Abstract
The chemoorganotrophic, aerobic, facultatively anaerobic, non-motile strain MWH-C5T isolated from the water column of oligo-mesotrophic Lake Mondsee (Austria) was phenotypically, phylogenetically, and chemotaxonomically characterized. The predominant fatty acids of the strain were C16:1ω7c/ω6c, C16:0, C12:1, and C8:0-3OH, the major quinone was ubiquinone Q-8, and the G+C content of the DNA of the strain was 55.5 mol%. The 16S rRNA gene similarity to the closest related type species were 96.6% (Curvibacter delicatus) and 95.7% (Rhodoferax fermentans). The phylogenetic analysis of the 16S rRNA gene sequences revealed the affiliation of the strain with the family Comamonadaceae (Betaproteobacteria), however, the revealed phylogenetic position of the strain did not indicate the affiliation to any previously described genus within this family. A family-wide comparison of traits revealed that the strain possesses a unique combination of G+C value, major fatty acids, and major 3-hydroxy fatty acid. Furthermore, the strain differs in several traits from the closest related genera. Based on the phylogeny of the strain and the differences to the closest related genera, we propose to establish the genus Limnohabitans gen. nov. to accommodate this strain, and to place the strain in the new species Limnohabitans curvus sp. nov., with the type strain MWH-C5T (DSM 21645T, = CCUG 56720 T). The type strain is closely related to a large number of uncultured bacteria detected by cultivation-independent methods in various freshwater systems.
The bacterioplankton of freshwater habitats is mainly composed of phylogenetic groups absent in marine bacterioplankton and in terrestrial habitats (Zwart et al., 2002; Hahn, 2006). Investigations with cultivation-independent methods demonstrated that the majority of taxa dwelling in the water column of freshwater lakes and ponds represent uncultured and undescribed taxa (Crump et al., 1999; Zwart et al., 2002; Eiler & Bertilson, 2004). In this paper, we characterize a strain isolated from the pelagic zone of a freshwater lake, which is closely related to uncultured bacteria numerously detected in freshwater samples, and propose to establish for this strain the novel genus Limnohabitans sp. nov. and the novel species Limnohabitans curvus sp. nov. within the family Comamonadaceae (Betaproteobacteria).
Strain MWH-C5T was isolated from deep, oligo-mesotrophic Lake Mondsee (47°50′2.92″N; 13°22′25.98″E) located in Austria. The strain was obtained by using the dilution-acclimatization method (DAM) and Nutrient broth soytone yeast extract (NSY) medium (Hahn et al., 2004; Hahn et al., 2005). The isolated bacterium grows on a variety of solidified complex media, e.g. Luria-Bertani agar (Difco BD), casitone agar (Difco BD), R2A agar (Remel), and NSY agar (Hahn et al., 2004), forming unpigmented, smooth, convex colonies.
Tests on growth of the strain on single carbon sources resulted only in weak growth. In order to avoid false negative results in such tests caused by too weak growth response on single substrate sources, tests on substrate utilization were performed in the presence of complex substrate mixtures as described previously (Hahn et al., in press). Briefly, growth enabled by utilization of specific substrates was determined by comparison of OD (575 nm) established in liquid one-tenth-strength NSY medium (0.3 gL−1) with and without 0.5 g L−1 test substance. OD differences of <10%, of 10-50%, and of >50% of the OD established on the medium without test substance were scored after 10 days of growth as no utilization (−), weak utilization (w) and good utilization (+), respectively. The analysis of the phylogenetic positions of the strain was performed by means of 16S rRNA gene sequence analysis. Sequences were obtained, aligned and analyzed by the neighbour-joining approach as described previously (Hahn et al., in press). In addition, trees were constructed by using the maximum-likelihood and the Bayesian inference tree-building methods. For the maximum-likelihood tree calculation and bootstrap analysis we used the genetic algorithm GARLI (Zwickl, 2006) and a grid computing system (Cummings & Huskamp, 2005). A bayesian inference tree was calculated by using the program Mr Bayes (Ronquist & Huelsenbeck, 2003) performing one millions generations and subsequently explored by the AWTY system (Nylander et al., 2008). The determination of the G+C content of DNA and the analyses of major respiratory lipoquinone analyses were both carried out by the Identification Service and Dr.B.J.Tindall, DSMZ, Braunschweig, Germany. Fatty acid profiles of strain MWH-C5T, as well as of the type strains of Curvibacter gracilis and Rhodoferax fermentans were characterized by using the MIS Sherlock automatic identification System (MIDI, Inc., Newark, DE) and the Sherlock Aerobic Bacterial Database (TSBA60) as described by Greenblatt et al. (1999). For each strain, biomass of replicated cultures obtained by growing the strains in NSY (3 g l−1) for 2 days at 21 °C was analyzed.
