-30 microconcentrators (Millipore, Billerica, MA). Fab fragments were generated by digestion of whole IgG with immobilized papain, and purified by removal of undigested IgG and Fc fragments by protein-A (Thermo Fisher Scientific, Rockford, IL). Antibody integrity and Fab fragments were confirmed by denaturing and Native-PAGE, and IgG concentration was assayed using a human IgG-capture ELISA.
Quantitative Immunofluorescence
AQP4-expressing U87MG cells were incubated for 20 min in live-cell blocking buffer (PBS containing 6 mm glucose, 1 mm pyruvate, 1% bovine serum albumin, 2% goat serum) and then for 30 min with NMO patient serum or recombinant NMO-IgG in blocking buffer. Cells were then rinsed extensively with PBS, fixed in 4% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100. Cells were then blocked again and incubated for 30 min with 0.4 μg/ml polyclonal, C-terminal specific rabbit anti-AQP4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and then rinsed with PBS. Finally, cells were incubated for 30 min with 4 μg/ml goat anti-human IgG-conjugated Alexa Fluor 488 and goat anti-rabbit IgG-conjugated Alexa Fluor 555 (Invitrogen) in blocking buffer. After incubation with secondary antibodies, cells were rinsed extensively in PBS, and coverglasses were mounted with VectaMount hard-set medium (Vector Laboratories, Burlingame, CA). In some experiments, U87MG cells were labeled as described above, but with a monoclonal mouse anti-Myc IgG (Covance, Emeryville, CA) or purified Fab fragments instead of whole NMO-IgG. Anti-Myc was stained with goat anti-mouse IgG-conjugated Alexa Fluor 488, whereas Fab fragments were stained with Dylight 488-linked F(ab′)2-specific secondary antibodies (Jackson Immunoresearch, West Grove, PA). Quantitative analysis of antibody binding was done on a Nikon Eclipse TE2000S inverted epifluorescence microscope (Nikon, Melville, NY) equipped with a Nikon 10 × air objective (numerical aperture 0.3). Green and red dyes were excited and observed through Chroma filter sets 41001 and 42001 (Chroma, Rockingham, VT), respectively. Images were recorded by a CCD camera (Hamamatsu Orca, Bridgewater, NJ), and intensities were determined using custom software. For determination of binding constants of monoclonal NMO-IgG to AQP4 isoforms, measured green/red fluorescence intensity ratios (G/R) were background-subtracted and normalized to the saturation level of the strongest binding antibodies (G/R ∼ 0.8) to give the fraction bound antibody. Nonspecific binding signal was determined by linear regression to measured G/R of a non-NMO monoclonal IgG (rAb-2B4) and subtracted. G/R values were fitted to the single-site binding model: F = [rAb]n/(KD + [rAb]n), where F is the fraction of bound rAb, KD the dissociation constant, and n the Hill coefficient.
Total Internal Reflection Fluorescence Microscopy
TIRFM was done using a Nikon Eclipse TE2000E microscope with a through-objective TIRF attachment and a 100× TIRF oil immersion objective (numerical aperture 1.49) mounted on a perfect focus module (Nikon). Alexa Fluor 555-labeled AQP4 was excited using an argon ion laser through a Z514/10 × excitation filter and Z514RDC dichroic mirror and detected through an ET605/40m emission filter (Chroma). Images were acquired using a QuantEM 512SC deep-cooled CCD camera (Photometrics, Tucson, AZ).
Single Particle Tracking
Prior to labeling of AQP4 with quantum dots (Qdots), cells expressing Myc-tagged AQP4 were washed with 2 ml of PBS containing 6 mm glucose and 1 mm pyruvate (GP buffer) and incubated for 5 min in blocking buffer. Cells were then incubated for 5 min with 70 ng/ml mouse anti-Myc antibody (Covance) in blocking buffer, rinsed, and incubated for 5 min with 0.1 nm goat F(ab′)2 anti-mouse IgG-conjugated Qdot 655 (Invitrogen) in blocking buffer. Cells were rinsed extensively and maintained throughout experiments in GP buffer. SPT was performed on a Nikon Eclipse TE2000S inverted epifluorescence microscope equipped with a Nikon 100× TIRF oil immersion objective (numerical aperture 1.45) and a deep-cooled CCD camera (Hamamatsu EM-CCD). Qdot fluorescence was excited using an E460SPUV excitation filter and 475DCXRU dichroic mirror and detected through a D655/40m emission filter (Chroma). Data were acquired continuously at 11 ms/frame (91 Hz) for 6 s. Image sequences were analyzed and trajectories constructed as described in detail previously (32). Diffusion data are reported in the form of cumulative distributions of ranges at 1 s, where P(range) is defined as the probability that the range of a particle is ≤ a given distance at t = 1 s.
