Abstract
Background
Making an informed decision about treating a prostate cancer detected following a routine prostate-specific antigen (PSA) test requires knowledge about disease natural history, such as the chances that it would have been clinically diagnosed in the absence of screening and that it would metastasize or lead to death in the absence of treatment.
Methods
We use three independently developed models of prostate cancer natural history to project risks of clinical progression events and disease-specific deaths for PSA-detected cases assuming they receive no primary treatment.
Results
The three models project that 20–33% of men have preclinical onset; of these 38–50% would be clinically diagnosed and 12–25% would die of the disease in the absence of screening and primary treatment. The risk that men under age 60 at PSA detection with Gleason score 2–7 would have been clinically diagnosed in the absence of screening is 67–93% and would die of the disease in the absence of primary treatment is 23–34%. For Gleason score 8–10 these risks are 90–96% and 63–83%.
Conclusions
Risks of disease progression among untreated PSA-detected cases can be nontrivial, particularly for younger men and men with high Gleason scores. Model projections can be useful for informing decisions about treatment.
Impact
This is the first study to project population-based natural history summaries in the absence of screening or primary treatment and risks of clinical progression events following PSA detection in the absence of primary treatment.
Keywords: Comparative modeling, natural history, prostatic neoplasm, PSA screening
1. Introduction
Choosing the optimal management strategy for a newly diagnosed localized prostate cancer is based, at least in part, upon our understanding of the natural history of the disease in the absence of any aggressive treatment intervention. Useful information about disease natural history for men clinically diagnosed and initially untreated before the widespread adoption of the prostate-specific antigen (PSA) test is available from population-based cohort studies (1, 2), yet relatively little information is available for men diagnosed following a routine PSA test (3). While the prognosis for screen-detected tumors appears to be better than that for clinically diagnosed tumors (4), predicting the natural history of a specific PSA-detected tumor is complicated by overdiagnosis and by the lead time associated with the test.
A large proportion of PSA-detected cancers are overdiagnosed, i.e., they would never have progressed to a symptomatic state or been clinically diagnosed in the absence of the test. By definition, an overdiagnosed tumor has an entirely different prognosis than a non-overdiagnosed one. The clinical challenge is to determine whether a given case is overdiagnosed at the time of diagnosis. Further complicating the situation, even if we are able to identify a non-overdiagnosed cancer, prognosis depends critically on the lead time, which is the time by which diagnosis is advanced by screening. Lead times can be highly variable across patients primarily due to heterogeneity of the disease.
Unfortunately, there is no sure way to assess whether a tumor is overdiagnosed or to predict its lead time in clinical practice. Consequently, once a cancer has been detected by screening, it is typically treated, altering its natural history. We can then no longer observe whether or when it would have progressed in the absence of treatment. This data limitation has spawned the development of model-based approaches for inferring lead time, overdiagnosis, and future survival from observed data on disease-specific incidence and deaths. For example, Nicholson and Harland (5) and Parker et al. (6) projected prostate cancer survival for PSA-detected cases using epidemiologic models based on published incidence and survival data. While these studies provided important insights about prognosis, they relied on simplifying assumptions about the populations under study, screening practices, and/or lead time distributions.
An alternative modeling approach is provided by Etzioni et al. (7), Draisma et al. (8), and Tsodikov et al. (9), who developed more biological models of prostate cancer natural history. These models consist of a series of transitions between a healthy state and the clinico-pathologic stages of disease. While these transitions are generally unobservable, the models quantitatively link the respective transition probabilities with resulting observable rates of stage- and grade-specific disease incidence. Calibration of the models to observed disease incidence data allows for estimation of these transition probabilities. Superimposing screening on the calibrated models allows for projection of overdiagnosis frequencies, probabilities of disease progression, and lead time distributions.
With the goals of making transparent all assumptions underlying these more complicated models, strengthening the robustness of modeling methods, and coordinating common input datasets, these three groups joined together to form the prostate working group of the Cancer Intervention and Surveillance Modeling Network (CISNET). In addition to facilitating deeper insights about disease natural history, the comparative modeling approach provides a degree of robustness to model specification since each modeling group makes different assumptions about the mechanisms of disease progression.
The CISNET prostate working group previously collaborated to quantify the contribution of screening to the population declines in prostate cancer mortality (10) and to reconcile differing estimates of overdiagnosis rates and mean lead times (11). In this paper, we briefly review these models and use them to examine lifetime risks of, mean ages at, and mean years between key disease progression events in the absence of screening and primary treatment. We then project risks of clinical progression events for PSA-detected localized prostate cases who do not receive curative treatment. In addition, we project 20-year prostate cancer and non-prostate cancer survival by age and Gleason score for these cases. Few studies have published detailed information about prostate cancer natural history in the PSA era absent screening and primary treatment. And, to our knowledge, no study has systematically projected population-based risks of clinical progression outcomes following PSA detection or uncertainty in associated survival were the disease to be left untreated.
2. Materials and Methods
To facilitate comparison, each natural history model was calibrated to the same incidence data: men aged 50–84 in years 1975–2000 in the core 9 registries of the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program (12). Missing stage and grade information was imputed assuming that it was missing completely at random. To address the upward drift in Gleason scoring of well to moderately differentiated disease during these years (13), final projections were reported using Gleason score categories 2–7 and 8–10. To disentangle incidence of clinical and screen diagnoses, each model relied on a common retrospective reconstruction of PSA screening patterns in the US population (14). Also, each model used US life tables to generate non-prostate cancer survival and a common Poisson regression model for prostate cancer survival (adapted from 2) fit to data from men clinically diagnosed in 1983–1986, just prior to the widespread dissemination of PSA screening. Each model guarantees individual survival during his lead time, and actual dates and causes of death are assigned based on the earlier of disease-specific and other-cause survival times. By explicitly standardizing these common elements, differences across models are due entirely to differences in the conceptual mechanisms used to represent the development and progression of the disease.
