Abstract
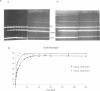
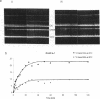
The hsdR, hsdM and hsdS genes coding for R.EcoK restriction endonuclease, both with and without a temperature sensitive mutation (ts-1) in the hsdS gene, were cloned in pBR322 plasmid and introduced into E.coli C3-6. The presence of the hsdSts-1 mutation has no effect on the R-M phenotype of this construct in bacteria grown at 42 degrees C. However, DNA sequencing indicates that the mutation is still present on the pBR322-hsdts-1 operon. The putative temperature-sensitive endonuclease was purified from bacteria carrying this plasmid and the ability to cleave and methylate plasmid DNA was investigated. The mutant endonuclease was found to show temperature-sensitivity for restriction. Modification was dramatically reduced at both the permissive and non-permissive temperatures. The wild type enzyme was found to cleave circular DNA in a manner which strongly suggests that only one endonuclease molecule is required per cleavage event. Circular and linear DNA appear to be cleaved using different mechanisms, and cleavage of linear DNA may require a second endonuclease molecule. The subunit composition of the purified endonucleases was investigated and compared to the level of subunit production in minicells. There is no evidence that HsdR is prevented from assembling with HsdM and HsdSts-1 to produce the mutant endonuclease. The data also suggests that the level of HsdR subunit may be limiting within the cell. We suggest that an excess of HsdM and HsdS may produce the methylase in vivo and that assembly of the endonuclease may be dependent upon the prior production of this methylase.
Full text
PDF

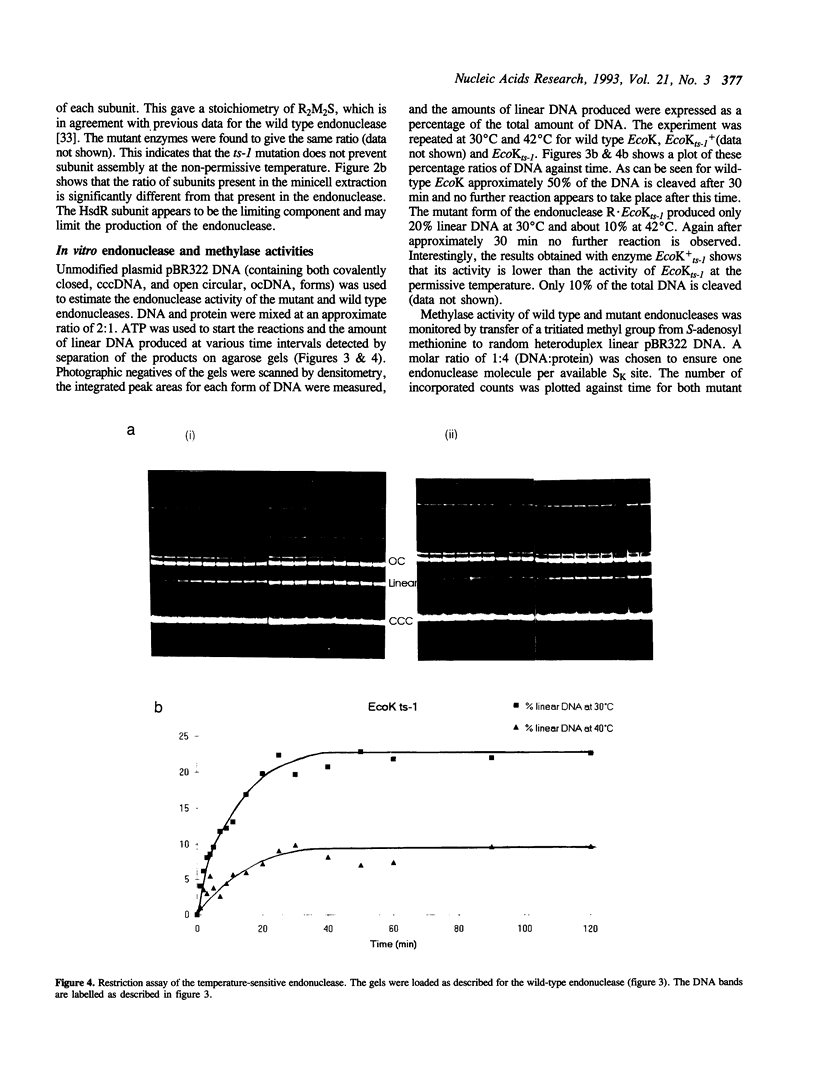
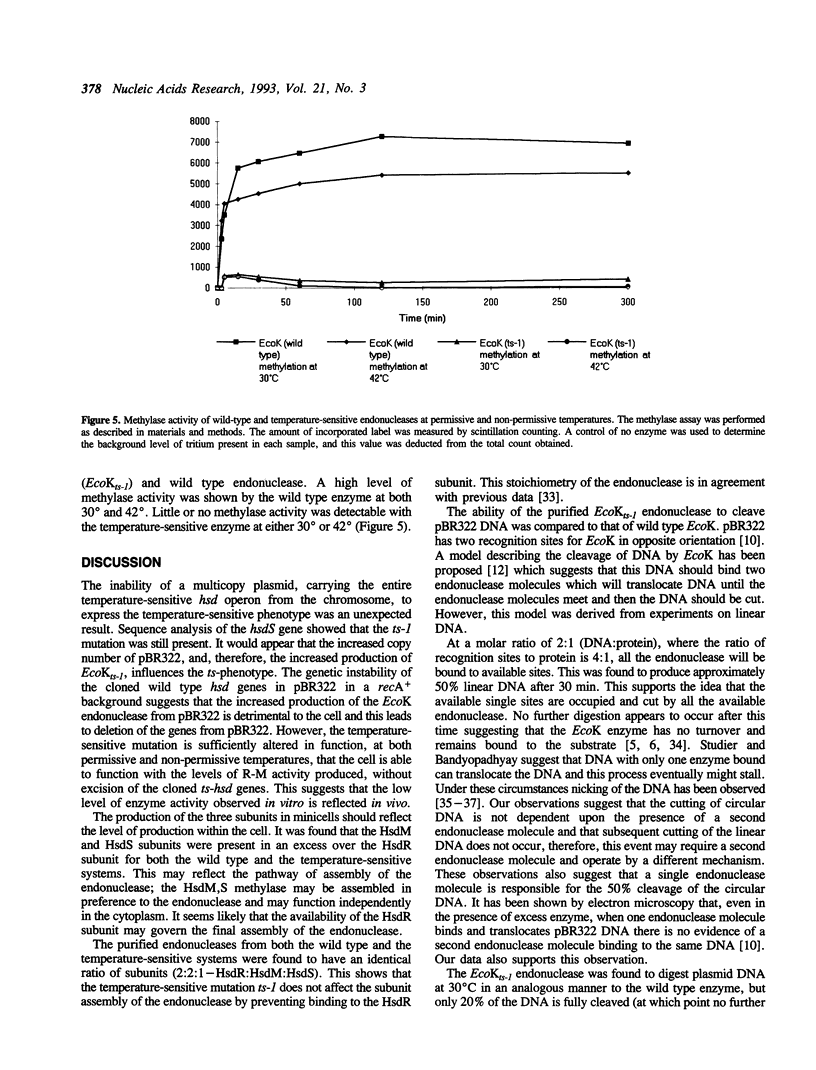

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler S. P., Nathans D. Studies of SV 40 DNA. V. Conversion of circular to linear SV 40 DNA by restriction endonuclease from Escherichia coli B. Biochim Biophys Acta. 1973 Mar 19;299(2):177–188. doi: 10.1016/0005-2787(73)90340-7. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI G., WEIGLE J. J. Host controlled variation in bacterial viruses. J Bacteriol. 1953 Feb;65(2):113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Burckhardt J., Weisemann J., Hamilton D. L., Yuan R. Complexes formed between the restriction endonuclease EcoK and heteroduplex DNA. J Mol Biol. 1981 Dec 5;153(2):425–440. doi: 10.1016/0022-2836(81)90287-4. [DOI] [PubMed] [Google Scholar]