Electrophoresis and Immunoblotting
Cell cultures were lysed with Native-PAGE sample buffer (Invitrogen) containing 1% dodecyl-β-d-maltoside (EMD Chemicals, Gibbstown, NJ) for 30 min on ice. Lysates were centrifuged at 20,000 × g for 30 min at 4 °C and the pellet discarded. For Blue-Native-PAGE, polyacrylamide native gradient gels (3–9%) were prepared as previously described (33). 10 μg of protein was mixed with 5% Coomassie Blue G-250 (Invitrogen) and loaded in each lane. Ferritin was used as the molecular mass standard (440 and 880 kDa). Running buffers were 25 mm imidazole, pH 7 (anode buffer), and 50 mm Tricine, 7.5 mm imidazole, 0.02% Coomassie Blue G-250, pH 7 (cathode buffer). Tricine SDS-PAGE was performed as previously described (34) with a 12% running gel and 3% stacking gel. Samples were not heated prior to loading. SeeBlue Plus2 Pre-Stained Standard (Invitrogen) was used as a molecular mass marker. Proteins were blotted onto polyvinyl difluoride membranes (Bio-Rad). For immunoblot analysis, membranes were blocked with 3% BSA and incubated with rabbit anti-AQP4 primary antibodies (Santa Cruz Biotechnology) for 2 h. Membranes were then rinsed and incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) and rinsed extensively, and labeled proteins were detected using the ECL Plus enzymatic chemiluminescence kit (Amersham Biosciences).
RESULTS
Approach for Quantitative Analysis of NMO-IgG Binding to AQP4
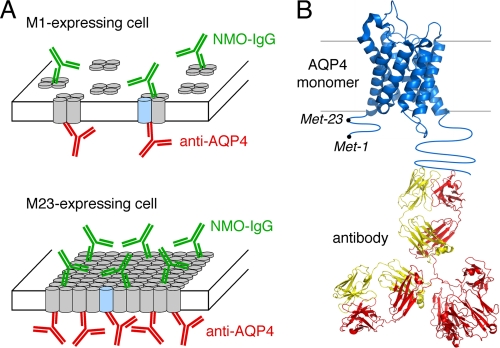
Fig. 1 diagrams the approach used for quantitative analysis of NMO-IgG binding to AQP4. Cells expressing specified AQP4 isoform(s) at their plasma membrane were incubated with NMO-IgG (NMO patient serum or recombinant monoclonal antibody), fixed, permeabilized, and then incubated with anti-AQP4 antibody (Fig. 1A). The anti-AQP4 antibody recognizes the AQP4 C terminus, which is common to all AQP4 isoforms (Fig. 1B). Fluorescent secondary antibodies were used to detect NMO-IgG by green fluorescence and AQP4 by red fluorescence. NMO-IgG binding to AQP4 was quantified by G/R fluorescence ratios. Binding affinity and stoichometry were determined by titration with increasing NMO-IgG concentration. In contrast to measurements done at a single NMO-IgG concentration, measurement of full, concentration-dependent binding provides a quantitative, unbiased description of NMO-IgG binding to AQP4.
FIGURE 1.
Schematic of the two-color ratio imaging method for quantitative measurement of NMO-IgG binding to AQP4 isoforms. A, AQP4 monomers (cylinders) are shown as assembling into tetramers (top) or OAPs (bottom). NMO-IgG (green) binds AQP4 at an extracellular domain, and a reference AQP4 antibody (red) binds on the cytoplasmic side. B, reference AQP4 antibody binds to the C terminus of AQP4, independent of the AQP4 N-terminal isoform and OAP formation.