The results of the phenotypic and chemotaxonomic investigations are presented in Tables 1 and 2. Strain MWH-C5T is an aerobic, chemoorganotrophic, facultatively anaerobic, non-motile bacterium, and possess a cell morphology of small curved rods (Fig. 1). The predominant (> 5% of total) fatty acids of the strain were C16:1ω7c/ω6c and C16:0, the sole detected 3-hydroxy fatty acid was C8:0-3OH, the major quinone was ubiquinone Q-8, and the G+C content of the DNA of the strain was 55.5 mol%.
Table 1.
Phenotypic traits of Limnohabitans curvus sp. nov. strain MWH-C5T, and the type strains of the type species of the genera Curvibacter and Rhodoferax (C. gracilis strain CCUG 49445T and R. fermentans strains CCUG 45364T). The substrate utilization tests were performed for all three strains under the same conditions. All three strains assimilated acetate, pyruvate, fumarate, succinate, citrate, gluconate, and glucose, none of the four strains assimilated L-carnitine. All three strains are oxidase and catalase positive. −, negative; +, positive; w, weakly positive; n.d., not determined.
| MWH-C5T | C. gracilis | R. fermentans | |
|---|---|---|---|
| Cell morphology | curved rods | curved rods | curved rods |
| Cell length (μm) | 1.0 -1.5 | 1.1 - 2.8$ | 1.5 - 3.0$$$ |
| Cell width (μm) | 0.4 - 0.5 | 0.4 - 0.5$ | 0.6 - 0.9$$$ |
| Min temp. of growth (°C) | 4 | 9§ | n.d.$$ |
| Max temp. of growth (°C) | 34 | 40§ | n.d.$$ |
| Max NaCl concentration (% NaCl) | 0.5 | < 3% $ | n.d.$$ |
| Anaerobic growth | + | n.d.$ | + $$ |
| Ethanol | w | − | w |
| Glycerol | − | + | + |
| Glyoxylate | − | w | − |
| Gycolate | − | + | − |
| D-Glycerate | + | + | w |
| Oxalate | − | − | w |
| DL-Lactate | − | + | − |
| Propionate | w | + | w |
| DL-Malate | + | + | w |
| Malonate | − | w | − |
| alpha-Ketoglutarate | + | + | w |
| Oxaloacetate | − | + | w |
| L-Arginine | − | − | + |
| L-Glutamate | − | + | + |
| L-Glutamine | − | + | w |
| L-Histidine | − | + | − |
| L-Phenylalanine | − | + | − |
| L-Proline | − | + | + |
| L-Serine | − | + | + |
| L-Tryptophan | − | + | w |
| D-Ribose | w | w | + |
| L-Sorbose | − | w | − |
| D-Galactose | w | w | + |
| D-Mannose | + | w | + |
| D-Saccharose | w | + | w |
| N-acetyl-glucosamine | − | w | + |
| Betaine | − | − | w |
| Spermidine | − | − | w |
| Quinone type | Q8 | Q8$ | Q8 + RQ8 $$ |
| DNA G+C content (mol%) | 55.5 | 66.0$ | 60 $$ |
data from Ding and Yokota (2004)
data from Hiraishi et al. (1991)
data presented by Ding and Yokota (2004) in the description of the genus Curvibacter
Table 2.
Whole cell fatty acid composition of Limnohabitans curvus sp. nov. strain MWH-C5T and the type strains of the type species of Curvibacter and Rhodoferax (C. gracilis strain CCUG 49445T and R. fermentans strains CCUG 45364T). All strains were cultivated under identical conditions (NSY medium (3 g l−1) at 21 °C for 2 days). The presented data were obtained from replicated cultures. nd, not detected.
