Abstract
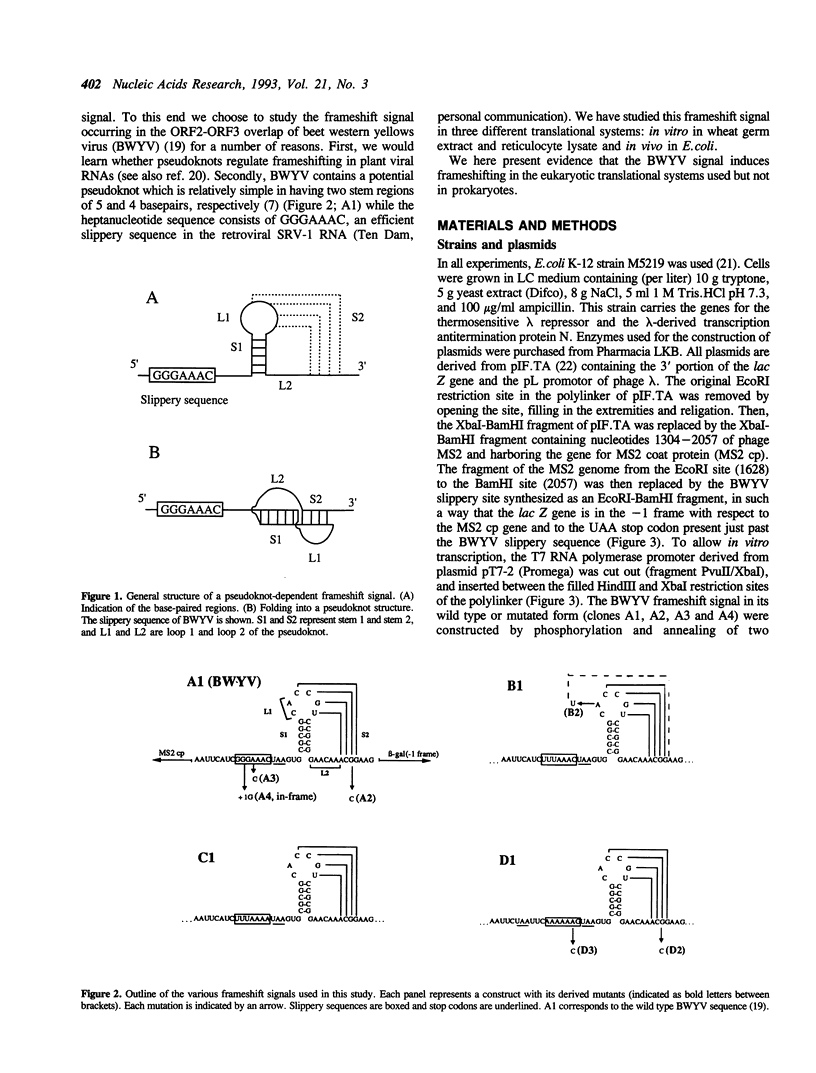
The genomic RNA of beet western yellows virus (BWYV) contains a potential translational frameshift signal in the overlap region of open reading frames ORF2 and ORF3. The signal, composed of a heptanucleotide slippery sequence and a downstream pseudoknot, is similar in appearance to those identified in retroviral RNAs. We have examined whether the proposed BWYV signal functions in frameshifting in three translational systems, i.c. in vitro in a reticulocyte lysate or a wheat germ extract and in vivo in E. coli. The efficiency of the signal in the eukaryotic system is low but significant, as it responds strongly to changes in either the slip sequence or the pseudoknot. In contrast, in E. coli there is hardly any response to the same changes. Replacing the slip sequence to the typical prokaryotic signal AAAAAAG yields more than 5% frameshift in E. coli. In this organism the frameshifting is highly sensitive to changes in the slip sequence but only slightly to disruption of the pseudoknot. The eukaryotic assay systems are barely sensitive to changes in either AAAAAAG or in the pseudoknot structure in this construct. We conclude that eukaryotic frameshift signals are not recognized by prokaryotes. On the other hand the typical prokaryotic slip sequence AAAAAAG does not lead to significant frameshifting in the eukaryote. In contrast to recent reports on the closely related potato leafroll virus (PLRV) we show that the frameshifting in BWYV is pseudoknot-dependent.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blinkowa A. L., Walker J. R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990 Apr 11;18(7):1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A. J., Inglis S. C. Mutational analysis of the "slippery-sequence" component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992 Sep 20;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990 May;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. C., Martin C. T., Pham T. T., Coleman J. E. Transcription by T7 RNA polymerase is not zinc-dependent and is abolished on amidomethylation of cysteine-347. Biochemistry. 1986 Jan 14;25(1):36–40. doi: 10.1021/bi00349a006. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987 Feb;61(2):480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Bishop D. H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992 Feb;186(2):389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z., Brown P. O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992 Mar;6(3):511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991 May 11;19(9):2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng T. H., Tu C. L., Bruenn J. A. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J Virol. 1992 Feb;66(2):999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. Retroviruses. Science. 1988 Jun 10;240(4858):1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- Veidt I., Lot H., Leiser M., Scheidecker D., Guilley H., Richards K., Jonard G. Nucleotide sequence of beet western yellows virus RNA. Nucleic Acids Res. 1988 Nov 11;16(21):9917–9932. doi: 10.1093/nar/16.21.9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]