Abstract
Genome-wide association studies (GWAS) have identified SNPs associated with breast cancer. However, they offer limited insights about the biological mechanisms by which SNPs confer risk. We investigated the association of GWAS information with a major oncogenic pathway in breast cancer, the Notch signaling pathway. We first identified 385 SNPs and 150 genes associated with risk for breast cancer by mining data from 41 GWAS. We then investigated their expression, along with 32 genes involved in the Notch signaling pathway using two publicly available gene expression data sets from the Caucasian (42 cases and 143 controls) and Asian (43 cases and 43 controls) populations. Pathway prediction and network modeling confirmed that Notch receptors and genes involved in the Notch signaling pathway interact with genes containing SNPs associated with risk for breast cancer. Additionally, we identified other SNP-associated biological pathways relevant to breast cancer, including the P53, apoptosis and MAP kinase pathways.
Keywords: GWAS, gene expression, Notch signaling pathway
Introduction
The lifetime risk of developing breast cancer is about one in eight for women, with around 192,370 new invasive cases, 62,280 new in situ cases being diagnosed and 40,170 deaths in the United States each year.1 Breast cancer is the second leading cause of cancer death among women in the US. While major inroads have been made in reducing mortality rates due to increased screening, digital mammography, specialized care, and the widespread use of therapeutic agents such as aromatase inhibitors, trastuzumab and others, defining the genetic architecture of breast cancer remains an important long-term goal for the development of more effective therapeutic strategies and early interventions. Recent advances in microarray technology and reduction in genotyping costs have made possible genome-wide association studies to identify genetic variants associated with risk for breast cancer.2–6 Although these studies are providing valuable clues about the broad patterns of genetic susceptibility to breast cancer, the ultimate goal of SNP and gene discovery is to identify and characterize the biological pathways and the molecular mechanisms underlying the disease. This is especially important in breast cancer, a group of biologically and genetically heterogeneous diseases with distinct oncogenic pathways and therapeutic targets. To date, there is little information associating GWAS data to known oncogenetic pathways involved in breast cancer. This knowledge gap is hindering translation of discoveries from GWAS into clinical practice to develop clinically useful genetic tests as well as early therapeutic interventions and new targeted drugs.
The objective of this study was to investigate the association of GWAS information with the Notch signaling pathway. The rationale for choosing the Notch signaling pathway in this study was that apart from its involvement in breast cancer, the Notch signaling pathway is involved in many types of cancer including breast cancer, lung cancer, neuroblastomas, skin cancer, cervical cancer, and prostate cancer.7 However, it is less well characterized compared to other biological pathways involved in breast cancer such as the estrogen, kinase, apoptosis and P53 pathways which are enriched with SNPs associated with risk for breast cancer. Consequently, association of GWAS information with the Notch signaling pathway may provide proof of concept that this approach could work and provide insights about the putative functional bridges between GWAS information and biological pathways that are less characterized and may not contain genes harboring mutations or SNPs associated with cancer. The Notch signaling pathway is extremely contextual-dependent, meaning that crosstalk and interaction with other pathways including those enriched by SNPs associated with risk for breast cancer would be very important in determining outcomes. With the exception of T-ALL (T-cell acute lymphoblastic leukemia),7 there are very few instances where mutations have been detected in solid tumors in Notch pathway genes, despite solid evidence that the pathway itself is very important to the biology of tumors.8–13 Thus, it is conceivable that the genes in the Notch signaling pathway may be regulated in trans by genetic variants located in genes in other biological pathways that crosstalk with the Notch signaling pathway. Therefore, modeling gene regulatory networks using GWAS information, gene expression data and genes involved in the Notch signaling pathway provides the best mechanism for understanding the potential molecular mechanisms underlying Notch dysregulation in breast cancer.
Our group8 among others9,10 have shown that the Notch signaling pathway plays a critical role in the development of breast cancer. Numerous cellular functions and microenvironment cues associated with tumorigenesis are modulated by Notch signaling, including cell fate, proliferation, apoptosis, adhesion, and angiogenesis.11,12 Additionally, Notch signaling plays an important role in the maintenance of breast tumor-initiating cells.13 Of the four known Notch receptors, three have been implicated in breast oncogenesis (Notch-1, -3, and -4) while one (Notch-2) has been suggested to have opposite roles and have a positive prognostic significance.14 Notch-2 has recently been associated with ER-positive breast cancer tumors.15 Both pan-Notch inhibitors and specific monoclonal antibodies (mAb) to individual Notch receptors are being developed for breast cancer. However, the molecular mechanisms underlying dysregulation and aberrant expression of Notch receptors and other genes involved in the Notch signaling pathway leading to breast cancer remain poorly understood. The association between the Notch signaling pathway and genes containing SNPs associated with risk for breast cancer could provide putative functional bridges between GWAS information with an oncogenic pathway that does not harbor mutations, but is involved in cancer development and progression.8–13 Therefore, elucidating the association of GWAS information with the Notch signaling pathway may help to determine which patients may benefit from Notch inhibitors and to explore the role of Notch transmembrane receptors as potential drug targets and predictive markers.
We hypothesized that genes containing SNPs with large (P ≤10−5) and small to moderate (P∼10−2–10−4) effects associated with risk for breast cancer directly or indirectly interact with the 4 Notch family members, other genes in the Notch signaling pathway, and potentially other biological pathways relevant to breast cancer. To formally test this hypothesis, we mined data from 41GWAS for SNPs and genes associated with risk for breast cancer, and two publicly available gene expression data sets derived from the Caucasian (42 cases and 143 controls)16 and Asian (43 cases and 43 controls)17 populations. Throughout this study, we have defined genes containing SNPs associated with risk for breast cancer as candidate genes, and assumed the gene and pathway as the units of association. We have assumed the P-value and correlation as measures of effect size for both GWAS and gene expression analysis.
Methods
Data sources
We mined the literature through PubMed searches and websites containing supplementary data on 41GWAS to identify SNPs and genes associated with risk for breast cancer. The search included terms (GWAS, GWA, WGAS, WGA, genome-wide, genomewide, whole genome, all terms + association, or + scan) in combination with breast cancer from the primary published reports through July 2010. All the reports were read and information was manually extracted and entered into the database. The inclusion criterion was that the study must include a sample size of ≥500 cases and ≥500 controls. We catalogued SNPs with large (P ≤10−5) and small to moderate (P∼10−2–10−4) effect sizes. We chose this liberal statistical threshold to allow examination of genes containing borderline SNPs with small effect sizes and to include GWAS of various sizes accommodating publication bias while maintaining a consistent approach. SNPs mapping to intergenic regions were not used in this study. SNP locations and gene names were verified using the dbSNP database and the chromosome report build 3.71. The Human Genome Nomenclature (HUGO) database was used to further check the authenticity of gene names and their aliases. The list of genes (gene symbols, full names), number of SNPs per gene, along with references of primary reports from which the GWAS information was derived are summarized in Table A in the Appendix, provided as supplementary data to this manuscript.