| Fatty acid | MWH-C5T | R. fermentans | C. gracilis |
|---|---|---|---|
| C8:0-3OH | 2.7 | 3.9 | 5.1 |
| C9:0-3OH | nd | 0.1 | nd |
| C10:0-3OH | nd | nd | 0.2 |
| C12:0 | 4.5 | nd | 5.6 |
| C12:0-3OH | nd | nd | nd |
| C14:0 | 1.0 | 0.2 | 1.2 |
| C14:1ω5c | 0.4 | nd | 0.3 |
| C15:0 | nd | 2.0 | nd |
| C15:1ω6c | nd | 0.1 | 0.1 |
| C15:1ω8c | nd | 0.2 | nd |
| C16:0 | 14.0 | 46.3 | 15.2 |
| C16:1ω5c | 0.2 | 0.3 | 0.8 |
| C16:1ω7c/ω6c | 76.7 | 44.1 | 49.2 |
| C17:0 | nd | 0.4 | nd |
| C17:0 cyclo | nd | nd | nd |
| C17:1ω6c | nd | 0.6 | 0.6 |
| C17:1ω8c | nd | 0.2 | nd |
| C18:0 | 0.3 | nd | nd |
| C18:1ω7c 11Me | 0.3 | 0.2 | 0.7 |
| C18:1ω7c/ω6c | 1.8 | 1.3 | 20.7 |
| C18:1ω9c | 0.2 | nd | nd |
| C19:1ω6c/C19:0 cyclo | nd | 0.1 | nd |
Fig. 1.
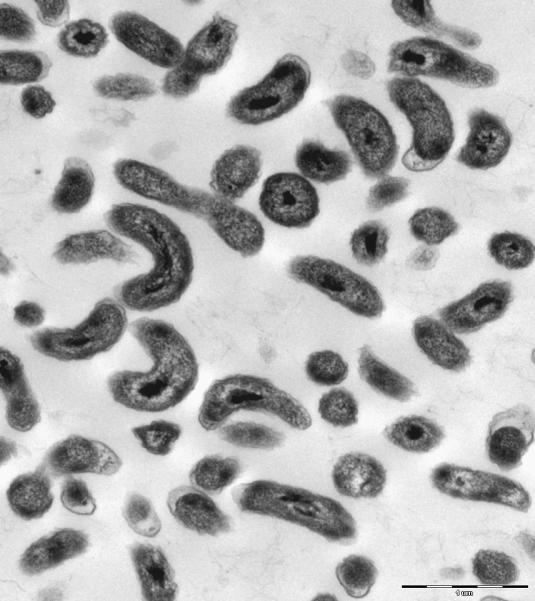
Electron microscopy image illustrating the cell morphology and size of the strain MWH-C5T. Cells of a liquid culture were concentrated by centrifugation and fixed with 2.5% glutaraldehyde, post-fixed with OsO4 and embedded with Spurr resin. Ultra-thin sections were counterstained with uranyl acetate and lead citrate. The image was obtained by transmission electron microscopy at a 20000-fold magnification.
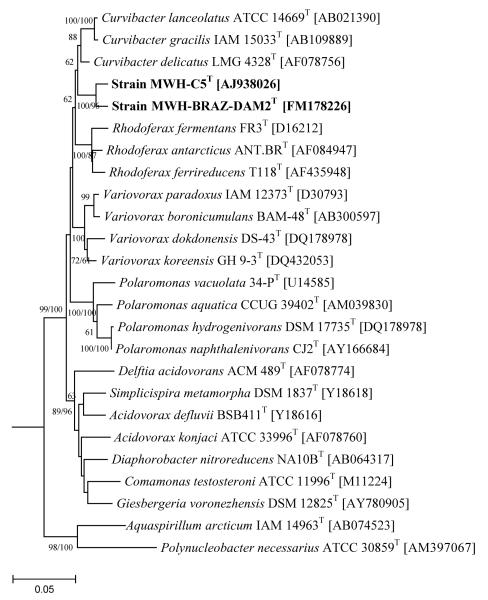
Phylogenetic analyses with all three tree-building methods revealed consistently the affiliation of strain MWH-C5T with the family Comamonadaceae (Fig. 2). The sequence of the strain did not cluster within any previously described genus in any tree generated with the three methods. The phylogenetic relationships reconstructed by using the three algorithms indicated in all three cases a close relationship of the strain with the genera Rhodoferax, Pseudorhodoferax, Curvibacter Polaromonas, Variovorax, Ramlibacter, and Caenimonas. Regarding the phylogenetic relationships of the strain MWH-C5T to these seven genera, the topologies and branching orders of the three trees was almost identical (data not shown). The trees only differed in the position of the type species of the genus Caenimonas, which clustered in the maximum-likelihood (ML) and bayesian inference (BI) trees with the genus Ramlibacter, while it clustered in the neighbour-joining (NJ) tree with the genus Curvibacter. Pair-wise sequence similarity values of 16S rRNA genes of strain MWH-C5T and type strains of these seven most closely related genera were in the range of 94.4% - 96.2% (average 95.3), which is very similar to the range of 93.0% – 96.5% (average 95.4) observed for pair-wise comparisons among the type species of the seven genera (Supplementary Table S1).