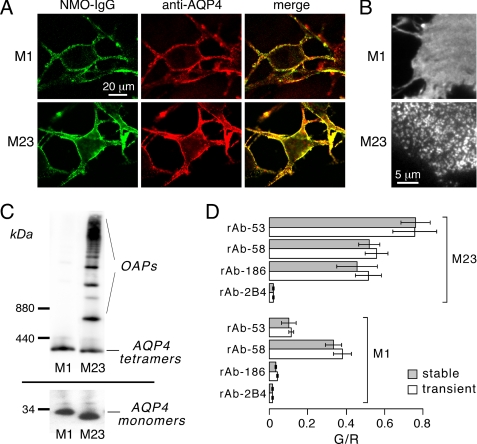
A cell line for analysis of NMO-IgG binding to AQP4 was selected that showed efficient plasma membrane targeting of AQP4 isoforms following stable or transient transfections, as well as human glial cell origin, rapid growth, and strong adherence to coverglass supports. The selected cell line, U87MG, was originally derived from a human astrocytoma (35). Fig. 2A shows high magnification confocal microscopy of U87MG cells stably expressing M1 or M23 AQP4 and stained with a recombinant monoclonal NMO-IgG (green) and an anti-AQP4 antibody (red). AQP4 was localized to the cell plasma membrane, with little intracellular red fluorescence. Fig. 2B shows TIRFM of the anti-AQP4 antibody-stained AQP4-expressing cells. A smooth pattern of fluorescence was seen for M1 AQP4 and a punctate pattern for M23 AQP4, as found in other cell types (36), confirming that M23 AQP4 forms OAPs in transfected U87MG cells whereas M1 AQP4 does not. Immunoblot analysis of cell homogenates showed the expected molecular sizes of the M1 and M23 isoforms of AQP4 (Fig. 2C, bottom). Blue-Native-PAGE of cells expressing M23 alone showed multiple high molecular mass bands corresponding to the expected formation of large supramolecular aggregates (OAPs), whereas M1-expressing cells showed only the expected ∼300 kDa band corresponding to individual AQP4 tetramers (Fig. 2C, top). Fig. 2D shows examples of measured G/R ratios after binding recombinant monoclonal NMO-IgGs (rAb-53, rAb-58, and rAb-186) or control antibody (rAb-2B4) (each at 20 μg/ml) to M1 or M23 AQP4-expressing cells. These initial measurements show considerable diversity in NMO-IgG binding to M1 versus M23 AQP4, with similar results found in both stably and transiently transfected U87MG cells.
FIGURE 2.
Characterization of stably transfected, AQP4-expressing U87MG cells. A, confocal fluorescence images show U87MG cells stably expressing M1 (top) or M23 (bottom) and labeled with NMO-IgG (green) and C-terminal anti-AQP4 antibody (red). B, TIRF images show distinct OAPs in M23-expressing cells (bottom) and a smooth fluorescence staining pattern in M1-expressing cells (top). C, AQP4 immunoblot following Blue-Native-PAGE (top) and Tricine SDS-PAGE (bottom) of stable AQP4-expressing U87MG cell lysates is shown. D, measured G/R fluorescence ratios in U87MG cells after stable (gray) or transient (white) transfection with M1 or M23 AQP4 and labeled with the indicated recombinant monoclonal NMO-IgG (mean ± S.E. (error bars), n = 4) are shown.
Heterogeneous NMO-IgG Binding to AQP4 Isoforms in NMO Serum Samples
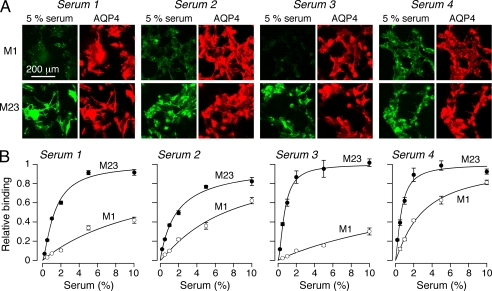
Binding of NMO-IgG in human NMO sera to the M1 and M23 AQP4 isoforms was measured by the fluorescence ratio imaging method. Initial measurements done on 10 serum specimens from NMO patients studied at a 1:100 dilution showed a wide range of relative M1:M23 binding from 0.14 to 0.67. Fig. 3A shows representative fluorescence micrographs for serum specimens from four NMO patients. Each serum specimen showed strong NMO-IgG binding to M23 AQP4 but variable binding to M1 AQP4. Fig. 3B shows concentration-dependent NMO-IgG binding in which background-corrected G/R ratios were determined as a function of serum concentration. NMO-IgG binding to AQP4 was saturable, with a wide variation in relative M23:M1 dissociation constants of ∼0.11 (serum 1), ∼0.26 (serum 2), ∼0.03 (serum 3), and ∼0.21 (serum 4). Interestingly, the binding data fitted well to a single-site, saturable binding model with near unity Hill coefficient, despite the presumed polyclonal composition of NMO serum. It is not possible to determine absolute binding affinity of NMO-IgG to AQP4 using serum because the fraction of NMO-IgG in total serum IgG is not known nor is the polyclonal NMO-IgG composition.
FIGURE 3.
Differential binding of NMO-IgG in NMO patient serum to M1 versus M23 AQP4. A, M1- and M23-expressing U87MG cells stained with 5% NMO serum (green) from four patients and with reference AQP4 antibody (red). B, binding curves for the NMO patient sera to M1 versus M23 AQP4 (mean ± S.E. (error bars), n = 5). Curves represent fits to single-site binding model.
