Abstract
The genetic architecture of the craniofacial complex has been the subject of intense scrutiny because of the high frequency of congenital malformations. Numerous animal models have been used to document the early development of the craniofacial complex, but few studies have focused directly on the genetic underpinnings of normal variation in the human craniofacial complex. The current study examines 80 quantitative traits derived from lateral cephalographs of 981 participants in the Fels Longitudinal Study, Wright State University, Dayton, Ohio. Quantitative genetic analyses were conducted using the SOLAR analytic platform, a maximum-likelihood variance components method that incorporates all familial information for parameter estimation. Heritability estimates were significant and of moderate to high magnitude for all craniofacial traits. Additionally, significant quantitative trait loci (QTL) were identified for 10 traits from the three developmental components (basicranium, splanchnocranium, and neurocranium) of the craniofacial complex. These QTL were found on chromosomes 3, 6, 11, 12, and 14. This study of the genetic architecture of the craniofacial complex elucidates fundamental information of the genetic architecture of the craniofacial complex in humans.
Keywords: QTL, genome-wide linkage analysis, Human, Craniofacial complex
Recent advances in developmental biology have led to the discovery of a number of genes and gene products important in the development of the skull (e.g., Mina et al., 1991; Brown et al., 1993; Mina et al., 1995; Mina, 2001; Abzhanov et al., 2006; Mina and Havens, 2007). However, our primary understanding of the specific genes related to human craniofacial morphology comes from genetic disorders associated with craniofacial anomalies (e.g., Slavkin, 1983; Olsen, 1998; Gorlin, 1998; Slavkin, 2001; Brennan and Pauli, 2001; Riise et al., 2002; Helms and Schneider, 2003; Shieh et al., 2006). Many of these anomalies affect multiple tissue types and often are not restricted to craniofacial structures. Examples include Axenfeld-Rieger syndrome, which is characterized by ocular and dental features, as well as hypospadias, anal stenosis and congenital heart defects (Idrees et al., 2006), or Williams syndrome with numerous neurologic and connective tissue deficits (Amenta et al., 2005).
While the advances made by examining the genetic etiology of orofacial pathogenesis are significant, they do not provide an adequate characterization of the genetic background to normal craniofacial development and morphology. The craniofacial complex is one of the primary foci for application of gene therapy and tissue engineering techniques (Dunn et al., 2005; Yelick and Vacanti, 2006; Hu et al., 2006; Garcia-Godoy and Murray, 2006). As it becomes increasingly possible to incorporate gene therapy and tissue engineering in the repair of craniofacial dysmorphology (congenital or acquired), studies elucidating the genetic underpinnings of continuous phenotypes typifying normal variation are of critical importance (Slavkin, 1983; Slavkin, 1995; Slavkin, 1996; Hu and Helms, 1999; Slavkin, 2001; Lindsey, 2001; Weng et al., 2001; Warren et al., 2003; Fong et al., 2003; MacArthur et al., 2004; MacArthur and Oreffo, 2005; Yelick and Vacanti, 2006). The current study examines fundamental features of the genetic architecture by assessing heritability and identifying QTL underlying normal variation of phenotypes throughout the craniofacial complex in participants from the Fels Longitudinal Study using modern variance components-based statistical genetic methods.
MATERIALS AND METHODS
Data used in the analyses presented here come from normal healthy individuals participating in the Fels Longitudinal Study. Initiated in 1929, the Fels Longitudinal Study is currently the world’s longest running study of human growth and body composition change over the lifespan (Roche, 1992). To date, there have been over 1,200 serial participants in the study, most of whom having participated in the study since birth. In addition, there are some 1,500 relatives from whom mixed cross-sectional and longitudinal data are available. This is a randomly ascertained cohort in that participating families were not selected for any specific feature or trait, and is, therefore, a study of normal variation in such traits. Most Fels Longitudinal Study participants live in or near southwest Ohio and neighboring states. Typically, participant examinations are scheduled at 1, 3, 6, 9, and 12 months of age, and then at 6-month intervals until age 18 years. Thereafter, most participants have biennial examinations.
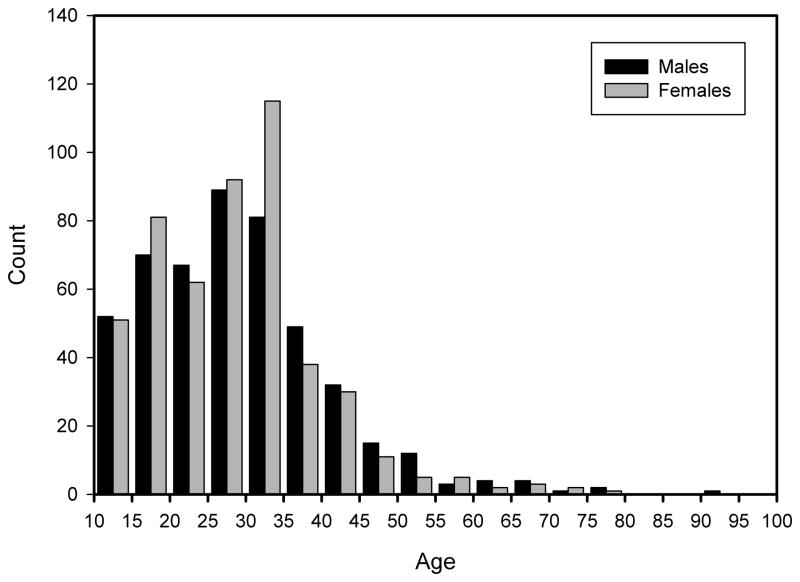
Data for the current study were obtained from lateral cephalographs of 981 participants (482 male, 498 female) ranging in age from 10.0 to 91.9 years (mean = 28.6 years; Figure 1) at the time of examination. Because the majority of participants have serial data, a single cephalograph was chosen for analysis. A minimum age was set at 10 years for the present study to minimize the effects of age.
Figure 1.
Age distribution of the total sample
The 981 individuals with cephalometric data come from 159 nuclear and extended families. In addition to parent-offspring and full sib-pairs, the larger pedigrees contain 16 other relative pair classes from which genetic information can be extracted in our analyses that use all familial information: e.g., avuncular (n pairs = 535), first cousins (n=435), second cousins (n=260), and so on. In all, there are a total of 3,620 relative pairings represented among the 981 participants in this study sample.
Radiography
Cranial radiography of Fels Longitudinal Study participants began in 1931 and ended in 1982. In such a long-term collection of radiographic data, quality and consistency in radiographic technique is particularly important. Consistency in positioning of the participant’s head, as well as the x-ray tube, helps to ensure the consistency of measurements taken from these radiographs. The radiographic protocol changed minimally over the 51 years that cranial radiographs were taken. The primary change was a shift in tube-to-film distance. Initially the tube-to-film distance was 36 inches; this was increased to 60 inches in 1948 and to 72 inches in 1953. Midline plane-to-film distance varied with head size until a standard 6 inches was implemented in 1965. Correction factors for radiographic enlargement have been calculated for all sets of distances allowing comparable measurements to be taken from radiographs of each time period. Most radiographs were taken without the use of a cephalostat but all participants were positioned by highly trained assistants (see Roche, 1992 for a review of research using this collection).
Phenotyping
Phenotyping was done using Nemoceph (CDIimaging), a commercially available software program designed for rapid and accurate collection of cephalometric data. Prior to measurement, each individual’s selected radiograph was first evaluated visually to ensure that correct positioning had occurred. Radiographs that demonstrated any feature that precluded accurate measurement, e.g., excessive rotation of the skull relative to the plate, were not used.
Quality of radiographic image does vary across the history of the study most notably as advances in film and radiographic screens became available. Each radiograph was assessed for overall quality (not only positioning, but also exposure, artifacts, etc.). Again, only those radiographs deemed acceptable by the assessor were included in the study.