Table 1 presents an at-a-glance comparison of the high-level natural history model features and implementation details of PSA screening and biopsy practices. In all models, the development of a new prostate cancer represents preclinical onset, i.e., the first point at which a properly directed needle biopsy would detect the cancer. And in all models the risk of onset depends on age. Given onset, the three models represent disease progression using different numbers of states and different assumptions about how disease can progress. In all models disease can progress from an organ-confined early stage to a distant metastatic stage. In all models disease can be clinically diagnosed, i.e., diagnosed in the absence of PSA screening. Clinical diagnosis could result from a digital rectal exam or from clinical manifestations of advanced disease, such as obstructive voiding symptoms. In the presence of PSA screening, disease can also be detected following a routine PSA test. At any screen event, the MISCAN and UMICH models estimate the probability of a positive biopsy given disease, while the FHCRC model decomposes this probability into three factors:
and combines external data for the first two factors with results from modeled PSA levels for the third factor.
Table 1.
Comparison of high-level features across models.
| Model feature | FHCRC | MISCAN | UMICH |
|---|---|---|---|
| Implementation | simulation | simulation | analytic |
| Disease states | (2 stages) × (2 grades) | (3 × 2 stages) × (3 grades) | (2 stages) × (2 grades) |
| local-regional, distant stage | T0 T3 local, distant stage | local-regional, distant stage | |
| low-moderate, high grade | low, moderate, high grade | low-moderate, high grade | |
| Progression depends on | current PSA level | current disease state | delay time and mode of detection |
| Stage progression | yes | yes | yes |
| Grade progression | no | yes | yes |
| PSA test sensitivity* | output of model | endogenous parameter | endogenous parameter |
| Biopsy compliance† | estimated from PLCO | combined with PSA sensitivity | combined with PSA sensitivity |
| Biopsy sensitivity‡ | based on literature review | combined with PSA sensitivity | combined with PSA sensitivity |
Pr(PSA positive|Disease)
Pr(Biopsy received|PSA positive, Disease)
Pr(Biopsy positive | Biopsy received, Disease)
The three natural history models are briefly reviewed below. Detailed descriptions of individual models and a joint report comparing the models are available on the CISNET website (15).
2.1. FHCRC
The FHCRC model assumes that a man’s PSA level (on a logarithmic scale) rises linearly with age and that it rises faster (i.e., it has a higher slope) beginning at onset of a biopsy-detectable preclinical tumor. In addition, disease grade is fixed at onset and post-onset PSA rises faster for Gleason score 8–10 than for Gleason score 2–7. The risk of disease onset is formalized as a hazard function that is proportional to age, while risks of transitioning from localized to metastatic states and from latent to symptomatic states are given by hazard functions that are proportional to the current PSA level (16–19). This dependence of disease progression on PSA levels implies that individuals with faster PSA growth will tend to have shorter intervals until the disease spreads beyond the prostate or becomes clinically diagnosed.
Grade-specific PSA slopes and variances in the FHCRC model were estimated using data from the control arm of the Prostate Cancer Prevention Trial, which conducted annual screening of 18,882 men for up to 7 years (20). PSA growth parameters were estimated by fitting random effects models to screened cases who had at least four tests. Given the estimated PSA growth parameters, we then estimated the disease transition hazards. To do this, we simulated natural histories and population disease trends under screening and identified the transition hazards that produced modeled disease incidence trends that best matched observed incidence trends by age, year, stage, and grade. Men with PSA levels of 4.0 ng/mL or greater at screening were assumed to receive a prostate biopsy based on age- and PSA-specific biopsy compliance rates observed in the Prostate, Lung, Colon, and Ovarian (PLCO) cancer screening trial, a randomized clinical trial involving PSA screening for 38,350 men (21). Biopsy sensitivity to detect occult tumors was allowed to improve over calendar years to reflect the dissemination of more extensive biopsy schemes in the late 1990s (18, 19). Given individual PSA trajectories, screening schedules, and biopsy compliance and sensitivity rates in the population, the hazard rate parameters were estimated using a simulated maximum likelihood algorithm to match model-projected incidence with SEER incidence.
Given the estimated PSA growth and hazards for disease transitions, the FHCRC model simulates complete disease histories for a population of individuals in the absence of screening and primary treatment. Natural history summaries of interest are then estimated empirically from this population. To project risks of clinical progression events for screen-detected cases, the model simulates another population of disease histories in the presence of screening but in the absence of curative treatment.
2.2. MISCAN
The MISCAN prostate cancer model also simulates individual life histories. The development of cancer in individuals is modeled as a sequence of tumor states, where prostate cancer develops from no prostate cancer through one or more screen-detectable preclinical states to a clinically diagnosed cancer. In each localized preclinical state, a tumor may grow to the next clinical T-stage (T1, impalpable; T2, palpable, confined to the prostate; and T3+, palpable, with extensions beyond the prostatic capsule), de-differentiate to a higher SEER histologic grade (well differentiated, Gleason score 2–6; moderately differentiated, Gleason score 7; and poorly differentiated, Gleason score 8–10), or give rise to symptoms and become clinically diagnosed. For these transitions, the time spent in the current state is generated from a Weibull distribution, where the parameters depend on the current state, and the choice of the next state is determined by transition probabilities. Additionally, there is a risk that a tumor in a SEER local-regional stage will develop into SEER distant stage disease. The transition to distant stage is modeled with a T-stage- and grade-specific hazard function. Consequently, the model includes 18 detectable preclinical states in the natural history that are derived from combinations of clinical T-stages, SEER histologic grades, and metastatic stages. The parameters for the natural history model were estimated using data from the Rotterdam section of the European Randomized study of Screening for Prostate Cancer (8, 22).