Fig. 2.
Neighbour-joining (NJ) tree (Kimura-2 correction) based on almost complete 16S rRNA gene sequences. The tree presents sequences of type species of almost all genera currently affiliated with the family Comamonadaceae. Only Caenibacterium thermophilum, which is proposed by Lütke-Eversloh et al. (2004) to be a later heterotypic synonym of Schlegelella thermodepolymerans (the type species of the genus Schlegelella) is not shown. The genera Polynucleobacter, Duganella, and, Herbaspirillum represent reference taxa not affiliated with the family Comamonadaceae. All Bootstrap values ≥ 60% (1000 iterations) are depicted. The tree was rooted by using archeal sequences (not shown). Bar, 5 nucleotide substitution per 100 nucleotides.
A family-wide comparison of traits of strain MWH-C5T and all type species of the 30 genera (including Pseudorhodoferax described by Bruland et al., in press) currently belonging to the family Comamonadaceae was performed. This comparative analysis was limited by the heterogeneity of data sets available for the different type species, because only very few traits were described for the overwhelming majority of strains and virtually no test on substrate utilization was performed for almost all strains. However, comparisons of chemotaxonomic traits, i.e. type of quinone, major fatty acids, major 3-hydroxy fatty acid, and G+C value, revealed a unique combination of these traits for strain MWH-C5T among all Comamonadaceae genera (Table S2). Strain MWH-C5T differs from all but two genera by its G+C content of DNA of 55.5 mol%, which is > 3 mol% lower in MWH-C5T than in other type strains. Among Comamonadaceae type strains only those of Gisbergeria species (Grabovich et al., 2006) and the type strain of the type species of the genus Polaromonas, i.e. P. vacuolata (Irgens et al., 1996), posses G+C values < 59 mol%. Interestingly, Polaromonas vacuolata differs significantly in this trait from the type strains of three other Polaromonas species, which have G+C content of 61.5 to 63.7 mol% (Weon et al., 2008). The G+C value of the fifth described Polaromonas species has not been determined (Kämpfer et al., 2006). Furthermore, the lack of C10:0-3OH is rare among Comamonadaceae bacteria, and all genera sharing this trait with strain MWH-C5T possess G+C values > 59 mol%.
A more detailed comparison with the seven most closely related genera (Fig. 2) revealed several differences to type species of these genera suitable for discrimination of the new taxon from these previously described taxa (Table 3). The G+C value is again an important discriminative trait, however the strain also differes in at least one more trait from all other type species. For instance by the absence of C10:0-3OH fatty acids from Caenimonas, Pseudorhodoferax, and Variovorax, by its ability to grow facultatively anaerobic from strictly aerobic Polaromonas, by a much lower salinity tolerance from Curvibacter type strains, by the ability to assimilate glucose and citrate from Ramlibacter, and by the ability to utilize alpha-ketoglutarate as a carbon source and the lack of rhodoquinone from Rhodoferax type strains.
Table 3.
Phenotypic and chemotaxonomic characteristics that differentiate Limnohabitans curvus gen. nov., sp. nov. strain MWH-C5T from the closest related genera as determined by the phylogenetic reconstruction presented in Fig. 2. Data for Caenimonas are from Ryu et al., (2008); for Curvibacter spp. are from Ding & Yokota (2004) and Tables 1 and 2; for Polaromonas spp. from Kämpfer et al. (2006), Sizova and Panikov (2007), Jeon et al. (2004), and Irgens et al. (1996); for Pseudorhodoferax are from Bruland et al., (in press); for Rhodoferax spp. from Madigan et al. (2000), Hiraishi et al. (1991), and Finneran et al. (2003), and from Tables 1 and 2; for Variovorax spp. from Yoon et al. (2006), Kim et al. (2006), and Miwa et al. (2008); for Ramlibacter from Heulin et al., (2003) and Ryu et al., (2008). Q, Quinone; RQ, rhodoquinone; +, positive; −, negative; NA, not available; d, variable; w, weak.