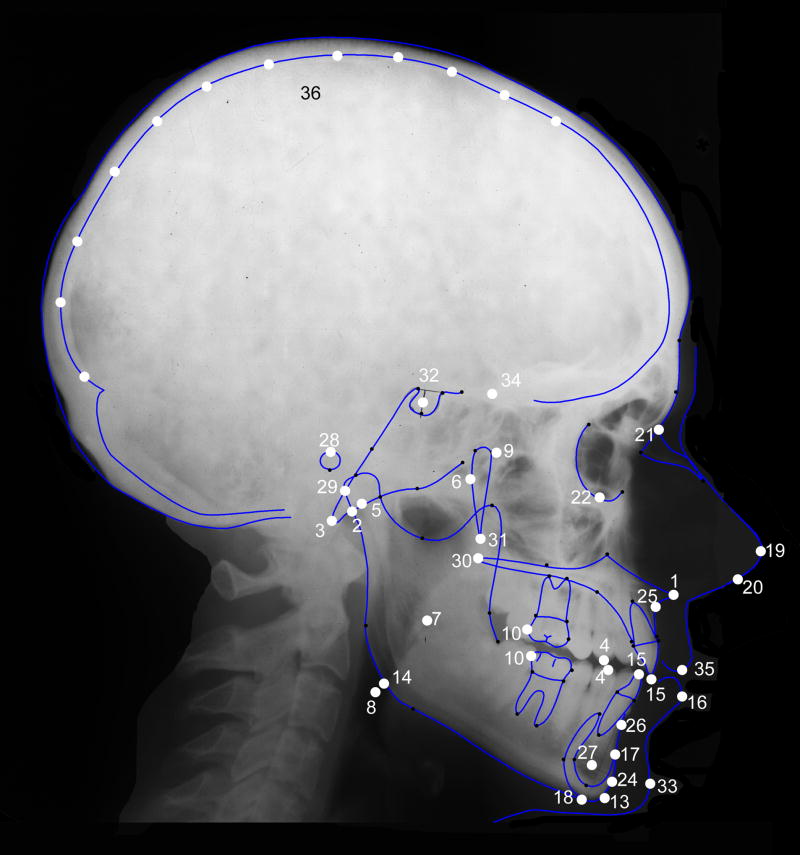
Following evaluation, radiographs were then scanned using an Epson Expression 10000XL scanner equipped with a transparency adapter. Radiographs were scanned directly into Nemoceph, which allows for adjustment of brightness and contrast as necessary, as well as the application of various filters (e.g., false color, inverse image) that assist in identification of cephalometric points. All radiographs were scanned along with a 10 cm ruler used to calibrate the images prior to measurement. Tracing of the radiograph begins with Nemoceph prompting the user to place markers on predefined cephalometric points (Table 1; Figure 2). Once these points have been placed, Nemoceph provides a rough outline of the external and internal aspects of the skull, central incisors, and first molars. These outlines are fit to the cranial contours and teeth by the user with standard computer drawing tools (e.g., handles and anchors) to provide an exact tracing of the craniofacial features. Once tracing is complete, the user has the option of collecting data based on several standard craniofacial analyses, such as a Rickett’s or Steiner’s analysis (Merow and Broadbent, Jr., 1990), or defining a unique set of measures. For our purposes, we have created a dataset that incorporates aspects of standard orthodontic analyses (the aforementioned Rickett’s and Steiner’s analyses) as well as a set of measures that, in our experience, play a frequent role in the comparative anatomical literature. As such, these measures should have a wide relevance across clinical and basic research.
Table 1.
Cephalometric points and planes identified on each lateral cephalograph. All points represent midline structures except where noted.
| Trait | Abbreviation | Description |
|---|---|---|
| 1. Anterior Nasal Spine | ANS | Tip of the anterior nasal spine |
| 2. Articulare | Ar | The intersection of the image of the posterior border of the ramus with the external surface of the basicranium |
| 3. Basion | Ba | The anterior margin of the foramen magnum |
| 4. Canine Tip | A3, B3 | Tip of canine (upper and lower respectively) |
| 5. Center of Condyle | Dc | Intersection point between the condyle axis (Xi-Cd) and the Ba-N plane. |
| 6. Center of Face | CF* | Posterior most point of the pterygomaxillary fissure |
| 7. Center of Mandible | Xi* | Point halfway from the height and depth of the mandibular ramus |
| 8. Cephalometric Gonion | Goc* | Intersection of tangent to the posterior and inferior borders of the mandible |
| 9. Cranial Centris | CC | The intersection of the facial axis and Ba-N plane |
| 10. Distal Molar | A6, B6 | Distal margin of first molar (upper and lower respectively) |
| 11. Facial Plane | FP | Line connecting nasion and pogonion |
| 12. Frankfort Horizontal | FH* | Plane defined by right and left porion and left orbitale |
| 13. Gnathion | Gn | The lowest, most anterior point on the mandibular symphysis |
| 14. Gonion | Go* | The external angle of the mandible |
| 15. Incisor Tip | A1, B1 | Tip of central incisor (upper and lower respectively) |
| 16. Lower Lip | LL | Vermillion border of lower lip |
| 17. Mental Protuberance | Pm | Point of inflection of the curvature at the profile of the chin (aka Suprapogonion) |
| 18. Menton | Me | The most inferior point on the mandibular symphysis |
| 19. Nasal Tip | NT | Most anterior point of the nose (soft tissue) |
| 20. Nasale Medium Columella | NM | The fleshy lower margin of the nasal septum (soft tissue) |
| 21. Nasion | N | The intersection of the nasal and frontal bones |
| 22. Orbitale | Or* | Inferior most point of the orbit |
| 23. Palatal Plane | ANS-PNS | Line connecting ANS and PNS |
| 24. Pogonion | Pog | The most anterior point of the mandibular symphysis |
| 25. Point A | Pt. A | Point in the median sagittal plane where the lower front edge of the anterior nasal spine meets the front wall of the maxillary alveolar process (aka subspinale). |
| 26. Point B | Pt. B | Deepest midline point on the mandible between infradentale and pogonion (aka supramentale). |
| 27. Point D | D | The center of the cross section of the mandibular symphysis |
| 28. Porion | Po* | Superior margin of external auditory canal |
| 29. Posterior Condyle | PCd* | The point of the condyle tangent to a perpendicular line extending from the S-N plane |
| 30. Posterior Nasal Spine | PNS | The posterior point of the hard palate |
| 31. Pterygomaxillary Fissure Inferior | PTM* | Teardrop shaped area between maxilla and pterygoid process of the sphenoid |
| 32. Sella | S | The pituitary fossa of the sphenoid bone |
| 33. Soft Tissue Pogonion | Pog(st) | The most anterior point of the chin (soft tissue) |
| 34. Sphenoethmoidal Suture | Se | Point identifying the sphenoethmoidal suture |
| 35. Stomion Superior | Sts | Lowest point of the vermillion border of upper lip (soft tissue) |
| 36. X### | Points defined along the endocranial surface of the neurocranium relative to the S-N plane (e.g., X100 = the point at which a line drawn 100° from the S-N plane intersects the endocranium). |
Points not on midline of the skull.
Figure 2.
Lateral cephalograph. Cephalometric points used in the analysis are identified (see Table 1 for key).
Two trained assessors measured all radiographs. To examine inter-observer reliability, a set of radiographs were traced and measured by both observers on a regular basis. In total, 331 radiographs were assessed by both individuals. Assessors were provided with the same scanned image, but all placement of points, and fitting of outlines, were conducted independently. Reliability was assessed using intra-class correlation and by calculation of average interobserver differences.
Interobserver reliability of measures was very high with an average intraclass correlation of 0.91 for angular metrics and 0.95 for linear metrics. The average interobserver difference for linear metrics was 1.12 mm, and the average difference for angular measures was 1.9 degrees (complete details are provided in supplemental material).