The MISCAN model represents the PSA test and subsequent biopsy as a single test with stage-specific sensitivities estimated from observed incidence data. For calibration to the US situation, we re-estimated these sensitivity parameters and estimated an additional stage-specific risk of clinical diagnosis to capture different pre-PSA disease diagnosis patterns in the US as compared with Europe. US-specific estimates for the parameters were obtained by calibrating the model to the observed age-specific incidence and age-specific SEER stage distribution (i.e., local-regional versus distant stage) using maximum likelihood (23).
As in the FHCRC model, natural history summary measures in the absence of screening and/or primary treatment are calculated empirically from simulated life histories in the absence of these interventions. And as in the FHCRC model, a new population of life histories is generated in the presence of screening to project risks of clinical progression events for PSA-detected cancers in the absence of primary treatment.
2.3. UMICH
The UMICH model of disease natural history consists of a sequence of analytical models rather than computer simulation algorithms. These models quantify the likelihood of observed disease incidence trends while averaging over distributions of unobserved factors influencing these trends.
The first component models disease incidence by age and calendar year. Disease incidence depends on time of disease onset, the time from onset to clinical diagnosis in the absence of screening (sojourn time), screening schedules, and test sensitivity. The model assumes distributions for the age at onset and the sojourn time with unknown parameters to be fit using population incidence data. As in the MISCAN model, test sensitivity reflects both the diagnostic properties of the test itself and the frequency and sensitivity of any subsequent biopsy. Sensitivity is modeled as increasing with time from disease onset. Screening schedules are random but are based on the reconstructed distribution of PSA screening patterns in the population (14). The unknown parameters are estimated by averaging over these distributions and calibrating the resulting marginal incidence against observed incidence (9).
The second component explicitly models disease grade (Gleason score 2–7 and Gleason score 8–10) and stage (SEER local-regional and distant) at diagnosis. These clinical characteristics depend on disease natural history through the time from disease onset to detection (delay time) and the mode of detection (screen or clinical). Given the distribution of age at diagnosis and calendar year of diagnosis output from the marginal incidence model, a Bayesian argument is used to derive a distribution for the delay time and the mode of diagnosis. The model for stage and grade at diagnosis is a multinomial logistic model, where delay time and mode of diagnosis are covariates (24). Putting this model together with the marginal incidence model produces a model for age-, stage-, and grade-specific incidence; calibrating this model to observed incidence allows for estimation of the parameters of the model for stage and grade at diagnosis.
An extension of the second component allows stage progression from screen detection to future (counterfactual) clinical detection. First, the fitted age-, stage-, and grade-specific model is run assuming zero test sensitivity to produce counterfactual “data” on the likely age-, stage-, and grade-specific incidence of disease in the absence of screening. Next, these “data” are used to estimate five unknown parameters representing allowable transition probabilities between stages and grades. Thus, under nonzero test sensitivity, the model is a joint model of age, stage, and grade at two points of diagnosis (real screen and counterfactual clinical). Model components are fit by maximum likelihood.
Given the estimated model components, we rely on analytic derivations to directly project probabilities of natural history summaries in the absence of screening and treatment. To project risks of clinical progression events for PSA-detected cases, we generate lead times from the fitted lead time distribution, assign stage and grade at clinical diagnosis conditional on lead time and age, stage, and grade at PSA detection, then generate prostate cancer survival from the common Poisson regression based on cases diagnosed in the pre-PSA era.
2.4. Model projections
We first use the three models to estimate lifetime risks of, mean ages at, and mean years between important prostate cancer natural history events in the absence of screening and primary treatment as follows.
Lifetime risks are calculated as proportions of individuals in the model populations with preclinical onset, clinical diagnosis, metastasis before clinical diagnosis, or prostate cancer death prior to non-prostate cancer death. To calculate risks conditional on onset, we restrict to individuals with onset in their lifetimes and calculate the proportion of these individuals that had the event in their lifetimes.
Mean ages at onset, clinical diagnosis, metastasis before clinical diagnosis, and prostate cancer death are projected in the presence of non-prostate cancer death. We average ages at each event among individuals that survive to the event in our model populations.
Mean years from onset to clinical diagnosis, to clinical diagnosis of metastatic disease, and to prostate cancer death are obtained by averaging time from onset to each endpoint among individuals in the model populations that reached that endpoint in their lifetimes. For example, we calculate mean years from onset to clinical diagnosis by summing these durations among men clinically diagnosed in their lifetimes and divide this total by the number of such men.
For reference, we also provide summary measures for PSA or clinical diagnosis in the presence of observed PSA screening patterns (14).
Next we project risks of clinical progression events for men who are PSA-detected in SEER local-regional stage in the model populations in the year 2000 (the most recent year to which the models are calibrated). Among these men, we estimate the proportion who would have gone on to be clinically diagnosed in the absence of screening. Note that by definition this proportion is one minus the fraction overdiagnosed. We also estimate the proportion who would have progressed to a metastatic stage prior to clinical diagnosis in the absence of screening and to prostate cancer death in the absence of any immediate or delayed primary treatment. Projections are tabulated by age and grade at PSA detection for all models and by age, grade, and PSA for the FHCRC model. At present, projections by PSA are only available from this model since it explicitly connects PSA growth and disease progression.
Finally, to provide a more complete picture of the risk of prostate cancer mortality if a PSA-detected tumor is left untreated, we project 20-year prostate cancer and non-prostate cancer survival by age and grade at PSA detection. The results reflect prostate cancer survival in the presence of other causes as in Nicholson and Harland (5), Albertsen et al. (2), and Parker et al. (6).