We used two publicly available gene expression data sets based on the case control design as in GWAS design to evaluate and establish the expression levels of candidate genes and genes involved in the notch signaling pathway. The first data set involved the Caucasian population, and consisted of 143 histologically normal breast tissues derived from patients harboring breast cancer who underwent curative mastectomy and 42 invasive ductal carcinomas of various histological grades obtained from breast cancer patients. The data set has been fully described by the originators.16 Briefly, this data set consisted of histological data. Histologically-normal breast has the potential to harbor pre-malignant changes at the molecular level and thus provides a boon for identifying risk markers. We postulated that a histologically-normal tissue with tumor-like gene expression patterns might harbor substantial risk for future cancer development. Thus genes associated with these high-risk tissues would be considered to be malignancy-risk genes. From this assumption, it follows that these genes could serve as potential molecular predictors of breast cancer. Normal breast cancer tissue included histologically normal and benign. All samples were assessed for global gene expression profiles using the Affymetrix platform on U133 Plus 2.0 Array. The tumors were not associated with any known genetic risk factors such as BRCA1 or BRCA2 mutations. The microarray data from these samples including the raw probe-level hybridization intensities were downloaded from the NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE10780.
Most GWAS have been performed on Caucasian populations. It remains unclear to what extent findings from these studies can be extrapolated to non-Caucasians. To determine whether results found using data from Caucasian population could be replicated in the Asian population, we used a second gene expression data set. The second data set involved a multi-ethnic Asian population, consisting of Malaysian breast cancer patients (Malays, Chinese and Indian). The data set has been described by.17 Briefly, the data set consisted of a total of 43 breast carcinomas and 43 patient-matched normal tissues collected from Kuala Lumpur, UKM and Putrajaya Hospitals in Malasia. The data set was generated using the Affymetrix platform’s U133A Chip and was downloaded from GEO accession number GSE15852. The two data sets contained similar information, both involved ductal carcinomas with same tumor grades. The clinical and histological characteristics of the two gene expression data sets used are summarized in Table 1.
Table 1.
Clinical and histological characteristics of Caucasian and Asian patients used in this study to generate gene expression data.
| ER, PR, Her2 and grade |
Caucasian population |
Asian population |
||||
|---|---|---|---|---|---|---|
| ER | PR | HER2/neu | ER | PR | HER2/neu | |
| Negative | 25 | 38 | 43 | 18 | 17 | 22 |
| Positive | 55 | 42 | 12 | 25 | 26 | 21 |
| Other | 10 | 10 | 35 | – | – | – |
| Total cases | 90 | 90 | 90 | 43 | 43 | 43 |
| Grade | Frequency | Frequency | ||||
| Well differentiated or 1 | 6 | 8 | ||||
| Moderately differentiated or 2 | 27 | 24 | ||||
| Poorly differentiated or 3 | 57 | 11 | ||||
| Total cases | 90 | 43 | ||||
In each of the two microarray data sets described above, entries in the data matrix were expression values generated by Affymetrix’s Microarray Analysis Suite 5.0 (MAS5) statistical algorithm.18 Following normalization and scaling, MAS5 signal values were summarized by Turkey’s biweight estimation of the probe level intensities within each probe set. This was followed by a global normalization (linear scaling) to give all chips the same average intensity. These procedures yield robust weighted means called average-scaled differences that are proportional to the amount of a particular RNA transcript present in the sample after background correction, which we used as the input in this analysis, after filtering out spiked control genes.
Data analysis
As a first step, we mapped SNPs to the genes by matching gene names, SNP IDs and positions using the information in the database (dbSNP). We then sorted and ranked the genes on the basis of P-values derived from GWAS, number of times the SNP in a particular gene has been replicated in multiple independent studies, and number of SNPs within each candidate gene. Genes containing SNPs with P-values P ≤ 10−5 were considered to have large effect size, whereas genes containing SNPs with P-values P<10−2–10−4 were considered to have small to moderate effect size. Relatively few SNPs mapped to candidate genes had P-values sufficiently small (P ≤ 10−5) or replicated in multiple independent studies to give conclusive evidence of association. Conversely, there were many genes containing several hundred SNPs with small to moderately significant P-values P∼10−2–10−4. These would likely contain several false positives, but may also contain genuine effects of small magnitude. Consequently, in our data analysis, we considered all the 150 candidate genes containing SNPs identified by GWAS. Our rationale was that, the presence of greater than expected number of associated SNPs in genes of similar biological functions interacting with each other and their downstream targets in biological pathways gives a degree of confidence that the associations are genuine, even if none is individually highly significant. The overall P-value for SNPs replicated in multiple independent studies was estimated using Fisher’s method.19
Briefly, we assumed that the P-values (Pi) are independent and uniformly distributed under their null hypotheses. Let Pi be the P-value for the corresponding statistic Pi where [Pi = P1, P2, … Pn]T is a vector of P-values obtained by performing independent test statistic [Ti = T1, T2, … Tn]T on individual SNPs [rsi = rs1, rs2, … rsn]T. Assuming H as a continuous monotonic function, a transformation of the P-value can be defined as Zi = H−1(1 − Pi).20 The statistics for combining K independent P-values or for combining information from K SNPs is given by the following equation.20
where, Z denotes the sum of Zi (Z-scores) of the transformed P-values for the K SNPs. The Z-scores were back transformed into P-values using Fisher’s method.19
The challenge was how to represent a gene containing multiple SNPs within the gene and how to account for correlations among those SNPs. Correlations among P-values of SNPs within a gene exist because of linkage disequilibrium among SNPs. Correlations among SNPs will invalidate the existing methods for combining independent P-values. Furthermore, the SNPs within a gene may have antagonistic functions which could not be captured by combining P-values. Therefore, the method for combining P-values in independent SNPs described above cannot be directly applied to combining P-values of SNPs within a gene. Wang et al,21 suggested choosing the most significant SNP from each gene as a representative. The limitation of that approach is that genes that contain a number of SNPs jointly having significant risk effects, but individually making only a small contribution, will be missed in such a representation. Therefore, in this study, we considered the gene and pathway containing SNPs as the units of association. This allowed us to holistically unravel the genetic susceptibility architecture of breast cancer by jointly considering all common variation within the gene and all the genes in the pathway through pathway prediction and modeling gene networks using candidate genes (ie, genes containing SNPs associate with risk for breast cancer) and genes involved in the Notch signaling pathway.
Next, we matched the 150 candidate genes containing SNPs along with genes involved in the Notch signaling pathway to probes on the U133 Plus 2.0 Chips and U133A Human Chips, representing gene expression from the Caucasian and Asian populations, respectively. The probes were extracted from the NetAffx Database using the batch query (Affymetrix Inc). We then used probes to extract the gene expression values for the candidate genes and genes involved in the Notch signaling pathway from gene expression data sets on Caucasian and the Asian populations, respectively.