Quantitative Binding of Recombinant Monoclonal NMO-IgGs to AQP4 Isoforms
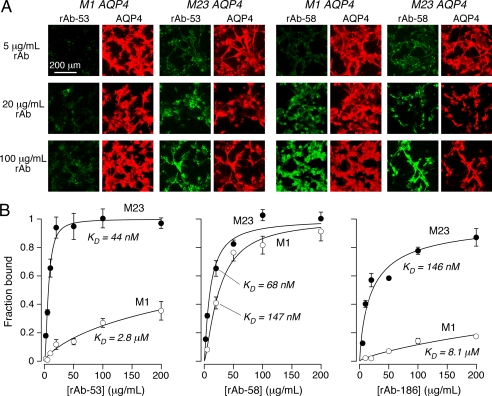
NMO-IgG in serum from a single NMO patient is polyclonal, consisting of a pool of monoclonal NMO-IgGs. A similar binding analysis was done using monoclonal recombinant NMO-IgGs derived from a single NMO patient to determine: (i) absolute binding affinities; (ii) whether differential M23:M1 binding is heterogeneous in a single NMO patient; and (iii) whether differential M23:M1 binding is due to differences in NMO-IgG binding affinity and/or stoichiometry. Fig. 4A shows fluorescence micrographs of two recombinant monoclonal NMO-IgGs (rAb-53 and rAb-58) from a single NMO patient. The monoclonal antibodies used in this study were derived from the cerebrospinal fluid of a single NMO patient (corresponding to Serum 1 in Fig. 3). Although strong monoclonal antibody binding to M23 AQP4 was seen in each case, binding to M1 AQP4 was variable. Fig. 4B summarizes concentration-dependent binding curves for three recombinant NMO-IgGs, together with curve fits for a single-site binding model. In each case the data fitted well to a single-site model with near unity Hill coefficient. The lowest dissociation constant was 44 ± 7 nm, which was found for binding of rAb-53 to M23 AQP4. Marked heterogeneity was found for monoclonal NMO-IgGs from a single NMO patient, with relative M23:M1 binding dissociation constants differing by a factor of ∼50 for rAb-53 and rAb-186, and ∼2 for rAb-58. The binding curves in Fig. 4B also support the conclusion that differences in M23:M1 binding are due to differences in binding affinity rather than to binding capacity.
FIGURE 4.
Differential binding of purified monoclonal NMO-IgGs to M1 versus M23 AQP4. A, representative fluorescence micrographs for binding of rAb-53 and rAb-58 (green) as a function of concentration, together with reference AQP4 antibody (red). B, binding curves for rAb-53 (left), rAb-58 (middle), and rAb-186 (right) to M1 versus M23 AQP4 (mean ± S.E. (error bars), n = 5). Curves represent fits to a single-site binding model, with fitted KD = 44 ± 7 nm and 2.8 ± 1 μm (rAb-53), 68 ± 9 nm and 147 ± 20 nm (rAb-58), and 146 ± 25 nm and 8.1 ± 3 μm (rAb-186), each for M23 and M1, respectively.
Mechanism of AQP4 Isoform-specific Binding of NMO-IgG
We investigated whether differences in NMO-IgG binding affinity to M23 versus M1 AQP4 are due to formation of OAPs by M23 AQP4 and/or to the different N termini of M23 versus M1. Measurements were made of NMO-IgG binding to cells expressing: (i) different ratios of M23:M1 AQP4; (ii) an M1 mutant that has a diminished ability to disrupt OAPs when co-expressed with M23; and (iii) M23 AQP4 mutants that have diminished ability to form OAPs. For these studies we used transiently transfected U87MG cells. The suitability of transient transfection was validated above by showing comparable binding of NMO-IgG in stably versus transiently transfected cells (Fig. 2D).
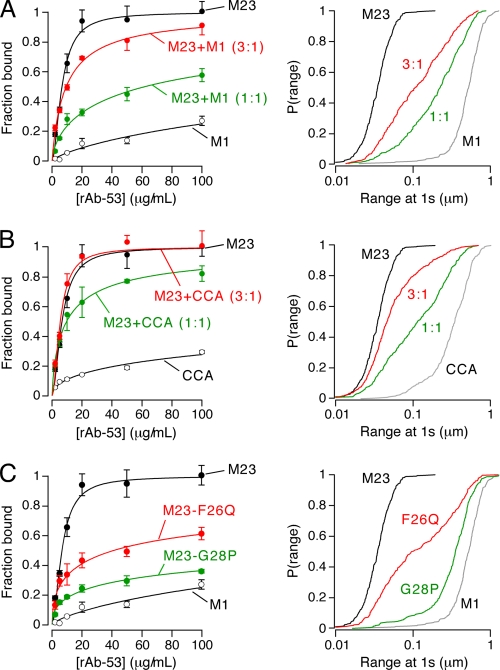
Fig. 5A shows concentration-dependent binding of NMO-IgG (rAb-53) to U87MG cells co-expressing different ratios of M23:M1 AQP4. The fraction of AQP4 in OAPs and average OAP size are directly related to the M23:M1 AQP4 ratio. Qdot single-particle tracking measurements were done to determine the characteristics of OAPs formed at different M23:M1 ratios. Fig. 5A (right) shows increased AQP4 diffusion with higher M1 content, as previously shown (26). This increase in AQP4 diffusion is due to active disruption of OAP growth by M1 AQP4. Not surprisingly, compared with binding of M23 alone, rAb-53 showed incrementally reduced AQP4 binding in 3:1 and 1:1 mixtures of M23:M1 (Fig. 5A, left).