3. Results
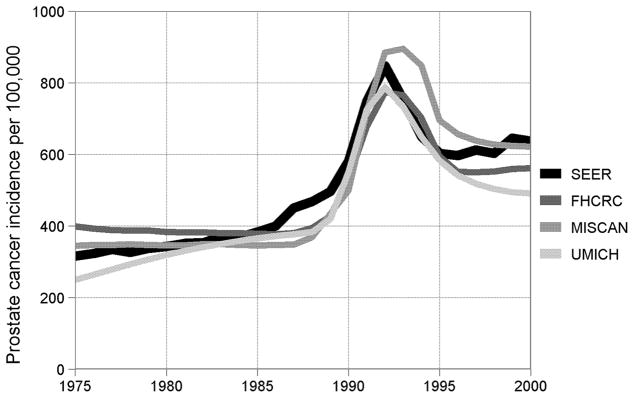
Figure 1 presents SEER observed and model-projected age-adjusted incidence per 100,000 men aged 50–84. Only the UMICH model captures the increasing trend in the pre-PSA years, and all models project a more sudden rise coincident with early PSA screening than was observed in SEER. Nonetheless, all models reproduce the scale of incidence in the pre-PSA period, the peak following the introduction of PSA screening in the late 1980s, and the re-stabilization at a higher level in the late 1990s.
Figure 1.
Age-adjusted prostate cancer incidence per 100,000 men aged 50–84 observed in the core 9 registries of the Surveillance, Epidemiology, and End Results program of the National Cancer Institute and corresponding projections by the three natural history models.
Table 2 presents natural history measures in the absence of screening or primary treatments projected by the three models. In general, the models are broadly consistent in the picture they present of prostate cancer natural history. The disease is widespread, progressing to a biopsy-detectable tumor in 20–33% of men. In the absence of early detection or primary treatments, 38–50% of these tumors go on to be clinically diagnosed, 5–9% metastasize by the time of clinical diagnosis, and 12–25% die of the disease. The preclinical period averages 7–14 years, permitting PSA testing to detect the disease often well in advance of clinical diagnosis (on average 4–9 years for men PSA detected in the year 2000 as shown in the last row of Table 2).
Table 2.
Natural history summary measures projected by the three models.
| Measure | FHCRC | MISCAN | UMICH |
|---|---|---|---|
| Lifetime risk of onset* | 33 | 27 | 20 |
| Lifetime risk of clinical diagnosis | 13 | 12 | 10 |
| Lifetime risk of metastasis by clinical diagnosis | 2 | 2 | 1 |
| Lifetime risk of prostate cancer death | 4 | 4 | 5 |
| Lifetime risk of clinical diagnosis given onset | 38 | 43 | 50 |
| Lifetime risk of metastasis by clinical diagnosis given onset | 7 | 9 | 5 |
| Lifetime risk of prostate cancer death given onset | 12 | 14 | 25 |
| Mean age at onset | 65 | 71 | 71 |
| Mean age at clinical diagnosis | 75 | 76 | 81 |
| Mean age at metastasis by clinical diagnosis | 74 | 78 | 81 |
| Mean age at prostate cancer death | 78 | 80 | 80 |
| Mean years from onset to clinical diagnosis | 14 | 9 | 7 |
| Mean years from onset to metastasis by clinical diagnosis | 13 | 13 | 4 |
| Mean years from onset to prostate cancer death | 18 | 15 | 8 |
| Lifetime risk of PSA or clinical diagnosis† | 14 | 20 | 15 |
| Lifetime risk of PSA or clinical diagnosis given onset† | 41 | 73 | 80 |
| Mean age at PSA or clinical diagnosis† | 74 | 74 | 75 |
| Mean years from PSA to clinical diagnosis† | 6 | 9 | 4 |
Onset represents the initial development of a biopsy-detectable preclinical tumor.
These measures are in the presence of observed PSA screening and are included for reference.
Table 3 presents risks of clinical progression events for PSA-detected cases who receive no primary treatment projected by the three models by age and grade. All results are based on modeled cases screen-detected in SEER local-regional stage in the year 2000. Note that projected risks of all clinical progression events decrease with age; this is to be expected since older men are more likely to die of other causes before the clinical progression event can occur. Also, projected risks tend to be higher when disease is more aggressive, indicated by higher disease grade. In general, even for Gleason score 2–7, all models estimate that at least two thirds of men under age 60 at PSA diagnosis would have gone on to be clinically diagnosed in the absence of screening. The risk that an untreated PSA-detected tumor would have metastasized before clinical diagnosis is 5–23% for men under 60 with Gleason score 2–7 and 22–35% for counterparts with Gleason score 8–10. The risk that an untreated PSA-detected tumor would lead to death in the absence of any primary treatment is 23–34% for men under 60 with Gleason score 2–7 and 63–83% for counterparts with Gleason score 8–10.
Table 3.
Projected frequencies (%) of clinical progression events for PSA-detected local-regional stage cases who receive no primary treatment by age and grade. Results from three models.
| Age group | Gleason grade | Clinical diagnosis |
Metastasis by clinical diagnosis |
Prostate cancer death |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FHCRC | MISCAN | UMICH | FHCRC | MISCAN | UMICH | FHCRC | MISCAN | UMICH | ||
| 50–54 | 2–7 | 92 | 71 | 93 | 12 | 23 | 5 | 25 | 29 | 34 |
| 55–59 | 2–7 | 85 | 67 | 90 | 11 | 19 | 5 | 23 | 26 | 33 |
| 60–64 | 2–7 | 77 | 58 | 86 | 11 | 17 | 5 | 20 | 22 | 31 |
| 65–69 | 2–7 | 68 | 49 | 80 | 9 | 14 | 5 | 16 | 17 | 28 |
| 70–74 | 2–7 | 56 | 40 | 74 | 8 | 10 | 4 | 12 | 12 | 23 |
| 75–79 | 2–7 | 44 | 32 | 66 | 6 | 8 | 4 | 8 | 10 | 18 |
| 80–84 | 2–7 | 36 | 24 | 59 | 5 | 6 | 3 | 6 | 6 | 13 |
| 50–54 | 8–10 | 96 | 96 | 93 | 35 | 23 | 26 | 77 | 83 | 68 |
| 55–59 | 8–10 | 90 | 95 | 90 | 33 | 22 | 25 | 69 | 71 | 63 |
| 60–64 | 8–10 | 84 | 92 | 86 | 30 | 19 | 24 | 60 | 66 | 56 |
| 65–69 | 8–10 | 76 | 89 | 80 | 27 | 21 | 23 | 49 | 55 | 49 |
| 70–74 | 8–10 | 66 | 81 | 74 | 23 | 20 | 21 | 39 | 45 | 41 |
| 75–79 | 8–10 | 53 | 72 | 66 | 20 | 19 | 19 | 29 | 33 | 32 |
| 80–84 | 8–10 | 46 | 64 | 59 | 16 | 17 | 16 | 21 | 23 | 24 |
Table 4 presents corresponding risks by age, grade, and PSA projected by the FHCRC model. We observe that the additional information provided by PSA at a screen detection is important, particularly for older men. For example, the risk that a cancer with Gleason score 2–7 would have gone on to be clinically diagnosed for men aged 65–69 increases from 58% for PSA 4–7 ng/mL at diagnosis to 80% for PSA 10 ng/mL at diagnosis. For the oldest age group, the risk that a cancer with Gleason score 2–7 would have gone on to be clinically diagnosed in the absence of screening, would have metastasized before clinical diagnosis in the absence of screening, or would have become terminal in the absence of primary treatment when PSA is 4–7 ng/mL at diagnosis is 37%, 43%, or 64% the risk faced when PSA is over 10 ng/mL at diagnosis. Corresponding results for a cancer with Gleason score 8–10 are 36%, 38%, and 35%.