On each data set containing candidate genes and genes involved in the Notch signaling pathway, we performed supervised analysis comparing mean gene expression profiles in cancer patients to mean gene expression profiles in cancer-free controls to identify significantly differentially expressed genes, which distinguished the two groups and were predictors of cancer, as demonstrated in Figures 1 and 2 for the Caucasian and Asian populations, respectively. We used the Benjamin and Hochberg22 procedure to correct for multiple testing. Genes were then ranked on estimated P-values and false discovery rate as shown in the Appendix Tables B and C for the Caucasians and Asians, respectively, and those passing a threshold (P ≤ 0.05) were selected. We performed correlation analysis using Pearson correlation (r) coefficient procedure to identify genes with similar expression profiles, and to assess the association between candidate genes and genes involved in the Notch signaling pathway. In correlation analysis, genes were treated as the variables and their expression values as the measurements. The correlation coefficient between the candidate gene X and Notchgene Y [and between candidate genes] was computed using the following equation.
where n is the sample size, Xi and Yj [X̄ and Ȳ] are the expression [mean] expression values for the candidate gene and the gene involved in the Notch signaling pathway, respectively. SX and SY are the standard deviations of the expression values for the candidate gene and the gene involved in the Notch signaling pathway, respectively. Both supervised and correlation analyses were performed using GenePattern.23
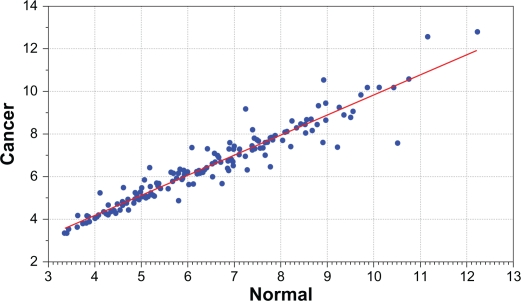
Figure 1.
Distribution of mean expression values for candidate genes between breast cancer patients (y-axis) and normal subjects (x-axis) in the Caucasian population. Blue dots significantly deviating from the red line indicate differential expression. The genes, estimates of P-values and false discovery rates for each gene are presented in Table B in the Appendix provided as supplementary data.
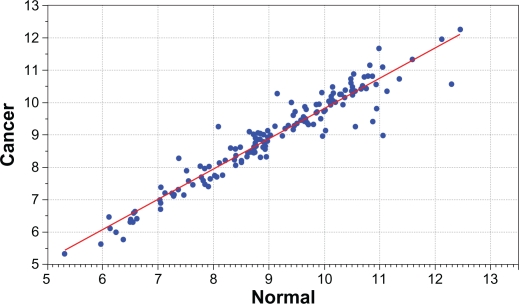
Figure 2.
Distribution of mean expression values for candidate genes between breast cancer patients (y-axis) and normal subjects (x-axis) in the Asian population. Blue dots significantly deviating from the red line indicate differential expression. The genes, estimates of P-values and false discovery rates for each gene are presented in Table C in the Appendix provided as supplementary data.
Finally, we performed pathway prediction and network modeling using the Osprey System24 to identify candidate genes which interact with genes involved in the Notch signaling pathway and other biological pathways relevant to breast cancer. The Osprey network modeling and visualization system is a very dynamic software platform which integrates experimental information from the literature with gene ontology information from the GO database about all the genes. Therefore, it allowed capturing all the genes that interact with the input genes (ie, candidate genes and genes in the Notch signaling pathway) that have been experimentally confirmed and are involved in the same biological process, which may have been missed during differential expression and co-expression analysis. Thus, is an optimal tool for pathway prediction, network modeling and in silico validation of predicted pathways and gene networks.
In pathway prediction and network visualization, we first performed pathway prediction using a set of candidate genes containing SNPs associated with risk for breast cancer and members of the Notch signaling pathway, which were differentially expressed between cases and controls in the Caucasian population. We repeated the same analysis for the Asian population. To determine whether genes containing SNPs with larger effects and SNPs replicated in multiple independent studies interact with genes in the Notch signaling pathway, we performed separate analysis for each set of genes. In pathway prediction, genes were represented by nodes and the interactions by vertices. Two genes were considered to share a genetic susceptibility architecture and network properties if they were interconnected as represented by the vertices and were correlated as determined by the correlation coefficient. To determine the functional relationships and biological properties of genes in the networks, we used the biological process category of the Gene Ontology classification built in the Osprey System to color-code the nodes (genes). We imposed level 3 filtering criteria to remove genes with spurious interactions, which could be less informative or could distort the reliability of network modeling. This approach also served as a validation step in that we randomly removed genes with fewer interactions and repeated the analysis.
Results
We investigated the association of GWAS information with the Notch signaling pathway. GWAS information included a total 497 SNPs associated with risk for breast cancer. The SNPs were derived from 41 GWAS, totaling more than 250,000 cases and 250,000 controls, mostly (99%) from the Caucasian populations. From the total, 112 SNPs were located in intergenic regions and were not used in the analysis. The remainder, 385 SNPs mapped to 150 genes, of which 130 candidate genes matched probes on the U133 Plus 2.0 Chip for data on Caucasian population and 111 candidate genes matched probes on the U133A Chip for data on Asian population, and were used in the analysis. The discrepancy in the number of genes in the two data sets is due to differences in Chip density (ie, difference in probes and unique number of genes represented on the U133 Plus 2.0 and U133A Human Chips). The list of gene symbols, SNP (rs_IDs), number of SNPs per gene along with the primary sources (ie, references) are provided as supplementary material in Table A in the appendix. Genes involved in the Notch signaling pathway included the 4 members of the Notch family of transmembrane receptors, NOTCH1, NOTCH2, NOTCH3, NOTCH4 and other genes involved in Notch signaling pathway including, PTEN, HES1, HES2, SKP2, DICER, XIAP, JAG1, JAG2, HES1, HES2, HEY1, HEY2, FBXW7, SKPIA, CCNA1, CCNA2, IAP, MYC, VEGFA, VEGFB, VEGFC and TP53, NUMB, DLL1, DLK2, DLL3 and DLL4. We hypothesized that genes containing SNPs associated with risk for breast cancer interact with the members of the Notch family of transmembrane receptors and other genes involved in the Notch signaling pathway and other pathways relevant to breast cancer.
As a first step, we evaluated the expression of candidate genes and genes involved in the Notch signaling pathway by comparing normal breast to breast tumors in Caucasian and Asian populations using publicly available gene expression data described in the methods section, as demonstrated in Figures 1 and 2 for the Caucasian and Asian populations, respectively. We sought to identify candidate genes and members of the Notch signaling pathway that were significantly differentially expressed between breast cancer and normal tissue. Such genes would serve as molecular predictors of breast cancer. We then used the identified differentially expressed genes as the input for pathway prediction and network modeling.