Seventy-seven measurements were made based on the craniometric points identified (see supplemental Table 1). Points and measurements chosen are designed to examine variation both within and between craniofacial components. For example, several measures are contained within their respective component, such as posterior base length (Ba-S), anterior base length (S-N), and angular measures such basicranial flexion (N-S-Ba), which are all contained within the basicranium. Similarly, there are measures isolated to the splanchnocranium including facial height (N-Pt. A) and palate length (PNS-Pt. A). Other measures span components, such as Ba-Pt. A, which incorporates both basicranial and splanchnocranial landmarks. Angular measures such as facial hafting (S-N-Pt. A) or the facial axis also span multiple cranial components. All linear measurements were corrected for radiographic enlargement using an established correction factor.
Because neurocranial landmarks are difficult to discern reliably on lateral cephalographs, a set of measurements were defined to capture the maximal amount of information from the neurocranium. First, a line from sella to nasion was identified. From this reference line, additional lines were placed every ten degrees up to 180 degrees (and Bastir et al., 2006 for a similar approach to quantifying craniofacial morphology; see, for example, Gonzalez et al., 2010). For each line, a measurement was taken from sella to the point of intersection with the endocranial surface. Because we start at nasion, the first couple of measures do not intersect the endocranial surface and, therefore, no measurement can be made. Measures up to S-X60 tend to be confounded by visualization problems on the radiographs. There are several structures, such as the crista galli and the frontal crest, which may lead to a reduced image quality in that region, with the lower measures more affected. Reliability coefficients of those measures are low but predictably rise with measures advancing along the neurocranial outline (S-X30 has an ICC of 0.2; S-X40 an ICC of 0.33; etc.). Starting with the distance S-X70, and continuing through the remainder of neurocranial measures, the reliability coefficients are consistently very high (ICC=0.99). As the first six such measures demonstrate less than acceptable reliability, these were not included in analyses; there are, therefore, 12 measures (S-X70 to S-X180) available to describe the morphology of the neurocranium.
As it could be argued that measurements defined in this manner are not necessarily homologous between individuals (Bastir and Rosas, 2006; see Gunz et al., 2008 for a discussion of analysis of semilandmark data), we also used a principal components analysis to extract latent variables describing the overall morphology of the neurocranium. Using a varimax rotation, three components were extracted that explained 45%, 26%, and 25% of the variance respectively (for a total of 96% of the variance explained by these three components). Factor patterns describing each component (Table 2) indicate that the first principal component (NeuroPC1) is heavily influenced by anterior neurocranial measurements (70 to 120 degrees from S-N), the second principal component (NeuroPC2) is influenced by posterior measures (150–180 degrees), and the third principal component (NeuroPC3) is influenced by intermediate measures (120–160 degrees). Thus, in general, individuals loading high on NeuroPC1 could be described as anteriorly elongate, individuals loading high on NeuroPC2 could be described as posteriorly elongated, and individuals loading high on NeuroPC3 could be described as possessing taller crania.
Table 2.
Factor loading scores for principal components decomposition of phenotypic correlation matrices.§
| Trait | NeuroPC1 | NeuroPC2 | NeuroPC3 |
|---|---|---|---|
| S-X70 | 97* | 9 | 10 |
| S-X80 | 98* | 4 | 13 |
| S-X90 | 97* | −1 | 21 |
| S-X100 | 91* | 0 | 36 |
| S-X110 | 83* | 7 | 51 |
| S-X120 | 72* | 21 | 64* |
| S-X130 | 55 | 40 | 72* |
| S-X140 | 39 | 52 | 75* |
| S-X150 | 27 | 60* | 73* |
| S-X160 | 17 | 72* | 63* |
| S-X170 | −4 | 91* | 34 |
| S-X180 | −3 | 96* | 9 |
Values are multiplied by 100 and rounded.
“Meaningful” factor loadings (loading >55).
Genotyping and the Whole Genome Linkage Map
Whole genome marker genotyping
Of the individuals with craniofacial phenotypic data, 618 had been genotyped for ~400 highly polymorphic autosomal genetic markers using the ABI PRISM Linkage Mapping Set-MD10 (Applied Biosystems, Foster City, CA). This mapping set consists of fluorescently labeled PCR primers that amplify dinucleotide repeat microsatellite loci (STRs) selected from the Genethon human linkage map (Weissenbach et al., 1992; Gyapay et al., 1994; Dib et al., 1996). This set is designed to create a map with markers spaced on average 10 cM apart (range 2.4 to 24.1 cM).
Genotyping error checking
Genotypes assigned to individuals were checked for Mendelian consistency using the PEDSYS (Dyke, 1994) program INFER. Discrepancies were initially referred to the lab for retyping of markers in order to resolve them. If discrepancies could not be resolved by re-typing, the program SimWalk2 (Sobel and Lange, 1996) was used to make decisions about the appropriate genotypes to exclude in that situation.
Estimation of allele frequencies and genetic marker maps
Estimates of allele frequencies for each genetic marker locus were obtained using SOLAR (Sequential Oligogenic Linkage Analysis Routines) (Almasy and Blangero, 1998). The linkage map was based on the deCODE map with non-deCODE markers added in by interpolation using physical location data.
Statistical Genetic Analyses
We used a maximum likelihood-based variance decomposition approach implemented in SOLAR (Almasy and Blangero, 1998) to estimate heritability for each craniofacial variable and to conduct multipoint linkage analysis of each cranial trait. Linkage analysis in SOLAR entails specification of the genetic covariance between arbitrary relatives as a function of the identity-by-descent (IBD) relationships at a given marker locus and models the covariance matrix for a pedigree as the sum of the additive genetic covariance attributable to the QTL, the additive genetic covariance due to the effects of loci other than the QTL, and the variance due to unmeasured environmental factors. The linkage analyses incorporated IBD allele sharing estimated from genotype data at the microsatellite markers in the human linkage map. Probabilities of IBD among relatives at marker loci in the human linkage map were estimated using Markov Chain Monte Carlo routines implemented in the computer package Loki (Heath, 1997).
The hypothesis of linkage for each trait was evaluated at 1 cM intervals along each chromosome using likelihood ratio tests, and likelihood ratio statistics were converted to the LOD score of classic linkage analysis (Ott, 1988).
The hypothesis of linkage was tested by comparing the likelihood of a restricted model in which variance due to the QTL equaled zero (no linkage) to that of a model in which it did not equal zero (i.e., is estimated). The LOD score of classical linkage analysis was obtained as the quotient of the difference between the two ln likelihoods (Ott, 1988).
To control for the genome-wide false positive rate, we calculated genome-wide P-values for each LOD score using a modification of a method suggested by Feingold et al. (1993) that takes into account pedigree complexity and the finite marker density of the linkage map. Accordingly, our threshold for significant evidence of linkage (corresponding to genome-wide α=0.05) was LOD=2.87, while suggestive evidence of linkage occurred at LOD=1.67. These values correspond to the expected false positive rates of once per 20 (“significant”) and once per 1 (“suggestive”) genome-wide linkage screens (Lander and Kruglyak, 1995).
Accounting for environmental contributions to the phenotypic variance can improve power to detect genetic effects. Prior to all analyses, we used likelihood ratio tests to screen each of the following variables for significant mean effects on the cephalographically-derived variables: age, sex, age2, age×sex, age2×sex, and head diameter (measured as the distance from Nasion to S-x180). After adjusting for the effects of all nominally significant (P≤0.10) covariates, we applied an inverse Gaussian transformation to the residuals to correct for departures from multivariate normality that might inflate evidence for linkage (Blangero et al., 2001). This transformation produces standardized traits with means and standard deviations approaching 0 and 1, respectively. All reported linkage analyses were conducted using these normalized residual data. The reported h2 is residual heritability (that part of the variance that is attributable to the additive effects of genes after covariate effects, including those of age and sex, are removed).