Table 4.
Projected frequencies (%) of clinical progression events for PSA-detected local-regional stage cases who receive no primary treatment by age, grade, and PSA at detection. Results from the FHCRC model.
| Age group | Gleason grade | Clinical diagnosis |
Metastasis by clinical diagnosis |
Prostate cancer death |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4–7 ng/mL | 7–10 ng/mL | > 10 ng/mL | 4–7 ng/mL | 7–10 ng/mL | > 10 ng/mL | 4–7 ng/mL | 7–10 ng/mL | > 10 ng/mL | ||
| 50–54 | 2–7 | 91 | 92 | 96 | 12 | 13 | 14 | 25 | 25 | 26 |
| 55–59 | 2–7 | 82 | 87 | 92 | 11 | 12 | 13 | 21 | 25 | 25 |
| 60–64 | 2–7 | 71 | 80 | 88 | 10 | 11 | 12 | 18 | 21 | 24 |
| 65–69 | 2–7 | 58 | 69 | 80 | 8 | 9 | 11 | 13 | 16 | 20 |
| 70–74 | 2–7 | 44 | 56 | 73 | 6 | 8 | 10 | 9 | 11 | 16 |
| 75–79 | 2–7 | 31 | 43 | 64 | 4 | 6 | 10 | 6 | 8 | 12 |
| 80–84 | 2–7 | 20 | 31 | 54 | 3 | 4 | 7 | 3 | 4 | 9 |
| 50–54 | 8–10 | 92 | 96 | 98 | 33 | 34 | 35 | 72 | 74 | 80 |
| 55–59 | 8–10 | 84 | 89 | 95 | 29 | 33 | 35 | 62 | 67 | 73 |
| 60–64 | 8–10 | 74 | 81 | 91 | 25 | 28 | 34 | 50 | 55 | 67 |
| 65–69 | 8–10 | 62 | 70 | 85 | 21 | 24 | 31 | 35 | 43 | 58 |
| 70–74 | 8–10 | 46 | 57 | 77 | 15 | 21 | 27 | 27 | 32 | 47 |
| 75–79 | 8–10 | 33 | 43 | 66 | 13 | 14 | 25 | 16 | 24 | 37 |
| 80–84 | 8–10 | 21 | 30 | 59 | 8 | 10 | 21 | 9 | 15 | 26 |
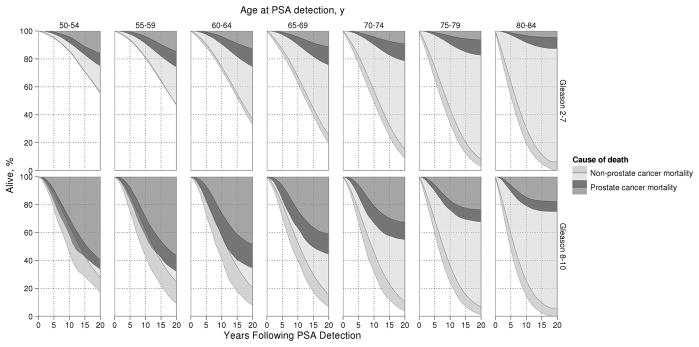
Complementing the projected risks that a PSA-detected cancer will lead to death if left untreated, Figure 2 presents projected 20-year survival for the same cohort of patients (men aged 50–84 screen detected in SEER local-regional stage in 2000 assuming no primary treatment) by age and grade at PSA detection. Ranges between the minimum and maximum values projected by the three models are represented in darker shaded areas for either cause of death; these regions can be interpreted as uncertainty due to model specification. Agreement across the three models is shown in lighter shaded areas for either cause of death. Among men under age 60 at PSA detection with Gleason score 2–7 disease, the three models project that 4–9% and 15–26% would die of their disease by 10 and 20 years after PSA detection in the absence of treatment. Corresponding projections for men with Gleason score 8–10 disease are 29–43% and 56–68%. Note that similar risks of prostate cancer and non-prostate cancer death and relatively greater inter-model uncertainty create overlapping projections for younger men with Gleason score 8–10.
Figure 2.
Prostate cancer and non-prostate cancer mortality following PSA detection for cases receiving no primary treatment projected by the three models by age and grade at PSA detection. For either cause of death, lighter areas reflect agreement by all three models and darker areas reflect inter-model uncertainty. Similar risks for either cause of death and relatively greater uncertainty about survival create substantial overlap for younger men with Gleason 8–10.