FIGURE 5.
Binding of NMO-IgG to mixtures of M1 and M23 AQP4 and to M23 mutants containing OAP-disrupting mutations. A, binding of rAb-53 (left) and cumulative distributions of diffusion range (right), measured by Qdot single-particle tracking, for M1 and M23 AQP4 mixtures at the indicated ratios (mean ± S.E. (error bars), n = 5). B, binding of rAb-53 (left) and diffusion range (right) for M23 AQP4 with M1 mutant CCA at the indicated ratios (mean ± S.E., n = 5). C, binding of rAb-53 (left) and diffusion range (right) for AQP4 mutants M23-F26Q (red) and M23-G28P (green) (mean ± S.E., n = 5).
Fig. 5B shows data from experiments similar to those in Fig. 5A, except that native M1 was replaced by the double cysteine mutant M1 C13A/C17A. CCA AQP4 does not form OAPs by itself, but when co-expressed with M23 it has greatly reduced ability to disrupt OAPs (26). Therefore, at the same M23:M1 ratio, cells expressing the CCA mutant in place of native M1 have greater OAP content, as confirmed by single-particle tracking (Fig. 5B, right). Concentration-dependent binding of rAb-53 showed greater binding to M23:CCA mixtures than to M23:M1 mixtures (Fig. 5B, left). Binding of rAb-53 to cells expressing a 3:1 ratio of M23 to CCA was identical to cells expressing M23 alone.
As an independent approach to address the binding specificity issue, we measured NMO-IgG binding to U87MG cells expressing M23 mutants containing an OAP-disrupting point mutation. The OAP-disrupting effect of these mutations was confirmed by Qdot single-particle tracking. Fig. 5C (right) shows that mutations F26Q and G28P in the M23 AQP4 N terminus greatly reduce OAP content, similar to our previous findings with corresponding rat isoforms of AQP4 (23). Fig. 5C (left) shows greatly reduced concentration-dependent binding of rAb-53 to cells expressing these M23 mutants compared with native M23. Together, the results in Fig. 5 indicate that OAP formation is responsible for the increased affinity of NMO-IgG to M23 versus M1 AQP4.
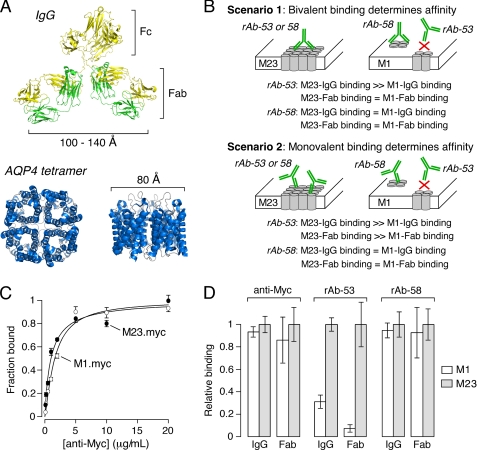
Two potential mechanisms, bivalent versus monovalent NMO-IgG binding, could account for the greater affinity of NMO-IgG to OAP versus non-OAP associated AQP4. Fig. 6A shows a comparison of the distance between adjacent Fab binding sites in IgG1 (37, 38) and the size of the AQP4 tetramer (39). Fig. 6B diagrams possible opposing binding mechanisms. (i) Bivalent binding: strong NMO-IgG binding requires a bivalent interaction in which both Fab sites must bind to AQP4 monomers or tetramers. For rAb-53, in which M23 binding is much stronger than M1 binding, the positions of the binding epitopes in AQP4 monomers are spaced such that a bivalent interaction between the Fab sites is not possible within a single tetramer, but is optimal for cross-linking of adjacent tetramers in OAPs. For rAb-58, the epitopes may be located at positions in which bivalent binding in a single tetramer can occur, resulting in similar binding to M1 and M23 AQP4. (ii) Monovalent binding: NMO-IgG binding involves classical monovalent interactions and is controlled primarily by affinities of individual Fabs to their respective epitopes. For rAb-53, a structural change in the epitope site upon OAP formation results in higher affinity. For rAb-58, the epitope site is not altered by OAP formation, resulting in similar affinity for M1 versus M23 AQP4.
FIGURE 6.