Using supervised analysis, we identified 71 candidate genes and 12 genes involved in the Notch signaling pathway, with significant differences in expression profiles between the cases and controls in the Caucasian population. The list of significantly differentially expressed genes involved in the Notch signaling pathways included HEY1 (P = 5.00E-06), HEY2 (P = 5.00E-06), JAG2 (P = 500E-06), MYC (P < 0.002), NOTCH1 (P = 3.00E-05), NOTCH2 (P = 2.00E-05), NOTCH4 (P = 5.00E-06), SP2 (P < 0.04), DICER1 (P < 0.0004) and FBXW7 (P = 5.00E-06). Repeating the same analysis using gene expression data from the Asian population, we identified 31 candidate genes and several genes involved in the Notch oncogenic pathway (PTEN, P5e-06; HEY2, P = 0.04; IAPP, P = 0.05) with significant differences in expression profiles between cases and controls. A full list of significantly differentially expressed (and non significantly differentially expressed) candidate genes between breast cancer patients and controls, their estimated P-values and false discovery rates for the Caucasian and Asian populations are provided in Tables B and C, respectively, provided as supplementary data. As expected, not all candidate genes exhibited differences in expression profiles between cases and controls. In addition, not all candidate genes differentially expressed in the Caucasian population were replicated in the Asian population.
This suggests that like GWAS results, gene expression can be heterogeneous among populations, making it difficult to replicate results. The observed differences in expression profiles between cases and controls in the two populations can be attributed to several factors; including the fact that gene expression varies among populations,25 differences in tissue procurement timing and storage, use of chips with different probe densities, as well as the genetic and phenotypic heterogeneity inherent in the GWAS data used in this study. Co-expression analysis however revealed that candidate genes that were not differentially expressed exhibited co-expression patterns with sets of genes distinguishing cancer from normal controls.
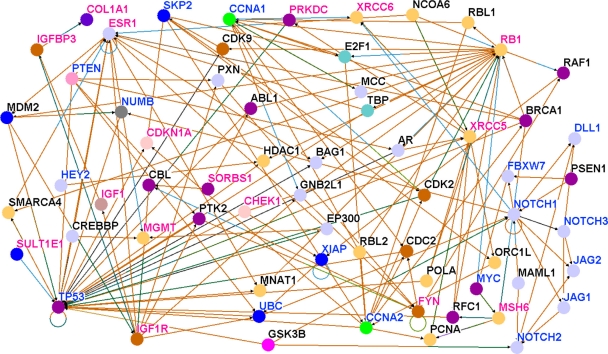
To formally test the hypothesis that candidate genes containing SNPs associated with risk for breast cancer interact with genes involved in the Notch signaling pathway, we performed pathway prediction and network modeling. As a first step, we performed pathway prediction and network modeling using the 71 candidate genes confirmed in the Causation population and all genes involved in the Notch signaling pathway. In addition we modeled the biological relationships of the genes in the predicted pathways and networks using Gene Ontology information and experimental information derived from the literature by text mining using the module built in the Osprey System. The key for GO information characterizing genes in the predicted pathways and networks according to the biological process in which they are involved is presented in Figure 3. The results of pathway prediction and network modeling for the Caucasian population are presented in Figure 4. For easy interpretation throughout the figures, names of candidate genes (ie, genes containing SNPs associated with risk for breast cancer) are shown in red. Names of genes involved in the Notch signaling pathway are shown in blue, while names of the new set of genes not identified by GWAS are shown in black. Nodes represent the genes and the vertices represent the interactions.
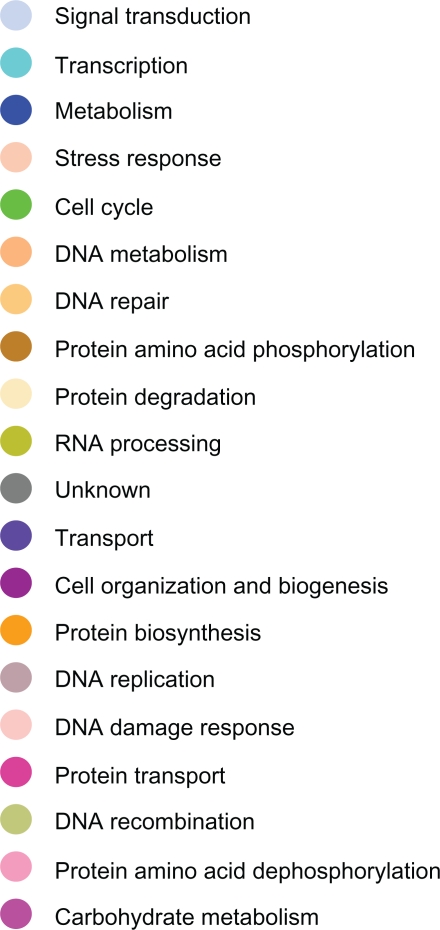
Figure 3.
Color codes indicating the biological process in which genes reported in figures 4–7 are involved.
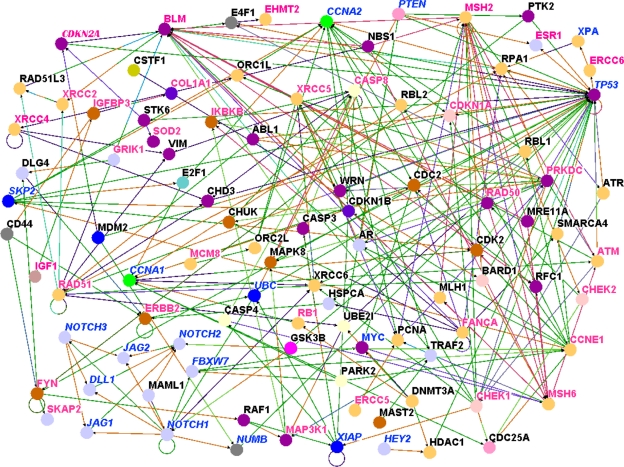
Figure 4.
Results of pathway prediction and network modeling showing interactions between genes containing SNPs associated with risk for breast cancer and genes involved in the Notch signaling and other biological pathways based on the Caucasian population. Nodes represent genes and vertices represent interactions. The color code denotes the biological process in which the genes are involved as defined in Figure 3. The color codes in the vertices indicate the experimental techniques or a combination thereof used to confirm the relationship between the genes as determined by the experiments reported in the literature. Candidate genes containing SNPs associated with risk for breast cancer are shown in red, genes involved in the Notch signaling pathway are shown in blue and new genes not reported in GWAS studies are shown in black. For the full names of genes and number of SNPs per gene including GWAS references, please refer Table A in supplementary data.
Members of the Notch family of transmembrane receptors NOTCH1, NOTCH2, were found to interact with genes containing SNPs associated with risk for breast cancer (Fig. 4). Interactions were found between NOTCH1 and several DNA repair genes, including XRCC6, XRCC5, and CHEK1; and between NOTCH2 and GSK3B, FANCA, MSH2. Also observed were interactions between other members of the Notch signaling pathway, for example, MYC, CNNA1, JAG1, FBXW7, CNNA2, XIAP, with genes containing SNPs associated with risk for breast cancer.