4. Discussion
In this article we use three independently developed models to project key events in the natural history of prostate cancer in the absence of screening and primary treatment, namely onset of preclinical biopsy-detectable disease, clinical diagnosis, transition to metastatic disease by the time of clinical diagnosis, and disease-specific mortality. The models also project frequencies of clinical progression events following PSA detection of local-regional stage disease in the year 2000. All models are estimated using common inputs for observed PSA screening patterns and are calibrated to reproduce US incidence trends among men aged 50–84 in the years 1975–2000. By using population incidence data to inform about underlying disease natural history, we provide unique insights regarding the distributions of unobservable events in disease progression.
Despite each model achieving projections that are reasonably consistent with observed incidence data, there are important differences in the projected courses of disease development and progression. For example, in all models, disease onset represents onset of biopsy-detectable tumors, so it is not surprising that projected risks of this event are lower than the estimated 36% of men with autopsy-detectable disease (25). However, the PSA test sensitivity in the FHCRC model is lower than that in the MISCAN and UMICH models; in other words, onset in the FHCRC model represents onset of disease that is harder to detect than that in the other models. Consequently, this model projects higher probabilities of onset and, given onset, lower probabilities of clinical diagnosis and prostate cancer death.
Another example of conceptual differences in disease natural history carrying implications for projected summary measures is manifested in projected mean years from onset to clinical diagnosis. The sequential stage progression formulation in the MISCAN model implies longer durations for cancers that are metastatic by the time of clinical diagnosis (mean 13 years) than for all cancers (mean 9 years). In contrast, the UMICH model implicitly assumes that tumors that become metastatic by the time of clinical diagnosis tend to grow faster (mean duration to diagnosis 4 years) than all cancers (mean duration to diagnosis 7 years).
All models project an unconditional lifetime risk of clinical diagnosis that is slightly higher than the 9% estimated in Ries et al. (26) based on incidence trends prior to the advent of PSA screening. Lifetime risks of dying from prostate cancer are close to the 3% obtained from Devcan (version 7) software for the pre-PSA era (26). Mean lead times associated with PSA screening range from 4 to 9 years and broadly agree with previously published estimates in the US setting (27, 5, 28).
The models also project risks of clinical diagnosis, metastasis by clinical diagnosis, and prostate cancer death in men aged 50–84 screen detected in SEER local-regional stage in 2000. Risks of these events are projected by age and grade for all three models and by age, grade, and PSA for the FHCRC model. As expected, risks fall with age (as non-prostate cancer death intervenes) and rise with more aggressive clinical characteristics.
It is worthwhile to observe three points related to these results. First, the risk that a PSA-detected patient would have gone on to be clinically diagnosed in the absence of screening is one minus the risk that he has been overdiagnosed. Thus, one minus the risk of clinical diagnosis reported in Tables 3 and 4 corresponds to the risk of overdiagnosis conditional on age and grade (and PSA) projected by the models. This complementary interpretation may help patients to make treatment decisions tailored to personalized clinical information known at diagnosis rather than based on broad estimates of overdiagnosis in the population as have been published previously (11).
Second, we find that even for men with Gleason score 2–7, the risks that the disease would lead to death in the absence of primary treatment (23–34% for men under age 60 at PSA detection) are nontrivial. Our estimates of disease-specific survival in this setting are based on men diagnosed in 1983–1986 without primary treatment, which we assume to be valid for contemporary cohorts. This assumption is imperfect because there have almost certainly been improvements in post-diagnosis monitoring of conservatively managed disease and a widened availability of more effective salvage therapies (29). However, because there is great uncertainty about how to correct for these improvements over time, we present the results without adjustment. Because these therapeutic advances have likely improved prostate cancer survival, the projections reported here may underestimate present-day prostate cancer survival in the absence of primary treatment.
Even if men are not treated at the time of screen detection, it is likely that intervention will occur at some later date should clinical events warrant it. To determine how our projections of disease-specific deaths would change in this setting, we assumed that all screen-detected cases received radical prostatectomy at their date of clinical diagnosis, and, accordingly, we inflated their post-lead-time survival by a factor of 0.65 (30). Under this assumption, the models projected that treatment reduced the risk of prostate cancer death by approximately 13–28% for men with Gleason score 2–7 and by approximately 13–20% for men with Gleason score 8–10. In absolute terms, for men under 60, the risk of prostate death decreased from 23–34% to 18–27% for Gleason score 2–7 disease and from 62–80% to 51–66% for Gleason score 8–10 disease.
These results have implications for management of PSA-detected prostate cancers. They suggest that, for younger men, the risks of progressing to lethal disease remain nontrivial even if curative treatment is pursued at clinical diagnosis. Since younger men are subject to low risks of overdiagnosis, these men would be well justified in considering primary treatment at PSA detection. However, we caution that the results presented here do not speak to the question of the optimal timing of primary therapy nor to the relative benefits of earlier versus later treatment. Instead, these results only provide alternative benchmark information for these treatment decisions. And as in other contexts, the optimal treatment decision should weigh any likely gains against known harms associated with treatments (for a review see 31).
Third, the FHCRC model implies that the PSA level carries important information about the risks of clinical progression events, particularly for older men. For example, for men aged 65–69 detected with Gleason score 2–7, these risks are 58%, 8%, and 13% when PSA is 4–7 ng/mL at diagnosis and 80%, 11%, and 20% when PSA is over 10 ng/mL at diagnosis. Consistent with findings in other studies (e.g., 32), these results indicate that PSA information stratifies risk. However, these results do not address whether early detection confers a survival benefit because any such benefit is contingent on treatment, which is disallowed by all models in this article. Despite this, the reader may be tempted to infer that PSA screening induces an implicit survival benefit because men with lower PSA at detection have a lower probability of dying of prostate cancer compared to men with higher PSA at detection. But these groups are not comparable due to selection artifacts. Specifically, the group of men with lower PSA at detection contains more overdiagnosed cases, and since overdiagnosed cases are not at risk of prostate cancer death, they artificially lower the risk of disease-specific mortality for this group as a whole. In addition, men with lower PSA at detection tend to have slower PSA growth in this model, which implies longer lead times and therefore also longer times to disease-specific mortality. This selection that occurs when a population is screened, and the artifacts that it generates, are key motivators for conducting randomized screening trials.