- Colson C., Glover S. W., Symonds N., Stacey K. A. The location of the genes for host-controlled modification and restriction in Escherichia coli K-12. Genetics. 1965 Nov;52(5):1043–1050. doi: 10.1093/genetics/52.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel A. S., Fuller-Pace F. V., Legge D. M., Murray N. E. Distribution and diversity of hsd genes in Escherichia coli and other enteric bacteria. J Bacteriol. 1988 Apr;170(4):1775–1782. doi: 10.1128/jb.170.4.1775-1782.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. II. Purification, subunit structure, and catalytic properties of the restriction endonuclease. J Biol Chem. 1972 Oct 10;247(19):6183–6191. [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Bickle T. A., Yuan R. The role of S-adenosylmethionine in the cleavage of deoxyribonucleic acid by the restriction endonuclease from Escherichia coli K. J Biol Chem. 1975 Jun 10;250(11):4159–4164. [PubMed] [Google Scholar]
- Hubacek J., Glover S. W. Complementation analysis of temperature-sensitive host specificity mutations in Escherichia coli. J Mol Biol. 1970 May 28;50(1):111–127. doi: 10.1016/0022-2836(70)90108-7. [DOI] [PubMed] [Google Scholar]
- Hubácek J., Weiserová M. DNA restriction and modification in Escherichia coli: functional analysis of the role of the dnaC(D) gene product. J Gen Microbiol. 1980 Jul;119(1):231–238. doi: 10.1099/00221287-119-1-231. [DOI] [PubMed] [Google Scholar]
- Hubácek J., Zinkevich V. E., Weiserová M. The location of a temperature-sensitive trans-dominant mutation and its effect on restriction and modification in Escherichia coli K12. J Gen Microbiol. 1989 Nov;135(11):3057–3065. doi: 10.1099/00221287-135-11-3057. [DOI] [PubMed] [Google Scholar]
- JACOB F., WOLLMAN E. L. Etude génétique d'un bactériophage tempéré d'Escherichia coli. l. Le système genétique du bactériophage. Ann Inst Pasteur (Paris) 1954 Dec;87(6):653–673. [PubMed] [Google Scholar]
- Kan N. C., Lautenberger J. A., Edgell M. H., Hutchison C. A., 3rd The nucleotide sequence recognized by the Escherichia coli K12 restriction and modification enzymes. J Mol Biol. 1979 May 15;130(2):191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S., Lautenberger J. A., Eskin B., Lackey D. Host-controlled restriction and modification enzymes of Escherichia coli B. Fed Proc. 1974 May;33(5):1128–1134. [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R., Heywood J. Restriction and modification of DNA. Annu Rev Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. Use of minicells for bacteriophage-directed polypeptide synthesis. Methods Enzymol. 1979;68:493–503. doi: 10.1016/0076-6879(79)68038-2. [DOI] [PubMed] [Google Scholar]