RESULTS
Basic quantitative genetic analyses using the complete data set of 618 phenotyped individuals reveal significant (p<0.001) additive genetic components to the variance (i.e., heritability or h2) for all 80 craniofacial traits measured in this study: h2 estimates ranged from 0.14 (for UI to NA) to 0.99 (for Molar Relation) with an average h2=0.43 across all traits.
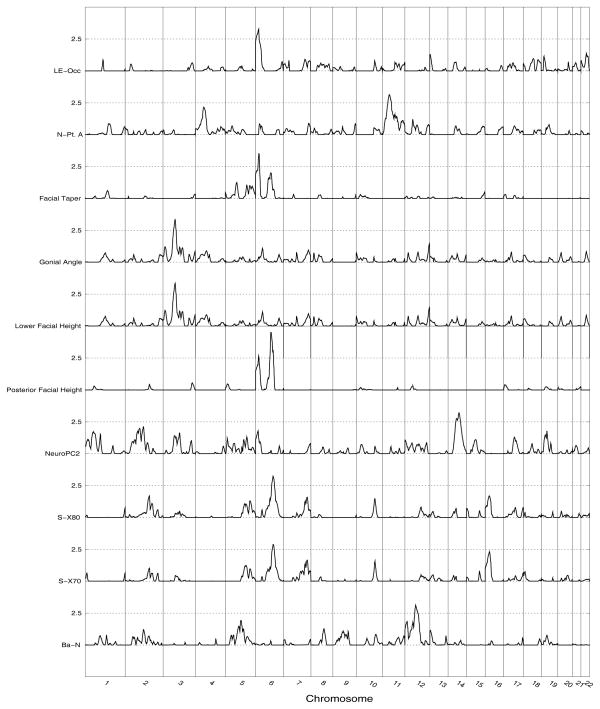
After confirming the reliability of the measurements and the identification of significant genetic effects on trait variation, we performed whole genome linkage analyses on the subset of 618 phenotyped and genotyped individuals to localize quantitative trait loci (QTL) accounting for these genetic effects. Linkage analyses identified several chromosomal regions harboring genes influencing craniofacial variation. Significant QTL (LOD>2.87) were identified for 10 craniofacial traits (Figure 3) and these results are presented in Table 3. One QTL was found on each of chromosomes 3, 11, 12, 14, and six QTL were found on chromosome 6 (Figure 4).
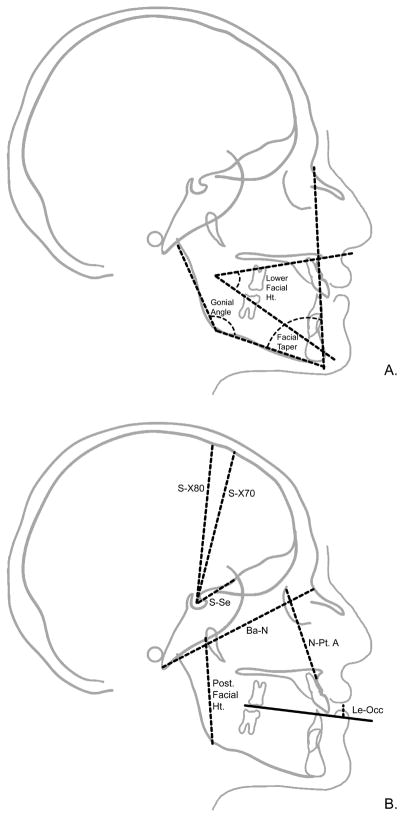
Figure 3.
Angular (A) and linear (B) measures collected from Lateral cephalographs. Measurements shown are those for which were detected one or more QTL accounting for a significant proportion of their variance.
Table 3.
Quantitative trait loci for craniofacial traits in Fels Longitudinal Study participants*.
| Trait | N | LOD | P** | Chromosome | cM*** | Cytogenetic location | #BP in 1-LOD Interval | #Genes in 1-LOD Interval | Component | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower facial height | 618 | 0.0+0.10**** | 0.553 | 3.35 | 0.011 | 3 | 84 | 3p21.31-p14.1 | 16,960,012 | 192 | Splanchnocranial | ||
| Gonial Angle | 573 | 0.0+0.10 | 0.506 | 3.37 | 0.014 | 6 | 22 | 6p25.1-p24.1 | 5,338,303 | 30 | Splanchnocranial | ||
| LE - Occ | 237 | 0.0+0.17 | 0.747 | 3.27 | 0.019 | 6 | 22 | 6p25.3-p24.1 | 12,718,164 | 71 | Soft tissue | ||
| Facial Taper | 654 | 0.0+0.09 | 0.496 | 3.52 | 0.007 | 6 | 23 | 6p25.1-p24.1 | 5,501,288 | 33 | Splanchnocranial | ||
| Post. Facial Ht | 573 | 0.0+0.11 | 0.563 | 4.52 | 0.0008 | 6 | 106 | 6q16.1-q21 | 16,146,101 | 47 | Splanchnocranial | ||
| S-X80 | 587 | 0.0+0.10 | 0.502 | 3.24 | 0.020 | 6 | 121 | 6q16.3-q22.33 | 26,595,994 | 119 | Neurocranial | ||
| S-X70 | 587 | 0.0+0.10 | 0.515 | 2.89 | 0.048 | 6 | 122 | 6q16.3-q23.2 | 27,699,629 | 124 | Neurocranial | ||
| N-Pt.A | 614 | 0.0+0.10 | 0.461 | 3.14 | 0.029 | 11 | 49 | 11p15.1-q12.1 | 58,193,54 | 221 | Splanchnocranial | ||
| Ba-N | 611 | 0.0+0.10 | 0.429 | 3.11 | 0.023 | 12 | 75 | 12q13.11-q21.31 | 35,563,317 | 363 | Basicranial | ||
| NeuroPC2 | 587 | 0.0+0.11 | 0.455 | 3.22 | 0.021 | 14 | 76 | 14q21.3-q31.3 | 39,665,099 | 255 | Neurocranial |
Maximum likelihood heritability estimates from linkage models: is the QTL-specific heritability, i.e., the proportion of the residual phenotypic variance due to the effect of the QTL, and is the residual heritability, i.e., the proportion of the phenotypic variance in the trait due to the effects of genes other than the QTL.
Genome-wide P-value associated with LOD score calculated using a modification of a method suggested by (Feingold et al. 1993) that takes into account pedigree complexity and the finite marker density of the baboon whole linkage map used in these analyses.
Location in centimorgans (cM) from pter-most marker locus in human genetic linkage map for that chromosome.
Standard errors for maximum likelihood estimates of residual that estimated on a boundary (e.g., 0) were obtained by interpolative maximization.
Figure 4.
Genomewide linkage results for craniofacial traits.
As noted, the cranium is frequently discussed in terms of developmental components; the basicranium (those structures supporting the brain), the splanchnocranium (the face and mandible), and the neurocranium (the bones surrounding the brain). Because traits within these components share similar developmental history and function, we investigated the significant QTL for each component for common patterns. Because some traits included aspects of multiple components, we investigated those separately.
Two significant QTL were found for basicranial measures, the first for the measure of total basicranial length (Ba-N). That QTL was localized to chromosome 12q21.33 and estimated to account for 43% of the detected residual phenotypic variance of variation in Ba-N. The second significant QTL found was for S-Se, a measure of anterior basicranial length. That QTL was localized to chromosome 2q24.3 and estimated to account for 46% of the detected residual phenotypic variance of variation in S-Se.
Our analyses yielded significant evidence for QTL affecting six splanchnocranial features including one soft-tissue measure. Four QTL were localized to chromosome 6 and one each to chromosome 3 and 11. The traits with QTL localized to chromosome 6 included three angular measures (Facial Taper, Posterior Facial Height, and the Gonial Angle), and the linear measure of lip embrasure to the occipital plane (LE-Occ). The QTL for the Gonial Angle, Facial Taper, and LE-Occ are in an overlapping region from 6p25-p24. They are estimated to account for 51%, 50%, and 75% of the detected residual phenotypic variance in their respective traits.