| Character | MWH-C5T | Caenimonas | Curvibacter | Polaromonas | Pseudorhodoferax | Ramlibacter | Rhodoferax | Variovorax |
|---|---|---|---|---|---|---|---|---|
| Cell morphology | Curved rods | Rods | Slightly curved rods | Rods | Short rods | Pleomorphic (rods to cocci(cysts)) |
Curved rods | Oval or rod shaped |
| Flagella | None | None | None or polar | None or single, polar |
Single, polar | None | Single, polar | Peritrichous |
| Psychrophilic growth | + | − | − | d | NA | − | − | − |
| Oxygen requirement | Facultatively anaerobic | Strictly aerobic | Aerobic or microaerophilic |
Obligately aerobic | Aerobic | Aerobic | Facultatively aerobic |
Strictly aerobic |
| Maximum salinity (%) | 0.5 | NA | 3 | 6 | NA | NA | 1 | 7 |
| Carbon source used for growth: |
||||||||
| alpha-Ketoglutarate | + | NA | + | + | NA | NA | − | NA |
| Citrate | + | − | + | + | d | − | + | NA |
| Gluconate (D−) | + | − | d | d | + | − | d | + |
| Glucose (D−) | + | − | d | d | − | − | d | + |
| Glutamate | − | NA | + | + | NA | NA | d | NA |
| Glutamine | − | NA | + | NA | NA | NA | − | NA |
| Glycerol | − | − | + | d | NA | NA | − | + |
| Glycolate | − | NA | + | NA | NA | NA | − | NA |
| Histidine (L−) | − | NA | + | − | NA | NA | − | NA |
| Lactate (DL−) | − | NA | + | d | NA | + | + | NA |
| Malate (DL−) | + | − | + | d | + | − | + | NA |
| Malonate | − | + | w | d | NA | NA | − | d |
| Mannose (D−) | + | − | d | − | − | − | d | + |
| N-acetyl-glucosamine | − | − | w | NA | NA | − | + | NA |
| Oxaloacetate | − | NA | + | d | NA | NA | w | NA |
| Phenylalanine (L−) | − | NA | + | NA | NA | NA | − | NA |
| Proline (L−) | − | NA | + | + | NA | NA | + | NA |
| Pyruvate | + | NA | + | + | NA | + | + | NA |
| Serine (L−) | − | NA | + | d | NA | NA | + | NA |
| Tryptophan (L−) | − | − | + | − | NA | NA | w | d |
| Quinones | Q-8 | Q-8 | Q-8 | Q-8, Q-9 | NA | NA | Q-8, RQ-8 | Q-8 |
| Major fatty acids | 16:0, 16:1 | 16:0,18:1, feat. 3 | 16:0, 16:1, 18:1 | 16:0, 16:1, 18:1 | 16:0,18:1, feat. 3 | NA | 16:0, 16:1 | 16:0, 16:1, 18:1 |
| Major 3-OH acid | 8:0 | 10:0 | 8:0 | 8:0, 10:0$ | 10:0 | NA | 8:0 | 10:0 |
| DNA G+C content (mol%) |
55.5 | 63 | 62 - 66 | 52 - 63$$ | 69 - 70 | 67 - 70 | 59 - 62 | 66 – 69 |
| Habitats | Freshwater | Activated sludge | Wellwater | Freshwater, marine | Activated sludge or soil |
Soil | Freshwater, activated sludge, marine |
Soil |
Only P. hydrogenivorans DSM 17735T and P. naphthalenivorans CJ2T contain C10:03-OH as the major hydroxylated fatty acid (Jeon et al., 2004; Sizova & Panikov, 2007)
Only the typestrain of Polaromonas vacuolata has a G+C vulue of 52 mol%, while the other type strains of Polaromonas species have G+C vulues of 62-64 mol%
BLAST searches with the 16S rRNA gene sequences of strain MWH-C5T and subsequent phylogenetic analyses revealed that strain MWH-C5T is closely related to a large number of uncultured strains (Supplemental Material Fig. S1) detected in a large number of cultivation-independent investigations on the diversity of freshwater bacteria (e.g., Zwart et al., 2002; Crump & Hobbie, 2005; Shaw et al., 2008). Some of these uncultured bacteria share 16S rRNA sequence similarities with strain MWH-C5T of 99.9%. The phylogenetic cluster formed by these uncultured organisms was previously designated the “Rhodoferax sp. BAL47” cluster (Zwart et al., 2002). The high number of sequences affiliated with this cluster, as well as their geographically widespread origin indicates a significant contribution of bacteria affiliated with this cluster to bacterioplankton in many freshwater lakes and ponds. This is also indicated by an investigation of 15 diverse lakes in northern Europe for the presence of bacteria affiliated with the “Rhodoferax sp. BAL47”cluster or 14 other clusters, which also contain taxa frequently detected in freshwater habitats, by a cultivation-independent method (Lindström et al., 2005; Zwart et al., 2003). The “Rhodoferax sp. BAL47”cluster and an actinobacterial cluster were the only clusters detected in all 15 investigated habitats. A prominent and well-investigated subgroup of the “Rhodoferax sp. BAL47” cluster is the so-called R-BT065 group (Šimek et al., 2001; Šimek et al., 2005), which can be detected by a specific FISH probe. Investigations of several freshwater ponds and lakes by using this FISH probe revealed that the targeted cells possess a planktonic lifestyle, and that this taxon comprises typically 5-30 % (maximum ~ 50 %) of total bacterioplankton cells in non-acidic stagnant freshwater habitats (Šimek et al., 2001; Šimek et al., 2005; Šimek et al., unpubl. data). The origin of strain MWH-C5T from the pelagic zone of a freshwater lake, as well as the close phylogenetic relationship with strains inhabiting the water column of freshwater systems indicates that this strain shares a planktonic lifestyle with R-BT065 bacteria and other members of the “Rhodoferax sp. BAL47” cluster.