To evaluate the strength of association between candidate genes and the Notch signaling pathway in the Caucasian population we estimated correlations between pairs of genes. Focusing on candidate genes with SNPs replicated in multiple GWAS studies. Significant correlations (P < 0.05) between Notch receptors and FGFR2 (NOTCH1, r = 0.20; NOTCH2, r = 0.37; NOTCH3, r = −0.23) were observed. Significant correlations between other genes involved in the Notch signaling pathway and FGFR2 were also observed, including with (MYC, r = 0.27; DICER, r = 0.36; HEY1, r = 0.44; HEY2, r = 0.57; JAG2, r = 0.47). Other candidate genes which exhibited significant correlations with genes involved in the Notch signaling pathway included (NOTCH1 versus PGR, r = −0.24; NOTCH4 versus PGR, r = −0.26; ESR1 versus SKP2, r = −0.46; FANCA versus FBXW7, r = −0.44; ERBB2 versus NOTCH3, r = 0.49; CDKN2A versus SKP2, r = 0.70; CDKN2A versus CHEK1, r = 0.68; CDKN2A versus CHEK2, r = 0.55; DICER versus BLM, r = −0.56; HES1 versus BLM, r = 47; HES2 versus BLM, r = 52, FBXW7 versus CHEK2, r = −0.49; FBXW7 versus ERBB2, r = −0.41 and NOTCH3 versus ERBB2, r = 0.49). This confirms our hypothesis that candidate genes are associated with Notch receptors and other genes involved in the Notch signaling pathway. Supporting the validity of our observations, in addition to the Notch signaling pathway, we identified other biological pathways relevant to breast cancer. Additional biological pathways enriched by SNPs and relevant to breast cancer identified in this analysis included the P53 pathway, the apoptosis control pathways, MAP kinase pathways, the estrogen receptor pathway and the insulin growth factor pathway.
In general, the interactions between candidate genes and Notch signaling appears to be complex involving multiple pathways, suggesting that multiple interacting pathways are likely involved in the development and progression of breast cancer. The involvement of multiple pathways also indicates that interactions between the Notch signaling pathway and candidate genes containing SNPs associated with risk for breast cancer may involve multi-pathway crosstalk. A clear example was the involvement of NUMB, which is involved in the Notch signaling pathway but also controls P53 tumor suppressor activity in breast cancer.26 NUMB is a cell fate determinant, which by asymmetrically partitioning at mitosis, controls cell fate choices by antagonizing the activity of the plasma membrane receptor of the Notch family.27
Of particular interest were the three-way interactions among genes containing SNPs with large (P ≤10−5) and small to moderate (P∼10−2–10−4) effects and genes involved in the Notch signaling pathway; and the interactions among candidate genes. This indicates that the genetic susceptibility architecture of breast cancer is complex and that even genes containing SNPs with small effects often considered as noise in GWAS analysis could potentially have significant effects on development and progression of cancer. These results demonstrate that pathway prediction and network modeling could potentially increase the power of GWAS analysis by taking into account complex interactions which could not be realized using GWAS alone.
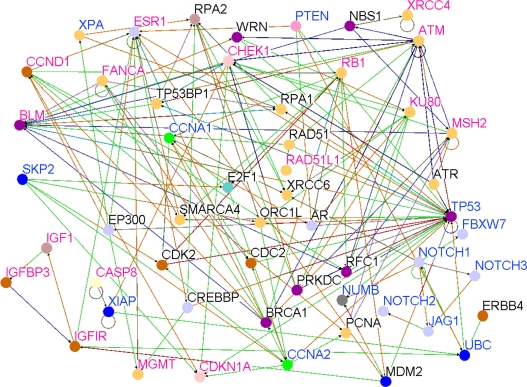
A major concern in genome-wide association studies is that majority of the GWAS studies ∼99% (based on this study) have been conducted on Caucasian populations. To determine whether results of pathway prediction and network modeling observed in the Caucasian population could be replicated in the Asian population, we performed pathway prediction using the 31 differentially expressed candidate genes identified using gene expression data derived from the Asian population and the set of genes involved in the Notch signaling pathway. The results showing pathway prediction and gene interaction networks for genes containing SNPs and members of the Notch signaling pathway based on the Asian population are presented in Figure 5. Genes involved in the Notch signaling pathway (NOTCH1, NOTCH2, NOTCH3, MYC, HEY2, FBXW7, PTEN, CCNA1, CCNA1, JAG2, JAG1), were found to interact directly or indirectly with genes containing SNPs associated with risk for breast cancer (ESR1, IGF1R, XRCC6, MGMT, CHEK1, MSH6, CDKN1A) (Fig. 5).
Figure 5.
Results of pathway prediction and network modeling showing interactions between genes containing SNPs associated with risk for breast cancer and genes involved in the Notch signaling and other biological pathways based on the Asian population. Nodes represent genes and vertices represent interactions. The color code denotes the biological process in which the genes are involved as defined in Figure 3. The color codes in the vertices indicate the experimental techniques or a combination thereof used to confirm the relationship between the genes as determined by the experiments reported in the literature. Candidate genes containing SNPs associated with risk for breast cancer are shown in red, genes involved in the Notch signaling pathway are shown in blue and novel genes are shown in black.
Like in the Caucasian population, genes containing SNPs associated with risk for breast cancer were also found to be associated with Notch signaling, P53, apoptosis and MAP kinase pathways. To assess the strength of association between Notch receptors and candidate genes in the Asian population, we estimated correlations. With the exception of NOTCH3 (r = 0.15), all the Notch receptors were significantly (P < 0.05) correlated with P53 (NOTCH1, r = 0.29; NOTCH2, r = 0.50; NOTCH4, r = 0.43). Significant correlations between the Notch receptors and IGF1-were also observed (NOTCH1, r = 0.47; NOTCH2, r = 0.53; NOTCH4, r = 0.29). Similar results were found between Notch receptors and the IGF1R (NOTCH1, r = 0.27; NOTCH2, r = 0.60; NOTCH4, r = 0.53) and between Notch receptors and the IGFBP3 (NOTCH1, r = 0.53; NOTCH2, r = 0.57; NOTCH3, r = 0.13; NOTCH4, r = 0.36). We also found the same results between Notch receptors and FYN (NOTCH1, r = 0.40; NOTCH2, r = 0.72; NOTCH3, r = 0.27; NOTCH4, r = 0.52). This confirms the association between candidate genes and the Notch signaling pathway. The P53, IGF1, IGFR1 and IGFBP3 and FYN genes have been reported in multiple independent GWAS studies. To the extent that these genes are also involved in different biological pathways, these results indicate crosstalk between the Notch signaling pathway and the P53 and insulin growth factor pathways. Additionally, significant correlations between IGF1 and PTEN (r = 0.65), and between PTEN and FYN (r = 0.61) were observed, yet another confirmation of association between the Notch signaling pathway and candidate genes. PTEN is a major regulator of the Notch signaling pathway. In general, the biological pathways identified in the Caucasian population were replicated in the Asian populations. This is an interesting result in that it demonstrates that although gene expression may vary across populations presumably due to environmental factors, the biological pathways underlying breast cancer development and progression could be the same in different populations. Environmental factors were not considered in this study.