Mechanism of increased NMO-IgG binding affinity to OAP-assembled AQP4. A, human IgG (Protein Data Bank ID code 1IGY) (38) and AQP4 (Protein Data Bank ID code 3GD8) (39) crystal structures showing relative size of the AQP4 tetramer compared with spacing between Fab binding sites in whole IgG. B, predictions of bivalent versus monovalent binding mechanisms. AQP4 monomers (cylinders) are shown as assembled in tetramers (M1) or OAPs (M23). NMO-IgG (green) binds either monovalently or bivalently to (unknown) extracellular domains on AQP4. C, binding of monoclonal mouse anti-Myc to cells expressing Myc-tagged M1 versus M23 AQP4. D, relative M1-to-M23 binding of whole IgG or purified Fab fragments of mouse anti-Myc (left), rAb-53 (middle), and rAb-58 (right) at a fixed concentration (mean ± S.E. (error bars), n = 5).
Fab binding to M1 and M23 AQP4 was measured to test these competing mechanisms. NMO-IgGs were digested with papain and purified to yield Fabs. The bivalent binding mechanism predicts little binding by Fabs and no difference between Fab binding to M1 versus M23 AQP4. The monovalent binding mechanism predicts that the increased binding of rAb-53 to M23 AQP4 would also be observed for its individual Fabs. As a control, we measured binding of Fabs generated from a mouse monoclonal anti-Myc antibody to external Myc-tagged AQP4 isoforms. As expected, whole anti-Myc IgG bound Myc-tagged M1 and M23 AQP4 equally, with KD ∼10 nm (Fig. 6C). Fig. 6D shows the binding of Fab fragments to M1 versus M23 AQP4. Whole IgG and Fabs from rAb-53 showed significantly greater binding to M23 AQP4, whereas whole IgG and Fabs from rAb-58 and anti-Myc showed similar M1 versus M23 binding. These data provide direct support for the second mechanism involving monovalent binding.
DISCUSSION
We previously showed that NMO patient serum and a recombinant monoclonal NMO-IgG were each able to bind to both the M23 and M1 isoforms of AQP4 (26), which contradicted an earlier study reporting undetectable binding to M1 AQP4 (28). Here, we used fluorescence ratio imaging to quantify the binding of NMO-IgG to AQP4 and to determine the role of AQP4 isoforms and OAPs in NMO-IgG binding. Our live cell system was developed out of a need for a robust method to characterize monoclonal NMO-IgGs and polyclonal NMO patient sera. We found that U87MG cells were suitable for quantitative binding measurements because they efficiently expressed AQP4 at the plasma membrane after stable or transient transfection, with little or no intracellular AQP4 expression. The excellent membrane expression of AQP4 in U87MG cells is likely due to their astrocyte origin and hence their expression of the same trafficking machinery as that in native human astrocytes. Immunoblot analysis of the stably transfected clones used in this study showed exclusive expression of individual AQP4 isoforms, with no detectable M23 in the M1 cell line (Fig. 2C). U87MG cells grow rapidly and adhere well to culture supports, making them suitable for automated and high throughput assays.
For all monoclonal and polyclonal NMO antibodies tested, NMO-IgG binding to M23-expressing cells was comparable with or greater than to M1-expressing cells, although measurable binding to M1 was found in all cases. NMO-IgG was found to bind to each AQP4 isoform in a concentration-dependent manner that fitted well to a single-site, saturable binding model with near unity Hill coefficient, consistent with apparent single-site, noncooperative binding. Preferential binding of NMO-IgG to M23 AQP4 was found to be a consequence of greater binding affinity rather than to greater binding capacity. Differences in the binding affinity of monoclonal NMO-IgG rAb-53 to the M23 and M1 isoforms of AQP4 were found to be a consequence of OAP formation by M23 AQP4 rather than to differences in the N-terminal sequences. This was proven using mutants of M1 and M23 AQP4 with altered abilities to form and disrupt OAPs. Using a two-color single-particle tracking approach, we recently showed that co-expressed AQP4 isoforms M1 and M23 co-assemble in AQP4 tetramers with differential abilities to assemble into OAPs. We showed that OAP size and content could be altered by changing the M1:M23 ratio or by altering the palmitoylation state of M1 AQP4 (26). We found here that co-expression of M1 and M23 also affects NMO-IgG binding (Fig. 5A). However, by reducing the ability of M1 to disrupt OAPs by expression of a palmitoylation-null M1 mutant, we found significantly increased NMO-IgG binding in mixtures with identical M23:M1 ratio (Fig. 5B). We previously showed that OAP formation by M23 requires N-terminal hydrophobic interactions at residues 24–26 just downstream of Met-23 (23), and we discovered M23 mutants with greatly reduced ability to form OAPs (M23-F26Q and M23-G28P). These OAP-disrupting M23 AQP4 mutants greatly reduced NMO-IgG binding (Fig. 5C). In contrast to isotherms for binding of NMO-IgG to individual M1 and M23 isoforms, binding to AQP4 mutants M23-F26Q and M23-G28P produced isotherms that did that not fit well to a single-site binding model, probably because expression of these mutants produces heterogeneous populations of OAP-assembled AQP4 and individual AQP4 tetramers.