Randomized screening trials provide the opportunity to evaluate the benefits of screening in comparable groups and in the presence of treatment. Thus, these trials have best chance of identifying a survival benefit if one exists. The fact that the US and European prostate cancer screening trials did not show an unequivocal benefit (33, 34) has led many to doubt whether such a benefit exists, but it has yet to be verified whether the conflicting results are due to lack of benefit or to differences in settings, protocols, and implementations.
Survival projections are relatively consistent across the models despite variability in the modeled rates of disease progression events. As for risks of prostate cancer death, it is important to note that the survival model is based on pre-PSA data and hence projections in the absence of primary treatment are also in the absence of present-day disease management practices. We compared our survival results with those of Nicholson and Harland (5) and Parker et al. (6). Projected 15-year prostate cancer survival rates appear to be consistent with Nicholson and Harland (Table 4 in 5); averages across the three CISNET models are 87% for men aged 50–54 to 92% for men aged 80–84 at PSA detection for Gleason scores 2–7, while they project 88% for men aged 50 to 96% for men aged 80 at PSA detection for Gleason score 6 (a complete list of age- and grade-specific projections could not be obtained with their model). In contrast, because Parker et al. (6) use grade categories 2–6, 7, and 8–10, we compare survival projections for Gleason score 8–10 only. We project survival probabilities of 50% for men aged 55–59 to 64% for men aged 70–74 at PSA detection, while they project 28% for men aged 55–59 to 72% for men aged 70–74 at PSA detection. Our projections, particularly those for younger men, may be more consistent with their alternative scenario which assumes shorter lead times and lower overdetection rates than were reported in Draisma et al. (8). These assumptions may be more realistic in the US setting (11).
Finally, it is important to recognize that projected risks of clinical progression and disease-specific mortality for PSA-detected cases are based on the coarse categorization of Gleason scores 2–7 into a single category. This categorization was adopted to overcome the tendency of Gleason scores to migrate from well to moderately differentiated grades over calendar time, which is difficult to model adequately. As a consequence, the model projections cannot distinguish cancers with Gleason scores 2–6 from the more aggressive cancers with Gleason scores 3+4 or 4+3. Therefore, the models may overstate the risks of clinical progression for men with lower Gleason scores in this category.
While there is broad qualitative agreement in the projections across models, there is also quantitative variability. This variability reflects real uncertainty about the natural history of the disease despite the successful calibration of each model to observed incidence data. Yet despite this uncertainty, the models provide important answers to questions that patients with PSA-detected prostate cancers consistently ask when faced with determining their optimal treatment course. The models provide estimates of the risks that their cancer has been overdiagnosed and that it would progress to a metastatic state or lead to death if left untreated. These risks are estimated based on factors that have been shown to be most predictive of prognosis, namely, age, grade, and PSA. If the projected risks of clinical progression outcomes are low, then patients can be reassured of the advisability of a conservative treatment approach. More generally, these tailored risks of disease progression events should be useful when selecting treatment, but it should be recognized that the extent to which treatment will alter these risks is not clear and is not addressed by the present manuscript.
Further work will examine how survival projections change if treatment is instituted some time after PSA detection but before clinical diagnosis, when there is evidence of disease progression. This future work will be contingent on obtaining reliable estimates for treatment efficacy conditional on its timing following PSA detection. Ongoing studies of active surveillance will hopefully provide us with this information, which will then be incorporated into the models to provide patients with even more tailored information about their likely outcomes under different treatment options.
Acknowledgments
Financial support: This work was supported by the National Cancer Institute of the National Institutes of Health [U01CA88160, R01CA131874]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
The authors are grateful to John L. Gore for comments on an earlier draft.
References
- 1.Johansson JE, Andren O, Andersson SO, et al. Natural history of early, localized prostate cancer. J Am Med Assoc. 2004;291:2713–9. doi: 10.1001/jama.291.22.2713. [DOI] [PubMed] [Google Scholar]
- 2.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. J Am Med Assoc. 2005;293:2095–101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 3.Albertsen P. Predicting survival for men with clinically localized prostate cancer. Cancer. 2008;112:1–3. doi: 10.1002/cncr.23107. [DOI] [PubMed] [Google Scholar]
- 4.Paquette EL, Sun L, Paquette LR, Connelly R, Mcleod DG, Moul JW. Improved prostate cancer-specific survival and other disease parameters: Impact of prostate-specific antigen testing. Urology. 2002;60:756–9. doi: 10.1016/s0090-4295(02)01960-x. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson PW, Harland SJ. Survival prospects after screen-detection of prostate cancer. BJU Int. 2002;90:686–93. doi: 10.1046/j.1464-410x.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 6.Parker C, Muston D, Melia J, Moss S, Dearnaley D. A model of the natural history of screen-detected prostate cancer, and the effect of radical treatment on overall survival. Br J Cancer. 2006;94:1361–8. doi: 10.1038/sj.bjc.6603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etzioni R, Legler JM, Feuer EJ, Merrill RM, Cronin KA, Hankey BF. Cancer surveillance series: Interpreting trends in prostate cancer–Part III: Quantifying the link between population prostate-specific antigen testing and recent declines in prostate cancer mortality. J Natl Cancer Inst. 1999;91:1033–9. doi: 10.1093/jnci/91.12.1033. [DOI] [PubMed] [Google Scholar]