- Rosamond J., Endlich B., Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J Mol Biol. 1979 Apr 25;129(4):619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Bandyopadhyay P. K. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri B., Nagaraja V., Bickle T. A. Bacterial DNA modification. Curr Top Microbiol Immunol. 1984;108:1–9. doi: 10.1007/978-3-642-69370-0_1. [DOI] [PubMed] [Google Scholar]
- Tal M., Weissman I., Silberstein A. A new method for stoichiometric analysis of proteins in complex mixture--reevaluation of the stoichiometry of E. coli ribosomal proteins. J Biochem Biophys Methods. 1990 Sep-Oct;21(3):247–266. doi: 10.1016/0165-022x(90)90018-8. [DOI] [PubMed] [Google Scholar]
- Van Pel A., Colson C. DNA restriction and modification systems in Salmonella. II. Genetic complementation between the K and B systems of Escherichia coli and the Salmonella typhimurium system SB, with the same chromosomal location. Mol Gen Genet. 1974;135(1):51–60. doi: 10.1007/BF00433901. [DOI] [PubMed] [Google Scholar]
- Vogel J. L., Li Z. J., Howe M. M., Toussaint A., Higgins N. P. Temperature-sensitive mutations in the bacteriophage Mu c repressor locate a 63-amino-acid DNA-binding domain. J Bacteriol. 1991 Oct;173(20):6568–6577. doi: 10.1128/jb.173.20.6568-6577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980 May;20(1):237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- Zinkevich V. E., Solonin A. S., Bogdarina I. G., Taniashin V. I., Baev A. A. Klonirovanie i restriktsionnyi analiz BamHI-EcoRI fragmenta DNK, soderazhashchego geny hsd-oblasti Escherichia coli. Dokl Akad Nauk SSSR. 1981;259(1):216–218. [PubMed] [Google Scholar]
- Zinkevich V. E., Weiserová M., Kryukov V. M., Hubácek J. A mutation that converts serine340 of the HsdSK polypeptide to phenylalanine and its effects on restriction and modification in Escherichia coli K-12. Gene. 1990 May 31;90(1):125–128. doi: 10.1016/0378-1119(90)90447-y. [DOI] [PubMed] [Google Scholar]
- Zinkevich V., Heslop P., Glover S. W., Weiserova M., Hubácek J., Firman K. Mutation in the specificity polypeptide of the type I restriction endonuclease R.EcoK that affects subunit assembly. J Mol Biol. 1992 Oct 5;227(3):597–601. doi: 10.1016/0022-2836(92)90210-b. [DOI] [PubMed] [Google Scholar]