The mechanism for higher affinity NMO-IgG binding to OAP-assembled AQP4 was examined by comparing whole (bivalent) NMO-IgG with its monovalent Fabs. OAP assembly resulted in a greater affinity for individual Fab binding sites rather than enhancement by bivalent IgG binding. We propose, therefore, that the binding epitope for many of the IgGs found in NMO may be located at the tetramer/tetramer interface created upon OAP assembly. However, differences in binding affinities vary, likely depending on the epitope structure, and some NMO-IgGs such as rAb-58 have high affinity for unassembled M1 AQP4.
The widely used assay for serum NMO-IgG immunofluorescence is performed in M1 AQP4-expressing HEK293 cells (1). Our data here indicate that M23 AQP4-expressing cells are superior because NMO-IgG binding to M23 AQP4 is as good as and generally much better than binding to M1 AQP4. The difference in binding is seen at all NMO-IgG concentrations and can be quite marked at low concentrations, as often found in human serum. Up to 30% of serum from patients with NMO, as defined by established clinical criteria, is found to be seronegative as assayed using the conventional method (29). This value is likely substantially lower utilizing more sensitive assays such as the imaging assay established here with a human astrocyte cell line strongly expressing the OAP-forming M23 isoform of AQP4. Indeed, a recent study of human serum samples reported an improvement from 70% to 97% sensitivity for NMO-IgG when using M23-expressing cells instead of M1-expressing cells (31). Our quantitative binding assay established here should also be useful in correlating serum NMO-IgG binding affinity and AQP4 isoform specificity with NMO clinical parameters, such as disease activity, treatment status, and patient characteristics. Base-line differences in NMO-IgG binding to OAP versus non-OAP-associated AQP4 may have diagnostic significance, and spontaneous and treatment-associated changes in binding may have prognostic significance.
This work was supported, in whole or in part, by National Institutes of Health Grants EY13574, EB00415, DK35124, HL73856, DK86125, and DK72517 (to A. S. V.). This work was also supported by grants from the Guthy-Jackson Charitable Foundation (to A. S. V. and J. L. B.) and Grant NMSSRG4320 from the National Multiple Sclerosis Society (to J. L. B.).
- NMO
- neuromyelitis optica
- AQP4
- aquaporin-4
- CCA
- C13A/C17A
- G/R
- green-to-red
- NMO-IgG
- serum autoantibody
- OAP
- orthogonal array of particles
- Qdot
- quantum dot
- rAb
- recombinant antibody
- TIRFM
- total internal reflection fluorescence microscopy.
REFERENCES
- 1. Lennon V. A., Kryzer T. J., Pittock S. J., Verkman A. S., Hinson S. R. (2005) J. Exp. Med. 202, 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matiello M., Lennon V. A., Jacob A., Pittock S. J., Lucchinetti C. F., Wingerchuk D. M., Weinshenker B. G. (2008) Neurology 70, 2197–2200 [DOI] [PubMed] [Google Scholar]
- 3. Jarius S., Aboul-Enein F., Waters P., Kuenz B., Hauser A., Berger T., Lang W., Reindl M., Vincent A., Kristoferitsch W. (2008) Brain 131, 3072–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett J. L., Lam C., Kalluri S. R., Saikali P., Bautista K., Dupree C., Glogowska M., Case D., Antel J. P., Owens G. P., Gilden D., Nessler S., Stadelmann C., Hemmer B. (2009) Ann. Neurol. 66, 617–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradl M., Misu T., Takahashi T., Watanabe M., Mader S., Reindl M., Adzemovic M., Bauer J., Berger T., Fujihara K., Itoyama Y., Lassmann H. (2009) Ann. Neurol. 66, 630–643 [DOI] [PubMed] [Google Scholar]
- 6. Kinoshita M., Nakatsuji Y., Kimura T., Moriya M., Takata K., Okuno T., Kumanogoh A., Kajiyama K., Yoshikawa H., Sakoda S. (2010) Biochem. Biophys. Res. Commun. 394, 205–210 [DOI] [PubMed] [Google Scholar]
- 7. Saadoun S., Waters P., Bell B. A., Vincent A., Verkman A. S., Papadopoulos M. C. (2010) Brain 133, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manley G. T., Fujimura M., Ma T., Noshita N., Filiz F., Bollen A. W., Chan P., Verkman A. S. (2000) Nat. Med. 6, 159–163 [DOI] [PubMed] [Google Scholar]