The initial whole genome screen for three splanchnocranial traits, Posterior Facial Height, Facial Taper, and Lower Facial Height found evidence for two QTL for each trait. The QTL detected for the trait Posterior Facial Height included one on chromosome 3 with LOD = 3.75 and the other on chromosome 6 with LOD = 4.52. For Facial Taper the two QTL detected in the initial scan were on chromosome 5 (LOD = 2.94) and chromosome 6 (LOD= 3.52). And, for Lower Facial Height, the two QTL were on chromosome 3 (LOD = 3.35) and chromosome 12 (LOD = 2.94). For these traits, we subsequently performed a second, sequential, whole genome linkage screen conditional on the QTL with the highest LOD score. For all three traits, the evidence for a second QTL in that second (conditional) linkage screen was neither significant nor suggestive.
The QTL localized to chromosome 6 for Posterior Facial Height is at 6q16.1-q21 and accounts for 56% of the residual phenotypic variance. The Lower Facial Height QTL localized to 3p21.31-p14.1 and accounts for 56% of the detected residual phenotypic variance. Finally, the facial length, defined as N-Pt. A, QTL localized to 11p15.1-q12.1 and accounts for 46% of the residual phenotypic variance.
Finally, three significant QTL were found for neurocranial traits. Two of these, influencing variation in the highly correlated S-X70 and S-X80 variables, mapped to the same location (i.e., 6q16.3). The estimated effect of these QTL on these two correlated measures was essentially identical, accounting for approximately 50–52% of the residual phenotypic variance. The third QTL was detected when we analyzed a synthetic variable, NeuroPC2, obtained from a principal components analysis of all three neurocranial metrics. The QTL for NeuroPC2, which (as noted earlier) emphasizes the posterior measures of the neurocranium, is localized to 14q21.3 and accounts for approximately 46% of the residual phenotypic variance on variation in this principal component.
DISCUSSION
To our knowledge, this study is the first to identify QTL influencing normal variation in the human craniofacial complex. This examination of genetic influences on the human craniofacial complex was conducted with an age-restricted sample of the Fels Longitudinal Study to minimize the effects of the substantial growth during early childhood. Subsequent studies will interrogate the QTL further in order to nominate positional candidate genes for further intensive study. Techniques available for this include fine-mapping of the QTL with high density SNP typing or examination of transcriptional profile data, both in conjunction with bioinformatic searches such as those provided by Ingenuity Systems.
Although each of the eleven detected QTL is estimated to account for all the additive genetic effects on the variance of the analyzed craniofacial phenotype (i.e., the residual heritability estimate in the maximized model equals 0), we do not contend that genetic effects on any of these traits are limited only to genes and variants under the QTL. There is, of course, a standard error about each maximum likelihood parameter estimate, including the residual heritability estimates; and other loci, which exert relatively weak additive genetics effects on these traits in these families, may be better detectable in studies of other pedigreed data sets and/or other primate species. Further, because data and theory point primarily to additive genetic effects on complex phenotypes (Hill et al., 2008), we have not accounted explicitly for dominance or epistatic interactions in these genome screens. Doing so, as well as testing for genotype-by-environment interactions (for example), in the future may identify other loci of importance. Nonetheless, we are confident that the detected QTL in this study are responsible for sufficiently substantive proportions of the observed quantitative variation in these craniofacial traits to warrant their continued study.
Data from Lateral Cephalographs
The data for this study come from lateral cephalographs. Lateral cephalographs have been a standard diagnostic tool for dentists and orthodontists and have served as the basis for basic and clinical research into growth and development of the craniofacial complex for decades (e.g., Lewis and Roche, 1972; Lewis and Roche, 1974; Riolo et al., 1974; Roche and Lewis, 1976; Lewis and Roche, 1977; Roche et al., 1977; Lewis et al., 1982; Ohtsuki et al., 1982; Lewis et al., 1985). While new imaging modalities are becoming more common, lateral cephalographs continue to be the standard diagnostic tool in most orthodontic practices.
Because plane film radiography reduces a three-dimensional structure to a two-dimensional image, quantitative data derived from lateral cephalographs is largely restricted to measures of midline structures (with some off-midline points and planes projected onto the midline). As described, linear metrics derived from lateral cephalographs must be corrected for the enlargement created by the radiographic protocol; angular data do not require correction. Because the radiographic protocol changed over the course of the 50+ year study it is likely that some error has been introduced. All changes in protocol and date of change are well-documented in the FLS archives and correction factors specific to the protocol used have been applied to all measurements, thus minimizing that error.
Recent years have seen a rise in the use of advanced medical and computer-aided technology designed to characterize morphology in complex structures such as the craniofacial complex. Technologies range from systems designed to identify XYZ coordinates from surface topologies such as the Microscribe 3D (Immersion Corporation) or the 3dMD system (www.3dmd.com) to medical imaging modalities such as CT or MRI. The former techniques provide accurate identification of points from the surface but are not able to characterize internal morphology in living participants. Additionally, the 3dMD system is limited in the ability to characterize areas covered by hair and may have error introduced due to variation in soft-tissue thicknesses. Medical imaging is very effective at providing an accurate means to assess three-dimensional morphology but can be expensive, uncomfortable for the participant and, in the case of CT, exposes the participant to x-irradiation.
Given the success of the current project to identify QTL of midline cranial structures, it is important to consider approaches allowing the examination of the complete morphology of the craniofacial complex. All the modalities described above are most often used to extract simple linear and angular measures similar to the procedures described for the current study (i.e., craniometric points are identified with lengths or angles measured between those points). Augmenting the current dataset with measures describing, for example, facial widths would provide additional phenotypes with relevance to clinical research of conditions such as orofacial clefting, or to comparative anatomical research in primate and human evolution.
There are additional techniques such as Fourier analysis or geometric morphometric approaches that may provide a more comprehensive means to evaluate shape in two or three dimensions. We chose to begin with simple metrics, because not only is there a wealth of both clinical and comparative anatomical literature with similar datasets, but, because characterizing large-scale craniofacial shape variation may introduce difficulties when applied to genetic analyses. For this paper we have made the assumption that localized traits are influenced by a small set of genes (i.e., are oligogenic in nature) and that these genes are characterized by a sufficient effect size to allow for detection with genetic epidemiologic techniques (Towne et al., 2002). If, as morphological analysis becomes more and more comprehensive, the number of genes involved increases, and effect size for individual genes decrease, the ability to accurately detect and localize specific genes is impaired. To test the feasibility of comprehensive geometric analyses, we have begun to examine the utility of a geometric morphometric approach in genetic analyses (McNulty et al., 2009; Sherwood et al., 2009).
Candidate genes
To investigate potential positional candidate genes under the QTL regions identified in our analyses, the current literature was searched for known genes affecting craniofacial structures. Several of the identified QTL harbor genes implicated in the growth and development of cranial structures. These include BMP6 for our Facial Taper QTL, and several members of the WNT family, WNT1, WNT10B for our Ba-N trait, and WNT5A for our Lower Facial Height trait. Kettunen et al (2006) have identified the role of BMP6 in early mesenchymal condensations and hypertrophic chondrocytes of the mouse cranial base. Wnt signals have been shown to promote growth of facial prominences by regulating the Bmp pathway (Brugmann et al., 2010). Specifically, Geetha-Loganathan et al (2009) have identified the expression of WNT5A in craniofacial mesenchyme of the chicken and Warner et al (2009) have identified the expression of WNT5A in the mouse secondary palate.