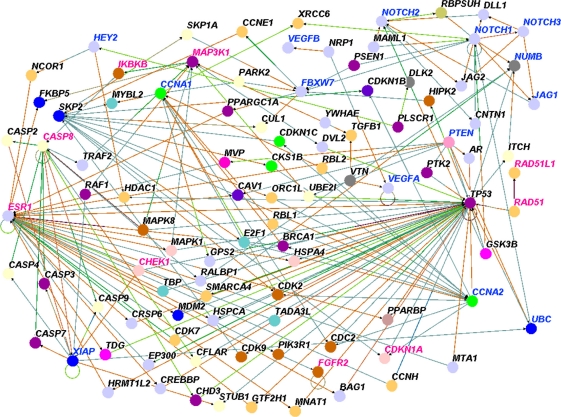
Both the validity and reproducibility of results from GWAS studies particularly the ones with small to moderate effect sizes (P∼10−2–10−4) have been challenged.28 Therefore, to determine whether genes involved in the Notch signaling pathway interact with genes that contain SNPs with good evidence of association (ie, “winner” SNPs) (P ≤10−5), we removed genes containing SNPs with small effects. We then repeated pathway prediction combining genes involved in the Notch signaling pathways and genes containing SNPs with large effects. Genes containing SNPs with larger effects (P ≤10−5) included ABCC4, BTNL8, CASP8, COLIA1, ECHDC1, ESR1, FBN1, FGFR2, GRIK1, MLK4, LOC643714, LSP1, NEKIO, PPP2R2B, RAD51L1, RFN146, SLC4A7, TOX3, STXBP4, TGFB1, MAP3K1, H19. The results of genes containing SNPs with large effects are presented in Figure 6. We found that genes containing SNPs with large effects interact with each other and with genes involved in the Notch signaling pathway and other biological pathways involved in breast cancer. In addition, we also identified genes with small effects, for example, the IKBKB (P = 0.002).
Figure 6.
Results of pathway prediction and network modeling showing interactions between genes containing SNPs associated with risk for breast cancer and genes involved in the Notch signaling and other biological pathways. The results are based on genes containing SNPs replicated in multiple independent GWAS. Nodes represent genes and vertices represent interactions. The color code denotes the biological process in which the genes are involved as defined in Figure 3. The color codes in the vertices indicate the experimental techniques or a combination thereof used to confirm the relationship between the genes as determined by the experiments reported in the literature. Candidate genes containing SNPs associated with risk for breast cancer are shown in red, genes involved in the Notch signaling pathway are shown in blue and novel genes are shown in black.
To further address the problem of reliability of GWAS data, we performed additional analyses combining genes containing SNPs replicated in multiple independent studies with genes involved in the Notch Signaling pathway. Genes containing SNPs reported in multiple independent studies included A2BP1, ADH1B, ALPL, ATM, BLM, CASP8, CCND1, CDKN1A, CHEK1, CSB, PMBT1, ECHDC1, EHMT, ERBB4, ESR1, FANCA, FGFR2, HCN1, ICAM5, IGF1, IGFIR, IGFBP3, KU80, LSP1, MGMT, MSH2, NEIL2, POLB, POLK, RAD51L1, RB1, RNF146, RPA1, RPA2, SKAP2, SOD2, MAP3K1, H19, TOX3, TGFB1, TNP1, XPA, XPC, XRCC3, XRCC4. The results are shown in Figure 7. We found that genes containing SNPs replicated in multiple independent studies interact with each other and with genes involved in the Notch signaling pathway, and genes involved in other breast cancer-relevant biological pathways. In addition, we identified novel genes not yet identified by GWAS (genes shown in black).
Figure 7.
Results of pathway prediction and network modeling showing interactions between genes containing SNPs associated with risk for breast cancer and genes involved in the Notch signaling and other biological pathways. The results are based on genes containing SNPs with the largest effect size (P < 10−5). Nodes represent genes and vertices represent interactions. The color code denotes the biological process in which the genes are involved as defined in Figure 3. The color codes in the vertices indicate the experimental techniques or a combination thereof used to confirm the relationship between the genes as determined by the experiments reported in the literature. Candidate genes containing SNPs associated with risk for breast cancer are shown in red, genes involved in the Notch signaling pathway are shown in blue and novel genes are shown in black.
To address the problem of publication bias and to determine whether members of the Notch signaling pathway interact with candidate genes with small to moderate effects, we performed further analyses combining the genes involved in the Notch signaling pathway and candidate genes containing SNPs with small effects. We found that genes containing SNPs with small effects interact with genes involved in the Notch signaling pathway (results not presented because we captured the same results in the four figures reported above). In additional, we identified novel genes not reported in GWAS studies, including MDM2, CHUK, AR, WRN, XRCC6, ABL1, HDAC1 and RAF1.
Overall, in all the analysis, we confirmed our hypothesis that genes containing SNPs associated with risk for breast cancer (regardless of effect size) interact with genes involved in the Notch signaling pathway as well as other biological pathways known to be relevant to breast cancer. Additionally, we identified novel genes not yet reported by GWAS. These results demonstrated that GWAS information can be leveraged with biological knowledge and gene expression data to infer the association between gene expression and breast cancer. The association of the Notch oncogenic pathway with genes containing SNPs associated with risk for breast cancer demonstrates that integrative analysis combing GWAS information, gene expression data and biological knowledge is a powerful approach to identifying molecular markers underlying GWAS findings.
Discussion
In this study, we provide evidence of association between breast cancer risk candidate genes and the Notch signaling pathway, an important oncogenic pathway involved in many aspects of tumor development, growth and progression, and a potential therapeutic target. Additionally, we identified other biological pathways including the ESR1 pathway, IGF pathway, the Map kinase pathway, the apoptosis pathway and the P53 pathway enriched by SNPs associated with risk for breast cancer. This suggests that regulation of the Notch pathway by candidate genes from GWAS is complicated and potentially involves multi-dimensional crosstalk between the Notch signaling pathway and other oncogenic pathways. Our results tend to agree with those in a recent association study, Fu et al,15 which showed the association between NOTCH2 and the P53 pathway in ER-positive breast tumors.