Based on the reconstructed phylogenetic position of strain MWH-C5T (Fig. 2), the phylogenetic distances to type species of closest related genera (Table S1), and the phenotypic and chemotaxonomic differences between the strain and type species of related genera (Table 3 and Table S2) we propose to establish the new genus Limnohabitans gen. nov., and the new species Limnohabitans curvus sp. nov. with strain MWH-C5T as type species and type strain of the new genus and the new species, respectively.
Description of Limnohabitans gen. nov.
Limnohabitans (Lim.no.ha.bi’tans. Gr. n. limne, lake; L. part. adj. habitans, inhabiting; N.L. part. adj. used as a masc. n. Limnohabitans, lake dweller, referring to the type of ecosystem inhabited by these bacteria).
Aerobic, facultatively anaerobic, chemo-organotrophic, oxidase and catalase positive bacteria. Cells are non-pigmented, non-motile curved rods. Not halotolerant, do not grow at NaCl concentrations > 0.5%. Mesophilic. Major fatty acids (constituting >5 % of total fatty acids) are C16:0 and C16:1ω7c/ω6c. The major quinone is ubiquinone Q-8. Isolated from the water column of freshwater habitats. The genus is affiliated to the class Betaproteobacteria and to the family Comamonadaceae. The type species is Limnohabitans curvus.
Description of Limnohabitans curvus sp. nov.
Limnohabitans curvus (cur.vus. L. masc. adj. curvus curved or crooked).
Cell morphology of curved rods, 1.0–1.5 μm in length and 0.4–0.5 μm in width. Chemoorganotroph, aerobic, facultatively anaerobic, oxidase and catalase positive. Colonies grown on NSY agar are unpigmented, circular and convex with smooth surface. Growth occurs at 4–34 °C, and with 0–0.5 % (w/v) NaCl. Assimilates acetate, glycerate, alpha-ketoglutarate, pyruvate, fumarate, citrate, malate, succinate, gluconate, glucose, and mannose. Week assimilation was observed for several substances (Table 1). No assimilation of glycerol, glyoxylate, glycolate, oxalate, lactate, malonate, oxaloacetate, arginine, glutamate, glutamine, histidine, phenylalanine, proline, serine, tryptophan, sorbose, N-acetyl-glucosamine, betaine, spermidine, and carnitine. Major cellular fatty acids (> 5% of total) are C16:0, and C16:1ω7c/ω6c. The major quinone is ubiquinone Q-8, and the G+C value of the DNA is 55.5%. The type strain is MWH-C5T (DSM 21645T, = CCUG 56720 T) isolated from Lake Mondsee, Austria.
Supplementary Material
Acknowledgements
D. Elhottová and J. Petrásek are acknowledged for determination of fatty acids profiles supported by the project ASCR - ISB No. AV0Z 60660521, and P. Masarova and L. Bucinska are acknowledged for performing electron microscopy analyses. P. Schumann is acknowledged for determination of G+C values, and B. Tindall is acknowledged for the analyses of quinones. This study was supported by the Czech-Austrian KONTAKT project MEB 060702 / CZ 05/2007 (granted to KS and MWH), by the Austrian Science Fund (FWF) project P19853 (granted to MWH), by the Grant Agency of the Czech Republic under research grant 206/08/0015 (granted to KS), and by the institutional project of the ASCR No. AV0Z 60170517. The authors also benefited from participation in ALTERnet (A Long-Term Biodiversity, Ecosystem and Awareness Research Network), an EU Network of Excellence (GOCE-CT-2003-505298).