Several studies report that mutations in some of our identified regions of interest are associated with dysmorphic craniofacial patterns. For example, several of the traits we examine link to two regions on chromosome six (6p25-p24 and 6q16-q21). There are numerous reports of mutations on in these regions associated with various forms of craniofacial dysmorphology including craniometaphyseal dysplasia (Lughetti et al., 2000), and oculodentodigital dysplasia (Boyadjiev et al., 1999) or other facial dysmorphologies (Pazooki et al., 2007).
Systems Biology and the Craniofacial Complex
The current literature contains a wealth of detailed genetic information on human dental and cranial disorders (e.g., Brennan and Pauli, 2001; Cohen, 2002; Riise et al., 2002; Mulliken, 2002; Shieh et al., 2006; Hennekam et al., 2010). It is also clear, however, that the advances made in the genetics of craniofacial disorders do not provide unambiguous answers to questions of causation. For instance, Maclean et al., (2005) lists at least five separate mutations associated with Axenfeld-Rieger malformations. Similarly, Bardet-Biedl syndrome has also been linked to mutations in at least six different genes (Riise et al., 2002). It has been suggested that this heterogeneity results from underlying variability in the phenotypic syndrome. In other words, disorders are not distinct entities or syndromes, but instead are variable manifestations along a continuous scale. While phenotypic expression varies within each syndrome, most would agree the presentation of each syndrome is sufficiently unique to warrant distinct and separate classification (Cohen, 2002; Mulliken, 2002).
In discussing the problems associated with genetic heterogeneity Cohen (2002) states that “other factors are involved that are not understood at the present time.” There are two especially prominent candidates for these other factors: 1) the environment; or 2) other, presently unknown, genes and their possible interactive effects. Environmental insults resulting in growth perturbation or gross anatomical deformities are relatively commonly encountered in utero and range from mechanical disruptions such as amniotic bands, to complications based on placental-cord insufficiencies, to the introduction of teratogenic substances (Cohen, 1990; Sherwood et al., 1992; Sherwood et al., 1997; Moss, 1997; Cox, 2004). The subtle effects of a “normal” environment (acknowledging the extreme heterogeneity of any individual’s environment) on variability are less easily characterized.
The other potential confounding factor in understanding the genetics of dysmorphology is the relationship of mutated genes with other genes. While it is readily acknowledged that complex traits are often oligogenic in nature, there persists an expectation that a given mutation will produce a singular outcome. Even if the (non-genetic) environment were held constant, this expectation would not be warranted. The cumulative pleiotropic effects of genes and gene-gene interactions would be expected to produce a wide range of phenotypes proportional to the number of genes involved. In other words, variability among normal genes would be expected to produce variable phenotypes when acting in concert with a mutated gene.
One approach to the characterization of the cumulative pleiotropic effects of genes that is becoming increasingly popular is that of systems biology (Nadeau et al., 2003; Ideker, 2004a; Ideker, 2004b; e.g., Ideker et al., 2006). The systems biology approach stresses the hierarchical nature of biological information and prioritizes the elucidation of gene networks and signaling pathways to characterize the system under investigation. As the interaction between genes is identified, it becomes clear the extent to which a pathway may be affected, not only by a mutation-bearing gene, but also by the variation in all genes involved in the pathway.
With regard to craniofacial biology, a well-developed pathway model has been developed relative to the disorder holoprosencephaly (HPE). This disorder had been characterized as genetically heterogeneous with at least eight genes identified as etiologic factors. Phenotypically, presentation of the condition ranges from the extreme morphology of an alobar cerebrum with cyclopia, to the microform condition of hypotelorism and single midline incisor (Ming and Muenke, 2002). Several genes have been identified as causative including Sonic Hedgehog (SHH), Patched1 (PTCH), and GLI2 (Ming and Muenke, 2002). Elucidation of the signaling pathways linking these genes has identified that all are components of the SHH signaling pathway. It is not surprising, therefore, that a disruption in the pathway, resulting from a mutation in any of those genes, may produce a similar phenotype and that mutations in multiple genes accounts for, in part, the variation in phenotypes characterizing the condition (Gripp et al., 2000; Orioli et al., 2001; Ming et al., 2002; Ming and Muenke, 2002; Roessler et al., 2003). Identification of additional such signaling pathways and gene networks is critical for a complete understanding of normal craniofacial development and the etiology of dysmorphologies.
CONCLUSION
This is the first paper, to the best of our knowledge, to report a large-scale genetic epidemiologic analysis of the human craniofacial complex. While significant advancements to understanding the genetic influences on the craniofacial complex have been made through studies of dysmorphic syndromes, or through the use of animal models, the examination of the genetic architecture underlying normal human variation will provide information useful across multiple disciplines. Our current work has successfully localized chromosomal regions influencing craniofacial variation with future efforts set to identify specific genes operating within those regions. Following that, the next step is to identify the gene networks and pathways in which those genes operate. While the current study seeks to characterize genetic influences on variation of a discreet morphological region, specifically midline cranial structures, the QTL identified, and the genes ultimately identified, are likely to represent components of gene networks and signaling pathways with effects throughout the craniofacial complex. In other words, while genes identified via genetic epidemiologic analysis of metrics derived from lateral cephalographs are, by definition, those affecting midline measures, it is unlikely that those gene effects are uniquely restricted to that anatomical region. Future work is likely to demonstrate that genes identified through our efforts play a role in gene networks throughout the cranium.
Supplementary Material
Acknowledgments
The authors are indebted to the participants of the Fels Longitudinal Study. In addition, we are grateful to those members of the research team of the Fels Longitudinal Study who established and maintained the craniofacial program, most notably Arthur Lewis and Alex Roche. We express our sincere gratitude to Kimberly Lever, Rebecca Junker, and Joe Wagner for phenotyping and database assistance. This work was supported in part by the National Institute of Dental and Craniofacial Research (National Institutes of Health) DE016692, DE016408 to R.J. Sherwood. Primary support for the Fels Longitudinal Study was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development HD012252. Development and implementation of the SOLAR statistical genetics analysis package is supported by grant MH059490 from the U.S. National Institute of Mental Health. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grants C06 RR017515 and C06 RR013556 from the National Center for Research Resources (NCRR), NIH. The supercomputing facilities used in this work at the AT&T Genomics Computing Center were supported by a gift from the SBC Foundation. Two anonymous reviewers provided comments that greatly improved the manuscript, we appreciate their advice as well as that of the editors.
Grant sponsors: National Institute of Dental and Craniofacial Research (NIH), Grant Numbers DE016692, DE016408.