- 8.Draisma G, Boer R, Otto SJ, et al. Lead times and overdetection due to prostate-specific antigen screening: Estimates from the European Randomized study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 9.Tsodikov A, Szabo A, Wegelin J. A population model of prostate cancer incidence. Stat Med. 2006;25:2846–66. doi: 10.1002/sim.2257. [DOI] [PubMed] [Google Scholar]
- 10.Etzioni R, Gulati R, Falcon S, Penson D. Impact of PSA screening on the incidence of advanced stage prostate cancer in the US: A surveillance modeling approach. Med Decis Making. 2008;28:323–31. doi: 10.1177/0272989X07312719. [DOI] [PubMed] [Google Scholar]
- 11.Draisma G, Etzioni R, Tsodikov A, et al. Lead times and overdiagnosis in prostate-specific antigen screening: Importance of methods and context. J Natl Cancer Inst. 2009;101:374–83. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surveillance, Epidemiology, and End Results (SEER) Program SEER*Stat Database: Incidence–SEER 9 Registries [Internet] Bethesda, MD: National Cancer Institute, Surveillance Research Program, Cancer Statistics Branch; released April 2004, based on the November 2003 submission [cited 2010 Dec 22]. Available from: http://seer.cancer.gov. [Google Scholar]
- 13.Albertsen PC, Hanley JA, Barrows GH, et al. Prostate cancer and the Will Rogers phenomenon. J Natl Cancer Inst. 2005;97:1248–53. doi: 10.1093/jnci/dji248. [DOI] [PubMed] [Google Scholar]
- 14.Mariotto AB, Etzioni R, Krapcho M, Feuer EJ. Reconstructing PSA testing patterns between black and white men in the US from Medicare claims and the National Health Interview Survey. Cancer. 2007;109:1877–86. doi: 10.1002/cncr.22607. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Intervention and Surveillance Modeling Network (CISNET) Prostate Working Group. Joint Profile [Internet] Bethesda, MD: National Cancer Institute; 2009. [cited 2010 Dec 22]. Available from: http://cisnet.cancer.gov/prostate/profiles.html. [Google Scholar]
- 16.Inoue LY, Etzioni R, Slate EH, Morrell C, Penson DF. Combining longitudinal studies of PSA. Biostatistics. 2004;5:483–500. doi: 10.1093/biostatistics/5.3.483. [DOI] [PubMed] [Google Scholar]
- 17.Inoue LYT, Etzioni R, CM, PM Modeling disease progression with longitudinal markers. J Am Stat Assoc. 2008;103:259–70. doi: 10.1198/016214507000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etzioni R, Tsodikov A, Mariotto A, et al. Quantifying the role of PSA screening in the us prostate cancer mortality decline. Cancer Causes Control. 2008;19:175–81. doi: 10.1007/s10552-007-9083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati R, Inoue LYT, Katcher J, Hazelton W, Etzioni R. Calibrating disease progression models using population data: A critical precursor to policy development in cancer control. Biostatistics. 2010;11:707–19. doi: 10.1093/biostatistics/kxq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349:215–24. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]
- 21.Pinsky PF, Andriole GL, Kramer BS, Hayes RB, Prorok PC, Gohagan JK. Prostate biopsy following a positive screen in the Prostate, Lung, Colorectal and Ovarian cancer screening trial. J Urol. 2005;173:746–50. doi: 10.1097/01.ju.0000152697.25708.71. [DOI] [PubMed] [Google Scholar]
- 22.Draisma G, Postma R, Schröder FH, van der Kwast T, de Koning HJ. Gleason score, age and screening: Modeling dedifferentiation in prostate cancer. Int J Cancer. 2006;119:2366–71. doi: 10.1002/ijc.22158. [DOI] [PubMed] [Google Scholar]
- 23.Wever EM, Draisma G, Heijnsdijk EA, et al. Prostate-specific antigen screening in the United States vs in the European Randomized Study of Screening for Prostate Cancer—Rotterdam. J Natl Cancer Inst. 2010;102:352–5. doi: 10.1093/jnci/djp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chefo S, Tsodikov A. Stage-specific cancer incidence: An artificially mixed multinomial logit model. Stat Med. 2009;28:2054–76. doi: 10.1002/sim.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etzioni R, Cha R, Feuer EJ, Davidov O. Asymptomatic incidence and duration of prostate cancer. Am J Epidemiol. 1998;148:775–85. doi: 10.1093/oxfordjournals.aje.a009698. [DOI] [PubMed] [Google Scholar]
- 26.Ries LAG, Melbert D, Krapcho M, et al., editors. SEER Cancer Statistics Review, 1975–2005. Bethesda MD: National Cancer Institute; 2008. [Google Scholar]
- 27.Gann PH, Hennekens CH, Stampfer MJ. A prospective evaluation of plasma prostate-specific antigen for detection of prostatic cancer. J Am Med Assoc. 1995;273:289–94. [PubMed] [Google Scholar]
- 28.Telesca D, Etzioni R, Gulati R. Estimating lead time and overdiagnosis associated with PSA screening from prostate cancer incidence trends. Biometrics. 2008;64:10–9. doi: 10.1111/j.1541-0420.2007.00825.x. [DOI] [PubMed] [Google Scholar]
- 29.Klein EA, Platz EA, Thompson IM. Epidemiology, etiology, and prevention of prostate cancer. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA, editors. Campbell-Walsh Urology. 9. Philadelphia: Saunders; 2007. pp. 2854–73. [Google Scholar]
- 30.Bill-Axelson A, Holmberg L, Ruutu M, et al. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med. 2005;352:1977–84. doi: 10.1056/NEJMoa043739. [DOI] [PubMed] [Google Scholar]
- 31.Wilt TJ, MacDonald R, Rutks I, Shamliyan TA, Taylor BC, Kane RL. Systematic review: Comparative effectiveness and harms of treatments for clinically localized prostate cancer. Ann Intern Med. 2008;148:435–48. doi: 10.7326/0003-4819-148-6-200803180-00209. [DOI] [PubMed] [Google Scholar]
- 32.Kattan MW, Cuzick J, Fisher G, et al. Nomogram incorporating PSA level to predict cancer-specific survival for men with clinically localized prostate cancer managed without curative intent. Cancer. 2008;112:69–74. doi: 10.1002/cncr.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andriole GL, Grubb RL, Buys SS, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]