- 9. Papadopoulos M. C., Manley G. T., Krishna S., Verkman A. S. (2004) FASEB J. 18, 1291–1293 [DOI] [PubMed] [Google Scholar]
- 10. Saadoun S., Bell B. A., Verkman A. S., Papadopoulos M. C. (2008) Brain 131, 1087–1098 [DOI] [PubMed] [Google Scholar]
- 11. Li J., Verkman A. S. (2001) J. Biol. Chem. 276, 31233–31237 [DOI] [PubMed] [Google Scholar]
- 12. Lu D. C., Zhang H., Zador Z., Verkman A. S. (2008) FASEB J. 22, 3216–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Binder D. K., Yao X., Verkman A. S., Manley G. T. (2006) Acta Neurochir. Suppl. 96, 389–392 [DOI] [PubMed] [Google Scholar]
- 14. Padmawar P., Yao X., Bloch O., Manley G. T., Verkman A. S. (2005) Nat. Methods 2, 825–827 [DOI] [PubMed] [Google Scholar]
- 15. Saadoun S., Papadopoulos M. C., Watanabe H., Yan D., Manley G. T., Verkman A. S. (2005) J. Cell Sci. 118, 5691–5698 [DOI] [PubMed] [Google Scholar]
- 16. Auguste K. I., Jin S., Uchida K., Yan D., Manley G. T., Papadopoulos M. C., Verkman A. S. (2007) FASEB J. 21, 108–116 [DOI] [PubMed] [Google Scholar]
- 17. Hasegawa H., Ma T., Skach W., Matthay M. A., Verkman A. S. (1994) J. Biol. Chem. 269, 5497–5500 [PubMed] [Google Scholar]
- 18. Jung J. S., Bhat R. V., Preston G. M., Guggino W. B., Baraban J. M., Agre P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 13052–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang B., Ma T., Verkman A. S. (1995) J. Biol. Chem. 270, 22907–22913 [DOI] [PubMed] [Google Scholar]
- 20. Lu M., Lee M. D., Smith B. L., Jung J. S., Agre P., Verdijk M. A., Merkx G., Rijss J. P., Deen P. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10908–10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landis D. M., Reese T. S. (1974) J. Cell Biol. 60, 316–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolburg H. (1995) J. Hirnforsch. 36, 239–258 [PubMed] [Google Scholar]
- 23. Crane J. M., Verkman A. S. (2009) J. Cell Sci. 122, 813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neely J. D., Christensen B. M., Nielsen S., Agre P. (1999) Biochemistry 38, 11156–11163 [DOI] [PubMed] [Google Scholar]
- 25. Furman C. S., Gorelick-Feldman D. A., Davidson K. G., Yasumura T., Neely J. D., Agre P., Rash J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13609–13614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crane J. M., Bennett J. L., Verkman A. S. (2009) J. Biol. Chem. 284, 35850–35860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tajima M., Crane J. M., Verkman A. S. (2010) J. Biol. Chem. 285, 8163–8170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nicchia G. P., Mastrototaro M., Rossi A., Pisani F., Tortorella C., Ruggieri M., Lia A., Trojano M., Frigeri A., Svelto M. (2009) Glia 57, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 29. Wingerchuk D. M., Lennon V. A., Pittock S. J., Lucchinetti C. F., Weinshenker B. G. (2006) Neurology 66, 1485–1489 [DOI] [PubMed] [Google Scholar]
- 30. Hinson S. R., Pittock S. J., Lucchinetti C. F., Roemer S. F., Fryer J. P., Kryzer T. J., Lennon V. A. (2007) Neurology 69, 2221–2231 [DOI] [PubMed] [Google Scholar]
- 31. Mader S., Lutterotti A., Di Pauli F., Kuenz B., Schanda K., Aboul-Enein F., Khalil M., Storch M. K., Jarius S., Kristoferitsch W., Berger T., Reindl M. (2010) PLoS One 5, e10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crane J. M., Verkman A. S. (2008) Biophys. J. 94, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 34. Schägger H. (2006) Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 35. Pontén J., Macintyre E. H. (1968) Acta Pathol. Microbiol. Scand. 74, 465–486 [DOI] [PubMed] [Google Scholar]
- 36. Crane J. M., Van Hoek A. N., Skach W. R., Verkman A. S. (2008) Mol. Biol. Cell 19, 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sosnick T. R., Benjamin D. C., Novotny J., Seeger P. A., Trewhella J. (1992) Biochemistry 31, 1779–1786 [DOI] [PubMed] [Google Scholar]
- 38. Harris L. J., Skaletsky E., McPherson A. (1998) J. Mol. Biol. 275, 861–872 [DOI] [PubMed] [Google Scholar]
- 39. Ho J. D., Yeh R., Sandstrom A., Chorny I., Harries W. E., Robbins R. A., Miercke L. J., Stroud R. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7437–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]