Grant sponsor: Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIH), Grant number HD012252
Grant sponsor: National Institute of Mental Health (NIH), Grant number: MH059490
Grant sponsor: National Center for Research Resources (NIH), Grant numbers: C06 RR017515, C06 RR013556
LITERATURE CITED
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta S, Sofocleous C, Kolialexi A, Thomaidis L, Giouroukos S, Karavitakis E, Mavrou A, Kitsiou S, Kanavakis E, Fryssira H. Clinical manifestations and molecular investigation of 50 patients with Williams syndrome in the Greek population. Pediatr Res. 2005;57:789–795. doi: 10.1203/01.PDR.0000157675.06850.68. [DOI] [PubMed] [Google Scholar]
- Bastir M, Rosas A. Correlated variation between the lateral basicranium and the face: a geometric morphometric study in different human groups. Arch Oral Biol. 2006;51:814–824. doi: 10.1016/j.archoralbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Bastir M, Rosas A, O’Higgins P. Craniofacial levels and the morphological maturation of the human skull. J Anat. 2006;209:637–654. doi: 10.1111/j.1469-7580.2006.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- Boyadjiev SA, Jabs EW, LaBuda M, Jamal JE, Torbergsen T, Ptacek LJ, Rogers RC, Nyberg-Hansen R, Opjordsmoen S, Zeller CB, Stine OC, Stalker HJ, Zori RT, Shapiro RE. Linkage analysis narrows the critical region for oculodentodigital dysplasia to chromosome 6q22-q23. Genomics. 1999;58:34–40. doi: 10.1006/geno.1999.5814. [DOI] [PubMed] [Google Scholar]
- Brennan AM, Pauli RM. Hajdu--Cheney syndrome: evolution of phenotype and clinical problems. Am J Med Genet. 2001;100:292–310. doi: 10.1002/1096-8628(20010515)100:4<292::aid-ajmg1308>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Brown JM, Wedden SE, Millburn GH, Robson LG, Hill RE, Davidson DR, Tickle C. Experimental analysis of the control of expression of the homeobox-gene Msx-1 in the developing limb and face. Development. 1993;119:41–48. doi: 10.1242/dev.119.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Powder KE, Young NM, Goodnough LH, Hahn SM, James AW, Helms JA, Lovett M. Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. Hum Mol Genet. 2010;19:920–930. doi: 10.1093/hmg/ddp559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM. Anomalies, syndromes, and dysmorphic growth and development. In: Enlow DH, editor. Facial Growth. Philadelphia: W.B. Saunders Co; 1990. pp. 331–345. [Google Scholar]
- Cohen MM. Perspectives on craniofacial anomalies, syndromes, and other disorders. In: Lin KY, Ogle RC, Jane JA, editors. Craniofacial Surgery: Science and Surgical Technique. Philadelphia: W.B. Saunders Company; 2002. pp. 3–38. [Google Scholar]
- Cox TC. Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin Genet. 2004;65:163–176. doi: 10.1111/j.0009-9163.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Jin Q, Taba M, Jr, Franceschi RT, Bruce RR, Giannobile WV. BMP gene delivery for alveolar bone engineering at dental implant defects. Mol Ther. 2005;11:294–299. doi: 10.1016/j.ymthe.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke B. Populations Genetics Laboratory Technical Report No 2. 2. Southwest Foundation for Biomedical Research; 1994. PEDSYS, a pedigree data management system user’s manual. [Google Scholar]
- Feingold E, Brown PO, Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993;53:234–251. [PMC free article] [PubMed] [Google Scholar]
- Fong KD, Nacamuli RP, Song HM, Warren SM, Lorenz HP, Longaker MT. New strategies for craniofacial repair and replacement: a brief review. J Craniofac Surg. 2003;14:333–339. doi: 10.1097/00001665-200305000-00011. [DOI] [PubMed] [Google Scholar]
- Garcia-Godoy F, Murray PE. Status and potential commercial impact of stem cell-based treatments on dental and craniofacial regeneration. Stem Cells Dev. 2006;15:881–887. doi: 10.1089/scd.2006.15.881. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Antoni L, Fu K, Whiting CJ, Francis-West P, Richman JM. Expression of WNT signalling pathway genes during chicken craniofacial development. Dev Dyn. 2009;238:1150–1165. doi: 10.1002/dvdy.21934. [DOI] [PubMed] [Google Scholar]
- Gonzalez PN, Perez SI, Bernal V. Ontogeny of robusticity of craniofacial traits in modern humans: a study of South American populations. Am J Phys Anthropol. 2010;142:367–379. doi: 10.1002/ajpa.21231. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ. Otodental syndrome, oculo-facio-cardio-dental (OFCD) syndrome, and lobodontia: dental disorders of interest to the pediatric radiologist. Pediatr Radiol. 1998;28:802–804. doi: 10.1007/s002470050469. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Gunz P, Mitteroecker P, Bookstein FL. Semilandmarks in three dimensions. In: Slice DE, editor. Modern Morphometrics in Physical Anthropology. New York: Plenum Publishers; 2008. pp. 73–98. [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. The 1993–94 Genethon human genetic linkage map. Nat Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JA, Schneider RA. Cranial skeletal biology. Nature. 2003;423:326–331. doi: 10.1038/nature01656. [DOI] [PubMed] [Google Scholar]
- Hennekam RCM, Krantz ID, Allanson JE. Gorlin’s Syndromes of the Head and Neck. 5. Oxford: Oxford University Press; 2010. [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Nadiri A, Kuchler-Bopp S, Perrin-Schmitt F, Peters H, Lesot H. Tissue engineering of tooth crown, root, and periodontium. Tissue Eng. 2006;12:2069–2075. doi: 10.1089/ten.2006.12.2069. [DOI] [PubMed] [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Ideker T. A systems approach to discovering signaling and regulatory pathways--or, how to digest large interaction networks into relevant pieces. Adv Exp Med Biol. 2004a;547:21–30. doi: 10.1007/978-1-4419-8861-4_3. [DOI] [PubMed] [Google Scholar]
- Ideker T. Systems biology 101--what you need to know. Nat Biotechnol. 2004b;22:473–475. doi: 10.1038/nbt0404-473. [DOI] [PubMed] [Google Scholar]
- Ideker T, Winslow LR, Lauffenburger DA. Bioengineering and systems biology. Ann Biomed Eng. 2006;34:1226–1233. doi: 10.1007/s10439-006-9119-3. [DOI] [PubMed] [Google Scholar]
- Idrees F, Bloch-Zupan A, Free SL, Vaideanu D, Thompson PJ, Ashley P, Brice G, Rutland P, Bitner-Glindzicz M, Khaw PT, Fraser S, Sisodiya SM, Sowden JC. A novel homeobox mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome associated with brain, ocular, and dental phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141:184–191. doi: 10.1002/ajmg.b.30237. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Nie X, Kvinnsland IH, Luukko K. Histological development and dynamic expression of Bmp2–6 mRNAs in the embryonic and postnatal mouse cranial base. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:1250–1258. doi: 10.1002/ar.a.20402. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lewis AB, Roche AF. Elongation of the cranial base in girls during pubescence. Angle Orthod. 1972;42:358–367. doi: 10.1043/0003-3219(1972)042<0358:EOTCBI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lewis AB, Roche AF. Cranial base elongation in boys during pubescence. Angle Orthod. 1974;44:83–93. doi: 10.1043/0003-3219(1974)044<0083:CBEIBD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lewis AB, Roche AF. The saddle angle: constancy or change? Angle Orthod. 1977;47:46–54. doi: 10.1043/0003-3219(1977)047<0046:TSACOC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lewis AB, Roche AF, Wagner B. Growth of the mandible during pubescence. Angle Orthod. 1982;52:325–342. doi: 10.1043/0003-3219(1982)052<0325:GOTMDP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lewis AB, Roche AF, Wagner B. Pubertal spurts in cranial base and mandible. Comparisons within individuals. Angle Orthod. 1985;55:17–30. doi: 10.1043/0003-3219(1985)055<0017:PSICBA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lindsey WH. Osseous tissue engineering with gene therapy for facial bone reconstruction. Laryngoscope. 2001;111:1128–1136. doi: 10.1097/00005537-200107000-00003. [DOI] [PubMed] [Google Scholar]