Abbreviations
- OD
optical density
- FISH
fluorescence in situ hybridization
- NJ
neighbour-joining
- ML
maximum-likelihood
- BI
bayesian inference
Footnotes
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of Limnohabitans curvus gen. nov., sp. nov. strain MWH-C5T is AJ938026.
References
- Bruland N, Bathe S, Willems A, Steinbüchel A. Pseudorhodoferax soli gen. nov., sp. nov., and Pseudorhodoferax caeni sp. nov., two novel Betaproteobacteria belonging to the Comamonadaceae family. Int J Syst Evol Microbiol. doi: 10.1099/ijs.0.006791-0. (in press) [DOI] [PubMed] [Google Scholar]
- Crump BC, Hobbie JE. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MP, Huskamp JC. Grid computing. Educause Review. 2005;40:116–117. [Google Scholar]
- Ding L, Yokota A. Proposals of Curvibacter gracilis gen. nov., sp. nov. and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of [Pseudomonas] huttiensis, [Pseudomonas] lanceolata, [Aquaspirillum] delicatum and [Aquaspirillum] autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov. and Herbaspirillum autotrophicum comb. nov. Int J Syst Evol Microbiol. 2004;54:2223–2230. doi: 10.1099/ijs.0.02975-0. [DOI] [PubMed] [Google Scholar]
- Eiler A, Bertilsson S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol. 2004;6:1228–1243. doi: 10.1111/j.1462-2920.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- Finneran KT, Johnsen CV, Lovley DR. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III) Int J Syst Evol Microbiol. 2003;53:669–673. doi: 10.1099/ijs.0.02298-0. [DOI] [PubMed] [Google Scholar]
- França L, Rainey FA, Nobre MF, da Costa MS. Tepidicella xavieri gen. nov., sp. nov., a betaproteobacterium isolated from a hot spring runoff. Int J Syst Evol Microbiol. 2006;56:907–912. doi: 10.1099/ijs.0.64193-0. [DOI] [PubMed] [Google Scholar]
- Grabovich M, Gavrish E, Kuever J, Lysenko AM, Podkopaeva D, Dubinina G. Proposal of Giesbergeria voronezhensis gen. nov., sp. nov. and G. kuznetsovii sp. nov. and reclassification of [Aquaspirillum] anulus, [A.] sinuosum and [A.] giesbergeri as Giesbergeria anulus comb. nov., G. sinuosa comb. nov. and G. giesbergeri comb. nov., and [Aquaspirillum] metamorphum and [A.] psychrophilum as Simplicispira metamorpha gen. nov., comb. nov. and S. psychrophila comb. nov. Int. J. Syst. Evol. Microbiol. 2006;56:569–576. doi: 10.1099/ijs.0.64027-0. 2006. [DOI] [PubMed] [Google Scholar]
- Greenblatt CL, Davis A, Clement BG, Kitts CL, Cox T, Cano RJ. Diversity of microorganisms isolated from amber. Microbiol Ecol. 1999;38:58–68. doi: 10.1007/s002489900153. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Pöckl M, Wu QL. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol. 2005;71:4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T. Emended description of the genus Polynucleobacter and the species P. necessarius and proposal of two subspecies, P. necessarius subspecies necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int J Syst Evol Microbiol. doi: 10.1099/ijs.0.005801-0. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pöckl M. The filtration-acclimatization-method for isolation of an important fraction of the not readily cultivable bacteria. J Microb Meth. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hahn MW. The microbial diversity of inland waters. Curr Opin Biotechnol. 2006;17:256–261. doi: 10.1016/j.copbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Heulin T, Barakat M, Christen R, Lesourd M, Sutra L, De Luca G, Achouak W. Ramlibacter tataouinensis gen. nov., sp. nov., and Ramlibacter henchirensis sp. nov., cyst-producing bacteria isolated from subdesert soil in Tunisia. Int J Syst Evol Microbiol. 2003;53:589–594. doi: 10.1099/ijs.0.02482-0. [DOI] [PubMed] [Google Scholar]
- Hiraishi A, Hoshino Y, Satoh T. Rhodoferax fermentans gen. nov., sp. nov., a phototrophic purple nonsulfur bacterium previously referred to as the “Rhodocyclus gelatinosus-like“ group. Arch Microbiol. 1991;155:330–336. [Google Scholar]