Several studies have now attempted pathway-based approaches to dissect the genetic susceptibility architecture of common diseases, for example, in inflammatory diseases,29 in bipolar disorder,30 in multiple sclerosis,31 in breast cancer,32,33 prostate cancer,34 and in seven common diseases.35 To our knowledge, this is the first study to associate GWAS information with the Notch signaling pathway. This is an important finding because although GWAS as demonstrated in this and other studies2–6 can effectively map loci contributing to phenotypes of interest in breast cancer, they offer limited insights about the biological mechanisms by which SNPs confer risk. Of particular interest is the association of Notch signaling with multiple DNA repair genes including the XRCC5 and XRCC6, which participate in non-homologous end joining for chromosomal DNA double-strand break repair, and they are essential for the efficient removal of apurinic/apyrimidinic sites near double-strand breaks.36 WRN, a member of the RecQ family of DNA helicases, interacts with XRCC5 and XRCC6 heterodimers (also known as Ku70/80).37 WRN and the serine/threonine protein kinase ATM, a master regulator of the cellular DNA damage response, cooperatively participate in an intra-S checkpoint in cells with collapsed replication forks.38 Protein kinase CHEK1 participates in all known cell cycle checkpoints.39 Checkpoint kinase 2 (CHEK2) is an important signal transducer of cellular responses to DNA damage, and it is considered a tumor suppressor; germ line defects of CHEK2 predispose to familial breast cancer and some other types of malignancies.40 FANCA participates in the repair of DNA interstrand crosslinks41 and MSH2, a gene frequently mutated in familial non-polylposis colon cancer,42 is a homolog of Drosophila MutS, a mismatch repair component.43
One possible explanation for these findings is that Notch signaling may be necessary for the survival of cells that are deficient in DNA repair. Also, DNA repair pathways are especially active in cells with stem-like phenotypes, potentially including tumor-initiating cells.44,45 Interestingly, an increase in Notch activity is part of the response to radiation in breast cancer-initiating cells,46 endothelial cells47 and glioma stem cells.48
A number of genes involved in cell proliferation and survival are represented in our analysis. Among them, GSK3B is a protein kinase involved in glycogen metabolism but also in proliferation, differentiation and survival that is downstream of the Wnt and AKT pathways49. GSK3B itself, as well as the WNT and AKT pathways are considered potential therapeutic targets in breast cancer.50 Interestingly, Drosophila Notch is known to interact genetically with the homolog of GSK3B.51 Additional interactions worth noting are those with IKKα (CHUK) and IKKβ (IKBKB). We have reported that Notch-1 interacts and cooperates with IKKα in cervical52 and breast cancer cells,53 and Vilimas et al have shown that it interacts with the IKK signalosome in T-ALL cells.54 These findings emphasize the potential cancer relevance of the Notch-NF-κB cross-talk. Similar considerations can be made for Raf-1, a key member of the Ras-Raf-MAP kinase pathway. We showed in 2002 that Ras-mediated transformation requires Notch-1.55 A number of cell cycle and proliferation-related genes containing breast cancer-related SNPs also show significant interaction with Notch pathway genes (eg, CDK2, CDC2, PCNA, cyclin E1), as does multi-functional oncogene c-Myc, which is known to be a direct Notch target and genetically interact with Notch-1 in T-ALL.56 Protein kinase c-Abl is another interesting Notch-interacting gene. In Drosophila, there is evidence of non-canonical interactions between Notch and Abl mediated by accessory protein Disabled.57 The anti-apoptotic mediator XIAP has been reported to physically interact with Notch-1, resulting in a direct interference with XIAP ubiquitination and degradation.58 Lipid phosphatase PTEN, which is frequently defective in breast cancer, has been shown to be regulated by Notch in other models.59,60 Histone deacetylase 1 (HDAC1) is displaced by Notch during the process of Notch-mediated transcriptional activation.61 We have recently revealed a two-way feedback between Notch-1 and the estrogen receptor alpha (ESR1) in breast cancer cells,8,53 as well as between Notch-1 and ERBB2.62
The E3 ubiquitin ligase MDM2, which targets tumor protein p53 for proteasomal degradation, was identified in our analysis as a novel candidate in both Caucasian and Asian populations. In a mouse model, it has been shown that Notch suppresses p53 in lymphomagenesis through repression of the ARF-MDM2-p53 tumor surveillance network.63 Interestingly, the endocytic protein NUMB, which is a negative regulator of Notch activity, can be found in a complex with p53 and MDM2, thereby preventing ubiquitination and degradation of p53.26
Thus, an analysis of interactions of genes that contain breast cancer-related SNPs with Notch pathway genes reveals genes and gene products that have been suggested to cross-talk with Notch signaling in other models, including invertebrate models,64 supporting the validity of our approach. Moreover, this analysis detected additional candidates for functional interactions with Notch, including numerous DNA repair and checkpoint genes, the androgen receptor (AR), free radical detoxifying enzyme superoxide dismutase 2 (SOD2) and other genes relevant to breast carcinogenesis and to responsiveness to therapeutic agents including chemotherapeutics and radiation. The association between the Notch signaling pathway and GWAS information is important in that numerous cellular functions and microenvironment cues associated with tumorigenesis are modulated by Notch signaling, including proliferation, apoptosis, adhesion, epithelial-to-mesenchymal transition and angiogenesis.11,64 Given that Notch signaling is activated in a wide variety of human breast cancer cases, components of the Notch pathway have been evaluated as prognosis markers and drug targets.14 In breast cancer clinical specimens, mRNA expression of Notch-1 and Notch ligand Jagged-1 have been shown to correlate strongly with poor prognosis.64–66 Loss of Notch-negative regulator NUMB, has been described in approximately 50% of human breast cancers.67 Notch-4 expression, as detected by immunohistochemistry, correlates with Ki67, a well-known proliferation marker in infiltrating breast carcinomas of ductal or lobular histologies.8 Conversely, and consistent with published in vitro data, expression of Notch-2 appears to have a positive prognostic significance.68
Although the results of this study offer valuable clues about association of GWAS information with the Notch signaling pathway and other pathways relevant to breast cancer, limitations in interpreting these results must be acknowledged. We have used results of genome-wide association studies and publicly available gene expression data in this analysis. Therefore, interpretation of our results is inherently subject to the constraints of such data. Key limitation include but are not limited to the fact that GWAS information was derived from results obtained from different studies conducted using different platforms, sample sizes, cryptic population stratifications, different phenotypes, and potentially different analysis techniques all of which could potentially affect our results.
However, the results presented in this study demonstrate conclusively that genes containing SNPs associated with risk for breast cancer interact with genes involved in the Notch signaling pathway and other biological pathways relevant to breast cancer. Important work remains to be done to determine how the SNPs disrupt the genes and pathways, leading to cancer development and progression. Such work is beyond the scope of this paper, but the work reported here is the first step in that direction.
Supplementary Data
Appendix
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Breast Cancer Facts and Figures 2009–2010. American Cancer Society, Inc. 250 Williams Street, NW, Atlanta, GA 30303.