- Lughetti P, Alonso LG, Wilcox W, Alonso N, Passos-Bueno MR. Mapping of the autosomal recessive (AR) craniometaphyseal dysplasia locus to chromosome region 6q21–22 and confirmation of genetic heterogeneity for mild AR spondylocostal dysplasia. Am J Med Genet. 2000;95:482–491. doi: 10.1002/1096-8628(20001218)95:5<482::aid-ajmg14>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Oreffo RO. Bridging the gap. Nature. 2005;433:19. doi: 10.1038/433019a. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Please CP, Taylor M, Oreffo RO. Mathematical modelling of skeletal repair. Biochem Biophys Res Commun. 2004;313:825–833. doi: 10.1016/j.bbrc.2003.11.171. [DOI] [PubMed] [Google Scholar]
- Maclean K, Smith J, St HL, Chia N, Williams R, Peters GB, Onikul E, McCrossin T, Lehmann OJ, Ades LC. Axenfeld-Rieger malformation and distinctive facial features: Clues to a recognizable 6p25 microdeletion syndrome. Am J Med Genet A. 2005;132:381–385. doi: 10.1002/ajmg.a.30274. [DOI] [PubMed] [Google Scholar]
- McNulty KP, Duren DL, Blangero J, Dyer T, Cole SA, Lee M, Siervogel RM, Towne B, Sherwood RJ. The geometry and architecture of craniofacial inheritance. Am J Phys Anthropol. 2009;138:289. [Google Scholar]
- Merow WW, Broadbent BH., Jr . Cephalometrics. In: Enlow DH, editor. Facial Growth. Philadelphia: W.B. Saunders Co; 1990. pp. 346–395. [Google Scholar]
- Mina M. Morphogenesis of the medial region of the developing mandible is regulated by multiple signaling pathways. Cells Tissues Organs. 2001;169:295–301. doi: 10.1159/000047894. [DOI] [PubMed] [Google Scholar]
- Mina M, Gluhak J, Upholt WB, Kollar EJ, Rogers B. Experimental analysis of Msx-1 and Msx-2 gene expression during chick mandibular morphogenesis. Dev Dyn. 1995;202:195–214. doi: 10.1002/aja.1002020211. [DOI] [PubMed] [Google Scholar]
- Mina M, Havens B. FGF signaling in mandibular skeletogenesis. Orthod Craniofac Res. 2007;10:59–66. doi: 10.1111/j.1601-6343.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- Mina M, Kollar EJ, Upholt WB. Temporal and spatial expression of genes for cartilage extracellular matrix proteins during avian mandibular arch development. Differentiation. 1991;48:17–24. doi: 10.1111/j.1432-0436.1991.tb00238.x. [DOI] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss ML. The functional matrix hypothesis revisited. 4. The epigenetic antithesis and the resolving synthesis. Am J Orthod Dentofacial Orthop. 1997;112:410–417. doi: 10.1016/s0889-5406(97)70049-0. [DOI] [PubMed] [Google Scholar]
- Mulliken JB. The craniofacial surgeon as amateur geneticist. J Craniofac Surg. 2002;13:3–17. doi: 10.1097/00001665-200201000-00002. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Burrage LC, Restivo J, Pao YH, Churchill G, Hoit BD. Pleiotropy, homeostasis, and functional networks based on assays of cardiovascular traits in genetically randomized populations. Genome Res. 2003;13:2082–2091. doi: 10.1101/gr.1186603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki F, Mukherjee D, Lewis AB, Roche AF. Growth of cranial base and vault dimensions in children. J Anthrop Soc Nippon. 1982;90:239–258. doi: 10.1002/ajpa.1330580305. [DOI] [PubMed] [Google Scholar]
- Olsen B. The genetic map of craniofacial anomalies. Harv Dent Bull. 1998;7:18–19. [PubMed] [Google Scholar]
- Orioli IM, Castilla EE, Ming JE, Nazer J, Burle de Aguiar MJ, Llerena JC, Muenke M. Identification of novel mutations in SHH and ZIC2 in a South American (ECLAMC) population with holoprosencephaly. Hum Genet. 2001;109:1–6. doi: 10.1007/s004390100537. [DOI] [PubMed] [Google Scholar]
- Ott J. Analysis of Human Genetic Linkage. Baltimore and London: The Johns Hopkins University Press; 1988. [Google Scholar]
- Pazooki M, Lebbar A, Roubergues A, Baverel F, Letessier D, Dupont JM. Pure familial 6q21q22.1 duplication in two generations. Eur J Med Genet. 2007;50:60–65. doi: 10.1016/j.ejmg.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Riise R, Tornqvist K, Wright AF, Mykytyn K, Sheffield VC. The phenotype in Norwegian patients with Bardet-Biedl syndrome with mutations in the BBS4 gene. Arch Ophthalmol. 2002;120:1364–1367. doi: 10.1001/archopht.120.10.1364. [DOI] [PubMed] [Google Scholar]
- Riolo ML, Moyers RE, McNamara JA, Hunter WS. Craniofacial Growth Series. An Atlas of Craniofacial Growth: Cephalometric Standards from the University School Growth Study, The University of Michigan. Ann Arbor: Center for Human Growth and Development, University of Michigan; 1974. [Google Scholar]
- Roche AF, Lewis AB. Late growth changes in the cranial base. In: Bosma JF, editor. Symposium on Development of the Basicranium. Bethesda, MD: U.S. Dept. Health, Education, and Welfare, NIH; 1976. pp. 221–239. [Google Scholar]
- Roche AF, Lewis AB, Wainer H, McCartin R. Late elongation of the cranial base. J Dent Res. 1977;56:802–808. doi: 10.1177/00220345770560071501. [DOI] [PubMed] [Google Scholar]
- Roche AF. Growth, Maturation and Body Composition: The Fels Longitudinal Study 1929–1991. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci U S A. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RJ, May RL, Meindl RS, Robinson HB. Growth alteration in the pathological human fetus. Am J Phys Anthropol Suppl. 1992;14:150. [Google Scholar]
- Sherwood RJ, McNulty KP, Duren DL, Mahaney MC, Williams-Blangero S, Siervogel RM, Towne B. Dissecting the genetic architecture of craniofacial shape. First International Conference on Biological Shape.2009. [Google Scholar]
- Sherwood RJ, Robinson HB, Meindl RS, May RL. Pattern and process of growth of the abnormal human fetus. Hum Biol. 1997;69:849–871. [PubMed] [Google Scholar]
- Shieh JT, Aradhya S, Novelli A, Manning MA, Cherry AM, Brumblay J, Salpietro CD, Bernardini L, Dallapiccola B, Hoyme HE. Nablus mask-like facial syndrome is caused by a microdeletion of 8q detected by array-based comparative genomic hybridization. Am J Med Genet A. 2006;140:1267–1273. doi: 10.1002/ajmg.a.31262. [DOI] [PubMed] [Google Scholar]
- Slavkin HC. Research on craniofacial genetics and developmental biology: implications for the future of academic dentistry. J Dent Educ. 1983;47:231–238. [PubMed] [Google Scholar]
- Slavkin HC. Recombinant DNA technology and oral medicine. Ann N Y Acad Sci. 1995;758:314–328. doi: 10.1111/j.1749-6632.1995.tb24836.x. [DOI] [PubMed] [Google Scholar]
- Slavkin HC. And the next 50 years? The future of recombinant DNA technology in oral medicine. J Public Health Dent. 1996;56:278–285. doi: 10.1111/j.1752-7325.1996.tb02452.x. [DOI] [PubMed] [Google Scholar]
- Slavkin HC. The human genome, implications for oral health and diseases, and dental education. J Dent Educ. 2001;65:463–479. [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Towne B, Demerath EW, Czerwinski SA. The genetic epidemiology of growth and development. In: Cameron N, editor. Human Growth and Development. San Diego: Academic Press; 2002. pp. 103–137. [Google Scholar]
- Warner DR, Smith HS, Webb CL, Greene RM, Pisano MM. Expression of Wnts in the developing murine secondary palate. Int J Dev Biol. 2009;53:1105–1112. doi: 10.1387/ijdb.082578dw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SM, Fong KD, Chen CM, Loboa EG, Cowan CM, Lorenz HP, Longaker MT. Tools and techniques for craniofacial tissue engineering. Tissue Eng. 2003;9:187–200. doi: 10.1089/107632703764664666. [DOI] [PubMed] [Google Scholar]
- Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Weng Y, Cao Y, Silva CA, Vacanti MP, Vacanti CA. Tissue-engineered composites of bone and cartilage for mandible condylar reconstruction. J Oral Maxillofac Surg. 2001;59:185–190. doi: 10.1053/joms.2001.20491. [DOI] [PubMed] [Google Scholar]
- Yelick PC, Vacanti JP. Bioengineered teeth from tooth bud cells. Dent Clin North Am. 2006;50:191–203. viii. doi: 10.1016/j.cden.2005.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.