- Irgens RL, Gosink JJ, Staley JT. Polaromonas vacuolata gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int J Syst Bacteriol. 1996;46:822–826. doi: 10.1099/00207713-46-3-822. [DOI] [PubMed] [Google Scholar]
- Jeon CO, Park W, Ghiorse WC, Madsen EL. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int J Syst Evol Microbiol. 2004;54:93–97. doi: 10.1099/ijs.0.02636-0. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, Busse H-J, Falsen E. Polaromonas aquatica sp. nov., isolated from tap water. Int J Syst Evol Microbiol. 2006;56:605–608. doi: 10.1099/ijs.0.63963-0. [DOI] [PubMed] [Google Scholar]
- Kim B-Y, Weon H-Y, Yoo S-H, Lee S-Y, Kwon S-W, Go S-J, Stackebrandt E. Variovorax soli sp. nov., isolated from greenhouse soil. Int J Syst Evol Microbiol. 2006;56:2899–2901. doi: 10.1099/ijs.0.64390-0. [DOI] [PubMed] [Google Scholar]
- Lindström ES, Kamst-Van Agterveld MP, Zwart G. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol. 2005;71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Elbanna K, Cnockaert MC, Mergaert J, Swings J, Manaia CM, Steinbüchel A. Caenibacterium thermophilum is a later synonym of Schlegelella thermodepolymerans. Int J Syst Evol Microbiol. 2004;54:1933–1935. doi: 10.1099/ijs.0.63204-0. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Jung DO, Woese CR, Achenbach LA. Rhodoferax antarcticus sp. nov., a moderately psychrophilic purple nonsulfur bacterium isolated from an Antarctic microbial mat. Arch Microbiol. 2000;173:269–277. doi: 10.1007/s002030000140. [DOI] [PubMed] [Google Scholar]
- Miwa H, Ahmed I, Yoon J, Yokota A, Fujiwara T. Variovorax boronicumulans sp. nov., a boron-accumulating bacterium isolated from soil. Int J Syst Evol Microbiol. 2008;58:286–289. doi: 10.1099/ijs.0.65315-0. [DOI] [PubMed] [Google Scholar]
- Nylander JA, Wilgenbusch J, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ryu SH, Lee DS, Park M, Wang Q, Jang HH, Park W, Jeon CO. Caenimonas koreensis gen. nov., sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol. 2008;58:1064–1068. doi: 10.1099/ijs.0.65416-0. [DOI] [PubMed] [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JB. It’s all relative: ranking the diversity of aquatic bacterial. Environ Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- Šimek K, Horňák K, Jezbera J, Mašín M, Nedoma J, Gasol JM, Schauer M. Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of b-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl Environ Microbiol. 2005;71:2381–2390. doi: 10.1128/AEM.71.5.2381-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Pernthaler J, Weinbauer MG, Horňák K, Dolan JR, Nedoma J, Mašín M, Amann R. Changes in bacterial community composition, dynamics and viral mortality rates associated with enhanced flagellate grazing in a meso-eutrophic reservoir. Appl Environ Microbiol. 2001;67:2723–2733. doi: 10.1128/AEM.67.6.2723-2733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova M, Panikov N. Polaromonas hydrogenivorans sp. nov., a psychrotolerant hydrogen-oxidizing bacterium from Alaskan soil. Int J Syst Evol Microbiol. 2007;57:616–619. doi: 10.1099/ijs.0.64350-0. [DOI] [PubMed] [Google Scholar]
- Weon HY, Yoo SH, Hong SB, Kwon SW, Stackebrandt E, Go SJ, Koo BS. Polaromonas jejuensis sp. nov., isolated from soil in Korea. Int J Syst Evol Microbiol. 2008;58:1525–1528. doi: 10.1099/ijs.0.65529-0. [DOI] [PubMed] [Google Scholar]
- Yoon J-H, Kang S-J, Oh T-K. Variovorax dokdonensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2006;56:811–814. doi: 10.1099/ijs.0.64070-0. [DOI] [PubMed] [Google Scholar]
- Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han SK. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]
- Zwart G, van Hannen EJ, Kamst-van Agterveld MP, Van der Gucht K, Lindström ES, Van Wichelen J, Lauridsen T, Crump BC, Han S-K, Declerck S. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl Environ Microbiol. 2003;69:5875–5883. doi: 10.1128/AEM.69.10.5875-5883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin; 2006. Ph.D. thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.