- 2.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature Genet. 2007;39(7):870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easton DF, Pooley KA, Dunning AM, Pharoah PD, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, et al. Common variants on Chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature Genet. 2007;39(7):865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 5.Commonly studied single-nucleotide polymorphisms and breast cancer: results from the breast cancer association consortium. J Natl Cancer Inst. 2006;98(19):1382–96. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- 6.Turnbull C, Ahmed S, Morrison J, Pernet D, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nature Genetics. 2010;42(6):504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allenspach EJ, Maillard I, Aster JC, Pear W. Notch signaling in cancer. Cancer Biol Therap. 2002;1(5):466–76. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo P, Miao H, D’Souza G, Osipo C, et al. Crosstalk between Notch and estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68(13):5226–35. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stylianou S, Clark RB, Brennan K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006;66(3):1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 10.Shi W, Harris AL. Notch signaling in breast cancer and tumor angiogenesis: Cross-talk and therapeutic potentials. J Mam Gland Biol Neopl. 2006;11:41–52. doi: 10.1007/s10911-006-9011-7. [DOI] [PubMed] [Google Scholar]
- 11.Leong KG, Karsan A. Recent insights into the role Notch signaling in tumorigenesis. Blood. 2006;107(6):2223–33. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 12.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009 Apr 17;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pannuti A, Foreman K, Rizzo P, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010 Jun 15;16(12):3141–52. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miele L. Rational targeting of Notch signaling in breast cancer. Expert Rev Anticancer Ther. 2008 Aug;8(8):1197–202. doi: 10.1586/14737140.8.8.1197. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Edvarden H, Kaushiva A, et al. Notch2 breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Molecular Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in historically-normal breast cancer. Breast Cancer Res Treat. 2009. [DOI] [PMC free article] [PubMed]
- 17.Ni IBP, Zakaria Z, Muhammad R, et al. Gene expression patterns distinguish breast carcinomas from normal breast tissues: The Malaysia context. Pathology-Res Pract. 2010;206:223–8. doi: 10.1016/j.prp.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Microarray Analysis Suite 5.0, Affymetrix Inc. Santa Clara, California.
- 19.Fisher RA. The distribution of the partial correlation coefficient. Metron. 1924;3(3–4):329–32. [Google Scholar]
- 20.Peng G, Luo L, Siu H, et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Europ J Hum Genet. 2010;18:111–7. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Bucan M. Pathway-based approaches to analysis of genome-wide association studies. Am J Hum Genet. 2007;81:1278–83. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Yosef Hochberg. Controlling the false discovery rat: a practical and powerful approach to multiple testing. J Royal Stat Society. Series B Methodology. 1995;57(1):289–300. [Google Scholar]
- 23.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P. GenePattern 2.0. Nature Genetics. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 24.Breitkreutz B, Stark C, Tyers M. Osprey: a network visualization system. BMC Genome Biology. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in Humans. PLoS Genetics. 2005;1(6):e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colaluca IN, Tosoni D, Nuciforo P, et al. Di Fiore PP. NUMB controls p53 tumor suppressor activity. Nature. 2008 Jan 3;451(7174):76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 27.Casanova JE. PARtitioning numb. EMBO Rep. 2007 Mar;8(3):233–5. doi: 10.1038/sj.embor.7400928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Nat Acad Sci U S A. 2009. [DOI] [PMC free article] [PubMed]
- 29.Eleftherohorinou H, Wright V, Hoggart C, et al. Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One. 2009;4(11):e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Askland K, Read C, Moore J. Pathway based analysis of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 31.Baranzini SE, Galwey NW, Wang J, et al. Pathway and network analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18(11):2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haiman CA, Hsu C, de Bakker PIW, et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17(6):825–34. doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 33.Menashe I, Maeder D, Garcia-Closas M, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;7(11):4453–9. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorlov IP, Gallick GE, Gorlova OY, et al. GWS meets microarray: Are the results of genome-wide association studies and gene expression profiling consistent? Prostate cancer as an example. PLoS One. 2009;4(8):e6511. doi: 10.1371/journal.pone.0006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–78. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts SA, Strande N, Burkhalter MD, et al. Ku is a 5’-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010 Apr 22;464(7292):1214–7. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comai L, Li B. The Werner syndrome protein at the crossroads of DNA repair and apoptosis. Mech Aging Dev. 2004 Aug;125(8):521–8. doi: 10.1016/j.mad.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Cheng WH, Muftic D, Muftuoglu M, et al. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell. 2008 Sep;19(9):3923–33. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010 Jan 15;16(2):376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevanlinna H, Bartek J. The CHEK2 gene and inherited breast cancer susceptibility. Oncogene. 2006 Sep 25;25(43):5912–9. doi: 10.1038/sj.onc.1209877. [DOI] [PubMed] [Google Scholar]
- 41.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007 Oct;8(10):735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 42.Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993 Dec 17;75(6):1215–25. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 43.Fishel R, Wilson T. MutS homologs in mammalian cells. Curr Opin Genet Dev. 1997 Feb;7(1):105–13. doi: 10.1016/s0959-437x(97)80117-7. [DOI] [PubMed] [Google Scholar]
- 44.Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R’s of radiobiology revisited. Stem Cells. 2010 Apr;28(4):639–48. doi: 10.1002/stem.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3522–7. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–85. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 47.Scharpfenecker M, Kruse JJ, Sprong D, Russell NS, Ten Dijke P, Stewart FA. Ionizing radiation shifts the PAI-1/ID-1 balance and activates notch signaling in endothelial cells. Int J Radiat Oncol Biol Phys. 2009 Feb 1;73(2):506–13. doi: 10.1016/j.ijrobp.2008.09.052. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010 Jan;28(1):17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel S, Doble B, Woodgett JR. Glycogen synthase kinase-3 in insulin and Wnt signaling: a double-edged sword. Biochem Soc Trans. 2004 Nov;32(Pt 5):803–8. doi: 10.1042/BST0320803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009 Jan 18;273(2):194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruel L, Bourouis M, Heitzler P, Pantesco V, Simpson P. Drosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch. Nature. 1993 Apr 8;362(6420):557–60. doi: 10.1038/362557a0. [DOI] [PubMed] [Google Scholar]
- 52.Song LL, Peng Y, Yun J, et al. Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene. 2008 Oct 2;27(44):5833–44. doi: 10.1038/onc.2008.190. [DOI] [PubMed] [Google Scholar]
- 53.Hao L, Rizzo P, Osipo C, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010 Jan 14;29(2):201–13. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilimas T, Mascarenhas J, Palomero T, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007 Jan;13(1):70–7. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 55.Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002 Sep;8(9):979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 56.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006 Aug 1;20(15):2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Gall M, De Mattei C, Giniger E. Molecular separation of two signaling pathways for the receptor, Notch. Dev Biol. 2008 Jan 15;313(2):556–67. doi: 10.1016/j.ydbio.2007.10.030. Epub 2007 Dec 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu WH, Hsiao HW, Tsou WI, Lai MZ. Notch inhibits apoptosis by direct interference with XIAP ubiquitination and degradation. EMBO J. 2007 Mar 21;26(6):1660–9. doi: 10.1038/sj.emboj.7601611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007 Oct;13(10):1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Graziani I, Eliasz S, De Marco MA, et al. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008 Dec 1;68(23):9678–85. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]
- 61.Kao HY, Ordentlich P, Koyano-Nakagawa N, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998 Aug 1;12(15):2269–77. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osipo C, Patel P, Rizzo P, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008 Aug 28;27(37):5019–32. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- 63.Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005 Aug 15;65(16):7159–68. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 64.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006 Dec;6(8):905–18. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 65.Reedijk M, Odorcic S, Chang L, et al. High-level co-expression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005 Sep 15;65(18):8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 66.Dickson BC, Mulligan AM, Zhang H, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007 Jun;20(6):685–93. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 67.Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004 Oct 25;167(2):215–21. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumor clinicopathological parameters in human breast cancer. Int J Mol Med. 2004 Nov;14(5):779–86. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix